Abstract
Aims
Components of bone morphogenetic protein (BMP) signalling have been implicated in both pathogenesis of pulmonary arterial hypertension (PAH) and endothelial-mesenchymal transition (EndoMT). In particular, the importance of BMP type 2 receptor in these processes has been extensively analysed. However, the contribution of BMP type 1 receptors (BMPR1s) to the onset of PAH and EndoMT remains poorly understood. BMPR1A, one of BMPR1s, was recently implicated in the pathogenesis of PAH, and was found to be down-regulated in the lungs of PAH patients, neither the downstream mechanism nor its contribution to EndoMT has been described. Therefore, we aim to delineate the role of endothelial BMPR1A in modulating EndoMT and pathogenesis of PAH.
Methods and results
We find that BMPR1A knockdown in endothelial cells (ECs) induces hallmarks of EndoMT, and deletion of endothelial Bmpr1a in adult mice (Bmpr1aiECKO) leads to development of PAH-like symptoms due to excessive EndoMT. By lineage tracing, we show that endothelial-derived smooth muscle cells are increased in endothelial Bmpr1a-deleted mice. Mechanistically, we identify ZEB1 as a primary target for BMPR1A in this setting; upon BMPR1A activation, ID2 physically interacts and sequesters ZEB1 to attenuate transcription of Tgfbr2, which in turn lowers the responses of ECs towards transforming growth factor beta (TGFβ) stimulation and prevents excessive EndoMT. In Bmpr1aiECKO mice, administering endothelial targeting lipid nanoparticles containing siRNA against Tgfbr2 effectively ameliorate PAH, reiterating the importance of BMPR1A-ID2/ZEB1-TGFBR2 axis in modulating progression of EndoMT and pathogenesis of PAH.
Conclusions
We demonstrate that BMPR1A is key to maintain endothelial identity and to prevent excessive EndoMT. We identify BMPR1A-induced interaction between ID2 and ZEB1 is the key regulatory step for onset of EndoMT and pathogenesis of PAH. Our findings indicate that BMPR1A-ID2/ZEB1-TGFBR2 signalling axis could serve as a potential novel therapeutic target for PAH and other EndoMT-related vascular disorders.
Keywords: EndoMT, Pulmonary arterial hypertension, Vascular remodelling, BMPR1A, BMP signalling
Graphical Abstract
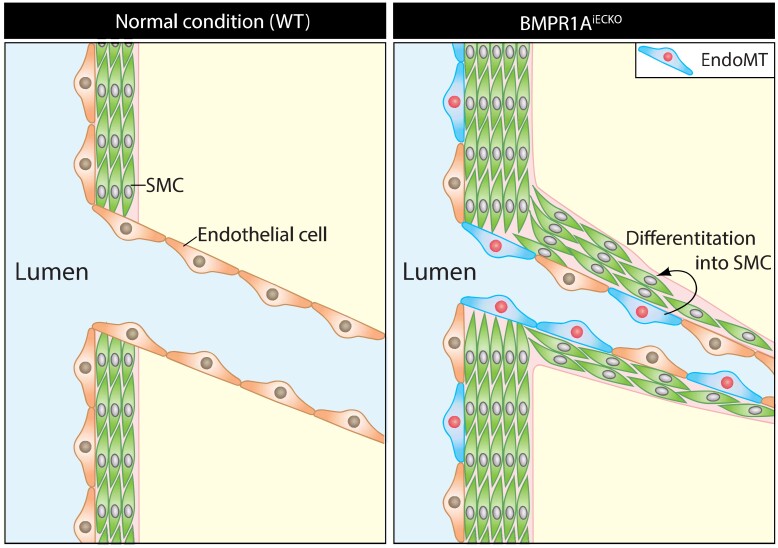
Graphical abstract.
Time of primary review: 53 days
1. Introduction
Pulmonary arterial hypertension (PAH) is a progressive disease with characteristic clinical hallmarks of excessive proliferation of pulmonary vascular cells and remodelling of distal pulmonary arterioles, which collectively leads to the obliteration of pulmonary arteries, increased pulmonary vascular resistance, and right ventricular failure.1–3 Dysregulation of bone morphogenetic protein (BMP) signalling has been implicated as a leading cause of PAH. In particular, mutations in BMP type 2 receptor (BMPR2), the obligatory component of BMP receptor complex, have been identified in both hereditary and idiopathic PAH patients, illustrating the importance of BMPR2 in the pathogenesis of PAH.4 In addition to BMPR2, recent studies in human subjects have implicated BMP receptor type 1A (BMPR1A/ALK3) in the pathogenesis of PAH. BMPR1A expression was found to be significantly decreased in the lungs of PAH patients.5 In addition, deleterious variants in BMPR1A were recently identified in four cases of PAH.6 Studies in mice, however, have not yielded a demonstrable role for BMPR1A in pulmonary vascular remodelling, in part due to embryonic lethality seen in the Bmpr1a−/− mice.7 Moreover, previous studies using BMPR1A deletion mice in smooth muscle cells led to no significant change in pulmonary pressure after chronic hypoxia exposure.8,9 Therefore, the contribution of endothelial BMPR1A on the pulmonary vasculature remains unknown.
In PAH, the vascular remodelling is multifactorial, including proliferation and differentiation of pre-existing vascular smooth muscle cell progenitors,10 and endothelial to mesenchymal transition (EndoMT) that may lead to loss of endothelial barrier function and further expansion of these cells into smooth muscle cell-like cells.11 Multiple mechanisms for EndoMT have been described to date, including loss of BMPR2 that promotes HMGA1 expression, and increased expression of Twist1 in pulmonary artery ECs (PAECs).12 EndoMT is a cellular trans-differentiation process reminiscent of epithelial to mesenchymal transition (EMT). EndoMT occurs when homeostatic signalling cascades that preserve the endothelial identity are disrupted.13–15 During EndoMT, hallmarks of endothelial characteristics such as expression of VE-Cadherin (CDH5) and stereotypic cellular morphology are lost in ECs with a concomitant gain of mesenchymal or fibroblast-like characteristics, including expression of α smooth muscle actin (αSMA) and type I collagen.16 While EndoMT is part of the physiological developmental process such as cardiac development,17,18 ectopic EndoMT has been implicated in a number of diseases in humans.
Mechanistically, it has been proposed that EndoMT is augmented by transforming growth factor (TGFβ) signalling, as shown in EMT.19,20 More recently, BMP signalling has also been closely tied to EndoMT in various vascular contexts.21,22 However, detailed molecular mechanism whereby BMP signalling promotes homeostasis of ECs and modulates EndoMT in the pulmonary vasculature remains to be fully defined. In this study, we analysed the role of BMPR1A in ECs using an inducible endothelial-specific Bmpr1a deletion mice (Bmpr1aiECKO) and cultured ECs, and found that attenuation of BMPR1A predisposed the onset of PAH-like symptoms in mice. Based on our data, we propose that BMPR1A is essential for the maintenance of endothelial fate by modulating Tgfbr2 expression in an ID2/ZEB1-dependent manner, and suggest the interface of ID2–ZEB1 interaction could serve as a novel target for PAH and other related diseases in humans.
2. Methods
2.1. Mice strains and animal husbandry
The Cdh5(PAC)CreERT2 transgenic mice (iEC Cre) were a kind gift from R. H. Adams.23 Bmpr1afl/fl mice have been previously described.24,25 The Rosa26mT/mG Cre reporter line26 was obtained from Jackson Laboratories (Stock number: 007676). All experimental animals were maintained on a C57BL/6J genetic background. Both male and female mice were used for the experiments, and littermate Cre-negative mice were used as control group. The experiments were conducted according to NIH guidelines on the use of laboratory animals, and the research protocols were approved by the Yale University IACUC. To induce Cre activity, fully adult mice (at least 2 months old) were injected with tamoxifen (2 mg/day) via intraperitoneal injection for 5 consecutive days. All mice after experiment were euthanized with carbon dioxide in a closed chamber of cervical dislocation method.
2.2. Right-ventricular systolic pressure measurement and hypertrophy
Right-ventricular systolic pressure (RVSP) measurements were performed at the designated time points after tamoxifen injection under isoflurane anaesthesia (3% for induction and 1% for maintenance of inhalation; generic name: Isothesia) by inserting a catheter (Millar Instruments, Houston, TX) into the right jugular vein as described previously.27 After RSVP measurement, mice were sacrificed and the heart was carefully dissected with micro-scissors under microscope to assess the weight ratio of the right ventricle/(left ventricles + septum).
2.3. Tissue preparation
For frozen sections, mice were anaesthetized by isoflurane inhalation (3%, Isothesia) and the lungs were flushed with phosphate-buffered saline (PBS) and 4% paraformaldehyde in PBS solution by inserting a needle connected to a reservoir into the right ventricle. Isolated lungs were fixed in freshly made 4% paraformaldehyde overnight at 4°C, rinsed with PBS at room temperature, incubated in 15% sucrose overnight at 4°C, and transferred to 30% sucrose at 4°C until the tissue sank. Fixed tissues were infiltrated with Tissue-Tek OCT embedding medium for 30 min at room temperature, transferred to an embedding mould filled with OCT, frozen on dry ice, and stored at −80°C. Frozen sections (10 μm thick) were cut at −20°C using Leica CM1510S-3 Cryostat (Leica, Wetzlar, Germany), and slides were kept at −80°C until used for immunostaining.
For vibratome sections, mice were anaesthetized by isoflurane inhalation (Isothesia), and the lungs were flushed with PBS by inserting a needle connected to a reservoir into the right ventricle. Animals were then tracheostomized with an angiocatheter, and the lungs were inflated with 2% low-melt agarose in PBS. Low-melt agarose-filled lungs were solidified in ice-cold PBS for 10 min. Then, lobes were separated, fixed in Dent’s fixative (4:1 methanol:DMSO) at 4°C for overnight, and rehydrated with PBS. Rehydrated lungs were cut using Leica VT1000 S vibratome (150 μm thickness) and incubated with PBS-T for 30 min at 37°C to remove residual agarose.
2.4. Immunohistochemistry and image analyses
In brief, the sections were permeabilized in ice-cold methanol for 10 min at −20°C and briefly air dried. OCT compound for cryosection was removed with flowing tap water. The specimens were first incubated in blocking buffer [1% foetal bovine serum (FBS), 3% bovine serum albumin, 0.5% Triton-X100, 0.01% sodium deoxycholate in Tris-buffered saline (TBS)] for 30 min at room temperature and subsequently incubated in blocking buffer containing antibodies overnight at 4°C. The next day, specimens were washed three times with 0.5% Triton-X100 in TBS (10 min/wash), incubated in secondary antibody at room temperature for 1 h. After they were washed three times with 0.5% Triton-X100 in the dark (10 min/wash), the specimens were mounted on glass slides with anti-fading mounting medium [10% 0.1 M Tris buffer (pH 9.0), 0.2 mg/mL P-phenylenediamine hydrochloride, and 4 mM sodium azide in glycerol]. Confocal microscopy was performed with Nikon ECLIPSE 80i fluorescence microscope, Leica spinning disk confocal microscope or a Leica SP5 confocal microscope. ImageJ (NIH) was used for the data analysis.
The following antibodies were used: anti-CD31 (1:100; BD), anti-αSMA (1:100; Sigma, St. Louis, MO), anti-SM22α (1:100; Abcam, Cambridge, UK), anti-green fluorescent protein (GFP; 1:100; Aveslab, Davis, CA), anti-FSP-1 (1:100; Calbiochem, St. Louis, MO), and anti-VE-cadherin (1:100; SantaCruz, Dallas, TX). Alexa Fluor 488, 555, and 647 donkey secondary antibodies were from Invitrogen. The histological samples for haematoxylin and eosin (H&E) staining were sectioned (10 μm thickness), mounted on slides, and stained with H&E staining kit (Vector Laboratories, Burlingame, CA). Elongation index for morphologic analysis of cultured cells was calculated as follows:
Elongation index = The longest axis of cell (a)/The shortest axis of cell (b).
2.5. Cell culture and siRNA treatment
Pulmonary artery ECs were purchased from ATCC (PCS-100-022). PAECs were grown on plates coated with 2% gelatin in EGM-2 media (Lonza, Basel, Switzerland) and used at passages between 2 and 7. Primary mouse lung endothelial cells (mLECs) were isolated from lungs of Bmpr1afx/fx (P5–7) mice. Briefly, dissected lungs were chopped with sterile scissors and digested in a solution of 2 mg/mL collagenase type I (Sigma-Aldrich, St. Louis, MO) in DMEM media (Invitrogen) containing 5% FBS, 100 IU/mL penicillin, and 100 mg/mL streptomycin for 1 h at 37°C. Thereafter, the cell suspension was immunomagnetically sorted in magnetic field using CD31 antibodies conjugated with dynabeads (Thermo Fischer Scientific, Waltham, MA). CD31+ sorted cells were seeded in EGM2 media. To delete Bmpr1a, control and Cre-expressing adenovirus were treated into isolated ECs.
For siRNA administration, transfection of siRNAs and plasmids were performed using Lipofectamine 2000 reagent (Invitrogen, Waltham, MA). siRNAs targeting BMPR1A (ID: s281), ID1 (ID: 11072), ID2 (ID: 11259), ID3 (ID: 115263), ZEB1 (ID: 109651), TCF3 (ID: s224712), and transforming growth factor beta receptor 2 (TGFBR2; ID: s14077) were purchased from Invitrogen (Silencer Select siRNA). Transfection efficiency was analysed by reverse transcriptase-polymerase chain reaction (RT–PCR) and Western blot at 48 h after transfection.
2.6. Western blot
The following primary antibodies were used for western blot: SM22α (Abcam, Cambridge, UK); αSMA (Sigma-Aldrich, St. Louis, MO); Flag (Sigma-Aldrich, St. Louis, MO); HA (Sigma-Aldrich, St. Louis, MO); pSMAD2 (Cell Signaling, Danvers, MA); pSMAD3 (Abcam, Cambridge, UK); totalSMAD2/3 (Cell Signalling); mouse TGFBR2 (SantaCruz); human TGFBR2 (SantaCruz); ID1 (Abcam, Cambridge, UK); ID2 (SantaCruz); ID3 (Abcam, Cambridge, UK); β-actin (SantaCruz); Zeb1 (Cell Signaling, Danvers, MA); and BMPR1A (MyBioSource, San Diego, CA). The secondary antibodies used were anti-mouse, rabbit, or goat Peroxidase AffiniPure antibodies from the Jackson Lab (catalogue: 715-035-150, 111-035-144, and 705-035-003, respectively).
2.7. Microarray data analysis
Data set associated with previously published report was used to determine expression level of BMPR1A in lung tissue specimens.28 An Agilent feature ID number (A_23_P1431) corresponding to BMPR1A was identified and extracted for comparison of BMPR1A expression in the two cohorts (control and PAH).
2.8. Plasmids and adenoviral constructs
Expression construct for Cre recombinase (addgene ID: #16583), TGFBR2 (addgene ID: #11766) was obtained from Addgene,29 and human ZEB1 and IDs construct was cloned from HUVEC cDNA. To generate the adenovirus, AdEasy™ Adenoviral Vector System (Agilent Technologies, Santa Clara, CA) was used. Each expression constructs were inserted into pShuttle-CMV vector. After DNA purification, those vectors were digested and linearized by Pme-I enzyme and electronically transformed into BJ5183 competent cells with pAd-Easy vector for homologous recombination. After colony screening, the final adenoviral vectors were linearized with Pac-I and transfected into HEK293A cells for viral packaging. Virus was titred using an Adeno-X Rapid Titer Kit (Clontech Laboratories, Mountain View, CA). The constructs were verified by sequencing analysis. Protein expression was confirmed by immunoblot analysis and infection efficiency was tested using immunostaining.
2.9. RNA extraction and real-time RT–PCR analysis
Total RNA was extracted from the samples using RNeasy mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions, and 1 μg of the RNA was reverse transcribed into cDNA using high capacity cDNA reverse transcription kit (Thermo Fisher). Quantitative RT–PCR was performed with FG Power SYBR Green PCR Master Mix (Thermofisher) and specific primers for each mRNA using Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The RT–PCR data were analysed with Bio-Rad CFX Manager software (Bio-Rad, Hercules, CA).
2.10. Transcription factor affinity prediction analysis
For promotor region analysis, a web-based transcription factor affinity prediction (TRAP) tool was used (http://trap.molgen.mpg.de/cgi-bin/trap_form.cgi).30 Human TGFBR2 promoter region sequence (−1670∼36) was used as input for TRAP analysis using JASPAR vertebrates as the matrix library, human promoters as the control, and Benjamini–Hochberg as the correction.
2.11. Site-directed mutagenesis and luciferase assay
The E-box mutation containing the construct for TGFBR2 were generated by site-directed mutagenesis from the wildtype construct using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). To generate the TGFBR2 promoter luciferase vector, the 1700 bp promoter region (−1670∼36) was synthesized (GeneArt Gene Synthesis; Life Technologies, Carlsbad, CA) and cloned into a pGL4-basic luciferase vector (Promega, Madison, WI). TGFBR2 luciferase activity was assayed 24 h after transfection using Dual-Luciferase reporter assay system (Promega, Madison, WI). HEK293 cells were used for the entire luciferase assay. TGFBR2 promoter luciferase vector (described above) and ZEB1 construct were transfected with Lipofectamine 2000 (Life Technologies, Carlsbad, CA) in combination with Renilla luciferase vector for normalization. To observe the interaction with ID proteins, ID1 or ID2 plasmids were added. Cells were collected 24 h after the transfection. Dual-Luciferase reporter system (Promega, Madison, WI) was used according to the manufacturer’s protocol. The luminescence was measured by a microplate reader (Bio-Tek, Winooski, VT). All experiments were performed three times in triplicates.
2.12. Chromatin immunoprecipitation
ChIP assays were performed using SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer’s protocol with some minor modifications. Cells cultured on 10 cm dishes were washed with ice-cold PBS for three times and fixed for 10 min by adding 1% paraformaldehyde solution. Fixation was quenched with glycine (0.12 M final concentration) for 5 min at room temperature. Cells were washed twice with ice-cold PBS, scraped into 1 mL PBS and centrifuged at 5000 r.p.m. for 10 min at 4°C. The cell pellets from two 10 cm dishes were lysed in 200 μL of lysis buffer. The lysate was then centrifuged at 5000 r.p.m. for 5 min at 4°C and the pellet was resuspended in 100 μL nuclease digestion buffer. The DNA was digested with 0.5 μL of micrococcal nuclease for 20 min at 37°C to a length of approximately 150–900 base pairs (checked by agarose gel electrophoresis). Lysates were centrifuged, and the pellet was resuspended in 500 μL ChIP buffer and sonicated for 3 × 30 s at power level 2 and 40% constancy. The solution was centrifuged at 10 000 r.p.m. for 10 min, and the supernatant was collected, which was the cross-linked chromatin. For ChIP, 150 μL of cross-linked chromatin was used for each immunoprecipitation and mixed with rabbit anti-Flag antibody (Sigma-Aldrich, 1:50) or the same amount of mouse IgG for control at 4°C overnight. Two per cent of cross-linked chromatin was saved as input control for later PCR. Thirty microlitres of protein G magnetic bead slurry were added to each immunoprecipitation reaction and incubated for 2 h at 4°C with rotation. The magnetic beads were washed three times with ChIP low salt buffer and once with ChIP high salt buffer. The bound chromatin on the beads was released in ChIP elution buffer by heating at 65°C for 30 min by vortexing at 1200 r.p.m. The chromatin was then digested with Protease K and purified using a spin column. The DNA was eventually eluted in 50 μL DNA elution buffer. The amount of precipitated DNA from each sample was measured with real-time PCR machine using primers flanking the −83 or −496 E-box motif in the TGFBR2 promoter region. The PCR primers (flanking the E-boxes) for the −83 E-box motif were 5′-ggaactcctgagtggtgtgg-3′ (forward) and 5′- ggggaaacaggaaactcctc-3′ (reverse), and for the −496 E-box motif were 5′-ttagaaatgcagaatctctg-3′ (forward) and 5′- ctttaggtcgaagtctagagg-3′ (reverse), respectively. The PCR primers for negative control (Lif) were 5′-caactggcacagctcaatgg-3′ (forward) and 5′-aaggtcccctcaacccagat-3′ (reverse).
2.13. In vivo siRNA delivery with 7C1 nanoparticle
Control and Tgfbr2-targeting nanoparticle were provided generated as previously described.31,32 After Bmpr1a deletion with tamoxifen injection, mice were injected with 7C1 control or TGFBR2 siRNA nanoparticle via tail vein (IV) every week for 3 weeks. RVSP and phenotypic analysis, including western blot and frozen section, were assessed 4 weeks after Bmpr1a deletion.
2.14. Statistics
All experiments were performed in triplicates (unless otherwise specified) in at least three independent experiments, and data shown are mean ± standard error of the mean. When only two groups were compared, statistical differences were assessed with unpaired two-tailed Student’s t-test. Otherwise, statistical significance was determined using one- or two-way analysis of variance followed by Bonferroni multiple comparison test. A P-value of <0.05 was considered statistically significant.
3. Results
3.1. Deletion of endothelial BMPR1A leads to pulmonary hypertension
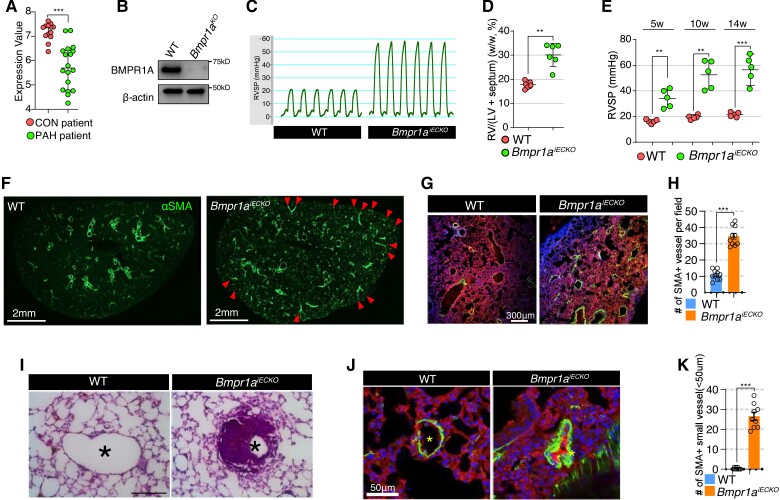
While previous report demonstrated decreased expression of BMPR1A in lungs of PAH patients in an angiopoietin-1-dependent manner,5 it only provided anecdotal evidence linking BMPR1A to PAH. To better understand the role of BMPR1A, we first reanalysed gene expression profile data set from a larger cohort of PAH subjects that included a control cohort (n = 13) and WHO Group 1 PAH cohort (n = 18).28 This analysis found significantly decreased expression of BMPR1A in PAH patient lungs compared with control lungs (Figure 1A). Given that expression of BMPR1A in the lungs was predominantly restricted to the pulmonary vascular endothelium5 and attenuated expression of BMPR1A in PAH patient (Figure 1A), we generated endothelial-specific, inducible Bmpr1a null mice (Bmpr1aiECKO) using Bmpr1afl/fl mice24 crossed to Cdh5(PAC)CreERT2 mice23 to investigate the role the potential contribution of endothelial BMPR1A to pulmonary vascular homeostasis and disease (see Supplementary material online, Figure S1A). As expected, Cre recombinase activation in mLECs from Bmpr1aiECKO mice dramatically reduced the expression of BMPR1A compared with control group (Figure 1B and Supplementary material online, Figure S1B and C).
Figure 1.
Loss of endothelial BMPR1A leads to pulmonary arterial hypertension. (A) Microarray-based gene expression profile of BMPR1A in human lungs from control and PAH subjects. ***P < 0.0001 (unpaired t-test). (B) Western blot showing BMPR1A expression level in lung ECs from Bmpr1aiECKO mice with (Bmpr1aKO) or without (WT) Cre activation. (C) Right ventricular systolic pressure in Bmpr1aiECKO mice in comparison with control mice (n = 6). (D) The weight of right ventricle is significantly increased in Bmpr1aiECKO mice (dots) compared with wildtype mice (dots). **P ≤ 0.005 (unpaired t-test). (E) The right ventricular systolic pressure is progressively elevated over 14 weeks in Bmpr1aiECKO mice (dots) after tamoxifen administration compared with wildtype mice (dots). [*** < 0.0001, **P ≤ 0.005 (unpaired t-test), n = 5]. (F) Immunostaining showing overall αSMA alteration with vibratome section (150 μm thickness). αSMA deposition was significantly elevated in Bmpr1aiECKO mice (left). Arrowheads indicate αSMA-positive pulmonary perivasculature (scale bar = 2 mm). (G) Confocal images showing CD31 and αSMA Immunostaining in the lung of control (left) and Bmpr1aiECKO (right) mice (CD31; αSMA; DAPI). Note that αSMA-positive vessels were increased in Bmpr1aiECKO mice compared with wildtype mice (scale bar = 300 μm). (H) Quantification of αSMA-positive arteries per field in the lungs of control and Bmpr1aiECKO mice [***P < 0.0001 (unpaired t-test), n = 10]. (I) Haematoxylin and eosin staining showing medial hypertrophy of pulmonary artery in Bmpr1aiECKO (right) mice (scale bar = 100 μm). (J) Confocal images showing the muscularized and occluded small (<50 μM) arteries (αSMA; CD31; DAPI) in Bmpr1aiECKO mice (scale bar = 50 μm). (K) Quantification of small αSMA-positive arteries (<50 μM) in the lungs of control and Bmpr1aiECKO mice [***P < 0.0001 (unpaired t-test), n = 9].
To investigate the phenotype of Bmpr1aiECKO mice, tamoxifen was administered to mice at 8 weeks of age (see Supplementary material online, Figure S1D) for the Cre recombinase activation. While systemic systolic blood pressure was not significantly changed at 5 weeks after endothelial Bmpr1a deletion (see Supplementary material online, Figure S2), Bmpr1aiECKO mice displayed spontaneous development of PAH as early as 5 weeks after deletion, as determined by increased RVSP (Figure 1C) and worsening right ventricular hypertrophy (Figure 1D), and RSVP became progressively worse with time (Figure 1E). Morphometric studies of the lungs identified significantly increased pulmonary vascular remodelling, as evidenced by a marked increase in perivascular staining of αSMA (Figure 1F–H and Supplementary material online, Figure S3 and S4A and B) and medial hypertrophy (Figure 1I) leading to occlusion of the vessel lumen in muscular pulmonary arteries compared with controls. In addition, immunohistochemistry showed that distal pulmonary vessels (<50 μm in size) were significantly more muscularized and occluded in Bmpr1aiECKO mice compared with controls (Figure 1J and K). Interestingly, endothelial deletion of a closely related receptor, namely ACVR1 (ALK2), did not lead to development of PAH (see Supplementary material online, Figure S5A–D), suggesting a unique role for BMPR1A in this context. Overall, these findings demonstrate a key role for BMPR1A, whose loss induces spontaneous pulmonary vascular remodelling and development of PAH.
3.2. Loss of endothelial BMPR1A induces endothelial to mesenchymal transition
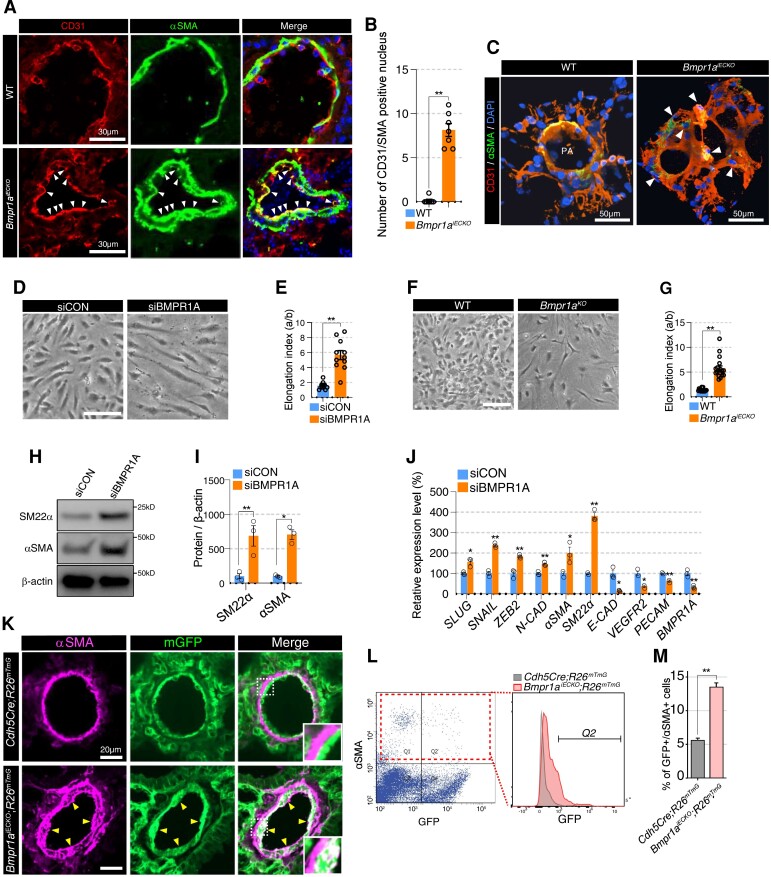
We sought to further investigate the cellular and molecular underpinning of vascular remodelling in response to the loss of endothelial Bmpr1a. Because endothelial loss of Bmpr1a lead to hypermuscularization in pulmonary arteries, we investigated whether endothelial to mesenchymal transition (EndoMT) caused the muscularization in Bmpr1aiECKO mice. First, we explored the pulmonary vasculature to detect endothelial marker (CD31) and αSMA double-positive cells which could be an evidence of EndoMT. Immunostaining with endothelial marker (CD31) and αSMA in pulmonary vasculature of Bmpr1aiECKO mice (Figure 2A and B), showed that ECs in pulmonary vasculature of Bmpr1aiECKO mice acquire mesenchymal identity, whereas in control mice CD31 and αSMA double-positive cells were relatively uncommon. Those CD31 and αSMA double-positive cells were also confirmed with three-dimensional reconstruction of the pulmonary vasculature with CD31 and αSMA immunostaining (Figure 2C and Supplementary material online, Videos S1 and S2). In addition, we also found that αSMA-positive ECs in retinal vasculature of Bmpr1aiECKO mice, indicating that the role of BMPR1A for maintaining endothelial identity is not limited in pulmonary vasculature (see Supplementary material online, Figure S6).
Figure 2.
Lack of BMPR1A in endothelial cells induces endothelial to mesenchymal transition. (A) Immunostaining showing CD31, αSMA, and DAPI in lung tissues from Bmpr1aiECKO (bottom) mice compared with control (top). White arrowheads indicate CD31 and αSMA double-positive endothelial cells (scale bar = 30 μm). (B)Quantification of CD31 and αSMA double-positive nucleus in the lung of control and Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 7]. (C) Snapshot of 3D reconstructed video (see Supplementary material online, Videos S1 and 2) with lung section of control (left) and Bmpr1aiECKO (right) mice (αSMA; CD31; DAPI). White arrowheads indicate αSMA/CD31 double-positive cells in Bmpr1aiECKO (right) mice (scale bar = 50 μm). (D) Representative images showing the morphology of control (left) or BMPR1A (right) siRNA-treated PAECs (scale bar = 50 μm). (E) Quantification of elongation index in control or BMPR1A siRNA-treated PAECs [**P ≤ 0.005 (unpaired t-test), n = 11]. (F) Representative images showing the morphology of control (left) and Bmpr1a-deleted (Bmpr1aKO, right) mouse lung ECs (scale bar = 50 μm). (G) Quantification of elongation index in control and Bmpr1a-deleted mouse lung ECs [**P ≤ 0.005 (unpaired t-test), n = 18]. (H) Western blot showing significantly increased expression of αSMA and SM22α in BMPR1A siRNA-treated PAECs compared with control siRNA-treated PAECs. (I) Quantification of expression of SM22α and αSMA in control or BMPR1A siRNA-treated PAECs [*P ≤ 0.05, **P ≤ 0.005 (unpaired t-test), n = 3]. (J) qRT–PCR showing the RNA expression of EndoMT and EC markers. Expression of transcripts associated with EndoMT (SLUG, SNAIL, ZEB2, and NCAD) is elevated with concomitant downregulation of endothelial transcripts (CDH5, VEGFR2, and PECAM) in BMPR1A siRNA-treated PAECs [**P ≤ 0.005, *P ≤ 0.05 (unpaired t-test), n = 3]. (K) Lineage tracing using ROSAmT/mG reporter mice showed that a subset of excessive αSMA expressing cells are descendants of endothelial cells. Arrowheads indicate αSMA and mGFP double-positive cells. The inset shows the area in white-dotted rectangle (scale bar = 20 μm). (L) Flow cytometry showing an increased number of αSMA/GFP double-positive cells in Cdh5(PAC)CreERT2; Bmpr1afl/fl; ROSA26mT/mG compared with Cdh5(PAC)CreERT; ROSA26mT/mG mice. (M) Quantification of the number of αSMA and GFP expressing cells (Q2 area) from control and Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 3].
Next, we sought to determine whether we can recapitulate this finding in vitro. We found that short interfering RNA (siRNA)-mediated knockdown of BMPR1A in human PAECs led to a dramatic change in cellular morphology from the typical cobblestone-like appearance of ECs to a spindle shape, reminiscent of mesenchymal cells (Figure 2D and E). Similarly, excision of Bmpr1a in pulmonary ECs isolated from Bmpr1afl/fl mice showed similar changes in cell morphology (Figure 2F and G). We measured the elongation index, which reflects the aspect ratio of cells,33,34 and found that it was significantly increased in PAECs subjected to BMPR1A knockdown and lung ECs from Bmpr1afl/fl mice subjected to Cre-induced excision (Figure 2E and G). Moreover, we found that the protein expression of αSMA and SM22α in BMPR1A siRNA-treated PAECs was significantly increased (Figure 2H and I). Interestingly, ECs with longer elongation index showed higher expression of EndoMT markers (FSP1, SM22α, and αSMA) and lower expression of EC marker (VECAD) than ECs in normal shape (see Supplementary material online, Figure S7A and B), suggesting that the morphological change in BMPR1A-deficient ECs is closely related with EndoMT process. As increased αSMA and SM22α expression is a phenomenon that is closely associated with EndoMT,14 we determined whether other EndoMT-related genes were also affected by BMPR1A knockdown. We found significant mRNA expression of multiple additional mesenchymal cell markers, including SLUG, SNAIL, ZEB2, and NCAD, while multiple endothelial markers, including CDH5, VEGFR2, and PECAM were significantly down-regulated (Figure 2J). Taken together, loss of endothelial BMPR1A led to EndoMT both in vivo and in vitro.
To further determine the involvement of EndoMT in the pulmonary vascular remodelling observed in the Bmpr1aiECKO mice, we bred Bmpr1aiECKO mice onto ROSA26mT/mG reporter mice (Bmpr1aiECKO; R26mT/mG), hence labelling pre-existing ECs with GFP at the time of Cre induction by tamoxifen injection (see Supplementary material online, Figure S1E). Immunohistochemistry on transverse section of lungs revealed that a significant fraction of αSMA staining cells also was GFP positive in Bmpr1aiECKO-ROSA26mT/mG mice, while GFP-positive ECs and αSMA-expressing SMCs were clearly separated in control mice (Figure 2K). We conducted flow cytometry analyses to determine the percentage of αSMA staining cells that were also GFP positive, and found a significantly higher percent of cells that were both αSMA and GFP positive in Bmpr1aiECKO mice compared with control (Figure 2L and M), further demonstrating that disruption of endothelial BMPR1A expression promotes EndoMT.
3.3. Lack of BMPR1A leads to increased TGFBR2 expression and activates TGFβ signalling
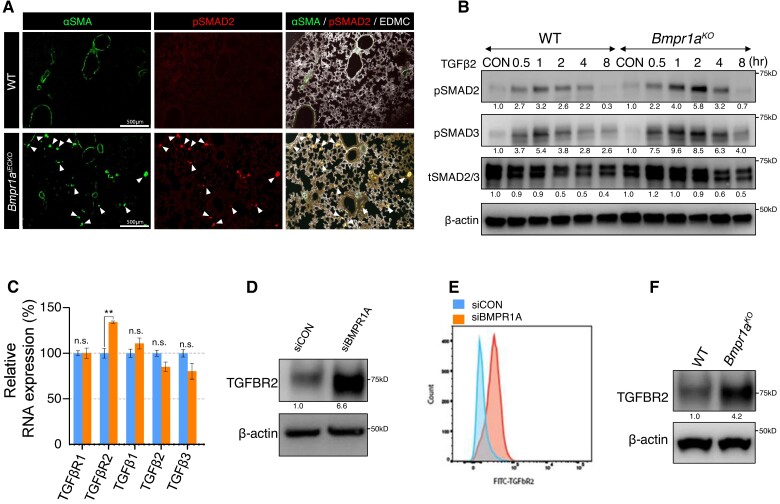
We next sought to further investigate the mechanism by which loss of BMPR1A promotes EndoMT. Given the known involvement of TGFβ signalling and its downstream targets in EndoMT, we investigated whether TGFβ signalling may be activated in Bmpr1aiECKO mice. We found a marked increase in staining for phosphorylated SMAD2 (pSMAD2), a well-established downstream target of TGFβ signalling, in the lungs of Bmpr1aiECKO mice compared with controls suggesting hyperactivated TGFβ signalling (Figure 3A). In addition, we found that ECs isolated from the lungs of Bmpr1aiECKO mice responded more robustly to TGFβ2 stimulation and showed increased SMAD2 and SMAD3 phosphorylation compared with those from control mice (Figure 3B).
Figure 3.
Increased TGFBR2 expression in the absence of endothelial BMPR1A. (A) Immunostaining showing Increased pSMAD2 deposition in lung tissues from Bmpr1aiECKO (bottom) mice compared with control (top). White arrows indicate pSMAD2 and αSMA double-positive cells. (scale bar = 500 μm). (B) Western blot showing that mouse lung endothelial cells with Bmpr1a deletion were more sensitive to TGFβ2 stimulation compared with those from control cells. (C) RNA expression of Tgfbr2 was significantly increased while the expression of other TGFβ signalling components was either slightly decreased or unaltered in BMPR1A siRNA-treated PAECs compared with control siRNA-treated PAECs. [**P ≤ 0.005 (unpaired t-test), n = 3]. (D) Western blot showing that TGFBR2 expression was increased in BMPR1A siRNA-treated PAECs compared with control siRNA-treated PAECs. (E) Flow cytometry showing a significant increase of TGFBR2 expression in BMPR1A siRNA-treated PAECs compared with control siRNA-treated PAECs. (F) Western blot showing increased TGFBR2 expression in mouse lung endothelial cells with Bmpr1a deletion compared with those from control cells.
To further elucidate the molecular mechanisms for augmented EndoMT in the absence of BMPR1A, we evaluated for any changes in the expression of TGFβ signalling components. We evaluated the expression levels of key components of TGFβ signalling in the context of BMPR1A knockdown in ECs, and found that the key TGFβ signalling receptor, namely TGFBR2, was significantly increased in response to BMPR1A knockdown, whereas multiple other components of TGFβ signalling were unaffected (Figure 3C). We further confirmed increased TGFBR2 protein expression in response to BMPR1A knockdown by western blot and flow cytometry (Figure 3D and E). Furthermore, western blots of lung ECs demonstrated an increased expression of TGFBR2 in Bmpr1aiECKO mice compared with control mice (Figure 3F). Taken together, our data demonstrate that lack of BMPR1A in ECs leads to a significant increase in TGFBR2 expression.
3.4. Endothelial TGFBR2 expression is regulated by interaction between ID2 and ZEB1
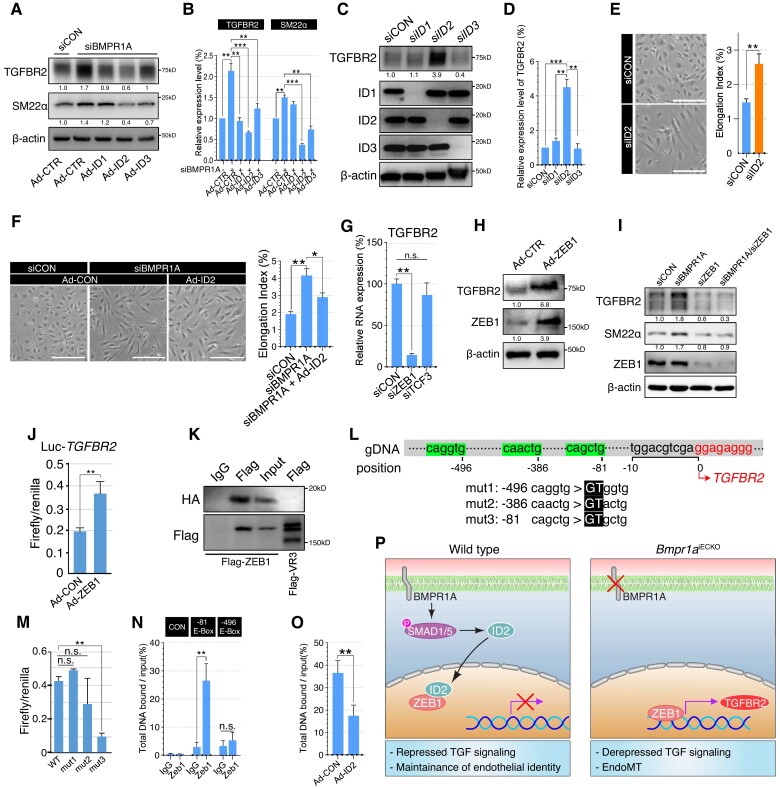
We further sought to define the molecular mechanism by which disrupted BMPR1A signalling results in increased TGFBR2 expression. Inhibitor of DNA-binding (ID) proteins are widely accepted as downstream transcriptional targets of BMP signalling, which serve as dominant-negative transcriptional regulators.35 ID proteins have been proposed to act as dominant-negative antagonists of the bHLH(basic helix-loop-helix) transcription family by inhibiting their binding to the E-box motif (CANNTG).36 We postulated that ID proteins could repress TGFBR2 expression in ECs in response to BMPR1A activity. In this scenario, lack of endothelial BMPR1A is likely to attenuate the activity of ID proteins, which could lead to de-repression of TGFBR2 expression and initiation of EndoMT. To test this possibility, we first sought to check whether the expression level of ID proteins is indeed reduced in response to BMPR1A knockdown. While ID4 was not decreased due to its lack of expression in ECs, other IDs expression level (ID1,2 and 3) was significantly lowered in BMPR1A siRNA-treated PAECs (see Supplementary material online, Figure S8).
Next, we sought to identify whether those ID proteins may be essential for mediating BMPR1A-dependent repression of TGFBR2 expression in ECs. Overexpression of ID genes in PAECs could decrease the expression of TGFBR2 and SM22α, with ID2 demonstrating the most potent inhibitory effects (Figure 4A and B). Since ID genes share common motifs,36 overexpression of one ID gene may inadvertently influence the function of the others. Therefore, we next treated PAECs with siRNAs targeting individual ID genes. While knockdown of ID1 or ID3 in PAECs did not alter the level of TGFBR2 expression, knockdown of ID2 resulted in a marked increase in TGFBR2 expression (Figure 4C and D). Taken together, ID2 appears to be the primary downstream factor which mediates the inhibitory effects of BMPR1A activity on TGFBR2 expression. In addition to its effects on TGFBR2 expression, knockdown of ID2 also affects the cell morphology of PAECs. It appears that ID2 siRNA-treated PAECs displayed more elongated shape compared with control siRNA-treated PAECs (Figure 4E). Conversely, overexpression of ID2 restored EC shape in BMPR1A siRNA-treated PAECs back to stereotypic cell morphology (Figure 4F), implying the role of ID2 in maintaining EC identity.
Figure 4.
ID2–ZEB1 interactions regulate transcription of TGFBR2. (A) Western blots showing TGFBR2 and SM22α expression in control or BMPR1A siRNA-treated PAEC cells with control, ID1, ID2, or ID3 overexpressing adenovirus infection. Note that ID2-overexpressed PAECs showed dramatic reduction of TGFBR2 expression. (B) Quantification of TGFBR2 and SM22α protein expression in C [**P ≤ 0.005 (unpaired t-test), n = 3]. (C) Western blots showing TGFBR2 expression in control, ID1, ID2, or ID3 siRNA-treated PAECs. Expression of TGFBR2 was significantly increased in ID2 siRNA-treated cells, but not in ID1 or ID3 siRNA. (D) Quantification of TGFBR2 protein expression in control, ID1, ID2, or ID3 siRNA-treated PAECs [***P < 0.0001, **P ≤ 0.005 (unpaired t-test), n = 3]. (E) Representative images showing the morphology of control or ID2 siRNA-treated PAECs (scale bar = 50 μm). Right panel shows quantification of elongation index [**P ≤ 0.005 (unpaired t-test), n = 27]. (F) Representative images showing the morphology of control or BMPR1A siRNA-treated PAECs with control or ID2 adenoviral overexpression (scale bar = 50 μm). Right panel shows quantification of elongation index [**P ≤ 0.005, *P ≤ 0.05 (unpaired t-test), n = 36]. (G) Quantification of TGFBR2 RNA expression in control, ZEB1 or TCF3 siRNA-treated PAECs. [**P ≤ 0.005 (unpaired t-test), n = 3]. (H) Western blots showing TGFBR2 expression in control or ZEB1 overexpressing PAECs. Overexpression of ZEB1 induces robust expression of TGFBR2 in PAECs. (I) Western blots showing the expression of TGFBR2, SM22α, and ZEB1 and BMPR1A in control, BMPR1A, ZEB1, or BMPR1A/ZEB1 siRNA-treated PAECs. Inhibiting ZEB1 effectively suppressed the ectopic expression of TGFBR2 and SM22α induced by BMPR1A siRNA treatment. (J) Luciferase assay using pGL4-luciferase reporter vector containing the TGFBR2 promoter region (−1670/+36) with control or ZEB1 overexpressing PAECs [**P ≤ 0.005 (unpaired t-test), n = 3]. (K) Co-immunoprecipitation using Flag-ZEB1 and HA-ID2-overexpressing PAECs showing that ID2 could physically interact with ZEB1. Flag-VEGFR3 infected cells were used as negative control for flag antibody. (L) Display of genomic sequence corresponding to the 5′-upstream and transcription initiation site (font) of TGFBR2. Three putative E-box motifs located at −496, −386, and −81 positions from the TGFBR2 transcription initiation site were highlighted. The mutated sequence with site-directed mutagenesis in pGL4- TGFBR2 promoter was highlighted in black box. (M) Luciferase assay using pGL4-basic Luc vector containing control, mut1, mut2, or mut3 TGFBR2 promoter region with ZEB1 overexpressing HEK293 cells. Note that an E-box motif at the −81 position within the TGFBR2 promoter confers ZEB1-mediated regulation of TGFBR2 expression [**P ≤ 0.005 (unpaired t-test), n = 3]. (N) Chromatin immunoprecipitation showing the interaction between ZEB1 and the E-box motif at the −81 position/−496 position within the TGFBR2 promoter or Lif (negative control). Note that only the E-box motif at the −81 position was pulled down with Flag-ZEB1-binding antibody [**P ≤ 0.005 (unpaired t-test), n = 3]. (O) Chromatin immunoprecipitation showing that ID2 overexpression inhibits the binding of ZEB1 to the TGFBR2 promoter (E-box motif at the −81 position) [**P ≤ 0.005 (unpaired t-test), n = 3]. (P) Our working model: In the presence of endothelial BMPR1A, ID2–ZEB1 interaction increases. We speculate that ID2 could prevent ZEB1 from binding to the promoter of TGFBR2, and inducing transcription by sequestering ZEB1, which helps effectively maintain endothelial fate (left). In Bmpr1aiECKO mice, lack of BMPR1A and ID2 allows ZEB1 to bind to the promoter of TGFBR2, and derepresses its expression, which leads to the loss of endothelial identity and EndoMT.
Since it has been proposed that ID2 functions as a transcriptional repressor by sequestering transcription factors,37 we explored the possibility that ID2 may interfere with a specific transcription factor to negatively regulate TGFBR2 expression. We first selected bHLH transcription factors which could bind to the TGFBR2 promoter using a web-based TRAP tool, and identified TCF3 and ZEB1 as potential TGFBR2 regulators.30 While TCF3 knockdown had minimal impact on TGFBR2 expression, we found that ZEB1 knockdown led to a robust decrease in TGFBR2 expression (Figure 4G). Overexpression of ZEB1 also robustly induced TGFBR2 expression (Figure 4H), further indicating that ZEB1 could modulate the transcription of TGFBR2. To test whether ZEB1 mediates BMPR1A knockdown-induced EndoMT, PAECs were concurrently treated with siRNAs targeting BMPR1A and ZEB1, and it attenuated the expression of TGFBR2 and SM22α induced by BMPR1A knockdown (Figure 4I).
Consistent with these findings, overexpression of ZEB1 was able to robustly induce the expression of a luciferase reporter driven by the TGFBR2 promoter construct containing 1670 bp upstream from the transcription start site (Figure 4J). Moreover, we found that ZEB1, which has been implicated in EMT,38 could form a complex with ID2 in PAECs (Figure 4K). We identified three putative ZEB1-binding E-box motifs located at −496, −386, and −81 positions from the TGFBR2 transcription initiation site and generated mutagenized vectors containing a point mutation in each E-box motif using site-directed mutagenesis (Figure 4L). Among the vectors we examined, only the mutation of the −81 positioned E-box motif (CAGCTG) failed to induce TGFBR2 promoter activity with ZEB1 overexpression, suggesting that ZEB1 binds to this E-box motif (Figure 4M). Furthermore, we conducted chromatin immunoprecipitation assay and found that ZEB1 was able to directly interact with the E-box motif at −81 position (Figure 4N) but not −496 from the TGFBR2 transcription initiation site or a promoter of unrelated gene (Lif). Moreover, this binding was significantly inhibited with ID2 overexpression (Figure 4O). Overall, our data demonstrate that ID2 mediates BMPR1A-induced repression of TGFBR2 expression by sequestering ZEB1 transcription factor to prevent its binding to the TGFBR2 promoter (Figure 4P).
3.5. Inhibition of TGFBR2 attenuates EndoMT and progression of PAH due to loss of endothelial BMPR1A
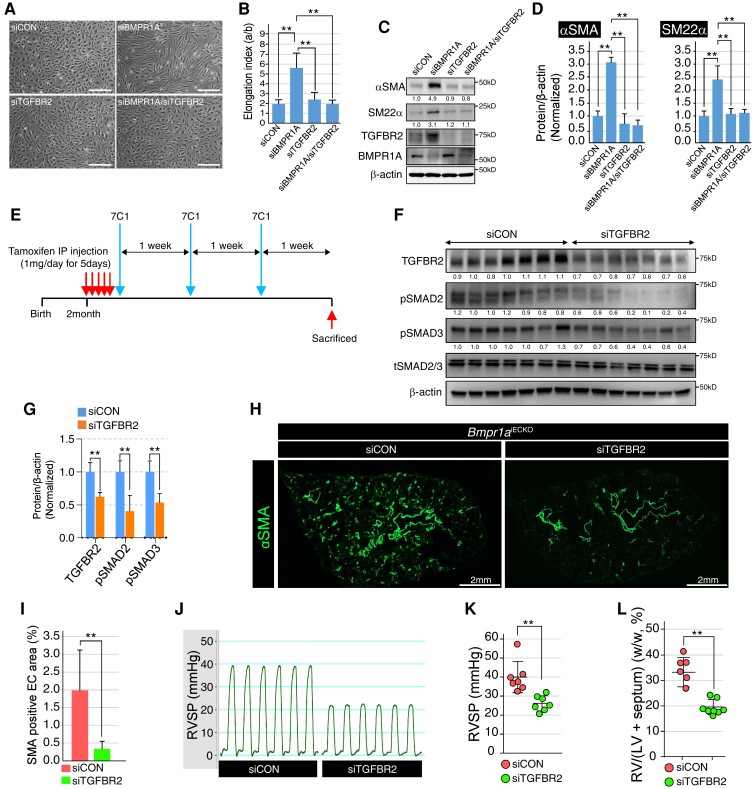
To investigate whether the EndoMT phenomenon driven by disrupted BMPR1A signalling can be rescued by concurrent silencing of TGFBR2 expression, we carried out simultaneous knockdown of BMPR1A and TGFBR2 in PAECs. We found restoration of the cellular morphology to a more cobblestone/endothelial-like appearance when TGFBR2 was concurrently knocked down with BMPR1A (Figure 5A and B). In addition, simultaneous knockdown of BMPR1A and TGFBR2 substantially attenuated the expression of mesenchymal cell markers induced by the absence of BMPR1A at both mRNA and protein levels (Figure 5C and D).
Figure 5.
Nanoparticle-mediated inhibition of TGFBR2 attenuates EndoMT and progression of PAH in vivo. (A) Representative images showing the morphology of control, BMPR1A, TGFBR2, or BMPR1A/TGFBR2 siRNA-treated PAECs. Morphological alteration in BMPR1A siRNA-treated PAECs was restored by concomitant inhibition of TGFBR2 (scale bar = 50 μm). (B) Quantification of elongation index in control, BMPR1A, TGFBR2, or BMPR1A/TGFBR2 siRNA-treated PAECs [**P ≤ 0.005 (unpaired t-test), n = 11]. (C) Western blots showing the expression of EndoMT markers (αSMA, SM22α, and TGFBR2) and BMPR1A in control, BMPR1A, TGFBR2, or BMPR1A/TGFBR2 siRNA-treated PAECs. Inhibiting TGFBR2 effectively suppressed the ectopic expression of EndoMT markers induced by BMPR1A siRNA treatment. (D) Quantification of αSMA (left) and SM22α (right) protein expression in control, BMPR1A, TGFBR2, or BMPR1A/TGFBR2 siRNA-treated PAECs [**P ≤ 0.005 (unpaired t-test), n = 3]. (E) Schematic illustration on the treatment regimen for 7C1 nanoparticles coated with either control or TGFBR2 siRNA. 7C1 nanoparticles were injected into the tail vein of Bmpr1aiECKO mice after tamoxifen administration. Three rounds of 7C1 nanoparticle injections were performed with a 1-week interval. (F) Western blot with whole lung lysate showed that treatment with TGFBR2 siRNA-coated 7C1 nanoparticle attenuated the ectopic expression of TGFBR2, pSMAD2, and pSMAD3 in Bmpr1aiECKO mice. (G) Quantification of TGFBR2, pSMAD2, and pSMAD3 protein level in control or TGFBR2 siRNA-coated 7C1 nanoparticle-injected Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 3]. (H) Immunostaining showed αSMA expression was significantly reduced in the lung of TGFBR2 siRNA-coated 7C1 nanoparticle-injected Bmpr1aiECKO mice (right) compared with control (left) (scale bar = 2 mm). (I) Quantification of αSMA-positive EC area in the lung of control or TGFBR2 siRNA-coated 7C1 nanoparticle-injected Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 14]. (J) Elevated right ventricle systolic pressure was restored to normal range in TGFBR2 siRNA-coated 7C1 nanoparticles-injected Bmpr1aiECKO mice. (K) Quantification of the right ventricle systolic pressure in control or TGFBR2 siRNA-coated 7C1 nanoparticles-injected Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 7]. (L) Quantification of the right ventricle heart wall thickness in control or TGFBR2 siRNA-coated 7C1 nanoparticles-injected Bmpr1aiECKO mice [**P ≤ 0.005 (unpaired t-test), n = 8].
Given the robust in vitro efficacy of TGFBR2 silencing in the rescue of BMPR1A knockdown phenotype, we sought to determine whether TGFBR2 silencing in vivo can also rescue the pulmonary vascular remodelling associated with PAH in Bmpr1aiECKO mice. While small molecule-based inhibition of TGFβ signalling is being tested in cancer clinical trials, systemic delivery of such inhibitors can be fraught with off-target toxicities, including cardiotoxicities.39 To achieve transcript and cell type-specific delivery of TGFBR2-targeting siRNA to the lung ECs in our Bmpr1aiECKO mice, we utilized TGFBR2-targeting siRNA formulated with the 7C1 nanoparticle, which has been previously found to effectively deliver siRNA to the lung endothelium when administered intravenously.31,32
After daily tamoxifen injection for 5 consecutive days to induce endothelial Bmpr1a deletion, 7C1 encapsulating TGFBR2-targeting siRNA was administered intravenously via tail vein for a total of 3 weekly doses (Figure 5E). We achieved robust silencing of TGFBR2 with the TGFBR2 siRNA-7C1 nanoparticle conjugate in the lungs of Bmpr1aiECKO mice compared with those mice receiving non-targeting control siRNA-7C1 nanoparticle conjugate (Figure 5F and G). Moreover, we found a significant reduction in SMAD2 and SMAD3 phosphorylation in the lungs of Bmpr1aiECKO mice treated with TGFBR2 siRNA, further suggesting that increased TGFBR2 expression is the key mechanism of EndoMT associated with BMPR1A disruption (Figure 5F and G). Suppressing TGFBR2 expression Bmpr1aiECKO mice resulted in a robust prevention of PAH, associated with a significant reduction of muscularization indicated by decreased staining for αSMA (Figure 5H and I) as well as RVSP (Figure 5J and K) and right ventricular hypertrophy (Figure 5L). Therefore, TGFBR2 appears to be the key downstream factor leading to ectopic EndoMT upon endothelial loss of BMPR1A, and inhibiting TGFBR2 may have therapeutic potential to ameliorate symptoms associated with PAH.
4. Discussion
Abnormal endothelial function, including contribution of EndoMT, has been described in multiple contexts of PAH.40,41 However, the molecular mediators of this process have not been fully elucidated. Our study demonstrates an essential role for endothelial BMPR1A in maintenance of endothelial identity. We find that loss of BMPR1A in ECs induces elevated expression of TGFBR2 which likely promotes spontaneous EndoMT, pulmonary vascular remodelling and PAH in mice. Therefore, our data support a model whereby BMPR1A functions a key factor mediating intricate interaction between BMP and TGFβ signalling, which balances the homeostasis and trans-differentiation of ECs. By systematic analyses, we demonstrated that BMPR1A-mediated maintenance of endothelial identity requires ID2 activity. In the process of EndoMT induced by endothelial loss of BMPR1A, ID2, but not other ID proteins, appears to induce ectopic transcriptional activation of TGFBR2. Considering that ID transcription modulators have been shown to relay BMP signalling collectively, but their individual functions in ECs have not been fully explored, our data illustrating the non-redundant and non-overlapping function of ID2 provide evidence that each ID protein may relay distinct aspects of BMP signalling in ECs.
For BMPR1A-mediated inhibition of TGFBR2 expression, we found that ZEB1, a well-characterized transcription factor which has been previously implicated in EndoMT, functions as a key downstream effectors. While BMPR1A modulates function of ZEB1 as a transcriptional factor, it does not appear to influence either transcription or translation of ZEB1 in ECs since we did not find any discernible changes in ZEB1 expression level in BMPR1A-deficient ECs. Rather, BMPR1A appears to modulate ZEB1 activity indirectly via ID2. Upon BMPR1A activation, ID2 expression becomes elevated, which in turn, facilitate the formation of ID2–ZEB1 complex. Therefore, we speculate that ID2-mediated sequestration of ZEB1 may prevent ZEB1 to induce expression of TGRBR2. As a consequence, loss of BMPR1A leads to de-repression of TGFBR2 expression in ECs, and subsequently promotes EndoMT and the onset of PAH. Further analyses on ZEB1, such as testing whether overexpression of ZEB1 could alleviate and partially rescue BMPR1A-deficiency induced PAH symptoms, are warranted to fully decipher the role of the role of ZEB1 in mediating BMPR1A activity in ECs.
Previously published work and our analyses of existing gene expression profiling data demonstrated decreased BMPR1A expression in the lungs of PAH subjects.5,28 In addition, deleterious mutations in BMPR1A have been recently identified in idiopathic PAH patients.6 However, its role in the pathogenesis of PAH has not been fully elucidated. Considering the incomplete penetrance of BMPR2 mutations in PAH (estimated to be ∼20% of mutation carriers), other molecules, including components of BMP signalling, are likely to be involved in driving the molecular pathogenesis of PAH. Our current findings provide key molecular evidence that implicates BMPR1A as a critical factor for endothelial preservation and vascular homeostasis, given the severe pulmonary vascular remodelling that occurs secondary to loss of BMPR1A, which leads to augmented TGFβ signalling, EndoMT, and spontaneous PAH.
While the role of BMPR2 in the pathophysiology of PAH has been well established, it is unclear which BMPR1 co-receptors are essential in this process. It has been reported that multiple BMP type 1 receptors are expressed in ECs.25 The most abundant BMPR1 in ECs is ACVRL/ALK1, which functions as a selective BMPR1 for BMP9/BMP10.25,42,43 The role of BMP9/ALK1 signalling in the pathophysiology of PAH remains a subject of controversy. For instance, Tu and colleagues44 reported that BMP9 inhibition could ameliorate the PAH symptoms in mice. Conversely, Long Morrell and colleagues45 demonstrated that BMP9 could provide a protective role for PAH patients. Therefore, it appears that BMP9/ALK1 signalling may provide a context-dependent input for the pathogenesis of PAH. Our analyses suggest that BMPR1A provides an indispensable role in ECs despite its relatively low expression. Moreover, considering that BMP9/ACVRL signalling is not affected by the loss of BMPR1A, our data suggest that BMP9/ACVRL signalling and BMP2/4/BMPR1A signalling provide non-redundant input for the pathogenesis of PAH.
Overall, our studies demonstrated that increased endothelial TGFBR2 expression could serve as a key molecular driver of EndoMT and PAH, and identified ZEB1 as a critical transcriptional regulator for TGFBR2 expression in ECs. BMPR1A appears to negatively modulate ZEB1 activity by inducing robust expression of ID2, which physically interacts with ZEB1 and prevents transcriptional activation of TGFBR2. Considering that ZEB1 has been previously implicated as a functional downstream of TGFβ signalling, facilitating EndoMT,46 our finding that ZEB1 functions as a critical regulator for TGFBR2 expression illustrates that EndoMT is regulated by complex feedback regulation of TGFβ signalling. Moreover, we demonstrated robust rescue of the phenotype through lung endothelial-targeted delivery of siRNA against TGFBR2, which resulted in decreased EndoMT both in vitro and in vivo, as well as amelioration of PAH in the Bmpr1aiECKO mice. Therefore, while there are remaining questions to fully address the implication of our findings in clinical settings, including the precise role of ZEB1 in modulating TGFBR2 expression, our study identified a potential therapeutic venue for selective augmentation BMPR1A signalling and/or ID2–ZEB1 interaction as robust, yet to be targeted therapeutic strategies in PAH.
Authors’ contributions
H.W.L., S.W.J., and H.J.C. design the experiments and wrote the manuscript. H.W.L, T.A., B.P., S.P., X.H., W.C., C.H.C., A.L., I.P., and O.H. performed the experiments. P.S.K., K.R.C., and D.G.A. provided key reagents. H.W.L., J.K., J.P., M.S., S.W.J., and H.J.C. analyse and interpreted the data.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgements
The authors thank Rita Webber for excellent animal care, and Drs Ralf Adams and Robert Behringer for providing mice strains.
Contributor Information
Heon-Woo Lee, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Takaomi Adachi, Division of Nephrology, Department of Medicine, Kyoto Prefectural University of Medicine, Kyoto, Japan.
Boryeong Pak, School of Life Sciences and Cell Logistics Research Center, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.
Saejeong Park, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Xiaoyue Hu, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Woosoung Choi, School of Life Sciences and Cell Logistics Research Center, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.
Piotr S Kowalski, David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02142, USA.
C Hong Chang, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Katharine R Clapham, Division of Pulmonary and Critical Care, Brigham and Women’s Hospital, Boston, MA 02127, USA.
Aram Lee, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA; Division of Biological Sciences, Sookmyung Women's University, Seoul 04310, Korea.
Irinna Papangeli, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Jongmin Kim, Division of Biological Sciences, Sookmyung Women's University, Seoul 04310, Korea.
Orjin Han, School of Life Sciences and Cell Logistics Research Center, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.
Jihwan Park, School of Life Sciences and Cell Logistics Research Center, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.
Daniel G Anderson, David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02142, USA.
Michael Simons, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA.
Suk-Won Jin, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA; School of Life Sciences and Cell Logistics Research Center, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.
Hyung J Chun, Yale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT 06511, USA; VA Connecticut Healthcare System, 950 Campbell Ave, 111B, West Haven, CT 06516, USA.
Funding
This work was supported by grants from National Institute of Health (HL142818 to HJC and HL114820 to S.W.J.), American Heart Association (Transformational Project Award to H.J.C.), and grants from National Research Foundation of Korea (2016R1A5A1007318 and 2019R1A2C2088125 to S.W.J.).
Data availability
The data described in this article will be available upon reasonable request to the corresponding authors.
Translational perspective.
The role of bone morphogenetic protein type I receptor A (BMPR1A) in repressing the expression of transforming growth factor beta receptor 2 by promoting ID2–ZEB1 interaction suggests that BMPR1A is a critical negative modulator for pathological endothelial to mesenchymal transition, and an essential factor for the maintenance of endothelial fate. Considering that endothelial to mesenchymal transition is a key underlying molecular mechanisms in a number of human cardiovascular diseases, our results suggest that BMPR1A could serve a novel candidate for successful therapy for the related diseases.
References
- 1. Farber HW, Loscalzo J. Pulmonary arterial hypertension. N Engl J Med 2004;351:1655–1665. [DOI] [PubMed] [Google Scholar]
- 2. Michelakis ED, Wilkins MR, Rabinovitch M. Emerging concepts and translational priorities in pulmonary arterial hypertension. Circulation 2008;118:1486–1495. [DOI] [PubMed] [Google Scholar]
- 3. Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 2008;118:2372–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans JD, Girerd B, Montani D, Wang XJ, Galie N, Austin ED, Elliott G, Asano K, Grunig E, Yan Y, Jing ZC, Manes A, Palazzini M, Wheeler LA, Nakayama I, Satoh T, Eichstaedt C, Hinderhofer K, Wolf M, Rosenzweig EB, Chung WK, Soubrier F, Simonneau G, Sitbon O, Graf S, Kaptoge S, Di Angelantonio E, Humbert M, Morrell NW. BMPR2 Mutations and survival in pulmonary arterial hypertension: an individual participant data meta-analysis. Lancet Respir Med 2016;4:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du L, Sullivan CC, Chu D, Cho AJ, Kido M, Wolf PL, Yuan JX, Deutsch R, Jamieson SW, Thistlethwaite PA. Signaling molecules in nonfamilial pulmonary hypertension. N Engl J Med 2003;348:500–509. [DOI] [PubMed] [Google Scholar]
- 6. Zhu N, Pauciulo MW, Welch CL, Lutz KA, Coleman AW, Gonzaga-Jauregui C, Wang J, Grimes JM, Martin LJ, He H. Novel risk genes and mechanisms implicated by exome sequencing of 2572 individuals with pulmonary arterial hypertension. Genome Med 2019;11:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mishina Y, Suzuki A, Ueno N, Behringer RR. BMPR encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev 1995;9:3027–3037. [DOI] [PubMed] [Google Scholar]
- 8. Vanderpool RR, El-Bizri N, Rabinovitch M, Chesler NC. Patchy deletion of BMPR1A potentiates proximal pulmonary artery remodeling in mice exposed to chronic hypoxia. Biomech Model Mechanobiol 2013;12:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El-Bizri N, Wang L, Merklinger SL, Guignabert C, Desai T, Urashima T, Sheikh AY, Knutsen RH, Mecham RP, Mishina Y, Rabinovitch M. Smooth muscle protein 22alpha-mediated patchy deletion of BMPR1A impairs cardiac contractility but protects against pulmonary vascular remodeling. Circ Res 2008;102:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sheikh AQ, Misra A, Rosas IO, Adams RH, Greif DM. Smooth muscle cell progenitors are primed to muscularize in pulmonary hypertension. Sci Transl Med 2015;7:308ra159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arciniegas E, Frid MG, Douglas IS, Stenmark KR. Perspectives on endothelial-to-mesenchymal transition: potential contribution to vascular remodeling in chronic pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol 2007;293:L1–L8. [DOI] [PubMed] [Google Scholar]
- 12. Mammoto A, Hendee K, Muyleart M, Mammoto T. Endothelial Twist1-PDGFB signaling mediates hypoxia-induced proliferation and migration of αSMA-positive cells. Sci Rep 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovacic JC, Dimmeler S, Harvey RP, Finkel T, Aikawa E, Krenning G, Baker AH. Endothelial to mesenchymal transition in cardiovascular disease: JACC state-of-the-art review. J Am Coll Cardiol 2019;73:190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piera-Velazquez S, Li Z, Jimenez SA. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am J Pathol 2011;179:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovacic JC, Mercader N, Torres M, Boehm M, Fuster V. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: from cardiovascular development to disease. Circulation 2012;125:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, Lui KO, Zhou B. Reassessing endothelial-to-mesenchymal transition in cardiovascular diseases. Nat Rev Cardiol 2018;15:445–456. [DOI] [PubMed] [Google Scholar]
- 17. von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 2012;110:1628–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 2005;132:5601–5611. [DOI] [PubMed] [Google Scholar]
- 19. Chen PY, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, Simons M. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest 2015;125:4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cooley BC, Nevado J, Mellad J, Yang D, St Hilaire C, Negro A, Fang F, Chen G, San H, Walts AD, Schwartzbeck RL, Taylor B, Lanzer JD, Wragg A, Elagha A, Beltran LE, Berry C, Feil R, Virmani R, Ladich E, Kovacic JC, Boehm M. TGF-beta signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci Transl Med 2014;6:227ra234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 2010;120:3493–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 2007;13:952–961. [DOI] [PubMed] [Google Scholar]
- 23. Wang YD, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD, Adams RH. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010;465:483–486. [DOI] [PubMed] [Google Scholar]
- 24. Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of BMPR/Alk3 conditional knockout mice. Genesis 2002;32:69–72. [DOI] [PubMed] [Google Scholar]
- 25. Lee HW, Chong DC, Ola R, Dunworth WP, Meadows S, Ka J, Kaartinen VM, Qyang YB, Cleaver O, Bautch VL, Eichmann A, Jin SW. Alk2/ACVR1 and Alk3/BMPR1A provide essential function for bone morphogenetic protein-induced retinal angiogenesis. Arterioscler Thromb Vasc Biol 2017;37:657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 27. Hwangbo C, Lee HW, Kang H, Ju H, Wiley DS, Papangeli I, Han J, Kim JD, Dunworth WP, Hu XY, Lee S, El-Hely O, Sofer A, Pak B, Peterson L, Comhair S, Hwang EM, Park JY, Thomas JL, Bautch VL, Erzurum SC, Chun HJ, Jin SW. Modulation of endothelial bone morphogenetic protein receptor type 2 activity by vascular endothelial growth factor receptor 3 in pulmonary arterial hypertension. Circulation 2017;135:2288–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, Ahmad F. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol 2010;298:H1235–H1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell 2001;105:445–457. [DOI] [PubMed] [Google Scholar]
- 30. Thomas-Chollier M, Hufton A, Heinig M, O'Keeffe S, Masri NE, Roider HG, Manke T, Vingron M. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc 2011;6:1860–1869. [DOI] [PubMed] [Google Scholar]
- 31. Khan OF, Kowalski PS, Doloff JC, Tsosie JK, Bakthavatchalu V, Winn CB, Haupt J, Jamiel M, Langer R, Anderson DG. Endothelial siRNA delivery in nonhuman primates using ionizable low-molecular weight polymeric nanoparticles. Sci Adv 2018;4:eaar8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dahlman JE, Barnes C, Khan O, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Sandu KS, Webber MJ, Novobrantseva T, Ruda VM, Lytton-Jean AKR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Schroeder A, Panigrahy D, Kotelianski V, Langer R, Anderson DG. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol 2014;9:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo-Azofeifa D, Delling M, Klein OD, Vale RD. Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. Elife 2018;7:e36739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loye AM, Kinser ER, Bensouda S, Shayan M, Davis R, Wang R, Chen Z, Schwarz UD, Schroers J, Kyriakides TR. Regulation of mesenchymal stem cell differentiation by nanopatterning of bulk metallic glass. Sci Rep 2018;8:8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J, Li X, Morrell NW. Id proteins in the vasculature: from molecular biology to cardiopulmonary medicine. Cardiovasc Res 2014;104:388–398. [DOI] [PubMed] [Google Scholar]
- 36. Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003;3:525–530. [DOI] [PubMed] [Google Scholar]
- 37. Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 1991;11:5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 2008;135:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderton MJ, Mellor HR, Bell A, Sadler C, Pass M, Powell S, Steele SJ, Roberts RR, Heier A. Induction of heart valve lesions by small-molecule ALK5 inhibitors. Toxicol Pathol 2011;39:916–924. [DOI] [PubMed] [Google Scholar]
- 40. Ranchoux B, Antigny F, Rucker-Martin C, Hautefort A, Pechoux C, Bogaard HJ, Dorfmuller P, Remy S, Lecerf F, Plante S, Chat S, Fadel E, Houssaini A, Anegon I, Adnot S, Simonneau G, Humbert M, Cohen-Kaminsky S, Perros F. Endothelial-to-mesenchymal transition in pulmonary hypertension. Circulation 2015;131:1006–1018. [DOI] [PubMed] [Google Scholar]
- 41. Good RB, Gilbane AJ, Trinder SL, Denton CP, Coghlan G, Abraham DJ, Holmes AM. Endothelial to mesenchymal transition contributes to endothelial dysfunction in pulmonary arterial hypertension. Am J Pathol 2015;185:1850–1858. [DOI] [PubMed] [Google Scholar]
- 42. Neal A, Nornes S, Payne S, Wallace MD, Fritzsche M, Louphrasitthiphol P, Wilkinson RN, Chouliaras KM, Liu K, Plant K, Sholapurkar R, Ratnayaka I, Herzog W, Bond G, Chico T, Bou-Gharios G, De Val S. Venous identity requires BMP signalling through ALK3. Nat Commun 2019;10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seki T, Yun J, Oh SP. Arterial endothelium-specific activin receptor-like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ Res 2003;93:682–689. [DOI] [PubMed] [Google Scholar]
- 44. Tu L, Desroches-Castan A, Mallet C, Guyon L, Cumont A, Phan C, Robert F, Thuillet R, Bordenave J, Sekine A, Huertas A, Ritvos O, Savale L, Feige JJ, Humbert M, Bailly S, Guignabert C. Selective BMP-9 inhibition partially protects against experimental pulmonary hypertension. Circ Res 2019;124:846–855. [DOI] [PubMed] [Google Scholar]
- 45. Long L, Ormiston ML, Yang X, Southwood M, Graf S, Machado RD, Mueller M, Kinzel B, Yung LM, Wilkinson JM, Moore SD, Drake KM, Aldred MA, Yu PB, Upton PD, Morrell NW. Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat Med 2015;21:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joseph JV, Conroy S, Tomar T, Eggens-Meijer E, Bhat K, Copray S, Walenkamp AM, Boddeke E, Balasubramanyian V, Wagemakers M, den Dunnen WF, Kruyt FA. TGF-beta is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis 2014;5:e1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data described in this article will be available upon reasonable request to the corresponding authors.