Abstract
The carboxyl-terminal domain of the small (S) envelope protein of hepatitis B virus was subjected to mutagenesis to identify sequences important for the envelopment of the nucleocapsid during morphogenesis of hepatitis delta virus (HDV) virions. The mutations consisted of carboxyl-terminal truncations of 4 to 64 amino acid residues and small combined deletions and insertions spanning the entire hydrophobic domain between residues 163 and 224. Truncation of as few as 14 residues partially inhibited glycosylation and secretion of S and prevented assembly or stability of HDV virions. Short internal combined deletions and insertions were tolerated for secretion of subviral particles with the exceptions of those affecting residues 164 to 173 and 219 to 223. However, mutants competent for subviral particle secretion had a reduced capacity for HDV assembly compared to that of the wild type. One exception was a mutant carrying a deletion of residues 214 to 218, which exhibited a twofold increase in HDV assembly (or stability), whereas deletions of residues 179 to 183, 194 to 198, and 199 to 203 were the most inhibitory. Substitutions of single amino acids between residues 194 and 198 demonstrated that HDV assembly deficiency could be assigned to the replacement of the tryptophan residue at position 196. We concluded that assembly of stable HDV particles requires a specific function of the carboxyl terminus of S which is mediated at least in part by Trp-196.
The assembly of hepatitis delta virus (HDV) particles depends upon the presence of hepatitis B virus (HBV) for the supply of its envelope proteins (23). The HDV particle consists of an outer envelope of HBV origin and an inner nucleocapsid made of a circular single-stranded RNA genome and two HDV-encoded proteins (p24 and p27) that bear the hepatitis delta antigen (HDAg) (29, 31). The viral envelope includes cell-derived lipids and the three HBV surface proteins designated large (L), middle (M), and small (S) (1). All three proteins are found at the surface of HBV virions, although S alone can be secreted as empty subviral lipoprotein particles through a nucleocapsid-independent mechanism (10). In addition, the mere presence of S is sufficient for assembly of HDV nucleocapsid-containing particles identical to mature HDV virions (30). However, in vitro infectivity requires the presence of the L protein in the viral envelope (27, 28). The 226 amino acid residues of the S protein sequence thus contain all the information necessary for its own secretion and that of HDV. Its synthesis occurs at the endoplasmic reticulum (ER) membrane as glycosylated (gp27) and nonglycosylated (p24) forms that anchor at the lipid membrane and dimerize. Then, they are transported via vesicles toward the Golgi compartment and secreted as empty 20-nm-diameter particles after oligomerization and budding at the pre-Golgi membrane into the lumen (15). The S protein contains at least two transmembrane domains the function of which has been verified experimentally (8, 9). The amino terminus (residues 1 to 3) is exposed at the luminal side of the ER membrane during synthesis; it is followed by a first transmembrane signal (signal I) located between residues 4 and 28, a cytosolic loop located between residues 28 and 80, and a second signal (signal II) that anchors the polypeptide chain into the membrane in the opposite direction with respect to signal I. The region located between residues 100 and 164 contains the major antigenic epitopes and a glycosylation site (11). It is translocated to the luminal compartment of the ER during synthesis and is found at the outside of secreted particles. It is referred to as an antigenic loop, and it was recently suggested that during synthesis, it may adopt an alternative cytoplasmic topology that corresponds to the nonglycosylated forms of S (22). The topology of the carboxyl-terminal domain located between residues 164 and 226 is not precisely known, but the major part, extending from residues 164 to 221, is predicted to be hydrophobic and to contain two transmembrane alpha-helices (21, 25).
Several recent studies have identified discrete regions or amino acid residues which are essential to S synthesis and/or secretion, including transmembrane signals and cysteine residues (3, 8, 9, 19, 20). More recently, we chose to examine the cytosolic loop of S, extending from residues 24 to 80, for its role in the envelopment of HDV nucleocapsid because its topology at the cytosolic side of the ER membrane appeared to be compatible with its binding to the nucleocapsid during virion assembly. We indeed identified a short sequence located at the carboxyl terminus of signal I, between residues 24 and 28, which had a critical influence on HDV assembly, but we could not demonstrate a direct interaction with the nucleocapsid (17). Prior to that study, Chen et al. (4) had reported that the carboxyl terminus of S was important for HDV virion assembly because its truncation by 50 residues was sufficient to abolish envelopment and secretion of coexpressed delta proteins. However, the capacity of this mutant for envelopment of an HDV RNA-containing nucleocapsid was not directly evaluated. In the present study, we examined the carboxyl terminus of S, extending from residues 162 to 226, for its function in HDV assembly. A series of truncations and combined deletion and insertion mutations in the DNA coding region for the S carboxyl terminus were constructed and examined for their effects on synthesis and secretion of subviral and HDV particles. The results indicated that the deletions (i) exerted a detrimental effect on subviral particle secretion (with the exception that one mutant retained wild-type [WT] properties) and (ii) reduced the capacity of S for HDV assembly (with the exception that one mutant showed facilitated assembly). Furthermore, we identified a short sequence located between residues 194 and 198 in which amino acid substitutions led to a drastic reduction of S capacity for HDV assembly without impairing its ability for subviral particle secretion.
MATERIALS AND METHODS
Production of HBV S envelope proteins in HuH-7 cells was achieved by using the expression vector p123 (17). Control plasmid pT7HB2.767, also referred to as env-negative (EN), is a derivative of p123 in which all of the three ATG start codons for the L, M, and S proteins were mutated to ACG to eliminate the expression of all three envelope proteins. Truncations were generated from plasmid p123 by using the PCR technique by amplification of a DNA fragment with a 5′ oligonucleotide spanning the XhoI restriction enzyme cleavage site (nucleotide 130) and a 3′ oligonucleotide at the position of the truncation that contained the TGA stop codon and the NsiI cleavage site. PCR-derived DNA fragments were then inserted into the corresponding XhoI to NsiI fragment in p123. Combined deletions and substitutions were carried out on plasmid p123 by using the PCR as described previously (17). The overlapping oligonucleotides contained a 15-nucleotide deletion corresponding to five amino acid residues in the S gene and a 6-nucleotide insertion for the HindIII cleavage site (corresponding to lysine and leucine residues) to provide easy detection and tracking of the mutation. The resulting S protein mutants thus contained a five-residue deletion that was replaced by insertion of the Lys-Leu sequence (see Fig. 1). They were designated by the numbers of the first and last residues of the deleted sequence and the letters K and L for the substitution with the Lys-Leu sequence. Single amino acid substitutions were also carried out by using the PCR overlap extension method. The mutations were designated by the one-letter code of the WT amino acid followed by its position in S and by the substituted amino acid. All PCR-generated fragments were cloned in p123 and sequenced by using the dideoxy method on double-stranded templates with Sequenase (Amersham). Clones containing the desired mutations within the PCR-generated fragments were selected and used for subsequent transfections. For production of HDAg proteins and replication of HDV RNA, we used the recombinant plasmid pSVLD3, which contains a head-to-tail trimer of full-length HDV cDNA for expression of HDV genomic RNA under the control of the simian virus 40 late promoter (18).
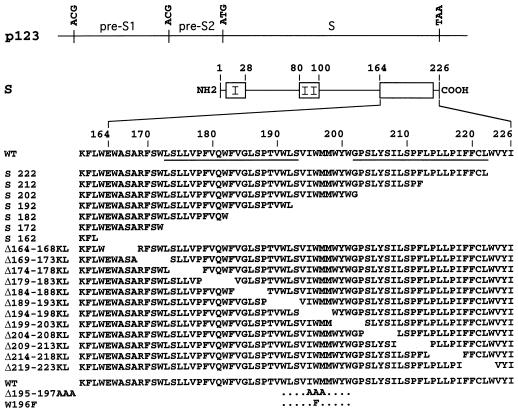
FIG. 1.
Schematic representations of S protein mutants. Plasmid p123 and S protein are depicted by horizontal thin lines. The HBV envelope protein open reading frame is divided into pre-S1, pre-S2, and S domains. Single base substitutions converted ATG (Met) start codons to ACG (Thr) codons for expression of the S protein only. Open boxes represent hydrophobic transmembrane regions in the S protein. Transmembrane signals I and II are indicated. The name and sequence of each mutant, including the positions of the first and last residues of the deleted sequence, are indicated. The letters KL correspond to an insertion of a Lys-Leu sequence at the site of deletion. The lower rows correspond to triple and single amino acid mutants. The position and nature of the substitutions are indicated. Underlined sequences represent putative transmembrane alpha-helices.
Cell culture and transfection of HuH-7 cells.
HuH-7 cells were maintained in Dulbecco modified Eagle medium/F12 supplemented with 5% fetal bovine serum. For production of HDV particles, cells were transfected with a mixture of (i) plasmid pSVLD3 for replication of HDV RNA and production of HDV proteins and (ii) the HBV recombinant plasmid p123 or derivatives for production of the WT or mutant S proteins, respectively. Transfection was carried out using Lipofectin (Life Technologies, Inc.) as described (17). For analysis of S protein expression by immunoprecipitation, transfections were carried out with 2 μg of p123 DNA or its derivatives per 0.5 × 106 cells per well and labeling was performed at day 6 posttransfection. For production of HDV particles, 0.5 × 106 cells were cotransfected with 0.6 μg of pSVLD3 DNA and 1.4 μg of p123 DNA or its derivatives. Culture medium was harvested on days 6, 9, 12, and 15 posttransfection and analyzed for the presence of envelope proteins and HDV RNA.
Preparation and characterization of particles produced by HuH-7 cells.
Culture fluids harvested on days 6, 9, 12, and 15 after transfection were used for purification of viral particles by precipitation in the presence of polyethylene glycol (PEG) as previously described (17). One half of the precipitate was resuspended in protein gel loading buffer and frozen at −70°C before protein analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting assay with anti-S antibodies. One half was resuspended in RNAble (Eurobio) for RNA extraction according to the guanidinium thiocyanate–acid phenol method (6).
For better characterization, PEG-precipitated particles were resuspended in 0.5 ml of 10 mM Tris-HCl (pH 7.4)–1 mM EDTA–150 mM NaCl (TNE), layered on the top of a 10 to 60% (wt/vol) cesium chloride gradient in TNE, and subjected to centrifugation at 35,000 rpm in an SW41 rotor (Beckman) for 16 h at 4°C. After centrifugation, fractions were collected from the bottom of the tube, and the density was determined by measurement of the refractive index. An aliquot of each fraction was used for detection of HDV RNA by agarose gel electrophoresis and blot hybridization and for detection of hepatitis B surface antigen (HBsAg).
Assays for viral RNA and envelope proteins.
Extraction of RNA from transfected cells or PEG-precipitated viral particles was carried out as described previously (17). Detection of HDV RNA was achieved after electrophoresis through a 1.2% agarose–2.2 M formaldehyde gel, transfer to a nylon membrane (Boehringer Mannheim) and hybridization to an HDV-specific RNA probe. A preparation of genomic HDV RNA standard was used to estimate the number of HDV molecules in each sample.
HBV envelope proteins were assayed as previously described (17). Briefly, particles sedimented from the culture medium were resuspended in disrupting buffer, submitted to SDS-PAGE, and immunodetected with rabbit R247 anti-S antibodies after transfer to a polyvinylidene difluoride membrane. Immunoblots were developed by chemiluminescence and exposure to Kodak films for detection of light emission.
Immunoprecipitation assay for S envelope proteins was carried out after metabolic labeling of transfected cells with [35S]Cys-[35S]Met (Amersham), immunoprecipitation of S proteins with R247 anti-S antibodies, SDS-PAGE, and autoradiography as described (17).
RESULTS
In an effort to understand the function of the hydrophobic carboxyl-terminal domain of HBV S protein in the morphogenesis of HDV particles, we constructed a series of mutants carrying mutations in the DNA coding region for S corresponding to progressive truncations of 4 to 64 amino acid residues. The mutant vectors designated by the position of the last residues in the S sequence are depicted in Fig. 1. We then constructed a series of progressive deletions, of five amino acid residues each, of amino acids located between positions 164 and 223. Each deleted sequence was replaced with a Lys-Leu coding sequence as described in the Materials and Methods section. The mutant vectors, referred to as KL mutants, were designated by the positions of the deleted residues in the S sequence and by the letters K and L for Lys and Leu, as indicated in Fig. 1. When needed, mutants referred to as GA mutants were generated. They consisted of a five-residue deletion and substitution with the Gly-Ala dipeptide. Mutant Δ195-197AAA contained a substitution of each of Ile-195, Trp-196 and Met-197 with Ala, and W196F was a single amino acid substitution of Trp-196 with Phe.
Effects of S protein carboxyl-terminal truncations on secretion of subviral and HDV RNA-containing particles.
Expression of WT and mutant S genes was examined by transient transfection of HuH-7 cells with p123 and each mutant expression vector as described previously (28). Transfected cells were labeled for 24 h with [35S]Cys and [35S]Met, and the envelope proteins from the cell lysates and supernatants were immunoprecipitated with anti-S antibodies (R247) and analyzed by SDS-PAGE and autoradiography. As reported previously, R247 antibodies recognize a linear epitope encompassing the Gln-54 to Ser-64 sequence of the S polypeptide (17).
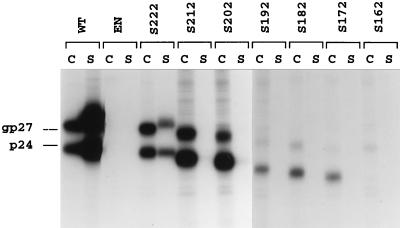
As shown in Fig. 2, transfection of HuH-7 cells with WT HBV DNA led to the synthesis of nonglycosylated (p24) and glycosylated (gp27) forms of the S protein. They were detected in both cell lysate and supernatant, indicating efficient synthesis and secretion. Mutant S222 had WT characteristics with regard to synthesis, whereas secretion was slightly affected. Mutants S212 and S202 exhibited a near-WT level of synthesis and stability, although the ratio of glycosylated to nonglycosylated forms was altered and their capacity for secretion was impaired. Interestingly, mutants S192, S182, and S172 were detected only in the cell lysates and predominantly as nonglycosylated polypeptides, although the N-glycosylation site is located at Asn-146. The deletion may have led to a protein topology incompatible with glycosylation and secretion as a result of misfolding and retention at the ER membrane. Finally, mutant S162, which lacks the entire carboxyl-terminal hydrophobic domain, failed to be detected under our experimental conditions.
FIG. 2.
Examination of WT and S protein truncated mutants expressed in HuH-7 cells. Six days after transfection of 0.5 × 106 HuH-7 cells with 2 μg of p123 DNA or its derivatives, transfected cells were labeled with 200 μCi of [35S]Cys-[35S]Met for 24 h. After labeling, cell lysates (C) and supernatants (S) were immunoprecipitated with rabbit R247 anti-S antibodies. One half of each immunoprecipitate was analyzed by SDS-PAGE. After electrophoresis, the gel was fixed, soaked in Amplify solution (Amersham), dried, and subjected to fluorography at −70°C for 48 h. The migration positions of glycosylated (gp27) and nonglycosylated (p24) S proteins are indicated. EN, env-negative plasmid.
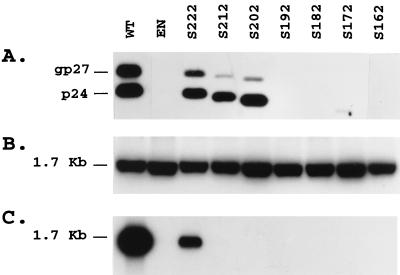
For production of enveloped HDV RNA-containing particles, HuH-7 cells were cotransfected with WT or mutant S expression vectors and the pSVLD3 HDV expression vector as described (27). Culture supernatant was harvested on days 6, 9, 12, and 15 posttransfection, and viral particles were precipitated in the presence of PEG before analyses for S protein by immunoblotting with anti-S antibodies and for HDV RNA by agarose gel electrophoresis and blot hybridization. Cells were also harvested on day 15 for analysis of intracellular HDV RNA. A control experiment was performed by cotransfection of HuH-7 cells with pSVLD3 and the env-negative HBV plasmid pT7HB2.767. As illustrated in Fig. 3 and as already demonstrated (24), there was no evidence for the release of enveloped HDV RNA containing particles in the absence of HBV envelope proteins. Transfection of HuH-7 cells with the mutants S222, S212, and S202 led to accumulation of subviral particles in the culture medium, as evidenced by the detection of nonglycosylated and glycosylated S polypeptides (Fig. 3A). The increased mobility of the S mutants corresponded to the expected molecular mass reduction. As shown in Fig. 3C, only mutant S222 was competent for HDV nucleocapsid envelopment, as indicated by the presence of viral RNA in PEG precipitate. Taken together, our results are consistent with previous studies (4) in demonstrating that the carboxyl terminus of S contains signal(s) important for glycosylation, stability, and secretion of subviral particles and for the envelopment of HDV RNA-containing nucleocapsid. Furthermore, a truncation by as few as 14 carboxyl-terminal residues was sufficient for partial loss of glycosylation and for inhibition of HDV assembly capability. This truncation may have led to the deletion of a specific sequence that is directly involved in the binding of S to the HDV nucleocapsid, or it may have affected the proper presentation of a nucleocapsid binding domain removed from the position affected by the deletion, by modifying the overall conformation of the S carboxyl terminus.
FIG. 3.
Detection of HDV RNA-containing particles in culture fluids from cells cotransfected with pSVLD3 HDV plasmid DNA and WT or S protein truncation mutant DNA. (A) Immunoblot analysis of S proteins extracted from culture fluids from 0.5 × 106 HuH-7 cells at days 6, 9, 12, and 15 after transfection with a mixture of 0.6 μg of pSVLD3 plasmid DNA and 1.4 μg of WT, env-negative (EN), or mutant HBV DNA. Particles sedimented from 200 μl of culture medium were disrupted in Laemmli sample buffer containing 2% SDS and 2% β-mercaptoethanol. Proteins were separated on a 12% acrylamide gel, transferred to a polyvinylidene difluoride membrane, and probed with anti-S antibody (1:2,000). (B) Cellular RNA was extracted from HuH-7 cells harvested at day 15 posttransfection. Five micrograms of total RNA was separated on agarose gel and analyzed for the presence of HDV RNA after transfer to a nylon membrane and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe. Following hybridization, filters were washed, dried, and autoradiographed at −70°C for 16 h with an intensifying screen. The size expressed in kilobases of HDV genomic RNA is indicated. (C) Particles sedimented from 500 μl of culture medium were disrupted in RNAble, and RNA was purified. RNA was analyzed by agarose gel electrophoresis followed by transfer to nylon membrane and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe as described for panel B.
Effects of combined deletion and substitution mutations on secretion of subviral and enveloped HDV RNA-containing particles.
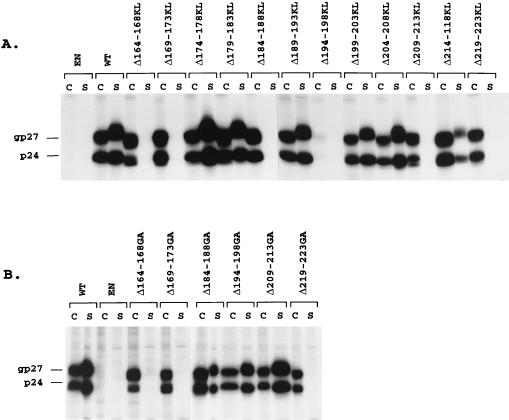
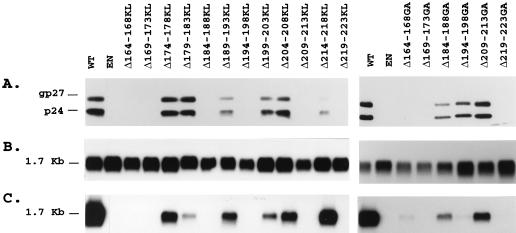
To further investigate the functional properties of the S carboxyl terminus in HDV assembly, a series of progressive deletions, of five amino acid residues each, of amino acids located between positions 164 and 223 was generated. Biosynthesis and secretion were examined by transient transfection of HuH-7 cells with p123 or derivative plasmids followed by immunoprecipitation assay with anti-S antibodies. As illustrated in Fig. 4A, mutants Δ174-178KL, Δ179-183KL, Δ189-193KL, Δ199-203KL, Δ204-208KL, and Δ214-218KL were competent for synthesis and secretion at levels comparable to that of the WT, whereas mutants Δ164-168KL, Δ169-173KL, Δ184-188KL, Δ209-213KL, and Δ219-223KL, detected at WT levels in the cell lysates, were deficient for secretion. Only traces of S proteins were visible in the lysate of cells transfected with mutant Δ194-198KL.
FIG. 4.
Examination of WT and combined deletion and insertion mutant S proteins expressed in HuH-7 cells. Each deletion and insertion consisted of a five-amino-acid-residue deletion and insertion of Lys-Leu (A) or Gly-Ala (B) residues as described in the Materials and Methods section. Six days after transfection of 0.5 × 106 HuH-7 cells with 2 μg of p123 DNA or its derivatives, transfected cells were labeled with 200 μCi of [35S]Cys-[35S]Met for 24 h. After labeling, cell lysates (C) and supernatants (S) were immunoprecipitated with anti-S antibodies. One half of each immunoprecipitate was analyzed as described in the legend for Fig. 2. The migration positions of glycosylated (gp27) and non-glycosylated (p24) S proteins are indicated. EN, env-negative.
To eliminate the possibility that the Lys-Leu substitution, rather than the deletion, had an inhibitory effect on synthesis or secretion of mutants Δ164-168KL, Δ169-173KL, Δ184-188KL, Δ194-198KL, Δ209-213KL and Δ219-223KL, we constructed a new panel of combined deletions and insertions in which the insertion of the Lys-Leu sequence was changed to that of Gly-Ala. Mutants designated Δ164-168GA, Δ169-173GA, Δ184-188GA, Δ194-198GA, Δ209-213GA and Δ219-223GA were analyzed for stability and secretion competence in HuH-7 cells by transient transfection, metabolic labeling and immunoprecipitation with anti-S antibodies. We observed that the substitution of Gly-Ala for Lys-Leu restored S synthesis and secretion in cells transfected with Δ194-198GA and restored secretion in cells transfected with Δ184-188GA and Δ209-213GA (Fig. 4B). It is thus likely that the introduction of the positively charged lysine residue in the Lys-Leu dipeptide at the position of the deletions caused the detrimental effects on S expression. It is noteworthy that residues 184 to 188 and 209 to 213 are located within alpha-helix transmembrane structures predicted by Persson and Argos (21). S proteins carrying the Gly-Ala dipeptide instead of the Lys-Leu dipeptide at deleted residues 164 to 168, 169 to 173, and 219 to 223 were deficient for secretion, although they were detected in cells at WT levels as nonglycosylated and glycosylated forms.
Production of HDV RNA-containing particles enveloped with KL or GA deletion mutants was attempted in HuH-7 cells by cotransfection with pSVLD3 as described above. Culture supernatant was harvested on days 6, 9, 12, and 15 posttransfection, and viral particles were precipitated in the presence of PEG before analysis for S protein and HDV RNA. HDV nucleocapsid envelopment was significantly reduced for all deletion mutants compared to that of the WT (by at least threefold) as measured by the amount of HDV RNA in the PEG precipitates normalized to that of S proteins (Fig. 5 and Table 1). One notable exception was mutant Δ214-218KL, which exhibited an apparent facilitation for nucleocapsid envelopment to a level equivalent to approximately twofold that for the WT. In contrast, mutants Δ179-183KL, Δ194-198GA, and Δ199-203KL exhibited the most-deleterious effect in HDV assembly and showed levels corresponding to 5, 1.5, and 10% of that of the WT, respectively. These estimations were conducted by comparisons of amounts of viral RNA and S proteins to known amounts of synthetic HDV RNA and purified HBV envelope protein preparations, respectively. Serial dilution (1/5) was made for each sample and standard of RNA or protein, and autoradiogram signal measurement was performed by using a densitometer. The amount of viral RNA was estimated at 1 pg (106 genome equivalents) per ml of culture medium for Δ194-198GA-coated particles and at 100 pg (108 genome equivalents) per ml for WT-coated particles, whereas Western blot analysis revealed that culture supernatants from WT- and Δ194-198GA-transfected cells contained approximately 80 ng of S proteins (2 × 1010 empty particle equivalents) per ml and 60 ng of S proteins (1.5 × 1010 empty particle equivalents) per ml, respectively. Overall, we observed that all deletion mutations had a negative effect on secretion of subviral particles except for Δ209-213GA (see Table 1) and to a greater extent on secretion of HDV nucleocapsid with the exception of Δ214-218KL. All remaining mutants, including carboxyl-terminal truncations, exhibited a greater than threefold reduction in their capacity for HDV assembly. Mutants Δ199-203KL and Δ179-183KL presented an inhibitory effect estimated at 10% and 5% of that of the WT, respectively, whereas the most-deleterious effect was observed with Δ194-198GA, which had efficiency in HDV envelopment 1.5% of that of the WT.
FIG. 5.
Detection of HDV RNA-containing particles in culture fluids from cells cotransfected with pSVLD3 HDV plasmid DNA and WT or S protein mutant DNA. (A) Immunoblot analysis of S proteins extracted from culture fluids from 0.5 × 106 HuH-7 cells at days 6, 9, 12, and 15 after transfection with a mixture of 0.6 μg of pSVLD3 plasmid DNA and 1.4 μg of WT, env-negative (EN), or mutant HBV plasmid DNA. Particles sedimented from 200 μl of culture medium were disrupted in Laemmli sample buffer containing 2% SDS and 2% β-mercaptoethanol. Proteins were separated on a 12% acrylamide gel, transferred to a polyvinylidene difluoride membrane and probed with anti-S antibodies (1:1,000). (B) Cellular RNA was extracted from HuH-7 cells harvested at day 15 posttransfection. Five micrograms of total RNA was separated on an agarose gel and analyzed for the presence of HDV RNA after transfer to a nylon membrane and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe. Following hybridization, filters were washed, dried, and autoradiographed at −70°C for 16 h with an intensifying screen. The size expressed in kilobases of HDV genomic RNA is indicated. (C) Particles sedimented by precipitation in the presence of 9% PEG from 500 μl of culture medium were disrupted in RNAble, and RNA was purified. RNA was analyzed by agarose gel electrophoresis followed by transfer at nylon membrane and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe as described for panel B.
TABLE 1.
S envelope protein mutant phenotypes
| WT and S mutant designation | Concn of:
|
||
|---|---|---|---|
| S protein (ng/ml of culture medium) | HDV RNA (pg/ml of culture medium) | HDV RNA (pg normalized to 80 ng of S protein) | |
| WT | 80 | 100 | 100 |
| EN | 0 | 0 | NAa |
| S-222 | 33 | 14 | 34 |
| S-212 | 7.5 | 0 | NA |
| S-202 | 6 | 0 | NA |
| S-192 | 0 | 0 | NA |
| S-182 | 0 | 0 | NA |
| S-172 | 1.5 | 0 | NA |
| S-162 | 0 | 0 | NA |
| Δ164-168KL | 0 | 0 | NA |
| Δ169-173KL | 0 | 0 | NA |
| Δ174-178KL | 63 | 17.5 | 22 |
| Δ179-183KL | 57 | 3.5 | 5 |
| Δ184-188GA | 32 | 11.5 | 29 |
| Δ189-193KL | 30 | 11.5 | 30 |
| Δ194-198GA | 60 | 1 | 1.5 |
| Δ199-203KL | 44 | 5.5 | 10 |
| Δ204-208KL | 64 | 15 | 19 |
| Δ209-213GA | 80 | 27 | 27 |
| Δ214-218KL | 17 | 39 | 183 |
| Δ219-223KL | 0 | 0 | NA |
| Δ195-197AAA | 80 | 1.5 | 1.5 |
| W196F | 80 | 1.5 | 1.5 |
NA, not applicable.
Effects of substitutions of amino acids between residues 194 and 198 of S on secretion of subviral and HDV particles.
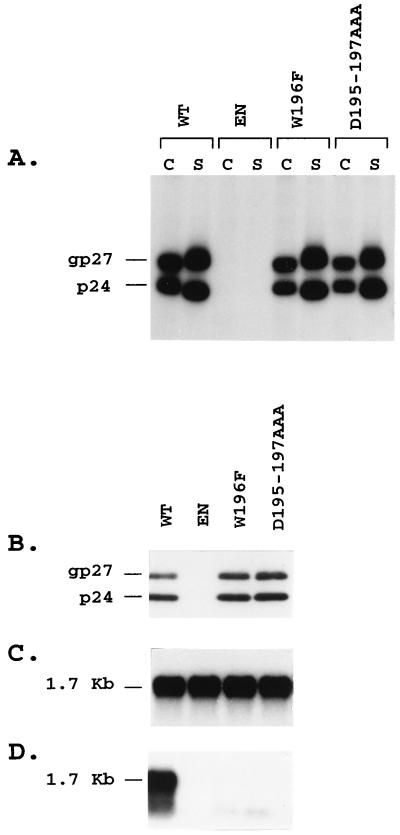
We then attempted to identify specific residues within the Val-Ile-Trp-Met-Met sequence corresponding to the deletion carried by the most-deleterious mutant, Δ194-198GA. The amino acid sequence at positions 193 to 202 has been predicted to form a turn between two alpha-helices located at positions 173 to 193 and 202 to 222 and to face the cytoplasm at the ER membrane. We first observed that the corresponding sequence on the woodchuck hepatitis B virus S protein, which is also competent for HDV nucleocapsid envelopment, contains a leucine residue instead of valine at position 194 and an isoleucine instead of methionine at position 198. We thus constructed a mutant, Δ195-197AAA, that carried a substitution of the Ala-Ala-Ala sequence for the Ile-Trp-Met tripeptide in order to change only conserved residues while strictly maintaining the length of the polypeptide chain. We observed that, after transfection in HuH-7 cells and immunoprecipitation with anti-S antibodies, Δ195-197AAA mutant had a WT phenotype with regard to synthesis and secretion of subviral particles but had lost its capacity for HDV assembly as judged by the absence of HDV RNA in PEG-precipitated particles from cells cotransfected with recombinant HDV plasmid pSVLD3 (Fig. 6).
FIG. 6.
Analysis of WT and W196F and Δ195-197AAA S protein mutants for subviral and HDV particle secretion. (A) HuH-7 cells transfected with WT or S protein mutants were labeled with 200 μCi of [35S]Cys-[35S]Met for 24 h, and cell lysates (C) and supernatants (S) were immunoprecipitated with rabbit R247 anti-S antibodies. (B) For expression of HDV RNA-containing particles, 0.5 × 106 HuH-7 cells were cotransfected with a mixture of 0.6 μg of pSVLD3 plasmid DNA and 1.4 μg of WT, env-negative (EN), or mutant DNA. Particles were harvested at days 6, 9, 12, and 15 posttransfection and sedimented by precipitation in the presence of 9% PEG from 200 μl of culture medium. Proteins were analyzed by SDS-PAGE and immunoblotting with anti-S antibodies (1:2,000). (C) Cellular RNA was extracted from HuH-7 cells harvested at day 15 posttransfection, and 5 μg was analyzed by gel electrophoresis and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe. (D) RNA purified from particles precipitated from 500 μl of culture medium was analyzed by gel electrophoresis and hybridization to a genomic strand-specific 32P-labeled HDV RNA probe. The gel migration positions of intracellular or extracellular genomic HDV RNA (1.7 Kb) are indicated in panels C and D, and those of S proteins (p24 and gp27) are indicated in panels A and B. EN, env-negative.
In an attempt to assign an essential function in HDV envelopment to a specific residue in the amino acid sequence from 194 to 198, we constructed a single amino acid substitution at Trp-196. It was targeted for specific mutagenesis because the indole ring of Trp is likely to protrude from the Val-Ile-Trp-Met-Met sequence in a position prone to establish hydrophobic binding in a protein-protein interaction. We decided to create a conservative mutation by substituting the indole ring of Trp with the phenyl ring of Phe in an attempt to generate the least structurally disturbing modification that maintained subviral particle secretion while preventing HDV assembly. The resulting mutant W196F had indeed a WT efficiency for subviral particle synthesis and secretion, and it appeared deficient for envelopment of HDV nucleocapsid as judged by the presence of only traces of HDV RNA in PEG-precipitated particles (Fig. 6). W196F S therefore represented the minimal mutant in the S carboxyl terminus that is secretion competent and HDV assembly deficient.
Characterization of HDV assembly-defective mutants.
Particles produced by HuH-7 cells cotransfected with HDV recombinant plasmid pSVLD3 and plasmids coding for Δ194-198GA, Δ195-197AAA, and W196F mutants were characterized by isopycnic centrifugation in a cesium chloride gradient. We sought to (i) verify the buoyant density of subviral and HDV particles, (ii) ascertain that traces of HDV RNA detected in the PEG precipitates of assembly-deficient mutants were indeed part of enveloped virions, (iii) better estimate the amount of viral RNA in virions, and (iv) visualize HDV proteins. After centrifugation and fractionation of the gradient, S proteins and genomic HDV RNA were detected in fractions with densities of about 1.20 g/cm3, consistent with the buoyant density of HDV (Fig. 7). As expected, there was no detectable HDV RNA or S proteins in the control experiment conducted with the env-negative plasmid pT7HB2.767, and only traces of viral RNA could be detected in the Δ194-198GA, Δ195-197AAA, and W196F particles. Since we did not have available a very sensitive antiserum, delta proteins were not easily resolved. However, both p24 and p27 forms were clearly visible in the fraction with a density of 1.20 g/cm3 of the WT control particles but were undetected in that of the HDV RNA-deficient particles (data not shown). By measurement of HDV RNA signals in WT particle extracts and Δ195-197AAA and W196F mutant particle extracts (normalized for S protein content) after blot hybridization and analysis with a densitometer, we confirmed the efficiency in envelopment of HDV nucleocapsid for these two mutants to be approximately 1.5% of that of the WT. We thus concluded that W196F S represents an HDV assembly-defective mutant with WT competence for subviral particle assembly and secretion but with a greater than 60-fold reduction in its capacity for production of stable HDV particles.
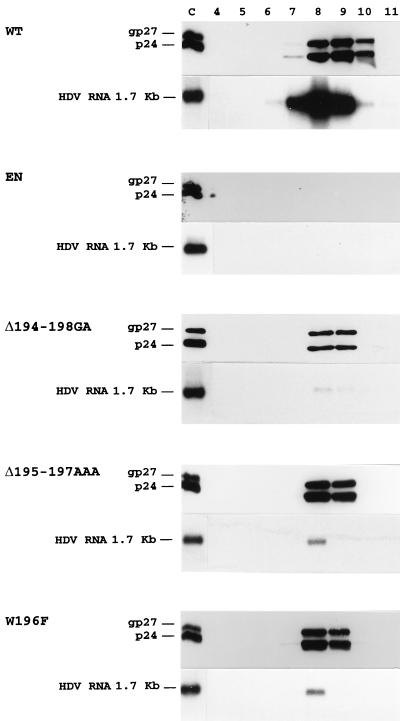
FIG. 7.
Characterization of HDV RNA-containing particles by isopycnic centrifugation in a cesium chloride gradient. Culture medium from HuH-7 cells was collected at days 6, 9, 12, and 15 after transfection of 0.5 × 106 cells with 0.6 μg of pSVLD3 plasmid DNA and 1.4 μg of WT, env-negative (EN), or S protein mutant DNA. Particles were concentrated from 5 ml of culture medium and subjected to centrifugation to equilibrium in 12-ml cesium chloride gradients. Fractions were collected from the bottom of the tube, and proteins from 1/4 of each 1-ml fraction were analyzed by immunoblotting with anti-S antibodies (1:2,000). RNA was extracted from each fraction for analysis by agarose gel electrophoresis and blot hybridization by using a genomic strand-specific 32P-labeled HDV RNA probe. The gel migration positions of genomic HDV RNA (1.7 kb) and S proteins (p24 and gp27) are indicated. The numbering (4 to 11) of each fraction is indicated. Fractions 8 and 9 correspond to fractions with densities of 1.22 and 1.18 g/cm3, respectively. Purified WT HDV particles were used as controls (C).
DISCUSSION
Unlike type D retroviruses, which possess a nucleocapsid that can bud at the host cell membrane in the absence of envelope proteins (14), or alphaviruses, for which budding is driven by a specific interaction between nucleocapsid and envelope proteins (26), hepadnaviruses use a nucleocapsid-independent budding mechanism where the S protein is the driving force (10). HDV takes advantage of the intrinsic capacity of S for lipoprotein particle formation to envelope its own nucleocapsid and exit the hepatocyte as a virion. The efficiency of HDV envelopment is likely to depend on the existence of a specific interaction between nucleocapsid and S polypeptide during maturation of HDV virions (16).
Recently, we identified a discrete sequence located at the carboxyl boundary of the transmembrane signal I for S, between Arg-24 and Ile-28, that is important for HDV particle assembly (17), and in a previous study, Chen et al. (4) reported that a carboxyl-terminal truncation of S by 50 residues led to inhibition of delta protein envelopment and secretion. Here, we demonstrate that the S carboxyl terminus indeed contains residues, in particular Trp-196, that are required for efficient assembly of stable HDV particles.
The carboxyl terminus of S is highly hydrophobic, and it is predicted to contain two transmembrane alpha-helices located at positions 173 to 193 and 202 to 222 (21). They bracket a short sequence (194 to 201) that presents a low degree of flexibility. Hydrophobicity and secondary structure predictions are compatible with an association of this region with the ER membrane through the two alpha-helices, placing the 194-201 turn sequence including Trp-196 at the cytoplasmic side of the ER membrane in a position potentially adequate for interaction with a nucleocapsid. However, a recent study aimed at mapping monoclonal anti-S antibodies using the phage display library technique suggested that the 187-207 sequence could be present at the outside of the S subvirus particles, a topology apparently incompatible with its binding to delta proteins during HDV maturation (5). Possibly, as suggested by Prange and Streeck (22) with regard to the antigenic loop, the entire carboxyl terminus of S could adopt the following two alternative topologies during synthesis: (i) a topology that positions Trp-196 at the cytosolic side of the ER membrane during maturation, and (ii) a topology where Trp-196 is at the luminal side of the ER membrane or at the outside of the particle as proposed by Chen et al. (5).
It is noteworthy that facilitation mutant Δ214-218KL, which presented an apparent twofold increase in HDV assembly or stability compared to that of WT S, was partially deficient for secretion of subviral particles. The 214-218KL mutation affects a domain which is predicted to reside at the center of a putative transmembrane alpha-helix structure (202 to 222). It thus is expected that a deletion and/or introduction of a positively charged Lys residue at this position could modify the transmembrane signal and thereby the secretion efficiency of the polypeptide.
Some of the HDV assembly-deficient mutants described here may have prevented nucleocapsid recognition without deleting a nucleocapsid binding motif per se, by simply inducing a steric hindrance that interferes with the proper presentation of the binding motif. Most of the deletions and insertions between residues 174 and 218 had the tendency to reduce synthesis or secretion of S proteins (Fig. 4) and to inhibit secretion or stability of HDV RNA-containing particles, suggesting that the length of the carboxyl-terminal polypeptide chain is very critical. Since the introduction of a positively charged Lys residue in the KL mutants is also a potential disrupting factor, the expression of additional mutants carrying substitutions that are conservative in terms of hybrophobicity should be very informative.
According to the present and preceding studies (17), it appears that envelopment of HDV nucleocapsid could involve several discrete regions that include at least one short sequence in the cytosolic loop (residues 24 to 28) and two short sequences in the carboxyl terminus (residues 179 to 183 and 194 to 203). Whether the antigenic loop contains additional HDV envelopment signals is currently under scrutiny.
The fact that residues essential to HDV assembly, such as Trp-196, are dispensable for subviral particle secretion and yet strictly conserved among HBV isolates or woodchuck hepatitis B virus suggests that the selection pressure that has led to their conservation concerns functions other than those involved in lipoprotein particle budding and secretion. We thus can speculate that HDV nucleocapsid envelopment relies upon S residues that are conserved because they are essential at the step of maturation or infectivity of HBV virions. It is not known precisely which specific residues of the L or S envelope proteins participate directly in the nucleocapsid envelopment during HBV virion maturation, but it is quite possible that in addition to the pre-S1 amino acid sequence at 103 to 124 already identified (2), the S polypeptide contains motifs or residues that are instrumental in this process. Whether mutations that are deleterious to HDV envelopment, such as the W196F substitution, are also inhibitory for HBV virion assembly remains to be determined.
Although the sequences at 219 to 222 and 164 to 173 were not tested in this study because stable mutants carrying deletion and insertion in these regions could not be generated, it is clear that discrete regions located at positions 179 to 183 and 199 to 203 and, more importantly, a Trp residue at position 196 participate in the HDV assembly process. The W196F mutation is conservative because it replaces the indole ring of tryptophan with the phenyl ring of phenylalanine, and it is predicted to increase flexibility while maintaining hydrophobicity (7). The exact function of Trp-196 and other motifs identified here as being important in the HDV assembly mechanism remains uncertain; in particular, direct binding is not demonstrated. Discrete regions or residues of S may be involved in interactions either intramolecularly, to establish a particular secondary structure in the S polypeptide, or intermolecularly in combination with membrane lipids, with the p27 HDV nucleocapsid protein including its farnesyl group, or with other S proteins.
Whatever mechanism is used for selective envelopment of HDV nucleocapsid by HBV envelope proteins, it must be a very specific process, because sera collected at the peak of an acute coinfection usually present a very high proportion of HDV nucleocapsid-containing particles to empty ones. The specificity for envelopment is likely mediated through specific interaction between S and delta proteins, which could be the target for the design of antiviral molecules.
ACKNOWLEDGMENTS
This work was supported in part by Association pour la Recherche contre le Cancer, Fondation pour la Recherche Médicale, La Ligue contre le Cancer, Conseil Régional de Languedoc-Roussillon, and Centre National de la Recherche Scientifique. S.J. was supported by a fellowship from Ministère de l’Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Bonino F, Heermann K H, Rizzetto M, Gerlich W H. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J Virol. 1986;58:945–950. doi: 10.1128/jvi.58.3.945-950.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss V, Ganem D. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J Virol. 1991;65:3813–3820. doi: 10.1128/jvi.65.7.3813-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P-J, Lai W-J, Wang C-J, Chen D-S. Hepatitis B surface antigen and large-form hepatitis delta antigen in HDV assembly: a further study. In: Hadziyannis S J, Taylor J M, Bonino F, editors. Hepatitis delta virus: molecular biology, pathogenesis, and clinical aspects. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 29–34. [PubMed] [Google Scholar]
- 5.Chen Y-C J, Delbrook K, Dealwis C, Mimms L, Mushahwar I K, Mandecki W. Discontinuous epitopes of hepatitis B surface antigen derived from a filamentous phage peptide library. Proc Natl Acad Sci USA. 1996;93:1997–2001. doi: 10.1073/pnas.93.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Davies D R, Padlan E A, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 8.Eble B E, Lingappa V R, Ganem D. Hepatitis B surface antigen: an unusual secreted protein initially synthesized as a transmembrane polypeptide. Mol Cell Biol. 1986;6:1454–1463. doi: 10.1128/mcb.6.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eble B E, MacRae D R, Lingappa V R, Ganem D. Multiple topogenic sequences determine the transmembrane orientation of hepatitis B surface antigen. Mol Cell Biol. 1987;7:3591–3601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem D. Hepadnaviridae and their replication. In: Fields B N, editor. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 11.Heermann K H, Gerlich W H. Surface proteins of hepatitis B viruses. In: McLachlan A, editor. Molecular biology of hepatitis B virus. Boca Raton, Fla: CRC Press; 1991. pp. 109–143. [Google Scholar]
- 12.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horten R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 14.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 15.Huovila A-P J, Eder A M, Fuller S D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang S B, Lai M M C. Isoprenylation mediates direct protein-protein interactions between hepatitis large delta antigen and hepatitis B virus surface antigen. J Virol. 1993;67:7659–7662. doi: 10.1128/jvi.67.12.7659-7662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenna S, Sureau C. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology. 1998;251:176–186. doi: 10.1006/viro.1998.9391. [DOI] [PubMed] [Google Scholar]
- 18.Kuo M Y-P, Chao M, Taylor J M. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangold C M T, Streeck R E. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993;67:4588–4597. doi: 10.1128/jvi.67.8.4588-4597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold C M T, Unckell F, Werr M, Streeck R E. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology. 1995;211:535–543. doi: 10.1006/viro.1995.1435. [DOI] [PubMed] [Google Scholar]
- 21.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 22.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzetto M, Hoyer B, Canese M G, Shih J W K, Purcell R H, Gerin J L. δ agent: association of δ antigen with hepatitis B surface antigen and RNA in serum of δ-infected chimpanzees. Proc Natl Acad Sci USA. 1980;77:6124–6128. doi: 10.1073/pnas.77.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu W-S, Bayer M, Taylor J M. Assembly of hepatitis delta virus particles. J Virol. 1992;66:2310–2315. doi: 10.1128/jvi.66.4.2310-2315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stirk H J, Thornton J M, Howard C R. A topological model for hepatitis B surface antigen. Intervirology. 1992;33:148–158. doi: 10.1159/000150244. [DOI] [PubMed] [Google Scholar]
- 26.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sureau C, Guerra B, Lanford R E. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J Virol. 1993;67:366–372. doi: 10.1128/jvi.67.1.366-372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sureau C, Guerra B, Lee H. The middle hepatitis B virus envelope protein is not necessary for infectivity of hepatitis delta virus. J Virol. 1994;68:4063–4066. doi: 10.1128/jvi.68.6.4063-4066.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor J M. Hepatitis delta virus and its replication. In: Fields B N, editor. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2809–2818. [Google Scholar]
- 30.Wang C-J, Chen P-J, Wu J-C, Patel D, Chen D-S. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J Virol. 1991;65:6630–6636. doi: 10.1128/jvi.65.12.6630-6636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K-S, Choo Q-L, Weiner A J, Ou J-H, Najarian R C, Thayer R M, Mullenbach G T, Denniston K J, Gerin J L, Houghton M. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]