Abstract
Glial cell line-derived neurotrophic factor (GDNF) signals through a multi-component receptor system predominantly consisting of glycosyl-phosphatidylinositol-anchored GDNF family receptor alpha-1 (GFRα1) and the Rearranged during transfection (RET) receptor tyrosine kinase. GDNF/RET signaling is vital to the central and peripheral nervous system, kidney morphogenesis, and spermatogenesis. In addition, the dysregulation of the GDNF/RET signaling has been implicated in the pathogenesis of cancers. Despite the extensive research on GDNF/RET signaling, a molecular network of reactions induced by GDNF reported across the published literature. However, a comprehensive GDNF/RET pathway resource is currently unavailable. We describe an integrated signaling pathway reaction map of GDNF/RET consisting of 1151 molecular reactions. These include information pertaining to 52 molecular association events, 70 enzyme catalysis events, 36 activation/inhibition events, 22 translocation events, 856 gene regulation events, and 115 protein-level expression events induced by GDNF in diverse cell types. We developed a comprehensive GDNF/RET signaling network map based on these molecular reactions. The pathway map was made accessible through WikiPathways database (https://www.wikipathways.org/index.php/Pathway:WP5143).
Graphical abstract
Biocuration and development of gene regulatory network map of GDNF/RET signaling pathway
Supplementary Information
The online version contains supplementary material available at 10.1007/s12079-023-00726-1.
Keywords: Multi-component receptor system, Receptor tyrosine kinases, Protein–protein interactions, Signaling network, Post-translational modifications
Introduction
Glial-cell-line-derived neurotrophic factor (GDNF) is a potent neurotrophic factor for the midbrain dopaminergic (DA) neurons (Lin et al. 1993). GDNF was first discovered in 1991 by researchers at Synergen, a biotechnology-based company in Boulder, Colorado, USA (Vastag 2010). It is a prominent member of the GDNF family of ligands (GFLs) that also comprises artemin (ARTN), neurturin (NRTN), and persephin (PSPN) (Zihlmann et al. 2005). A wide variety of cell types synthesize and secrete GDNF, including glial cells such as astrocytes, oligodendrocytes, Schwann cells, motor neurons (MNs), and skeletal muscles. GDNF is first synthesized as a precursor protein pro-GDNF. After a series of protein cleavage and processing, the 211 amino acid pro-GDNF is finally converted into the active and mature form of GDNF (Cintron-Colon et al. 2020).
The GDNF signals several intracellular pathways through receptors, including a unique multi-component receptor system, preferably the glycosyl-phosphatidylinositol (GPI)-linked cell surface co-receptor, GFRα1, and the signal transducer RET (REarrangement during Transfection) receptor tyrosine kinase. Besides GFRα1 and RET, GDNF also signals through neural cell adhesion molecule-1 (NCAM1), syndecan-3 (SDC3), and integrin-β1 (ITGB1), independent of RET. In mammals, two major alternate splice isoforms of RET that differ in their C-terminal tails (RET9 and RET51) are considered to exhibit different signaling properties in vivo (de Graaff et al. 2001). Cells that co-express both RET and GFRα1 receptors mediate cis signaling. The signaling mediated by soluble or immobilized GFRα1 undergoes ‘trans’-signaling, where RET is activated outside the lipid rafts by GFRα1 (Paratcha et al. 2001).
The GDNF/RET signaling is primary to the central and peripheral nervous system as a survival factor for multiple neuron types and a neuroprotective factor during many neurodegenerative disorders (Takahashi 2001). The RET signaling pathway is essential for the growth and formation of kidneys, as well as the maturation of spermatogonia, which are the precursors of sperm cells (Schuchardt et al. 1996; Meng et al. 2000). It is known that RET is an oncogenic driver in several cancers, including papillary thyroid carcinomas (PTCs), non-small cell lung cancers (NSCLCs), breast, ovarian, and head and neck tumors (Adashek et al. 2021; Thein et al. 2021; Regua et al. 2022). In addition, mutations in the RET gene have been associated with thyroid melanoma and multiple endocrine neoplasia (MEN) syndromes-2A and -2B (Adashek et al. 2021). Therefore, GDNF/RET signaling systems are being targeted with multiple therapeutic approaches in the treatment of diverse diseases, including neurological conditions and cancers (Plaza-Menacho et al. 2014; Mahato and Sidorova 2020).
However, the molecular events associated with GDNF/RET signaling are scattered across literature. They have not been assembled in a form that can be used for multi-omics level approaches. Hence, we report the assembly and development of a topology-oriented comprehensive GDNF/RET signaling network map. This GDNF/RET signaling pathway map forms part of the neurological system pathways such as BDNF/p75NTR (Sandhya et al. 2013), oxytocin-oxytocin receptor signaling (Chatterjee et al. 2016), galanin-galanin receptor (Gopalakrishnan et al. 2021), serotonin (Sahu et al. 2018), and corticotropin-releasing hormone (Subbannayya et al. 2013) developed by our group and submitted to NetPath (Kandasamy et al. 2010) and WikiPathways (Kutmon et al. 2016). Here, we describe the generation of an integrated pathway map of GDNF/RET signaling by manual curation. This comprehensive signaling pathway map can be used for knowledge-based analysis of multi-omics datasets for improvement of our understanding of fundamental biological processes associated with GDNF signaling.
Methodology
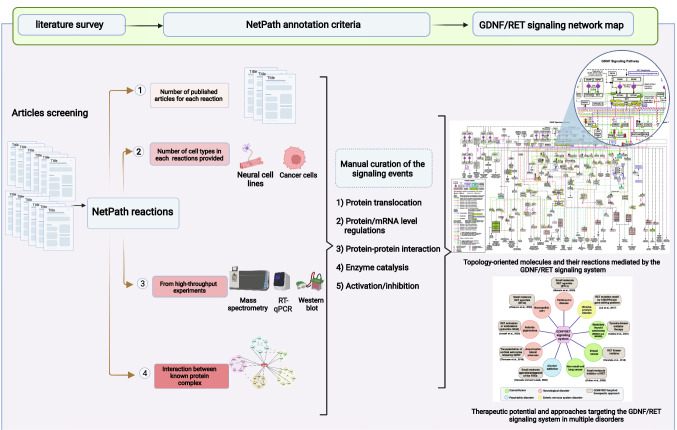
A literature survey was carried out in PubMed to gather the articles associated with GDNF signaling using the search term ‘("GDNF" OR "glial cell-derived neurotrophic factor" OR "ATF" OR "ATF1" OR "ATF2" OR "astrocyte-derived trophic factor") AND ("signaling" OR "signalling" OR "pathway")’. A manual screening of these articles containing relevant information of signaling events reported to be induced by GDNF was performed. Later, manual curation of the signaling events was carried out from the screened articles based on NetPath annotation criteria (Kandasamy et al. 2010). These events were categorized into five groups such as (a) molecular association (protein–protein interactions (PPIs)), (b) enzyme-catalysis (enzyme–substrate reactions and also the post-translational modifications), (c) activation/inhibition events, (d) protein translocation events, and e) gene regulation events at mRNA/protein level (Kandasamy et al. 2010). The curated signaling events were merged into a single pathway map and each curated reaction was hyperlinked to the PubMed identifier of the research article from which the reactions have been curated. The PathVisio tool was used for the representation and visualization of the network map (Kutmon et al. 2015). The GDNF pathway map was made freely accessible through WikiPathways database.
Results and discussion
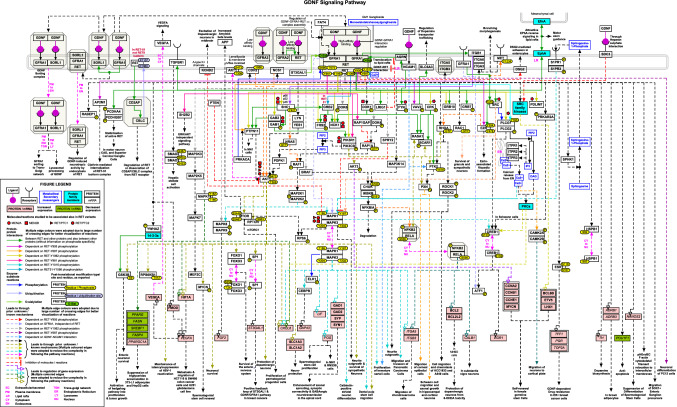
From 3,259 articles that were manually screened, 1061 articles which had relevant information pertaining to GDNF signaling were retrieved. A total of 1151 signaling events associated with GDNF stimulation were curated from these shortlisted articles (Supplementary Data S1). These events included categories of 52 molecular associations, 70 enzyme catalysis, 36 activation/inhibition reactions, and 22 translocation events. Furthermore, we also cataloged 115 protein and 856 mRNA expression regulation events modulated by GDNF stimulation in various mammalian cells/cell lines. A topology-oriented GDNF/RET signaling network map was manually drawn using PathVisio software and made freely accessible through WikiPathways database (https://www.wikipathways.org/index.php/Pathway:WP5143) (Fig. 1).
Fig. 1.
Schematic representation of GDNF signaling pathway. Schematic representation of GDNF-induced signaling reactions. The signaling pathway map represents molecules involved in ligand-receptor interactions and GDNF-activated downstream molecular events including molecular association, enzyme catalysis, translocation, and gene regulation events. Information regarding the post-translational modification site and the residue is also shown in the pathway
The signaling pathways via GDNF/GFRα1/RET complex
The GDNF/RET signaling pathway is a significant pathway to explore for its critical role in the development and maintenance of the nervous system (Takahashi 2001). It is also involved in the development of other organs, such as the kidneys (Costantini and Shakya 2006) and the pancreas (Lucini et al. 2008). Most of the GDNF effects are mediated by the receptor tyrosine kinase RET and GFRα1. The binding of GDNF to GFRα1/RET leads to auto-phosphorylation of tyrosine residues of RET including Tyr905, Tyr1015, Tyr1062, Tyr1096, Tyr687, Tyr981, Ser696, and Thr675. The former 4 residues are only specific to the RET51 isoform and eventually become the docking sites for adaptor proteins such as Grb7/Grb10, PLCγ, Shc/Enigma, and Grb2, respectively, leading to the activation of various signaling modules (Airaksinen et al. 1999; Takahashi 2001).
For instance, the phosphorylation of RET at Tyr1062 is found to be critical for the function of RET51, which is required for the activation of RAS/MAPK, p38MAPK, JNK and PI3K/AKT pathway (Besset et al. 2000; Segouffin-Cariou and Billaud 2000). Further, these pathways are vital for enteric neuroblasts differentiation and survival, and the branching morphogenesis of the ureteric bud (Fisher et al. 2001). Similarly, phosphorylated Tyr1015 residue activates PLCγ pathway, which regulates Ca2+ influx from the endoplasmic reticulum and thereby induces calcium signaling (Kawai and Takahashi 2020).
GDNF/RET signaling in cell-specific functions
The GDNF/RET signaling promote cell survival and/or neurite outgrowth in several neuronal subpopulations such as in Neuro2a cells via the activation of PI3K/AKT1 pathway (Encinas et al. 2001) and in sympathetic neurons via MAPK1/3 activation (Coulpier and Ibanez 2004). However, GDNF/SDC3 signaling is implicated in the regulation of neurite outgrowth in hippocampal neurons (Bespalov et al. 2011). Further, GDNF/GFRα1/NCAM1 signaling is reported to promote dendrite growth and spine formation via the activation of FAK, SRC and MAPK1/3 in the hippocampus (Irala et al. 2016). In TGW cell lines, the activated RET by Tyr1062 phosphorylation leads to the association between RET with GAB1 and SHC, which induces the activation of CREB and NF-kB through RAS/MAPK and PI3K/AKT pathways, respectively, which are involved in neuron survival (Hayashi et al. 2000). In adult rat substantia nigra, GDNF stimulates PI3K/AKT downstream signaling resulting in the translocation of NF-kB to nucleus and regulates the higher expression of Calbindin-D28K thereby protecting the neurons from degeneration, the main pathological change associated with Parkinson's disease (PD) (Howland and Wang 2008). In neuronal cell line RN33B, which is RET independent, GDNF promotes cell survival by activation of SRC-like kinase, phosphorylation of CREB, and protein up-regulation of FOS rather than activating RAS/MAPK pathway. In MN1 cells, GDNF stimulates the activation of RAS/MAPK and PI3K/AKT1 pathways, PLCγ phosphorylation, which results in the up-regulation of CREB and FOS at mRNA and protein level, respectively, which are essential for cell survival (Trupp et al. 1999).
The GDNF also plays an important role in spermatogonial stem cell (SSC) functions. In C18-4 cells, a mouse spermatogonial stem cell line, GDNF stimulation induces the phosphorylation and association of RET between SHC1 and GRB2 which in turn induces the phosphorylation of CREB1, ATF1, and TNF via the RAS/MAPK1/3 pathway that promotes the transcription of c-FOS, which leads to SSC proliferation (He et al. 2008). Similarly, GDNF/ GFRα1/RET induces spermatogenesis in mice spermatogonial progenitor cells (SPCs) by inducing the phosphorylation of RPS6 and RPTOR via MAPK1/3 kinase signaling, an important step in self-renewal and mitotic proliferation of SPCs (Wang et al. 2017).
GDNF/RET signaling in cancer
The GDNF/RET signaling is implicated in the development and metastasis of several cancers (Mulligan 2018; Subbiah and Cote 2020). In in vitro cell and animal models of breast cancer, GDNF/RET signaling induces MAPK and JNK activation that leads to tumor cell proliferation and survival suggesting the potential role of RET as a novel therapeutic target in breast cancer (Boulay et al. 2008). GDNF stimulates GFRα1 and induces the activation of NFKB1 which promotes the invasive potential in human pancreatic cancer cells (Takahashi et al. 2004). GDNF stimulates GFRα1 and induces the activation of AKT1, MAPK1, MAPK8, PIK3CA, AKT1, RHOA, NRAS, RAC1, and RNASE9, which are involved in tumor migration and invasion in pancreatic cancer PANC-1 cells (Veit et al. 2004). The exposure of cytotoxic agents to prostate cancer cells promotes the increased expression of GDNF which stimulates GFRα1 and induces the activation of SRC and MAPK1, which enhances prostate cancer resistance to genotoxic chemotherapy (Huber et al. 2015).
The GDNF secretion by nerve cells increases the PDL1 expression through JAK2-STAT1 signaling pathway contributing to immune surveillance escape in head and neck squamous cell carcinoma (HNSCC) (Lin et al. 2017). Activation of GDNF/RET/AKT signaling in neuroblastoma (NB) cells inhibits the GSK3β activity, resulting in increases in PTCH1, GLI2 and GLI1 and decreases in GLI3, which activates hedgehog signaling pathway and thereby promotes the proliferation of NB cells and tumor growth (Ruan et al. 2016). In breast cancer cells, GDNF stimulates GFRα1 and induces the phosphorylation of RET (Tyr 905), ESR1 (Ser 167 and 118), AKT1 (Ser 473), JUN (Ser 73), RPS6KB1 (Thr 389), MAPK1/3 (Tyr204/Thr202), MAPK8/9 (Thr183/Tyr185) and upregulation of PGR, TFF1, TOP2A at gene level in MCF7 cells which promoted the survival of aromatase inhibitor-resistant cells (Morandi et al. 2013). In human glioblastoma cells, GDNF stimulates GFRα1 and induces the upregulation of VEGFA and FMOD at mRNA and protein levels and subsequently, the transport of VEGFA from extracellular vesicle to cytoplasm which induces angiogenesis (Chen et al. 2018). A recent study has shown that RET and MEN2A molecular activities can be inhibited by the dephosphorylation of protein tyrosine phosphatase receptor-type-A (PTPRA) which in turn regulates GDNF-dependent RET-RAS-MAPK growth signaling and the oncogenic activity of RET (Yadav et al. 2020). Unlike the other RET-induced malignancies, which are caused due to RET gene mutation, papillary thyroid carcinoma (PTC), the most common form of thyroid cancer is mediated as a result of chromosomal translocation of RET gene (Santoro and Carlomagno 2013; Prescott and Zeiger 2015). The application of small molecule inhibitors capable of inhibiting RET protein kinase activity has been an effective therapeutic approach for PTCs (Prescott and Zeiger 2015).
Conclusions
We developed a comprehensive GDNF/RET signaling pathway map based on the information derived from the literature related to GDNF signaling. The diverse signaling modules and the interactions specific to the phosphosites in RET and also their information pertaining to distinct RET variants are distinguished in this pathway map. We believe that our map provide effective visualization of the pathway to derive novel insights into GDNF/RET signaling mechanisms by gene-set enrichment analysis approaches using future multiomic datasets. Information on the GDNF-mediated cellular responses in this map will facilitate effective integration and updation of the map with evolving information on GDNF/RET signaling as a platform for reference and analysis of GDNF associated physiological processes and pathological conditions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Department of Biotechnology (DBT), Government of India, for research support to the Institute of Bioinformatics (IOB), Bangalore. We thank the Yenepoya (Deemed to be University) for the facility support to the Centre for Integrative Omics data Science (CIODS). Praseeda Mol is a recipient of a Senior Research Fellowship from Inspire Fellowship from the Department of Science and Technology (DST), Government of India. Rajesh Raju is a recipient of the Young Scientist Award (YSS/2014/000607) from the Science and Engineering Research Board, Department of Science and Technology (DST), Government of India.
Abbreviations
- GDNF
Glial-cell-line-derived neurotrophic factor
- GFRα1
GDNF family receptor alpha-1
- GFRAs
GDNF family receptor alpha members
- GFLs
GDNF family ligands
- GPI
Glycosyl-phosphatidylinositol
- RET
Rearranged during transfection
- NCAM1
Neural cell adhesion molecule-1
- CNS
Central nervous system
- MAPK
Mitogen-activated protein kinase
- PLC
Phospholipase-C
- SDC3
Syndecan-3
- ITGB1
Integrin-β1
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Praseeda Mol, Email: prasy004@gmail.com.
Rex Devasahayam Arokia Balaya, Email: rexprem@yenepoya.edu.in.
Shobha Dagamajalu, Email: shobha_d@yenepoya.edu.in.
Sreeranjini Babu, Email: sreeranjinibabu611@gmail.com.
Pavithra Chandrasekaran, Email: cpavithrasekar01@gmail.com.
Reshma Raghavan, Email: reshmarechu1505@gmail.com.
Sneha Suresh, Email: sneha108ss@gmail.com.
Namitha Ravishankara, Email: namitharavi12@gmail.com.
Anu Hemalatha Raju, Email: anulatha279@gmail.com.
Bipin Nair, Email: bipin@am.amrita.edu.
Prashant Kumar Modi, Email: prashantmodi@yenepoya.edu.in.
Anita Mahadevan, Email: mahadevananita@gmail.com.
Thottethodi Subrahmanya Keshava Prasad, Email: keshav@yenepoya.edu.in.
Rajesh Raju, Email: rajeshraju@yenepoya.edu.in.
References
- Adashek JJ, Desai AP, Andreev-Drakhlin AY, Roszik J, Cote GJ, Subbiah V. Hallmarks of RET and Co-occuring genomic alterations in RET-aberrant cancers. Mol Cancer Ther. 2021;20(10):1769–1776. doi: 10.1158/1535-7163.MCT-21-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airaksinen MS, Titievsky A, Saarma M. GDNF family neurotrophic factor signaling: four masters, one servant? Mol Cell Neurosci. 1999;13(5):313–325. doi: 10.1006/mcne.1999.0754. [DOI] [PubMed] [Google Scholar]
- Bespalov MM, Sidorova YA, Tumova S, Ahonen-Bishopp A, Magalhaes AC, Kulesskiy E, Paveliev M, Rivera C, Rauvala H, Saarma M. Heparan sulfate proteoglycan syndecan-3 is a novel receptor for GDNF, neurturin, and artemin. J Cell Biol. 2011;192(1):153–169. doi: 10.1083/jcb.201009136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besset V, Scott RP, Ibanez CF. Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J Biol Chem. 2000;275(50):39159–39166. doi: 10.1074/jbc.M006908200. [DOI] [PubMed] [Google Scholar]
- Boulay A, Breuleux M, Stephan C, Fux C, Brisken C, Fiche M, Wartmann M, Stumm M, Lane HA, Hynes NE. The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res. 2008;68(10):3743–3751. doi: 10.1158/0008-5472.CAN-07-5100. [DOI] [PubMed] [Google Scholar]
- Chatterjee Oishi, Patil Krutika, Sahu Apeksha, Gopalakrishnan Lathika, Mol Praseeda, Advani Jayshree, Mukherjee Srabani, Christopher Rita, Prasad T. S. Keshava. An overview of the oxytocin-oxytocin receptor signaling network. Journal of Cell Communication and Signaling. 2016;10(4):355–360. doi: 10.1007/s12079-016-0353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ba H, Lu C, Dai J, Sun J. Glial cell line-derived neurotrophic factor (GDNF) promotes angiogenesis through the demethylation of the fibromodulin (FMOD) promoter in glioblastoma. Med Sci Monit. 2018;24:6137–6143. doi: 10.12659/MSM.911669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintron-Colon AF, Almeida-Alves G, Boynton AM, Spitsbergen JM. GDNF synthesis, signaling, and retrograde transport in motor neurons. Cell Tissue Res. 2020;382(1):47–56. doi: 10.1007/s00441-020-03287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. BioEssays. 2006;28(2):117–127. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- Coulpier M, Ibanez CF. Retrograde propagation of GDNF-mediated signals in sympathetic neurons. Mol Cell Neurosci. 2004;27(2):132–139. doi: 10.1016/j.mcn.2004.06.001. [DOI] [PubMed] [Google Scholar]
- de Graaff E, Srinivas S, Kilkenny C, D'Agati V, Mankoo BS, Costantini F, Pachnis V. Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 2001;15(18):2433–2444. doi: 10.1101/gad.205001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas M, Tansey MG, Tsui-Pierchala BA, Comella JX, Milbrandt J, Johnson EM., Jr c-Src is required for glial cell line-derived neurotrophic factor (GDNF) family ligand-mediated neuronal survival via a phosphatidylinositol-3 kinase (PI-3K)-dependent pathway. J Neurosci. 2001;21(5):1464–1472. doi: 10.1523/JNEUROSCI.21-05-01464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CE, Michael L, Barnett MW, Davies JA. Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development. 2001;128(21):4329–4338. doi: 10.1242/dev.128.21.4329. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan L, Chatterjee O, Raj C, Pullimamidi D, Advani J, Mahadevan A, Keshava Prasad TS. An assembly of galanin-galanin receptor signaling network. J Cell Commun Signal. 2021;15(2):269–275. doi: 10.1007/s12079-020-00590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Ichihara M, Iwashita T, Murakami H, Shimono Y, Kawai K, Kurokawa K, Murakumo Y, Imai T, Funahashi H, Nakao A, Takahashi M. Characterization of intracellular signals via tyrosine 1062 in RET activated by glial cell line-derived neurotrophic factor. Oncogene. 2000;19(39):4469–4475. doi: 10.1038/sj.onc.1203799. [DOI] [PubMed] [Google Scholar]
- He Z, Jiang J, Kokkinaki M, Golestaneh N, Hofmann MC, Dym M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26(1):266–278. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland JG, Wang YT. Synaptic plasticity in learning and memory: stress effects in the hippocampus. Prog Brain Res. 2008;169:145–158. doi: 10.1016/S0079-6123(07)00008-8. [DOI] [PubMed] [Google Scholar]
- Huber RM, Lucas JM, Gomez-Sarosi LA, Coleman I, Zhao S, Coleman R, Nelson PS. DNA damage induces GDNF secretion in the tumor microenvironment with paracrine effects promoting prostate cancer treatment resistance. Oncotarget. 2015;6(4):2134–2147. doi: 10.18632/oncotarget.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irala D, Bonafina A, Fontanet PA, Alsina FC, Paratcha G, Ledda F. The GDNF-GFRalpha1 complex promotes the development of hippocampal dendritic arbors and spines via NCAM. Development. 2016;143(22):4224–4235. doi: 10.1242/dev.140350. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11(1):R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K, Takahashi M. Intracellular RET signaling pathways activated by GDNF. Cell Tissue Res. 2020;382(1):113–123. doi: 10.1007/s00441-020-03262-1. [DOI] [PubMed] [Google Scholar]
- Kutmon Martina, van Iersel Martijn P., Bohler Anwesha, Kelder Thomas, Nunes Nuno, Pico Alexander R., Evelo Chris T., Murphy Robert F. PathVisio 3: An Extendable Pathway Analysis Toolbox. PLOS Computational Biology. 2015;11(2):e1004085. doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Melius J, Waagmeester A, Sinha SR, Miller R, Coort SL, Cirillo E, Smeets B, Evelo CT, Pico AR. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44(D1):D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260(5111):1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Lin C, Cao W, Ren Z, Tang Y, Zhang C, Yang R, Chen Y, Liu Z, Peng C, Wang L, Wang X, Ji T. GDNF secreted by nerves enhances PD-L1 expression via JAK2-STAT1 signaling activation in HNSCC. Oncoimmunology. 2017;6(11):e1353860. doi: 10.1080/2162402X.2017.1353860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucini C, Maruccio L, Facello B, Cocchia N, Tortora G, Castaldo L. Cellular localization of GDNF and its GFRalpha1/RET receptor complex in the developing pancreas of cat. J Anat. 2008;213(5):565–572. doi: 10.1111/j.1469-7580.2008.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato AK, Sidorova YA. Glial cell line-derived neurotrophic factors (GFLs) and small molecules targeting RET receptor for the treatment of pain and Parkinson's disease. Cell Tissue Res. 2020;382(1):147–160. doi: 10.1007/s00441-020-03227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, Saarma M, Sariola H. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287(5457):1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Morandi A, Martin LA, Gao Q, Pancholi S, Mackay A, Robertson D, Zvelebil M, Dowsett M, Plaza-Menacho I, Isacke CM. GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res. 2013;73(12):3783–3795. doi: 10.1158/0008-5472.CAN-12-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan LM. GDNF and the RET receptor in cancer: new insights and therapeutic potential. Front Physiol. 2018;9:1873. doi: 10.3389/fphys.2018.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratcha G, Ledda F, Baars L, Coulpier M, Besset V, Anders J, Scott R, Ibanez CF. Released GFRalpha1 potentiates downstream signaling, neuronal survival, and differentiation via a novel mechanism of recruitment of c-Ret to lipid rafts. Neuron. 2001;29(1):171–184. doi: 10.1016/S0896-6273(01)00188-X. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I, Mologni L, McDonald NQ. Mechanisms of RET signaling in cancer: current and future implications for targeted therapy. Cell Signal. 2014;26(8):1743–1752. doi: 10.1016/j.cellsig.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Prescott JD, Zeiger MA. The RET oncogene in papillary thyroid carcinoma. Cancer. 2015;121(13):2137–2146. doi: 10.1002/cncr.29044. [DOI] [PubMed] [Google Scholar]
- Regua AT, Najjar M, Lo HW. RET signaling pathway and RET inhibitors in human cancer. Front Oncol. 2022;12:932353. doi: 10.3389/fonc.2022.932353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Luo H, Wang J, Ji X, Zhang Z, Wu J, Zhang X, Wu X. Smoothened-independent activation of hedgehog signaling by rearranged during transfection promotes neuroblastoma cell proliferation and tumor growth. Biochim Biophys Acta. 2016;1860(9):1961–1972. doi: 10.1016/j.bbagen.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Sahu A, Gopalakrishnan L, Gaur N, Chatterjee O, Mol P, Modi PK, Dagamajalu S, Advani J, Jain S, Keshava Prasad TS. The 5-Hydroxytryptamine signaling map: an overview of serotonin-serotonin receptor mediated signaling network. J Cell Commun Signal. 2018;12(4):731–735. doi: 10.1007/s12079-018-0482-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya VK, Raju R, Verma R, Advani J, Sharma R, Radhakrishnan A, Nanjappa V, Narayana J, Somani BL, Mukherjee KK, Pandey A, Christopher R, Prasad TS. A network map of BDNF/TRKB and BDNF/p75NTR signaling system. J Cell Commun Signal. 2013;7(4):301–307. doi: 10.1007/s12079-013-0200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Carlomagno F. Central role of RET in thyroid cancer. Cold Spring Harb Perspect Biol. 2013;5(12):a009233. doi: 10.1101/cshperspect.a009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchardt A, D'Agati V, Pachnis V, Costantini F. Renal agenesis and hypodysplasia in ret-k- mutant mice result from defects in ureteric bud development. Development. 1996;122(6):1919–1929. doi: 10.1242/dev.122.6.1919. [DOI] [PubMed] [Google Scholar]
- Segouffin-Cariou C, Billaud M. Transforming ability of MEN2A-RET requires activation of the phosphatidylinositol 3-kinase/AKT signaling pathway. J Biol Chem. 2000;275(5):3568–3576. doi: 10.1074/jbc.275.5.3568. [DOI] [PubMed] [Google Scholar]
- Subbannayya T, Balakrishnan L, Sudarshan G, Advani J, Kumar S, Mahmood R, Nair B, Sirdeshmukh R, Mukherjee KK, Umathe SN, Raju R, Prasad TS. An integrated map of corticotropin-releasing hormone signaling pathway. J Cell Commun Signal. 2013;7(4):295–300. doi: 10.1007/s12079-013-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah V, Cote GJ. Advances in targeting RET-dependent cancers. Cancer Discov. 2020;10(4):498–505. doi: 10.1158/2159-8290.CD-19-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12(4):361–373. doi: 10.1016/S1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Funahashi H, Sawai H, Sakamoto M, Matsuo Y, Yamamoto M, Okada Y, Hayakawa T, Manabe T. Glial cell line-derived neurotrophic factor enhances nuclear factor-kappaB activity and invasive potential in human pancreatic cancer cells. Pancreas. 2004;29(1):22–27. doi: 10.1097/00006676-200407000-00051. [DOI] [PubMed] [Google Scholar]
- Thein KZ, Velcheti V, Mooers BHM, Wu J, Subbiah V. Precision therapy for RET-altered cancers with RET inhibitors. Trends Cancer. 2021;7(12):1074–1088. doi: 10.1016/j.trecan.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupp M, Scott R, Whittemore SR, Ibanez CF. Ret-dependent and -independent mechanisms of glial cell line-derived neurotrophic factor signaling in neuronal cells. J Biol Chem. 1999;274(30):20885–20894. doi: 10.1074/jbc.274.30.20885. [DOI] [PubMed] [Google Scholar]
- Vastag B. Biotechnology: crossing the barrier. Nature. 2010;466(7309):916–918. doi: 10.1038/466916a. [DOI] [PubMed] [Google Scholar]
- Veit C, Genze F, Menke A, Hoeffert S, Gress TM, Gierschik P, Giehl K. Activation of phosphatidylinositol 3-kinase and extracellular signal-regulated kinase is required for glial cell line-derived neurotrophic factor-induced migration and invasion of pancreatic carcinoma cells. Cancer Res. 2004;64(15):5291–5300. doi: 10.1158/0008-5472.CAN-04-1112. [DOI] [PubMed] [Google Scholar]
- Wang M, Guo Y, Wang M, Zhou T, Xue Y, Du G, Wei X, Wang J, Qi L, Zhang H, Li L, Ye L, Guo X, Wu X. The glial cell-derived neurotrophic factor (GDNF)-responsive phosphoprotein landscape identifies raptor phosphorylation required for spermatogonial progenitor cell proliferation. Mol Cell Proteomics. 2017;16(6):982–997. doi: 10.1074/mcp.M116.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav L, Pietila E, Ohman T, Liu X, Mahato AK, Sidorova Y, Lehti K, Saarma M, Varjosalo M. PTPRA phosphatase regulates GDNF-dependent ret signaling and inhibits the RET mutant MEN2A oncogenic potential. Science. 2020;23(2):100871. doi: 10.1016/j.isci.2020.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihlmann KB, Ducray AD, Schaller B, Huber AW, Krebs SH, Andres RH, Seiler RW, Meyer M, Widmer HR. The GDNF family members neurturin, artemin and persephin promote the morphological differentiation of cultured ventral mesencephalic dopaminergic neurons. Brain Res Bull. 2005;68(1–2):42–53. doi: 10.1016/j.brainresbull.2004.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.