Abstract
Background
Our understanding of the impact of copy number variants (CNVs) on psychopathology and their joint influence with polygenic risk scores (PRS) remains limited.
Methods
The UK Biobank recruited 502,534 individuals aged 37 to 73 living in the UK between 2006 and 2010. After quality control, genotype data from 459,855 individuals were available for CNV calling. A total of 61 commonly studied recurrent neuropsychiatric CNVs were selected for analyses and examined individually and in aggregate (any CNV, deletion or duplication). CNV risk scores (CRS) were used to quantify intolerance of CNVs to haploinsufficiency. Major depressive disorder (MDD) and generalized anxiety disorder (GAD) PRS were generated for white British individuals (N=408,870). Mood/anxiety factor scores was generated using item-level questionnaire data (N=501,289).
Results
CNV carriers showed higher mood/anxiety scores than non-carriers, with the largest effects seen for intolerant deletions. A total of 11 individual deletions, and 8 duplications were associated with higher mood/anxiety. Carriers of the 9p24.3 (DMRT1) duplication showed lower mood/anxiety. Associations remained significant for most CNVs when excluding individuals with psychiatric diagnoses. Nominally significant CNV×PRS interactions provided preliminary evidence that associations between select individual CNVs, but not CNVs in aggregate, and mood/anxiety may be modulated by PRS.
Conclusions
CNVs associated with risk for psychiatric disorders showed small to large effects on dimensional mood/anxiety scores in a general population cohort, even when excluding individuals with psychiatric diagnoses. CNV×PRS interactions showed that associations between select CNVs and mood/anxiety may be modulated by PRS.
Keywords: copy number variants, polygenic risk scores, psychopathology, depression, mood, anxiety
Introduction
Copy number variants (CNVs) are deletions or duplications of sections of DNA that often result in changes in gene dosage.(1–3) CNVs increase risk for neurodevelopmental and psychiatric disorders.(4–6) However, the full impact of CNVs on psychopathology remains unclear for several reasons.
First, low frequency of CNVs, especially those with large, pathogenic effects, means that previous studies have mostly focused on a single psychiatric disorder. For example, CNVs have been strongly implicated in autism spectrum disorder (ASD),(7, 8) which has led organizations such as the American Academy of Pediatrics to recommend genomic screening for all children experiencing ASD symptoms.(9, 10) CNVs are also associated with increased risk for schizophrenia.(11, 12) In particular, 22q11 deletion syndrome is now widely considered to be the single largest genetic risk factor for schizophrenia (11, 13, 14). At least 16 other CNVs have since been associated with increased schizophrenia risk.(11, 12) CNVs have also been implicated in mood disorders, such as bipolar disorder,(15) but less robustly, (16, 17) and with smaller effects, than in schizophrenia.(18) There have also been reports of CNVs being associated with increased risk for depression,(19) especially treatment-resistant major depression.(20) Thus, CNVs are associated with risk for psychiatric disorders with strong neurodevelopmental and genetic origins, such as ASD and schizophrenia. However, the role of CNVs in mood disorders is unclear.
Second, previous CNV studies have mostly used binary diagnostic categories to examine disease risk, and the behavioral dimensions underlying risk remain unknown. These studies were mostly conducted using samples ascertained from clinic populations, which may show limited generalizability. Moreover, even very large CNV studies have limited statistical power since many CNVs only occur in one or two patients, even in these very large samples. In general population studies of more prevalent psychiatric disorders, such as depression, statistical power remains limited due to possibly weaker effects of CNVs on psychiatric disorders without strong neurodevelopmental and genetic origins. Thus, using a dimensional psychiatric phenotype in a population-based sample may be an effective way to increase both generalizability and statistical power. Results from population-based samples may be more generalizable than those from psychiatric samples. Dimensional phenotypes circumvent the issue of CNVs occurring in only one or two patients since CNVs associations can also be examined in individuals with subclinical psychopathology, and thus, across the whole continuum of psychiatric symptomatology, beyond the most severe manifestations of disease.
Third, the joint influence of rare and common genetic variants on psychopathology remains unclear. In recent years, genome-wide association studies (GWAS) have been conducted for most major psychiatric disorders, finding hundreds of common loci that are associated with risk for schizophrenia,(21) ASD,(22) bipolar disorder,(23) major depression,(24) post-traumatic stress disorder,(25) and so on. Polygenic risk scores (PRS) that index cumulative effects of common variants from these GWAS are indeed associated with disease liability and widely used in research studies. However, how these common variants act together with CNVs is unclear, although efforts to delineate the combined effects of rare and common variants on neuropsychiatric phenotypes are underway(26–28). While CNVs are robustly associated with risk for psychiatric disorders, their outcomes vary widely. This variable expressivity may be, at least partly, due to differences in genetic background. Indeed, evidence suggests that PRS associations are greater in individuals with pathogenic CNVs, but carriers of large, rare CNVs have also been shown to have slightly lower PRS (i.e., lower predicted liability) than non-carriers.(29, 30) Outcomes of mutations being dependent on genetic background, a phenomenon known as genetic interaction or epistasis, has been detected in model organisms(31). While the molecular mechanisms underlying epistasis remain unclear, several models have been proposed(31). Examining potential interactions between common and rare variants in humans, although challenging(31), may provide additional insights into these underlying mechanisms. For example, 22q11.2 deletions are hypothesized to lower tolerance for expression of biological pathways involved in schizophrenia, with genes in this region possibly amplifying effects of common variants across the genome(32).
In the present study, we used data from the UK Biobank, a large population-based sample of over 500,000 adults aged 37 to 73, to derive dimensional scores of mood/anxiety using factor models of item-level data from mental health questionnaires. We then used genotype data to call CNVs and calculate psychiatric polygenic risk scores (PRS) to examine the effects of CNVs, PRS, and CNV×PRS interactions on these mood/anxiety factor scores. We hypothesized that 1) CNV carriers would show higher mood/anxiety scores than non-carriers; 2) PRS would be positively associated with mood/anxiety; and 3) associations between CNVs and psychopathology may be modulated by PRS.
Methods and Materials
Sample
The UK Biobank recruited 502,534 individuals (54% female) aged 37 to 73 living in the United Kingdom between 2006 and 2010. Phenotypic data were collected at assessment centers using touchscreen devices and through nurse-led interviews. Participants provided blood, urine, and saliva samples. Written informed consent was obtained from all participants. All procedures involving human participants were approved by the North West Multi-Centre Ethics Committee (approval number 11/NW/0382). Data were released under application number 40980.
Genotyping and CNV calling
DNA was extracted from whole blood and genotyped on two Affymetrix arrays: ~50,000 on the UK BiLEVE Array and ~450,000 on the UK Biobank Axiom Array. Quality control filters were: genotypic call rate >0.95; waviness factor >−0.05 and <0.05; log R ratio SD <0.35; BAF SD <0.08. Of the 488,377 individuals with genotypic data, 28,522 were excluded for failing these quality control filters, leaving 459,855 individuals for CNV calling.
CNVs were called with PennCNV(33) and QuantiSNP(34) using our previously published pipeline(35, 36) (https://martineaujeanlouis.github.io/MIND-GENESPARALLELCNV/#/) and the following parameters: number of consecutive probes for CNV detection ≥3; CNV size ≥1Kb; confidence scores ≥15. CNVs detected by both algorithms were merged using CNVision(37) to minimize false discoveries. We then used a CNV inheritance analysis algorithm to concatenate adjacent CNVs of the same type using these criteria: gap between CNVs ≤150 kb; CNV size ≥1000 bp; probes ≥3. CNVs were then selected for analysis using these criteria: confidence score ≥30 (with at least one detection algorithm), size ≥50 kb, unambiguous type (deletion or duplication), and overlap with segmental duplicates, HLA regions or centromeric regions <50%.
CNVs were annotated using Gencode V35 lifted to hg19 coordinates (https://www.gencodegenes.org/human/release_35lift37.html). We used bedtools (https://bedtools.readthedocs.io/en/latest/) to find gene components (UTRs, start and stop codons, exons and introns) that overlapped with CNVs. As detailed elsewhere(35), we selected 61 recurrent CNVs that have previously been associated with neuropsychiatric symptoms in multiple studies(11, 38–41) (eTable 1). These 61 recurrent CNVs were identified if they showed >40% overlap with a specific deletion or duplication, or if they disrupted a gene(s). All recurrent CNVs were verified visually. Specifically, 5,235 recurrent CNVs were verified due to low likelihood scores (<150). Only 0.8% (N=43) of these 5,235 recurrent CNVs were found to be false positives.
CNV Risk Scores
CNV risk scores (CRS) reflect the probability of intolerance to haploinsufficiency or triplosensitivity of each gene encapsulated in every CNV identified in an individual. In this study, the CRS was calculated as the sum of the inverse loss of function observed/expected upper bound fraction (1/LOEUF) for each gene encompassed in a deletion or duplication using our published annotation pipeline.(36) Briefly, each coding gene with all isoforms with at least one start and one stop codon fully encompassed in the filtered CNVs was identified using Ensembl map (Gencode V35lift37 (hg19))(42) and annotated using the inverse LOEUF (1/LOEUF) score (gnomAD version 2.1.1),(43) which is available for 19,197 genes and ranges from 0.5 (gene tolerant to haploinsufficiency) to 33.3 (gene intolerant to haploinsufficiency). A score of 0 was attributed to individuals with no coding genes encompassed in any CNV.
Polygenic Risk Scores
Our approach for calculating polygenic risk scores (PRS) has recently been described in detail (44). Briefly, we removed single nucleotide polymorphisms (SNPs) with >5% missingness, samples with >10% missingness, and samples in which genotyped sex was different from reported sex. The 1000 Genomes Project was used for imputation,(45) retaining polymorphic sites with imputation quality R2≥0.7 and MAF≥0.01. Given evidence that European-ancestry (EUR) GWAS do not yield accurate PRS for non-EUR individuals(44) and unavailability of non-EUR GWAS, PRS were only computed for the White British cohort (N=408,870). PRS-CS (a high-dimensional Bayesian regression approach that utilizes continuous shrinkage priors)(46) was used to infer posterior effect sizes of SNPs overlapping with the generalized anxiety disorder GWAS summary statistics (to calculate PRS-GAD)(47) or the major depressive disorder GWAS summary statistics (to calculate PRS-MDD)(48) and an external EUR linkage disequilibrium reference panel. PRS were standardized.
Mood/anxiety factor
We estimated one-factor confirmatory factor models(49) in Mplus(50) using mean- and variance-adjusted weighted least squared estimator (WLSMV) to generate factor scores of mood/anxiety in all individuals with available phenotype data. eTable 2 shows the 16 mood, anxiety, and neuroticism items that were entered into the confirmatory factor model and their factor loadings. These 16 items were collected for 501,289 individuals using touchscreen devices at the first assessment center. Fit indices suggested a fair model fit (RMSEA=0.091, TFI=0.907, CLI=0.892). Mood/anxiety factor scores were standardized, and their distribution is presented in eFigure 1. To validate this mood/anxiety factor, we used linear regression to compare factor scores between individuals with a primary or secondary International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10) diagnosis of 1) dementia, 2) substance use, 3) anxiety, 4) psychosis, 5) bipolar/mania, 6) depression, and 7) anxiety/depression (eTable 3) and individuals without a psychiatric diagnosis (N=451,978).
Statistical analyses
All statistics and graphics were generated using R.(51) Linear regression was used to test for main effects of CNVs and PRS, and CNV×PRS interaction effects on mood/anxiety factor scores. The 61 neuropsychiatric recurrent CNVs (eTable 1) were analyzed individually and in aggregate (i.e., any CNV; any deletion; any duplication) by comparing mood/anxiety scores between CNV carriers and non-carriers. Individual recurrent CNVs with fewer than five observations were excluded from individual CNV analyses. We conducted aggregate recurrent CNV analyses first including and then excluding carriers of the high frequency 2q13 (NPHP1), 15q11.2, 15q13.3 (CHRNA7), and ZNF92 because these CNVs account for more than half of all CNVs in the UK Biobank. Next, CNV risk scores (CRS) were categorized such that individuals with total 1/LOEUF >2.86 (i.e., carriers of CNVs including at least one intolerant gene, since 1/0.35=2.86 where 0.35 represents the cutoff for a gene’s intolerance to predicted loss of function (pLoF)) were compared to individuals with total 1/LOEUF = 0 (i.e., no coding genes encompassed in any CNV). We then examined the effect of intolerant recurrent CNVs by comparing recurrent CNV carriers with a CRS >2.86 to individuals with a CRS of 0. Thus, we tested for associations with mood/anxiety factor scores of aggregate CNVs (i.e., any CNV; any deletion; any duplication) categorized in five ways: 1) recurrent (61 neuropsychiatric recurrent CNVs in eTable 1); 2) intolerant (CRS >2.86 across the genome); 3) intolerant recurrent (recurrent CNVs with CRS >2.86); 4) recurrent rare (57 neuropsychiatric recurrent CNVs excluding carriers of the high frequency 2q13 (NPHP1), 15q11.2, 15q13.3 (CHRNA7), and ZNF92); 5) intolerant recurrent rare (52 neuropsychiatric recurrent CNVs with CRS >2.86 excluding carriers of the high frequency CNVs listed above and the ZMYM5, CRYL1, VPS13B, 16q23.3, 17q21.31 CNVs with a CRS>2.86). Finally, we tested for CNV×PRS interactions. Again, CNVs were analyzed individually and in aggregate (i.e., any CNV; any deletion; any duplication). Age, sex, and 10 ancestry principal components were included as covariates in all models. To control for multiple testing, the false discovery rate (FDR) was set at 5%(52).
Sensitivity analyses
CNV analyses were conducted including all participants (N=460,226) and only White British participants (N=386,963). Further sensitivity analyses were conducted excluding individuals with a primary or secondary ICD-10 diagnosis for any psychiatric disorder (N=50,501) (eTable 3).
Results
Individuals with psychiatric disorders show high mood/anxiety scores
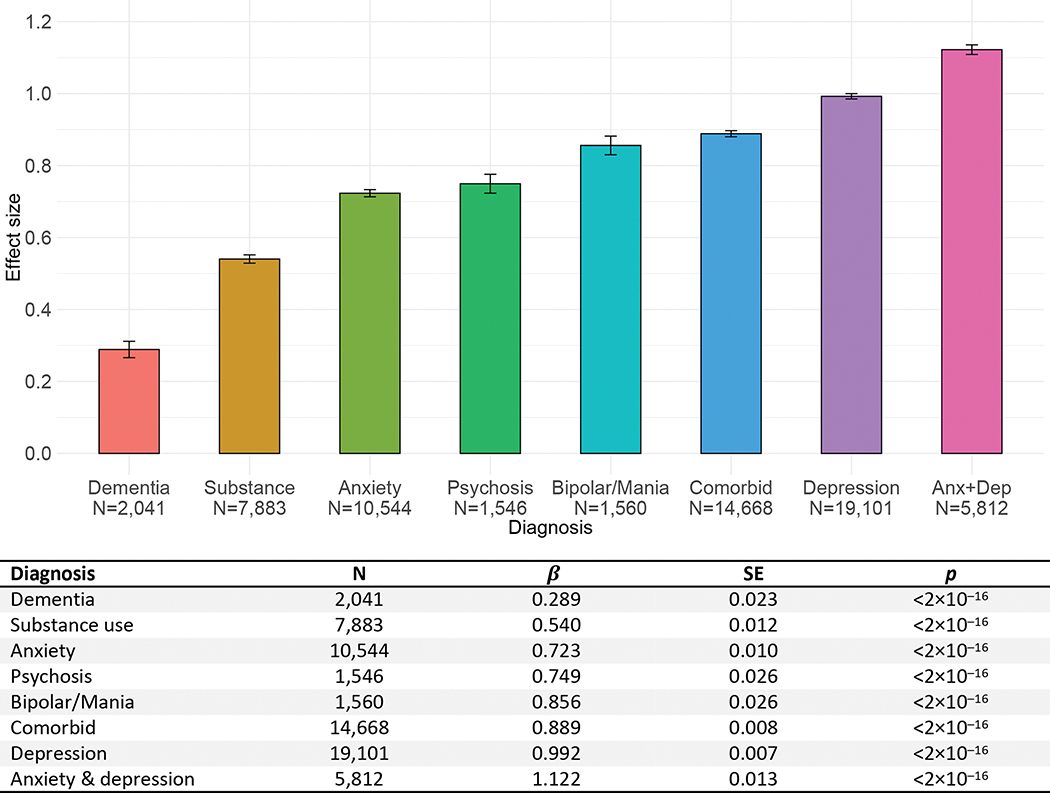
Distribution of mood/anxiety factor scores is presented in eFigure 1 and factor loadings are presented in eTable 2. Fit indices suggested a fair model fit (RMSEA=0.091, TFI=0.907, CLI=0.892). Figure 1 shows standardized mean differences in mood/anxiety scores between individuals with and without psychiatric diagnoses. Differences were significant for all diagnoses and of small to large effect size, such that individuals with diagnoses showed higher mood/anxiety scores. The largest effect size was seen for individuals with a diagnosis of mixed anxiety and depression, who scored more than one standard deviation (SD) above individuals without a psychiatric diagnosis (Figure 1). Large effect sizes were also seen for individuals with diagnoses of anxiety, psychosis, bipolar/mania, and depression.
Figure 1.
Standardized mean differences between individuals with and without psychiatric diagnoses on mood/anxiety factor scores.
Error bars represent standard errors.
β coefficients correspond to standardized effect sizes, with values of 0.2, 0.5, and 0.8 indicating small, medium, and large effects, respectively.
Diagnostic groups were compared to individuals without psychiatric diagnoses (N=451,978).
CNV carriers show higher mood/anxiety scores than non-carriers
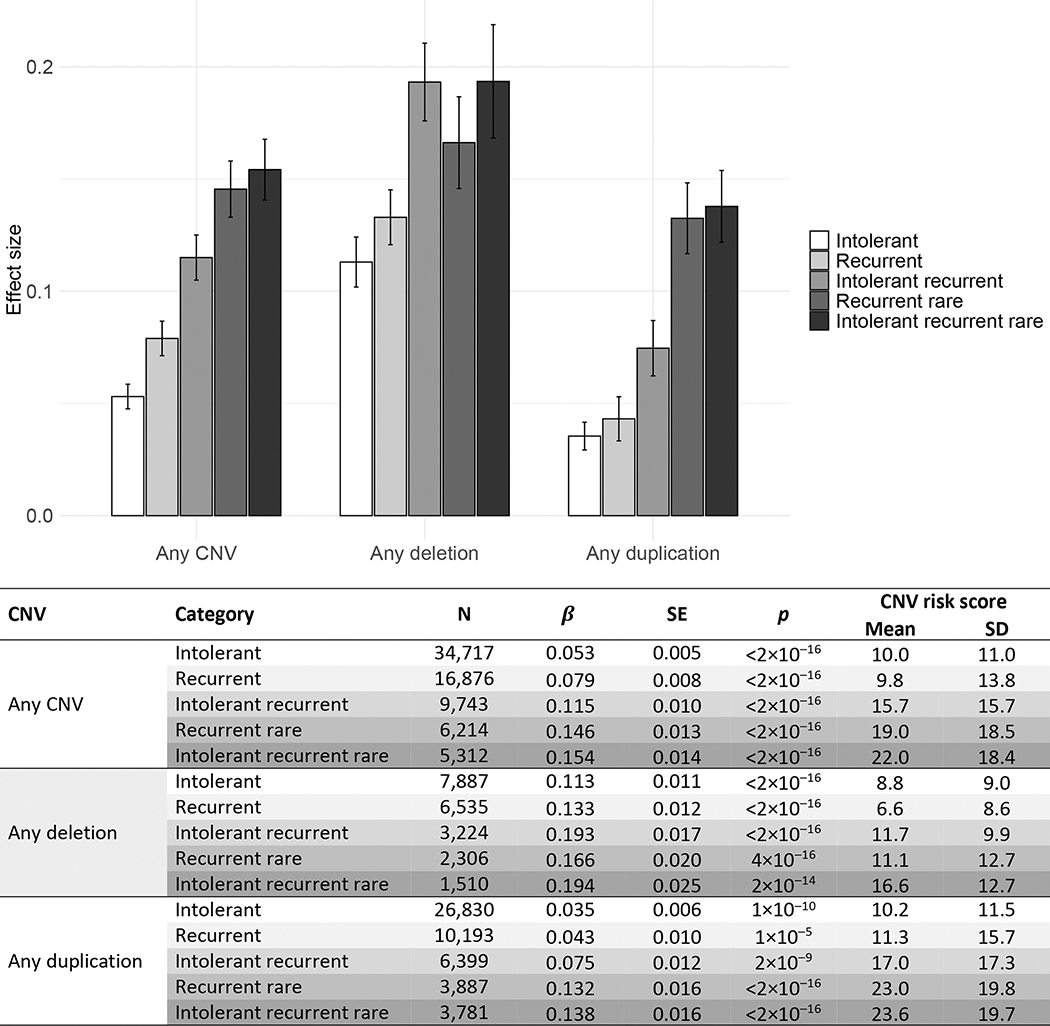
Figure 2 shows standardized mean differences in mood/anxiety factor scores between CNV carriers (intolerant; recurrent; intolerant recurrent; recurrent rare; intolerant recurrent rare) and non-carriers, as well as mean CNV risk scores (CRS) for all CNV categories. Overall, CNV carriers showed higher mood/anxiety scores than non-carriers, with very small to small effect sizes. Mood/anxiety scores were highest in individuals with intolerant recurrent rare deletions and lowest for individuals with intolerant duplications (Figure 2).
Figure 2.
Standardized mean differences between CNV carriers and non-carriers on mood/anxiety factor scores.
Error bars represent standard errors.
β coefficients correspond to standardized effect sizes, with values of 0.2, 0.5, and 0.8 indicating small, medium, and large effects.
Intolerant = CNV risk score (CRS)>2.86; Recurrent = any of the 61 neuropsychiatric recurrent CNVs; Intolerant recurrent = any of the 61 neuropsychiatric recurrent CNVs with a CRS>2.86; Recurrent rare = any of the 57 rare neuropsychiatric recurrent CNVs i.e., excluding the high frequency 2q13 (NPHP1), 15q11.2, 15q13.3 (CHRNA7), and ZNF92 CNVs; Intolerant recurrent rare = any of the 52 rare neuropsychiatric recurrent CNVs with a CRS>2.86 (i.e., excluding the high frequency CNVs listed above and the ZMYM5, CRYL1, VPS13B, 16q23.3, 17q21.31 CNVs with a CRS>2.86)
Results were similar when including only white British participants (eTable 4) and excluding participants with psychiatric diagnoses (eTable 5), such that CNV carriers showed higher mood/anxiety scores than non-carriers, with very small to small effect sizes.
Individual recurrent CNVs show small to large associations with mood/anxiety scores
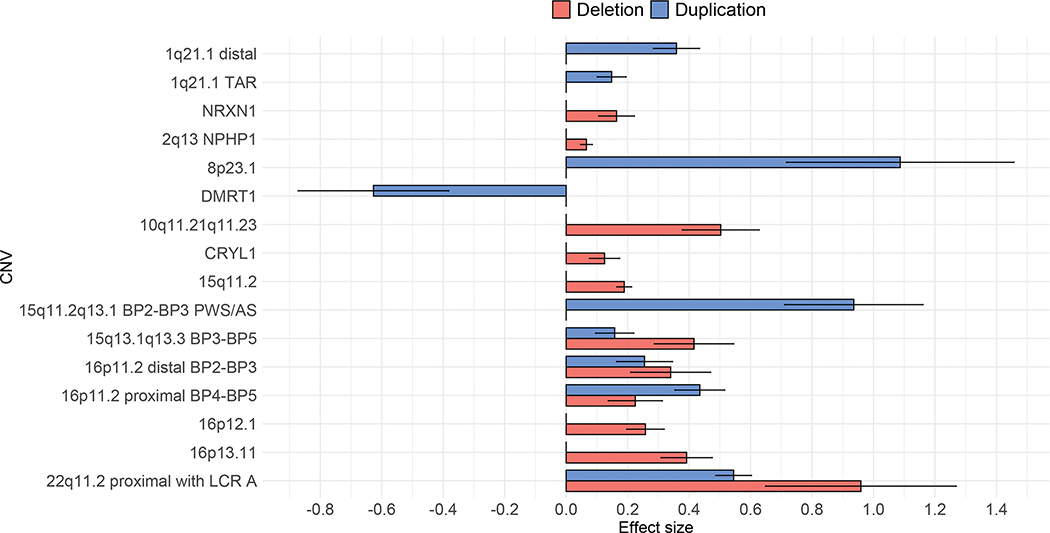
After FDR correction, 11 deletions (eTable 6) and 7 duplications (eTable 7) remained statistically significantly associated with higher mood/anxiety scores, with effect sizes ranging from small to large (Figure 3). Carriers of the DMRT1 duplication showed lower mood/anxiety scores, suggesting a protective effect of this CNV (Figure 3). Results were almost identical when including only individuals of white British ancestry (eTables 6–7). Excluding individuals with psychiatric diagnoses attenuated the large association seen for 22q11.2 deletion, resulting in an effect size of 0.29, which suggests a small subclinical effect (eTable 6). A total of 7 deletions and 7 duplications remained significantly associated with higher mood/anxiety scores when excluding individuals with psychiatric diagnoses. Interestingly, for CNVs no longer reaching statistical significance, effect sizes remained small to medium, suggesting that excluding individuals with psychiatric diagnoses may have reduced power to detect subclinical associations (eTables 6–7).
Figure 3.
Effect sizes and standard errors for CNVs with FDR corrected statistically significant associations with mood/anxiety factor scores
Effect sizes of 0.2, 0.5, and 0.8 indicate small, medium, and large effects, respectively. Error bars represent standard errors.
Psychiatric polygenic risk scores are positively associated with mood/anxiety scores
Major depressive disorder (MDD) (β=0.072, SE=0.002, pFDR<2×10−16) and generalized anxiety disorder (GAD) (β=0.069, SE=0.002, pFDR<2×10−16) polygenic risk scores (PRS) were positively associated with mood/anxiety scores.
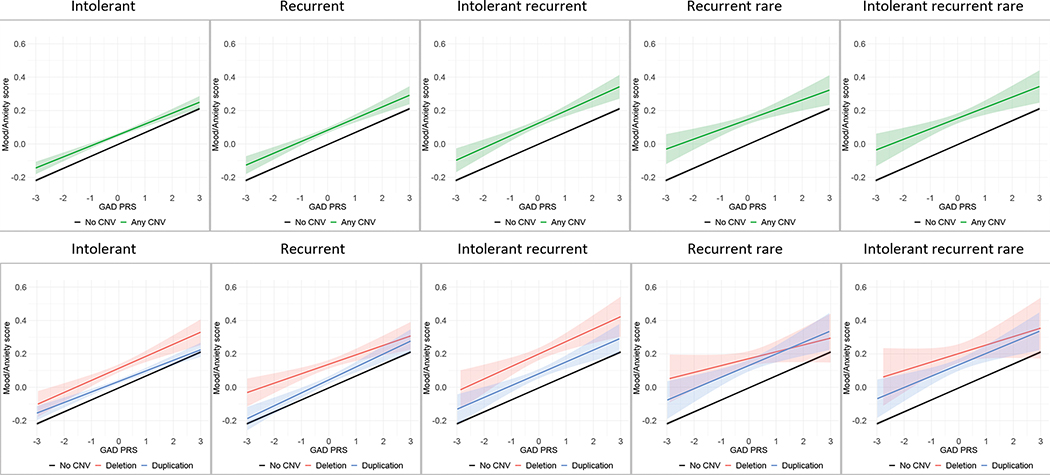
Aggregate CNVs do not show interactive effects with polygenic risk scores
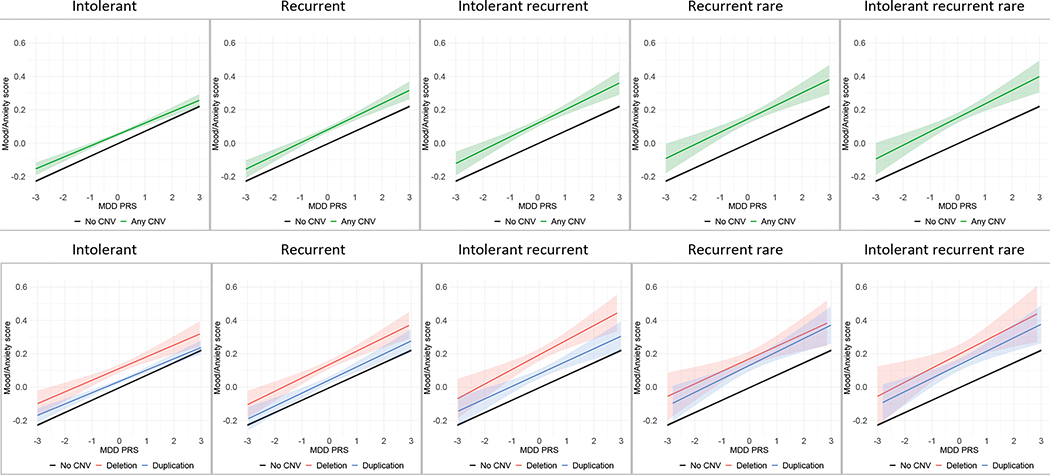
CNV×PRS interactions were not statistically significant for any of the CNVs categories (intolerant; recurrent; intolerant recurrent; recurrent rare; intolerant recurrent rare) or for either of the PRS (major depressive disorder or generalized anxiety disorder) (Table 1), suggesting that the effects of CNV burden and genetic background, as indexed by PRS, each independently influence mood/anxiety in an additive fashion (Figures 4 and 5).
Table 1.
CNV×PRS interactions on mood/anxiety factor scores
| CNV | Category | N | β | SE | P | |
|---|---|---|---|---|---|---|
| MDD PRS | Any CNV | Intolerant | 29,475 | −0.006 | 0.006 | 0.318 |
| Recurrent | 14,355 | 0.004 | 0.008 | 0.624 | ||
| Intolerant recurrent | 8,363 | 0.006 | 0.011 | 0.609 | ||
| Recurrent rare | 5,396 | 0.004 | 0.013 | 0.752 | ||
| Intolerant recurrent rare | 4,608 | 0.008 | 0.015 | 0.595 | ||
| Any deletion | Intolerant | 6,734 | −0.004 | 0.012 | 0.728 | |
| Recurrent | 5,666 | 0.006 | 0.013 | 0.677 | ||
| Intolerant recurrent | 2,808 | 0.013 | 0.019 | 0.489 | ||
| Recurrent rare | 1,980 | 0.001 | 0.022 | 0.971 | ||
| Intolerant recurrent rare | 1,284 | 0.010 | 0.027 | 0.705 | ||
| Any duplication | Intolerant | 22,741 | −0.007 | 0.007 | 0.322 | |
| Recurrent | 8,547 | 0.003 | 0.011 | 0.749 | ||
| Intolerant recurrent | 5,441 | 0.000 | 0.013 | 0.979 | ||
| Recurrent rare | 3,396 | 0.006 | 0.017 | 0.731 | ||
| Intolerant recurrent rare | 3,304 | 0.006 | 0.017 | 0.735 | ||
| GAD PRS | Any CNV | Intolerant | 29,475 | −0.006 | 0.006 | 0.324 |
| Recurrent | 14,355 | −0.002 | 0.008 | 0.835 | ||
| Intolerant recurrent | 8,363 | 0.002 | 0.011 | 0.868 | ||
| Recurrent rare | 5,396 | −0.012 | 0.014 | 0.370 | ||
| Intolerant recurrent rare | 4,608 | −0.008 | 0.015 | 0.588 | ||
| Any deletion | Intolerant | 6,734 | 0.001 | 0.012 | 0.953 | |
| Recurrent | 5,666 | −0.015 | 0.013 | 0.270 | ||
| Intolerant recurrent | 2,808 | 0.003 | 0.019 | 0.883 | ||
| Recurrent rare | 1,980 | −0.030 | 0.023 | 0.188 | ||
| Intolerant recurrent rare | 1,284 | −0.020 | 0.028 | 0.464 | ||
| Any duplication | Intolerant | 22,741 | −0.008 | 0.007 | 0.218 | |
| Recurrent | 8,547 | 0.006 | 0.011 | 0.590 | ||
| Intolerant recurrent | 5,441 | −0.001 | 0.014 | 0.953 | ||
| Recurrent rare | 3,396 | −0.002 | 0.017 | 0.886 | ||
| Intolerant recurrent rare | 3,304 | −0.004 | 0.017 | 0.832 |
β coefficients correspond to standardized effect sizes, with values of 0.2, 0.5, and 0.8 indicating small, medium, and large effects
CNV = copy number variant; PRS = polygenic risk score; MDD = major depressive disorder; GAD = generalized anxiety disorder
Figure 4.
CNV × major depressive disorder (MDD) PRS interactions on mood/anxiety factor scores
Intolerant = CNV risk score (CRS)>2.86; Recurrent = any of the 61 neuropsychiatric recurrent CNVs; Intolerant recurrent = any of the 61 neuropsychiatric recurrent CNVs with a CRS>2.86; Recurrent rare = any of the 57 rare neuropsychiatric recurrent CNVs i.e., excluding the high frequency 2q13 (NPHP1), 15q11.2, 15q13.3 (CHRNA7), and ZNF92 CNVs; Intolerant recurrent rare = any of the 52 rare neuropsychiatric recurrent CNVs with a CRS>2.86 (i.e., excluding the high frequency CNVs listed above and the ZMYM5, CRYL1, VPS13B, 16q23.3, 17q21.31 CNVs with a CRS>2.86)
Figure 5.
CNV × generalized anxiety disorder (GAD) PRS interactions on mood/anxiety factor scores
Intolerant = CNV risk score (CRS)>2.86; Recurrent = any of the 61 neuropsychiatric recurrent CNVs; Intolerant recurrent = any of the 61 neuropsychiatric recurrent CNVs with a CRS>2.86; Recurrent rare = any of the 57 rare neuropsychiatric recurrent CNVs i.e., excluding the high frequency 2q13 (NPHP1), 15q11.2, 15q13.3 (CHRNA7), and ZNF92 CNVs; Intolerant recurrent rare = any of the 52 rare neuropsychiatric recurrent CNVs with a CRS>2.86 (i.e., excluding the high frequency CNVs listed above and the ZMYM5, CRYL1, VPS13B, 16q23.3, 17q21.31 CNVs with a CRS>2.86).
Effects of select individual recurrent CNVs may be modulated by polygenic risk scores
CNV×PRS interactions between individual recurrent deletions and duplications and major depressive disorder (MDD) polygenic risk scores (PRS) are presented in eTables 8 and 9, respectively. Nominally significant CNV×PRS interactions were seen for 15q13.1q13.3 and 15q13.3 (CHRNA7) deletions, and 22q11.2 duplications. Specifically, carriers of 15q13.1q13.3 deletions and 22q11.2 duplications with low MDD PRS showed higher mood/anxiety scores than carriers of these CNVs with high MDD PRS, in contrast to non-carriers, where PRS was positively associated with mood/anxiety. On the other hand, carriers of the 15q13.3 (CHRNA7) deletion with high MDD PRS showed even higher mood/anxiety scores than non-carriers with high PRS (eFigure 2). However, these CNV×PRS interactions did not survive FDR correction.
CNV×PRS interactions between individual recurrent deletions and duplications and generalized anxiety disorder (GAD) PRS are presented in eTables 10 and 11, respectively. Nominally significant CNV×PRS interactions were seen for 7q11.23 and 15q13.3 duplications, and PAFAH1B1 deletions, whereby carriers of these CNVs with low GAD PRS showed higher mood/anxiety scores than carriers of these CNVs with high GAD PRS, in contrast to non-carriers, where PRS were positively associated with mood/anxiety (eFigure 2). Again, these CNV×PRS interactions did not survive FDR correction.
Discussion
Using data from nearly half a million individuals from a population-based cohort, we found associations between copy number variants (CNVs) and mood/anxiety factor scores, even when excluding individuals with psychiatric diagnoses. Effect sizes were largest for recurrent deletions encompassing genes intolerant to haploinsufficiency. Copy number variant-by-polygenic risk score (CNV×PRS) interactions were not significant when examining CNVs in aggregate, but preliminary evidence was found for select individual recurrent CNVs, such that associations with mood/anxiety scores may be modulated by genetic background indexed by the aggregate effects of common variants. Specifically, nominally significant CNV×PRS interactions suggested that differences in mood/anxiety scores between non-carriers and carriers of these select recurrent CNVs may also differ by major depressive disorder (MDD) and generalized anxiety disorder (GAD) polygenic risk score (PRS). These findings advance our understanding of the genetic underpinnings of psychopathology in several ways.
First, we used item-level data from mental health questionnaires to derive dimensional factor scores indexing mood/anxiety symptomatology, finding small to large associations with CNVs. These findings are in line with previous evidence for substantial associations between CNVs and risk for psychiatric disorders.(11, 15, 19) Our findings of higher mood/anxiety scores in carriers of Prader-Willi / Angelman syndrome and 16p11.2 duplications are directly in line with a previous UK Biobank study on CNVs and depression.(19) We also found significant effects of 6 additional duplications and 11 deletions. Our findings also advance knowledge by showing that CNVs associations manifest across the spectrum of symptom severity, not only at the most severe end of disease. Associations between 14 recurrent deletions and duplications remained significant when excluding individuals with psychiatric diagnoses, and for CNVs no longer reaching statistical significance, effect sizes remained small to medium. Overall, our findings demonstrate the utility of dimensional psychiatric phenotypes to examine CNV associations. Future studies that can derive factors measuring other domains of psychiatric symptomatology are needed to extend these findings. Such studies may also determine the specificity of CNV associations, namely whether the same CNVs are associated with different symptom profiles and severities.
Second, we found preliminary evidence for nominally significant CNV×PRS interactions. Specifically, carriers of the 15q13.3 (CHRNA7) deletion with high MDD PRS showed nominally higher mood/anxiety scores than non-carriers with high MDD PRS. On the other hand, carriers of 15q13.1q13.3 deletions and 22q11.2 duplications with low MDD PRS showed nominally higher mood/anxiety scores than carriers of these CNVs with high MDD PRS, in contrast to non-carriers, where PRS was positively associated with mood/anxiety. Similarly, carriers of 7q11.23 and 15q13.3 duplications, and PAFAH1B1 deletions with low GAD PRS showed nominally higher mood/anxiety scores than carriers of these CNVs with high GAD PRS, while in non-carriers PRS was positively associated with mood/anxiety. However, these preliminary, nominally significant findings warrant replication because sample sizes for most of these CNVs were small (N<60). Indeed, efforts to delineate the combined effects of rare and common variants on neuropsychiatric phenotypes using large-scale datasets are already underway(26–28).
While there is considerable evidence for the role of both common and rare genetic variants on psychiatric disorders, their joint effects on disease risk remain relatively unexamined. Moreover, the few studies that have investigated both common and rare variants have focused on neurodevelopmental disorders, namely schizophrenia,(30, 53, 54) autism spectrum disorders (ASD),(55) and attention deficit hyperactivity disorder (ADHD).(29, 55, 56). Thus, our preliminary findings of nominally significant CNV×PRS interactions are, to the best of our knowledge, the first to suggest these joint effects on dimensional scores of mood/anxiety, as well as in a population-based sample. Determining the molecular mechanisms underlying interactive effects of common and rare genetic is beyond the scope of this study, although several models of epistasis have been proposed(31). Nevertheless, our preliminary, nominally significant finding that, for most CNVs, PRS was negatively associated with mood/anxiety scores points to a complex interrelationship, possibly involving both amplification and attenuation of genes.
The utility of PRS for clinically relevant risk stratification in CNV carriers has been demonstrated in 22q11.2 deletion syndrome, where PRS are associated with clinical outcomes, (32) as well as cognitive functioning.(57) Our results build on previous findings by suggesting that associations between rare variants and psychopathology may be modulated by common variants, implying that PRS may explain some of the clinical and phenotypic variation observed among carriers of the same recurrent CNVs. However, these CNV×PRS interactions also suggest that PRS risk algorithms developed from standard GWAS populations, which likely include few CNV carriers, may show reduced prediction accuracy in CNV carriers, underestimating or overestimating the effect of high PRS in these individuals. Moreover, it is important to note that sample sizes for most CNVs were small (N<60). Thus, further work in even larger samples is needed to examine CNV×PRS interactions on psychopathology, as well as other medical outcomes(26–28), to determine the exact nature of the relationship between rare and common genetic variation. For example, joint effects of CNVs and PRS could also be the sum of interaction and conditional effects and should be tested in future studies with larger samples.
Moreover, while CNV×PRS interactions were not significant when examining CNVs in aggregate, we found preliminary evidence to suggest that associations between select individual recurrent CNVs and mood/anxiety may be modulated by PRS. These findings further highlight the importance of looking beyond aggregate effects of CNVs to examine individual recurrent CNVs. Nevertheless, our finding of small, yet robust, associations between aggregate CNVs and mood/anxiety are in line with our previous work showing associations between CNV risk scores (CRS) and autism spectrum disorder (ASD) risk (58), and cognitive functioning (36), as well as psychopathology, and brain structure (59). Moreover, given that the rarity of many CNVs makes it difficult to examine individual associations, CRS are a useful preliminary analytic tool. However, the exact interrelationship between common and rare variants will invariably depend on the specific variants under study, as well as the outcome, and may even vary by individual. Thus, future studies with even larger sample sizes, including other CNVs and PRS, are needed(26–28).
This study has limitations. While the use of a dimensional, data-driven mood/anxiety score increased power to detect CNVs associations, we were still underpowered to test many CNVs due to their rarity. Nevertheless, we found significant positive associations between 19 individual deletions and duplications and mood/anxiety. Second, PRS could only be calculated in White British individuals. The lack of racial diversity in psychiatric and genetics research is a larger issue that is not unique to our study, but the exclusion of other ancestry groups limits generalizability of our findings.
Supplementary Material
KEY RESOURCES TABLE.
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | ||||
| Bacterial or Viral Strain | ||||
| Biological Sample | ||||
| Cell Line | ||||
| Chemical Compound or Drug | ||||
| Commercial Assay Or Kit | ||||
| Deposited Data; Public Database | ||||
| Genetic Reagent | ||||
| Organism/Strain | ||||
| Peptide, Recombinant Protein | ||||
| Recombinant DNA | ||||
| Sequence-Based Reagent | ||||
| Software; Algorithm | R | http://www.R-project.org | ||
| Transfected Construct | ||||
| Other |
Acknowledgements
This work used the UK Biobank health resource under Application Number 40980 and was funded by the National Institute of Mental Health (NIMH) grant number U01-MH119690.
Footnotes
Disclosures
All authors reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, et al. (2006): Copy number variation: new insights in genome diversity. Genome research. 16:949–961. [DOI] [PubMed] [Google Scholar]
- 2.Zarrei M, MacDonald JR, Merico D, Scherer SW (2015): A copy number variation map of the human genome. Nature reviews genetics. 16:172–183. [DOI] [PubMed] [Google Scholar]
- 3.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, et al. (2010): Diversity of human copy number variation and multicopy genes. Science. 330:641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glessner JT, Li J, Wang D, March M, Lima L, Desai A, et al. (2017): Copy number variation meta-analysis reveals a novel duplication at 9p24 associated with multiple neurodevelopmental disorders. Genome medicine. 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merikangas AK, Corvin AP, Gallagher L (2009): Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends in genetics. 25:536–544. [DOI] [PubMed] [Google Scholar]
- 6.Asadollahi R, Oneda B, Joset P, Azzarello-Burri S, Bartholdi D, Steindl K, et al. (2014): The clinical significance of small copy number variants in neurodevelopmental disorders. Journal of Medical Genetics. 51:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. (2007): Strong association of de novo copy number mutations with autism. Science. 316:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. (2010): Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Council on Children With D, Section on Developmental Behavioral P, Bright Futures Steering C, Medical Home Initiatives for Children With Special Needs Project Advisory C (2006): Identifying infants and young children with developmental disorders in the medical home: an algorithm for developmental surveillance and screening. Pediatrics. 118:405–420. [DOI] [PubMed] [Google Scholar]
- 10.Hyman SL, Levy SE, Myers SM, Council On Children With Disabilities SOD, Behavioral P (2020): Identification, Evaluation, and Management of Children With Autism Spectrum Disorder. Pediatrics. 145. [DOI] [PubMed] [Google Scholar]
- 11.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. (2017): Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL, et al. (2014): The penetrance of copy number variations for schizophrenia and developmental delay. Biological psychiatry. 75:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy KC (2002): Schizophrenia and velo-cardio-facial syndrome. The Lancet. 359:426–430. [DOI] [PubMed] [Google Scholar]
- 14.Tsuang MT, Stone WS, Faraone SV (1999): Schizophrenia: a review of genetic studies. Harvard Review of Psychiatry. 7:185–207. [PubMed] [Google Scholar]
- 15.Green E, Rees E, Walters J, Smith K, Forty L, Grozeva D, et al. (2016): Copy number variation in bipolar disorder. Molecular psychiatry. 21:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charney AW, Stahl EA, Green EK, Chen C-Y, Moran JL, Chambert K, et al. (2019): Contribution of rare copy number variants to bipolar disorder risk is limited to schizoaffective cases. Biological psychiatry. 86:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozeva D, Kirov G, Conrad DF, Barnes CP, Hurles M, Owen MJ, et al. (2013): Reduced burden of very large and rare CNV s in bipolar affective disorder. Bipolar disorders. 15:893–898. [DOI] [PubMed] [Google Scholar]
- 18.Bergen S, O’dushlaine C, Ripke S, Lee P, Ruderfer D, Akterin S, et al. (2012): Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Molecular psychiatry. 17:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendall KM, Rees E, Bracher-Smith M, Legge S, Riglin L, Zammit S, et al. (2019): Association of Rare Copy Number Variants With Risk of Depression. JAMA psychiatry. 76:818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Dushlaine C, Ripke S, Ruderfer DM, Hamilton SP, Fava M, Iosifescu DV, et al. (2014): Rare copy number variation in treatment-resistant major depressive disorder. Biological psychiatry. 76:536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, et al. (2018): Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 173:1705–1715. e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. (2019): Identification of common genetic risk variants for autism spectrum disorder. Nature genetics. 51:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. (2019): Genome-wide association study identifies 30 loci associated with bipolar disorder. Nature genetics. 51:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. (2019): Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature neuroscience. 22:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, et al. (2019): International meta-analysis of PTSD genome-wide association studies identifies sex-and ancestry-specific genetic risk loci. Nature communications. 10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquemont S, Huguet G, Klein M, Chawner S, Donald KA, van den Bree MBM, et al. (2022): Genes To Mental Health (G2MH): A Framework to Map the Combined Effects of Rare and Common Variants on Dimensions of Cognition and Psychopathology. The American journal of psychiatry. 179:189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein M, Glahn D, Kendall K (2022): COPY NUMBER VARIATION AND THEIR INTERACTION WITH POLYGENIC RISK SCORES IN PSYCHIATRIC DISORDERS. European Neuropsychopharmacology. 63:e40–e41. [Google Scholar]
- 28.Sebat J (2022): CHARACTERIZATION OF THE COMBINED EFFECTS OF RARE VARIANTS AND POLYGENIC RISK BY WHOLE GENOME ANALYSIS OF PSYCHIATRIC DISORDERS AND QUANTITATIVE TRAITS. European Neuropsychopharmacology. 63:e315. [Google Scholar]
- 29.Martin J, O’Donovan MC, Thapar A, Langley K, Williams N (2015): The relative contribution of common and rare genetic variants to ADHD. Transl Psychiatry. 5:e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi S, Ninomiya K, Kushima I, Saito T, Shimasaki A, Sakusabe T, et al. (2020): Polygenic risk scores in schizophrenia with clinically significant copy number variants. Psychiatry and clinical neurosciences. 74:35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner B (2011): Molecular mechanisms of epistasis within and between genes. Trends in Genetics. 27:323–331. [DOI] [PubMed] [Google Scholar]
- 32.Cleynen I, Engchuan W, Hestand MS, Heung T, Holleman AM, Johnston HR, et al. (2021): Genetic contributors to risk of schizophrenia in the presence of a 22q11.2 deletion. Molecular psychiatry. 26:4496–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. (2007): PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome research. 17:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, et al. (2007): QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic acids research. 35:2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huguet G, Schramm C, Douard E, Jiang L, Labbe A, Tihy F, et al. (2018): Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA psychiatry. 75:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huguet G, Schramm C, Douard E, Tamer P, Main A, Monin P, et al. (2021): Genome-wide analysis of gene dosage in 24,092 individuals estimates that 10,000 genes modulate cognitive ability. Molecular psychiatry.1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. (2015): Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. (2011): A copy number variation morbidity map of developmental delay. Nature genetics. 43:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coe BP, Witherspoon K, Rosenfeld JA, Van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. (2014): Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature genetics. 46:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-De-Luca D, Sanders S, Willsey A, Mulle J, Lowe J, Geschwind D, et al. (2013): Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Molecular psychiatry. 18:1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. (2014): CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 505:361–366. [DOI] [PubMed] [Google Scholar]
- 42.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, et al. (2019): GENCODE reference annotation for the human and mouse genomes. Nucleic acids research. 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. (2020): The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz LM, Merikangas AK, Ruparel K, Jacquemont S, Glahn DC, Gur RE, et al. (2021): Stability of Polygenic Scores Across Discovery Genome-Wide Association Studies. bioRxiv.2021.2006.2018.449060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. (2016): Next-generation genotype imputation service and methods. Nature genetics. 48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW (2019): Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature communications. 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, et al. (2020): Reproducible genetic risk loci for anxiety: results from~ 200,000 participants in the Million Veteran Program. American Journal of Psychiatry. 177:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. (2021): Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in> 1.2 million individuals highlight new therapeutic directions. Nature neuroscience. 24:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reise SP, Moore TM, Haviland MG (2010): Bifactor models and rotations: Exploring the extent to which multidimensional data yield univocal scale scores. Journal of personality assessment 92:544–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muthén L, Muthén B (2015): Mplus - The comprehensive modelling program for applied researchers: user’s guide. 5. [Google Scholar]
- 51.Team RC (2013): R: A language and environment for statistical computing.
- 52.Benjamini Y, Yekutieli D (2001): The control of the false discovery rate in multiple testing under dependency. Annals of statistics.1165–1188. [Google Scholar]
- 53.Bergen SE, Ploner A, Howrigan D, Group CA, Consortium tSWGotPG, O’Donovan MC, et al. (2019): Joint contributions of rare copy number variants and common SNPs to risk for schizophrenia. American Journal of Psychiatry. 176:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tansey KE, Rees E, Linden D, Ripke S, Chambert K, Moran J, et al. (2016): Common alleles contribute to schizophrenia in CNV carriers. Molecular psychiatry. 21:1085–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LaBianca S, LaBianca J, Pagsberg AK, Jakobsen KD, Appadurai V, Buil A, et al. (2021): Copy Number Variants and Polygenic Risk Scores Predict Need of Care in Autism and/or ADHD Families. Journal of autism and developmental disorders. 51:276–285. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Neale BM, Liu L, Lee SH, Wray NR, Ji N, et al. (2013): Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: Genome-wide association study of both common and rare variants. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 162:419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies RW, Fiksinski AM, Breetvelt EJ, Williams NM, Hooper SR, Monfeuga T, et al. (2020): Using common genetic variation to examine phenotypic expression and risk prediction in 22q11. 2 deletion syndrome. Nature medicine. 26:1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douard E, Zeribi A, Schramm C, Tamer P, Loum MA, Nowak S, et al. (2021): Effect sizes of deletions and duplications on autism risk across the genome. American Journal of Psychiatry. 178:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander-Bloch A, Huguet G, Schultz LM, Huffnagle N, Jacquemont S, Seidlitz J, et al. (2022): Copy Number Variant Risk Scores Associated With Cognition, Psychopathology, and Brain Structure in Youths in the Philadelphia Neurodevelopmental Cohort. JAMA psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.