Abstract
Little is known about the mechanisms used by enveloped viruses to separate themselves from the cell surface at the final step of budding. However, small sequences in the Gag proteins of several retroviruses (L domains) have been implicated in this process. A sequence has been identified in the M proteins of rhabdoviruses that closely resembles the PPPPY motif in the L domain of Rous sarcoma virus (RSV), an avian retrovirus. To evaluate whether the PPPY sequence in vesicular stomatitis virus (VSV) M protein has an activity analogous to that of the retroviral sequence, M-Gag chimeras were characterized. The N-terminal 74 amino acids of the VSV (Indiana) M protein, including the PPPY motif, was able to replace the L domain of RSV Gag and allow the assembly and release of virus-like particles. Alanine substitutions in the VSV PPPY motif severely compromised the budding activity of this hybrid protein but not that of another chimera which also contained the RSV PPPPY sequence. We conclude that this VSV sequence is functionally homologous to the RSV L domain in promoting virus particle release, making this the first example of such an activity in a virus other than a retrovirus. Both the RSV and VSV motifs have been shown to interact in vitro with certain cellular proteins that contain a WW interaction module, suggesting that the L domains are sites of interaction with unknown host machinery involved in virus release.
The mechanism of assembly and budding for retroviruses is distinguished from that of the enveloped negative-strand RNA viruses in at least one very conspicuous way. The former is driven by a single large precursor protein, Gag (Fig. 1A), which upon budding from the plasma membrane is processed by the viral protease (PR) to form the structural proteins of the virion (MA, CA, and NC) (6, 27). In contrast, assembly and release of negative-strand RNA viruses such as vesicular stomatitis virus (VSV), a member of the Rhabdoviridae family, are driven by structural proteins (in this case, the M and N proteins) which are individually translated prior to assembling at the plasma membrane (28). In spite of this obvious difference, the assembly processes are quite similar in that the viral components are attached to the plasma membrane through a matrix protein (MA in retroviruses; M in rhabdoviruses) as they are assembled and the completed structures are released by budding through the membrane. However, the actual mechanism of virus-cell separation remains undefined for all enveloped viruses.
FIG. 1.
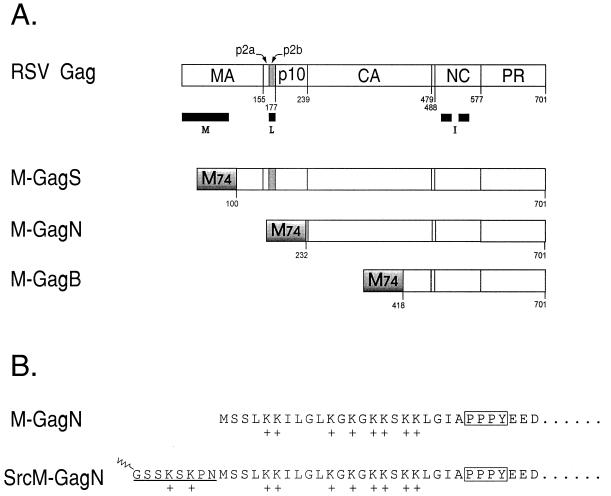
Structure of the Gag protein and M-Gag chimeras. (A) Structure of the wild-type Gag protein of RSV. The cleavage sites that are cut by the viral protease (PR) to generate the mature viral proteins are marked by the residue numbers. Black bars mark the positions of the three assembly domains in RSV Gag: M, membrane-binding domain; L, late domain; I, interaction domains. The PPPPY sequence that defines the core of the L domain lies within the p2b peptide (gray). The M-GagS, M-GagN, and M-GagN proteins are chimeras formed by the fusion of 74 amino acids from the N terminus of the VSV M protein (M74, shaded box) to different lengths of the Gag protein. The residue in Gag to which the M74 fragment is fused is indicated below each molecule. Foreign amino acids were inserted at the fusion junction in M-GagS (A-L-V) and in M-GagN (A-P-R-Q) but not in M-GagS. (B) Amino-terminal sequence of the M-GagN chimera and its Src derivative. The terminal sequence of M-GagN is identical to that of the VSV M protein. Eight lysine residues which have been implicated in membrane binding are indicated by plus signs. The PPPY motif is boxed. In the SrcM-GagN molecule, the amino-terminal sequence of the M protein is preserved. The addition of a sequence containing the first 10 codons of v-src to the chimeric M-gag gene in pSV.M-GagN is expected to yield the protein shown. The N-terminal methionine is predicted to be removed cotranslationally as myristic acid (indicated by the zigzag) is linked to the subterminal glycine in the Src peptide.
The minimal elements within the Rous sarcoma virus (RSV) Gag protein that are essential for assembly of budding-competent virus-like structures have been delineated by deletion analysis (Fig. 1A). These include a membrane targeting and binding signal (the M domain) within the matrix (MA) sequence at the very amino terminus of each Gag protein (25, 32, 37) and one or more regions for interaction between Gag proteins (the interaction or I domains) needed for the tight packing of molecules into high-density structures on the membrane. Efficient release of particles requires the action of a third element in Gag, a short sequence that has been termed the late (L) domain, because mutations within these regions cause a block late in the assembly-release pathway (18, 30).
The RSV sequence that is critical for late domain activity lies within the p2b region near the amino terminus of Gag where the most critical residues have been mapped to a PPPPY motif. Functional homologues of the RSV PPPPY sequence have been confirmed in the P(S/T)APP sequences of the human immunodeficiency virus type 1 (HIV-1) and visna virus Gag proteins, the YPDL sequence of equine infectious anemia virus (EIAV), as well as PPPY in Mason-Pfizer monkey virus (M-PMV) (10, 18, 21, 30, 33, 34). Mutations in each of these regions cause a dramatic reduction in particle release, though there is little sequence similarity to suggest a common function. Even more surprisingly, the L domains in different viruses are found at different locations within their respective Gag proteins: near the amino terminus in RSV and at the Gag carboxy terminus in HIV and EIAV. Yet, the functional similarity of the sequences from HIV, EIAV, and visna virus has been proven by showing that each can substitute for the L domain of RSV and allow the formation of budding-competent chimeric particles (18, 21, 33). Evidence is mounting which suggests that these late domains serve as binding sites for cellular factors that act to facilitate virus release (8, 11).
Until now there has been no confirmation of elements related to the retroviral L domains in other kinds of viruses. However, the expression of M in the absence of other VSV proteins results in the protein being packaged and released in low-density particles (12), similar to ones released by a truncated RSV Gag protein which contains little more than the membrane-binding and late domains (29). The M protein, therefore, is likely to possess functions equivalent to these retroviral assembly domains. Indeed, a homologue of the HIV-1 membrane-binding domain has been previously defined in the VSV M protein, and the two appear quite similar in function, utilizing both a hydrophobic region and a cluster of positively charged amino acids near the amino terminus (14, 15, 25, 35–37). Candidate rhabdovirus L domains have now been identified in the M proteins of VSV and several related viruses that are similar in sequence to the L domain of RSV (11). In this study, we demonstrate that a PPPY motif within the VSV M protein is indeed a functional homologue of the retroviral L domain, making this the first description of such an activity outside of the retroviruses. These findings imply that the mechanism of virus-cell separation is conserved between these two very different virus groups.
MATERIALS AND METHODS
M-Gag chimeras.
Vectors for the expression of chimeric proteins were constructed by modification of plasmid pSV.Myr0 (31) which is a simian virus 40-derived vector bearing the gag gene under control of the late promoter. To create pSV.M-GagS, a VSV (RSV) sequence containing the first 74 triplets of the M protein coding sequence was amplified by PCR. The 5′ primer UpNdeI (5′-GATCGATCGACATATGAGTTCCTTAAAGAAGATTCTCGGT; NdeI site underlined) introduced an NdeI site at the M initiator AUG; the downstream primer (primer M74SpeI, 5′-ACAAGACTTGACTAGTGCAGATCTAACCGTCATTTTCACTGT) inserted an SpeI site (underlined) after the 74 M codons. The amplified M74 sequence was cut with NdeI and ligated to a 130-nucleotide SstI-NdeI fragment bearing the 5′ untranslated region from pSV.Myr0. The ligated fragments were reamplified, using a 5′ primer which spans the upstream SstI site (dCA5; 5′-AGAAGGTACGAGCTCTACTGCAGGGA-3′; SstI site underlined) and the 3′ primer M74SpeI, and then cloned into vector pSV.ΔMA6 by using its SstI site and the unique SpeI site preceding codon 100 of gag (17). For the pSV.M-GagN vector, the downstream primer was M74NotI (5′-CATACAAGACTGCGGCCGCG CAGATCTAACCGTCATTTTCACTGT; NotI site underlined) and the final PCR product was cloned into pSV.Myr1RNot by using the SstI site and the unique NotI site in its p10 coding sequence (30). For pSV.M-GagB, the 3′ primer was M74BamHI (5′-TCTGAACGGAGGATCCGATCTAACCGTCATTTTCACTGT; BamHI site underlined). The SstI-BamHI fragment was then cloned into the SstI and BglII sites of pSV.Myr0. Each new vector was confirmed by DNA sequencing.
Point mutations in the PPPY motif.
The SstI-NotI fragment of pSV.M-GagN and the SstI-SpeI fragment of pSV.M-GagS were subjected to PCR by using mutagenic primers that modified the codons for the PPPY motif from CCACCCCCTTAT to CCACCCCCTGCT (for the PPPY to PPPA change) or to GCTCCCCCTTAT (PPPY to APPY). After recloning the mutated fragments into the pSV.M-GagN and pSV.M-GagS vectors, the presence of the mutations in each clone was confirmed by DNA sequencing.
Addition of the Src membrane-binding domain to M-GagN.
The M74 sequence from pSV.M-GagN was cloned into the vector pSV.Myr1RNot that bears a modified gag allele that differs from wild-type gag in two ways: replacement of the first 10 codons with a sequence encoding the first 10 residues of the v-Src oncoprotein (31) and placement of a unique NotI site in p10 (30). The vector was prepared by digestion with Bsu36I (which cuts after the ninth v-src codon), Klenow treatment to fill in recessed ends, and then NotI digestion. The M74-coding sequence (prepared by NdeI digestion, Klenow treatment, and NotI digestion) was inserted, and resulting clones were sequenced to confirm the in-frame fusion of the v-src, M74, and gag(p10) segments.
Budding assay.
The budding assay was performed in COS-1 cells as first described by Wills et al. (31). At 48 h posttransfection, cells were labelled for 2.5 h with [35S]methionine and Gag-related proteins in the medium and in cell lysates were analyzed by immunoprecipitation using anti-RSV or anti-CA sera, as indicated. For some experiments, the release of each chimeric protein was estimated by measuring the amount of released CA protein by PhosphorImager analysis of the dried gel. After correcting for minor differences in protein expression (evaluated by measuring the amount of labelled Gag protein present in cell lysates obtained from plates transfected in parallel but labelled for only 5 min) and for different numbers of methionine residues in the precursors, the release was expressed relative to that of authentic (wild-type) Gag protein.
RESULTS
A proline-rich sequence that closely resembles the core motif of the RSV L domain exists in the matrix (M) protein of VSV: LGIAPPPYEED in the VSV M protein; TASAPPPPYVG in the RSV p2b peptide. Related motifs exist in the M protein of rabies virus, as well as in Ebola and Marburg viruses (11). By analogy with the L domain of RSV Gag, a function for the rhabdovirus PPPY sequence in facilitating particle release appeared likely but could not be assumed for several reasons. PPPY motifs occur in nonviral proteins for which they clearly do not serve a role in budding. The VSV sequence is shorter than the RSV L domain by one proline, and the sequences flanking the proline motif are very different. Finally, no activity analogous to the L domain has been ascribed to any viral proteins other than those of retroviruses. To evaluate the possible L domain activity of the rhabdovirus PPPY sequence, we constructed chimeric proteins in which the first 74 amino acids of the VSV M protein (M74) replaced various portions of RSV Gag (Fig. 1A).
The M74 sequence contains both the PPPY motif and an amino-terminal basic region that has been implicated in membrane binding. Like the full-length M protein, M74 has been proven capable of causing the release of low-density, membrane-enclosed particles (12). Therefore, we hypothesized that the M74 fragment might substitute for both the M and L domains of the RSV Gag protein and support the assembly and release of virus-like particles of normal density. Luckily, this M74 fragment does not cause the cytopathic effects that are observed with the full-length M protein (35). The M74 sequence was inserted in place of the first 100, 232, and 418 residues of the RSV Gag protein to create the chimeras M-GagS, M-GagN, and M-GagB, respectively (Fig. 1A). In all three chimeras the RSV membrane-binding domain has been removed, making particle release dependent on any such activity that may reside within the M74 sequence. Release of particles containing the M-GagN and M-GagB proteins, which lack the RSV L domain, would be consistent with the presence in the M74 sequence of a functional element similar to the RSV L domain. Such particles would be expected to be of a density characteristic of retroviruses since the RSV I domains are retained in each case.
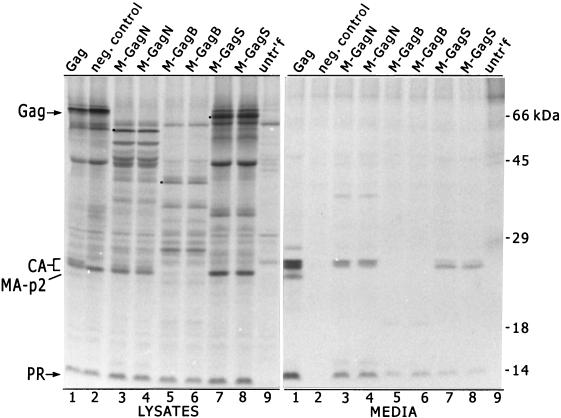
The assembly and budding abilities of each of the chimeric proteins were measured in a budding assay that has been used for studies of Gag protein assembly (31) (Fig. 2). COS-1 cells expressing either the Gag protein or one of the chimeras were labelled with [35S]methionine for 2.5 h, after which Gag-related proteins in the medium were evaluated by immunoprecipitation with an anti-RSV serum. In the case of wild-type Gag, the full-length protein and cleavage intermediates were detected in cell lysates (Fig. 2, lysates lane 1). Cleavage products formed by the action of the viral protease on Gag accumulated in the medium (Fig. 2, media lane 1). Such material has been shown to be contained in particles resembling authentic infectious virions in size, shape, density, rate of release, proteolytic maturation, and the ability to package viral RNA and reverse transcriptase when these are expressed in combination with the Gag protein (1, 5, 13, 23, 29, 31). As a negative control, we included a mutant Gag protein that bears a defective membrane-binding domain; it released no detectable material into the medium (media lane 2).
FIG. 2.
Release of particles from cells expressing M-Gag chimeras. Wild-type Gag protein and the three M-Gag chimeras were expressed in COS-1 cells (31). Two independent clones were tested for each of the chimeras. Gag-related proteins present in the cell lysates and in the medium of expressing cells were analyzed by radioimmunoprecipitation, using an anti-RSV serum which recognizes primarily the Gag protein and its cleavage products. The labelled proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% polyacrylamide gel. The positions of full-length Gag protein and the CA, MA, and PR proteins in cell lysates are marked on the left. The full-length M-Gag chimeras are marked by dots. A mutant Gag protein which has a defective membrane-binding domain serves as the negative control for budding (lanes 2). Samples from untransfected cells were loaded in lanes 9.
Each of the chimeric M-Gag proteins was readily detected in cell lysates where the uncleaved proteins (marked by dots in Fig. 2, lanes 3 to 8) as well as multiple cleavage products were observed. The electrophoretic mobilities of the full-length cell-associated proteins, analyzed after labelling for only 5 min, were consistent with the calculated masses of 73, 59, and 39 kDa for the M-GagS, M-GagN, and M-GagB proteins, respectively (not shown). Clearly, all three chimeras were capable of being released from cells (media lanes 3 to 8), indicating that L domain activity is present in each. Mature CA and PR proteins accumulated in the medium of cells expressing M-GagN and M-GagS (lanes 3, 4, 7, and 8). The patterns of cleavage by the viral protease appeared to be normal, although the proportion of the CA species in the M-GagS chimera appeared skewed in some experiments (Fig. 2, media lanes 7 and 8). This pattern has been previously noted with certain RSV mutants bearing altered L domains and indicates that the maturation of the CA protein by cleavage at its C terminus is slowed somewhat (33). PR but no CA protein was observed in the medium of M-GagB-expressing cells (lanes 5 and 6). This was expected, since most of the CA sequence is absent in this chimera. One predicted cleavage product, the fusion of M74 with the C-terminal fragment of MA, was not observed in the M-GagS lysate or medium samples (Fig. 2, lanes 7 and 8). This could be due to the presence of an unanticipated PR cleavage site at the fusion junction in this protein or by an interference with the MA epitope(s) recognized by the anti-RSV serum. No matrix-related proteins were seen in the M-GagN and M-GagB samples (Fig. 2, lanes 3 to 8) since the RSV MA region was completely deleted in these chimeras.
The proteolytic processing of the chimeric proteins suggested that they were packaged into actual virus-like particles. This was confirmed for the M-GagN chimera by the analysis of its buoyant density in isopycnic sucrose gradients, using previously published methods (1). The measured density of 1.15 g/ml for M-GagN particles (Fig. 3) overlaps the reported density range for authentic retrovirions and for particles produced by wild-type Gag expression in COS-1 cells (1.15 to 1.18 g/ml) (1, 7, 32). Thus, it is clear that the M74 sequence is able to replace the MA-p2b region of the Gag protein, compensating for the loss of both the membrane binding and late domain-like activities.
FIG. 3.
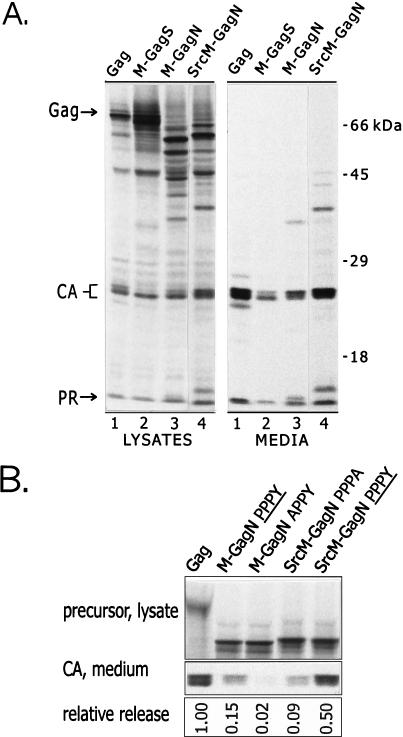
Enhancement of the budding activity of the M-GagN chimera by the addition of the Src membrane-binding signal. (A) Particle assembly and release by the SrcM-GagN protein were compared to those of the wild-type Gag protein and the M-GagS and M-GagN chimeras. Anti-RSV serum was used for immunoprecipitation. (B) The relative levels of particle release by the various proteins were compared. Expression and labelling were carried out as described in the legend for Fig. 2, except that an anti-CA serum was used for immunoprecipitation. The CA protein that accumulated in the medium during a 2.5-h labelling period (bottom panel) was measured by PhosphorImager analysis of the dried gel. These counts were normalized for the amount of Gag or M-Gag precursor present in lysates of parallel plates labelled for only 5 min (top panel) and for the number of methionine residues in the precursors. The release of each chimera was then expressed relative to that of the wild-type Gag protein. The gel shown represents one typical experiment. The chimeras which contain a wild-type VSV L domain are indicated by PPPY in the lane heading; the other chimeras carry the indicated mutant sequences. In three independent experiments, the average relative release (± standard error) was as follows: Gag, 1.00; M-GagN PPPY, 0.157 ± 0.052; M-GagN APPY, 0.014 ± 0.004; SrcM-GagN PPPA, 0.057 ± 0.016; SrcM-GagN PPPY, 0.348 ± 0.075.
Although the chimeras were released, the accumulation of extracellular CA protein was 6- to 10-fold lower for the M-GagN and M-GagS chimeras than for the wild-type Gag protein (Fig. 2, media lanes 1 and 3 to 8). The accumulation of PR protein in the medium of M-GagB-producing cells indicated that the release of M-GagB was similar to that seen with the other two chimeras (Fig. 2, media lanes 5 and 6). Since the M-GagS protein, which contains the proline motifs of both RSV and VSV, appeared to perform no better than M-GagN or M-GagB, we predicted that release of the chimeras was limited by suboptimal membrane-binding activity in M74. This interpretation is consistent with reports that the G glycoprotein (which is not expressed in our system) is needed for maximal budding activity in VSV and rabies virus (16, 24) and that regions of VSV M outside the M74 fragment contribute to hydrophobic interactions of M with the membrane (35).
To test whether the VSV membrane-binding activity was limiting in the chimeras, we attached the plasma membrane-binding signal of the viral v-Src (pp60v-src) oncoprotein onto M-GagN (Fig. 1B). The amino-terminal 8 to 10 amino acids of v-Src are known to confer strong plasma membrane binding even upon normally cytosolic proteins (19) and can substitute for the entire RSV M domain to enable particle assembly and release (32). When placed at the amino terminus of the SrcM-GagN chimera (Fig. 1B), the v-Src peptide did strongly boost particle release over that achieved by M-GagN (Fig. 3A). Release levels for SrcM-GagN were within threefold of that of the wild-type Gag protein on average and in some experiments were nearly indistinguishable from that of wild-type Gag (Fig. 3B). We conclude that the M74 sequence in the context of the M-Gag chimeras is not a fully efficient substitute for the RSV M domain. The discrepancy between Gag and the M-Gag chimeras may also indicate that the full function of the VSV sequence requires the presence of the G glycoprotein or else that the L domain is not fully accessible in the chimeras. Regardless, the potent release of SrcM-GagN, in the absence of the RSV proline motif, demonstrates that an effective L domain activity resides in the M74 sequence.
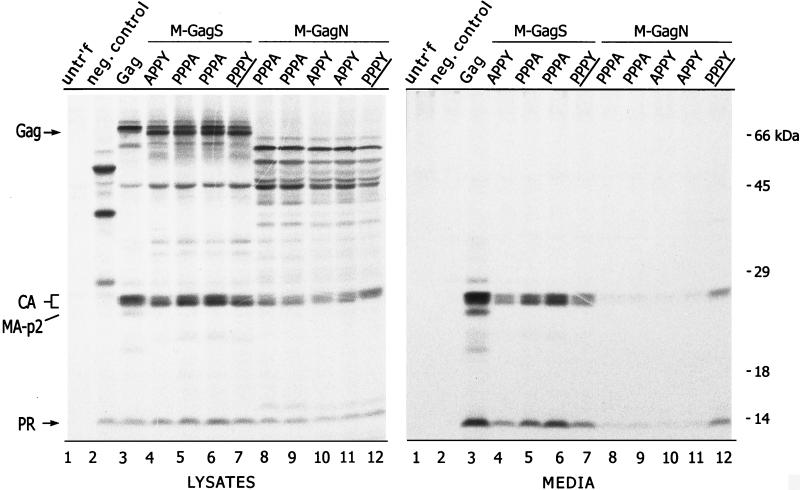
To test whether the particle release activity maps to the PPPY motif in M74, alanine substitutions were created within this sequence. Point mutations in the PPPY motif of M-GagN had drastic effects on budding, although none altered the levels of precursor proteins and cleavage intermediates in cell lysates (Fig. 4, compare lanes 8 to 11 with lane 12). Compared to the unmodified M-GagN protein (lane 12), alteration of the first proline residue (PPPY changed to APPY, lanes 10 and 11) or the tyrosine (PPPY to PPPA, lanes 8 and 9) in the M-GagN protein dropped particle release to nearly undetectable levels. On average in three experiments, the CA protein released into the medium by the M-GagN alanine mutants was only 2% of that seen with wild-type Gag protein, compared to 15% for the unmodified M-GagN (Fig. 3B). In sharp contrast, neither point mutation had any inhibitory effect on the release of the M-GagS protein, which retains the unaltered PPPPY sequence of RSV (Fig. 4, compare lanes 4 to 6 with lane 7).
FIG. 4.
Alterations to the PPPY motif of M-GagN cause a drastic reduction in budding activity. Particle release by the M-GagN chimera bearing the wild-type PPPY motif (distinguished by PPPY in the lane heading) was compared to those of mutants in which the peptide was altered by an alanine substitution at either the tyrosine (PPPA; two independent clones shown) or the first proline (APPY; two clones). Similar mutations in M74 were also evaluated in the context of the M-GagS protein. The negative control for budding is a mutant Gag protein that contains a large deletion spanning the p2b sequence (lanes 2). Samples from untransfected cells were loaded in lanes 1.
Finally, since the Src peptide cannot compensate for loss of L domain function in mutant RSV Gag proteins (30), we predicted that the budding of the SrcM-GagN chimera would be sensitive to alteration of the PPPY motif. Indeed, the mutation of the motif from PPPY to PPPA reduced SrcM-GagN particle release by sixfold, similar to the effect seen upon mutation of the motif in M-GagN (Fig. 3B). The magnitude of this effect was also comparable to that caused by similar point mutations in the RSV PPPPY motif of RSV Gag (33). Taken together the effects of point mutations on the budding of M-GagN, Src-MGagN, and M-GagS provide strong evidence that the VSV M protein has an activity that is functionally homologous to that of the retroviral late domain.
DISCUSSION
These studies have demonstrated that a sequence containing the PPPY motif of VSV M protein can substitute for the L domain of the RSV Gag protein to promote the release of chimeric particles. These findings support and extend the recent report that the integrity of the proline motif of the VSV M protein is important for the ability of the full-length protein to be released from cells in membrane-enclosed low-density particles (11). As a result, the M protein of VSV is now confirmed to possess the same three activities that have been defined as essential for Gag-directed budding: membrane binding activity (4), protein-protein interaction (9), and L domain function (this report). It is very likely that the proline motifs in the M proteins of other rhabdoviruses and in the presumptive matrix proteins of Marburg and Ebola viruses (11) will prove to have the same activity.
The idea that L domains function as binding sites for host factors to provide the machinery for budding was first presented (8) based on the sequence similarity between the RSV L domain and a protein interaction motif (XPPXY) recognized by proteins that contain a WW domain (3, 26). The WW domain is a protein interaction module found in a variety of proteins that are involved in cytoskeletal function, signal transduction, and regulatory events. One example, Yes-associated protein or YAP (2), a signal transduction protein, was proven to interact in vitro with the p2b region of RSV Gag protein (8). Similarly, the VSV PPPY and rabies virus PPEY sequences have now been shown by in vitro ligand blotting assay to bind the WW domains of YAP (11). Whether the functional in vivo ligand for the RSV and VSV L domains is YAP or perhaps another of the many and diverse cellular WW-containing proteins remains to be determined. The most compelling evidence to date for interactions between viral L domains and host factors comes from studies of the EIAV late domain (22). The YPDL sequence in EIAV p9 has now been shown to interact with the AP50 subunit of the adaptor protein complex 2 (AP2) both by in vitro binding assays and by in vivo colocalization.
Mutagenesis of the PPPY motif in the M-GagN and SrcM-GagN chimeras has demonstrated the importance of this motif in the budding activity of the M protein. However, none of the mutations tested completely abolished budding. This leaky phenotype is similar to what has been seen with RSV and M-PMV late domain mutants (18, 30, 33, 34). Perhaps some leakiness is not surprising if indeed the block to release occurs at a very late step, as some particles may spontaneously break off the cell surface. On the other hand, leakiness may reflect some redundancy within the viral proteins, providing multiple interaction sites for presumptive host factors. Alternatively, if only a very few molecules of the relevant factors are needed at the budding site, the reduced affinity of the mutant L domain for its ligand or simple trapping of a few host molecules by a nonspecific means may allow the inclusion of sufficient factors to drive some budding activity.
Whether all these different viruses function through the very same cellular machinery but do so by interacting with different components within it remains to be elucidated. The striking sequence differences between the EIAV late domain and those of RSV, VSV, HIV-1, and visna virus certainly imply that the latter ones do not bind directly with AP50. It is also possible that the proline motifs from RSV and VSV, though very similar in sequence, work through different WW domain proteins, given the observation that the most potent chimeric protein (SrcM-GagN) could not equal the release efficiency of the authentic Gag protein. This idea is consistent with findings that cellular XPPXY motifs with slight sequence differences in and around the motif itself react with quite different arrays of cellular WW domains (20). Clearly, the elucidation of the mechanism of virus budding will require a fuller accounting of the host machinery involved. A comparison of host factors that interface with the different retrovirus and rhabdovirus proteins is likely to be a fruitful approach to this goal.
ACKNOWLEDGMENTS
This work was funded by grants from the National Institutes of Health (CA47482) and the American Cancer Society (FRA-427) to J.W.W. and from the National Institutes of Health to P.P.
Special thanks are due to Laurence Garnier for her assistance with the buoyant density analysis.
REFERENCES
- 1.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H I, Einbond A, Kwak S-J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 3.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong L D, Rose J K. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craven R C, Bennett R P, Wills J W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991;65:6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. In: Krausslich H-G, editor. Morphogenesis and maturation of retroviruses. Berlin, Germany: Springer-Verlag; 1996. pp. 65–94. [DOI] [PubMed] [Google Scholar]
- 7.Dickson C, Eisenman R, Fan H, Hunter E, Teich N. Protein biosynthesis and assembly. In: Weiss R, Teich N, Varmus H, Coffin J M, editors. RNA tumor viruses. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 513–648. [Google Scholar]
- 8.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 9.Gaudin Y, Barge A, Ebel C, Ruigrok R W H. Aggregation of VSV M protein is reversible and mediated by nucleation sites: implications for viral assembly. Virology. 1995;206:28–37. doi: 10.1016/s0042-6822(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 10.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harty R N, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 15.Lenard J, Vanderoef R. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J Virol. 1990;64:3486–3491. doi: 10.1128/jvi.64.7.3486-3491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mebatsion T, Konig M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 17.Nelle T D, Wills J W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellman D, Garber E A, Cross F R, Hanafusa H. An N-terminal peptide from p60src can direct myristylation and plasma membrane localization when fused to heterologous proteins. Nature (London) 1985;314:374–377. doi: 10.1038/314374a0. [DOI] [PubMed] [Google Scholar]
- 20.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 21.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puffer B A, Watkins S C, Montelaro R C. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakalian M, Wills J W, Vogt V M. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spearman P, Wang J-J, van der Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 27.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 28.Wagner R R, Rose J K. Rhabdoviridae: the viruses and their replication. In: Fields B N, Knipe D, Howley P, et al., editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1121–1135. [Google Scholar]
- 29.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Z, Sun W, Suryanarayana K, Justice P, Robinson D, Wagner R R. Membrane-binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J Virol. 1994;68:7386–7396. doi: 10.1128/jvi.68.11.7386-7396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zakowski J J, Petri W A, Jr, Wagner R R. Role of matrix protein in assembling the membrane of vesicular stomatitis virus: reconstitution of matrix protein with negatively charged phospholipid vesicles. Biochemistry. 1981;20:3902–3907. doi: 10.1021/bi00516a037. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]