Abstract
The H5N1 avian influenza virus that killed 6 of 18 persons infected in Hong Kong in 1997 was transmitted directly from poultry to humans. Viral isolates from this outbreak may provide molecular clues to zoonotic transfer. Here we demonstrate that the H5N1 viruses circulating in poultry comprised two distinguishable phylogenetic lineages in all genes that were in very rapid evolution. When introduced into new hosts, influenza viruses usually undergo rapid alteration of their surface glycoproteins, especially in the hemagglutinin (HA). Surprisingly, these H5N1 isolates had a large proportion of amino acid changes in all gene products except in the HA. These viruses maybe reassortants each of whose HA gene is well adapted to domestic poultry while the rest of the genome arises from a different source. The consensus amino acid sequences of “internal” virion proteins reveal amino acids previously found in human strains. These human-specific amino acids may be important factors in zoonotic transmission.
At irregular intervals, influenza viruses that contain avian virus-derived genes cause pandemics that vary in severity from serious (Asian influenza [1957] and Hong Kong influenza [1968]) (39) to catastrophic (Spanish influenza [1918]). The molecular events that permit an avian influenza virus to transfer to and replicate in humans are largely unknown, as the precursor viruses have heretofore not been available for study. The H5N1 influenza viruses that infected 18 humans in Hong Kong in 1997 and caused the deaths of six (13, 35, 41) had been isolated 2 months earlier from chickens with lethal influenza (10). Thus, the putative progenitor virus was available for molecular analysis. There was no convincing evidence of human-to-human transmission; instead, the available evidence indicated that each case had been caused by independent transmission of virus to humans from birds in poultry markets.
Avian influenza viruses have a receptor specificity unlike that of human strains; they bind preferentially to terminal SAα2,3-galactose determinants, whereas human strains preferentially bind to terminal SAα2,6-galactose sequences (20, 25). The direct transmission of H5N1 viruses from chickens to humans in Hong Kong indicates that receptor specificity is not a definitive host restriction factor and that an intermediate host (17, 29) is not necessarily required for the first stage of transmission to humans. Previous studies indicate that the viral nucleoprotein (NP) and PB2 and M2 proteins are important in determining host range (2, 8, 28, 34), but the specific domains that affect host range involved have not been identified.
Circumstantial evidence has indicated that avian influenza viruses can directly infect humans (3, 5, 9, 31, 38, 42). In 1983, a highly lethal influenza outbreak in chickens in Pennsylvania was caused by an avian influenza virus of the H5 subtype;efforts to isolate the virus from poultry workers during that outbreak were unsuccessful (4). The H5N1 viruses isolated from humans in Hong Kong represent the first known direct transmission of avian influenza virus that has caused severe respiratory disease and death in humans. The present study characterized the genomes of H5N1 avian influenza viruses isolated from poultry markets in Hong Kong and compared them with virus isolated from the index human case. We established that a rapidly evolving reassortant influenza virus was present in Hong Kong poultry markets, and we propose that the presence of specific amino acids typical of human influenza viruses permitted direct transmission to humans.
MATERIALS AND METHODS
Viruses.
The viruses used in this study are listed in Table 1 (Table 1 also lists abbreviations used below for some viruses). The viruses were isolated in chicken embryos as described previously (30) and were grown in chicken embryos. Since these viruses are lethal for chicken embryos, the replication time was optimized and virus was harvested just before the death of the embryos. All viruses were handled in a biosafety level 3+ facility approved for use by the U.S. Department of Agriculture; the staff wore fitted HEPA-filtered masks and took prophylactic rimantadine.
TABLE 1.
Influenza A viruses sequenced in this study
| Pathogenicity and host | Virus strain | Subtype | Abbreviation | Source | Pathogenicity in chickens | Connecting peptide | Carbohydrate at residue 154 of HA |
|---|---|---|---|---|---|---|---|
| Pathogenic | |||||||
| Human | A/Hong Kong/156/97 | H5N1 | HK156-97 | Index human infection | + | TPQRERRRKKR | − |
| Chicken (Hong Kong) | A/Chicken/Hong Kong/220/97 | H5N1 | CHK220-97 | Farm, New Territories, organs | + | TPQRERRRKKR | + |
| A/Chicken/Hong Kong/258/97 | H5N1 | CHK258-97 | Farm, New Territories, organs | + | APQRERRRKKR | + | |
| A/Chicken/Hong Kong/786/97 | H5N1 | CHK786-97 | Market, New Territories, organs | + | TPQRERRRKKR | + | |
| A/Chicken/Hong Kong/y385/97 | H5N1 | CHK385-97 | Tsuen Wan Market, Hong Kong, feces | + | TPQRERRRKKR | + | |
| A/Chicken/Hong Kong/y388/97 | H5N1 | CHK388-97 | Tsuen Wan Market, Hong Kong, feces | + | TPQRERRRKKR | + | |
| A/Chicken/Hong Kong/728/97 | H5N1 | CHK728-97 | Market, New Territories, organs | + | TPQRERRRKKR | − | |
| A/Chicken/Hong Kong/915/97 | H5N1 | CHK915-97 | Market, New Territories, organs | + | TPQRERRRKKR | − | |
| A/Chicken/Hong Kong/p17/97 | H5N1 | CHK17-97 | Central Market, Hong Kong, feces | + | TPQRERRRKKR | − | |
| A/Chicken/Hong Kong/1203/97 | H5N1 | CHK1203-97 | Market, Hong Kong, organs | + | TPQRERRRKKR | − | |
| A/Chicken/Hong Kong/976/97 | H5N1 | CHK976-97 | Apleichau Market, Hong Kong, feces | + | TPQRERRRKKR | − | |
| A/Chicken/Hong Kong/990/97 | H5N1 | CHK990-97 | Apleichau Market, Hong Kong, feces | + | TPQRERRRKKR | − | |
| Duck | A/Duck/Hong Kong/p46/97 | H5N1 | DHK46-97 | Central Market, Hong Kong, feces | + | TPQRERRRKKR | − |
| A/Duck/Hong Kong/y283/97 | H5N1 | DHK283-97 | Tsuen Wan Market, Hong Kong, feces | + | TPQRERRRKKR | − | |
| Goose | A/Goose/Hong Kong/w355/97 | H5N1 | GHK355-97 | Cheung Sha Wan Market, Hong Kong, cloacal swab | + | TPQRERRRKKR | − |
| Chicken (Mexico) | A/Chicken/Puebla/8623-607/94 | H5N2 | CPUE1-94 | NSa | NDb | VPQRKRKTR | ND |
| A/Chicken/Puebla/14585-622/94 | H5N2 | CPUE2-94 | NS | ND | VPQRKRKTR | ND | |
| A/Chicken/Queretaro/14588-19/95 | H5N2 | CQUE95 | NS | ND | VPQRKRKTR | ND | |
| Nonpathogenic | |||||||
| Duck | A/Duck/Hong Kong/205/77 | H5N3 | DHK205-77 | NS | ND | VPQRETR | − |
| A/Duck/Hong Kong/698/79 | H5N3 | DHK698-79 | NS | ND | VPQRETR | − | |
| A/Duck/Minnesota/1525/81 | H5N1 | DMN81 | NS | ND | VPQRETR | − | |
| A/Duck/Potsdam/2216-4/84 | H5N6 | DP84 | NS | ND | VPQRETR | − | |
| A/Duck/Potsdam/1402-6/86 | H5N2 | DP86 | NS | ND | VPQRETR | − | |
| A/Duck/Nanchang/266/98 | H4N6 | DNC98 | NS | ND | ND | ND | |
| Aquatic bird | A/Aquatic bird/Hong Kong/m603/98 | H11N1 | ABHK603-98 | Nature, New Territories, feces | ND | ND | ND |
| Chicken (Mexico) | A/Chicken/Hidalgo/26654-1368/94 | H5N2 | CHID94 | NS | ND | VPQRETR | ND |
| A/Chicken/Mexico/26654-1374/94 | H5N2 | CMEX94 | NS | ND | VPQRETR | ND | |
| Turkey | A/Turkey/Ramon/73 | H5N2 | TRAM73 | NS | ND | VPQRETR | − |
| Gull | A/Gull/Pennsylvania/4175/83 | H5N1 | GPA83 | NS | ND | VPQRETR | − |
NS, not specified.
ND, not determined in this study.
Sequencing and analysis.
The viruses sequenced in this study are listed in Table 1. Viral RNA was extracted from infected allantoic fluid with the RNeasy extraction kit (Qiagen, Santa Clara, Calif.) and amplified by reverse transcription-PCR (32). PCR products were purified with the QIAquick PCR purification kit (Qiagen), sequenced with synthetic oligonucleotides by using rhodamine Dye-Terminator Cycle Sequencing Ready Reaction kits with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystems Inc. Foster City, Calif.), and electrophoresed on model 377 (Perkin-Elmer, Applied Biosystems Inc.) DNA sequencers by the Center for Biotechnology at St. Jude Children’s Research Hospital.
Analysis of sequence data was performed with Wisconsin Package version 9.1-Unix (Genetics Computer Group, Madison, Wis.) and GeneDoc version 2.3.000 (24) software. Phylogenetic analysis was done with PHYLIP version 3.57C software (14) to implement a neighbor-joining algorithm.
Analysis of nucleotide and amino acid changes in avian influenza viruses.
A defined region was sequenced for each of six viral genes (the genes for polymerases [PA, PB1, and PB2], nucleoprotein [NP], hemagglutinin [HA], and neuraminidase [NA]) in groups of two to six related viruses, and the resulting nucleotide and amino acid sequences were compared within groups (see Table 3). A matrix analysis was performed with GeneDoc version 2.3.000 (24) to quantitate the nucleotide and amino acid changes in each gene of each isolate, and the values were averaged for each group. The percentage of nucleotide changes that resulted in amino acid changes was calculated for each gene of each virus in a group, and the values were averaged.
TABLE 3.
Comparison of nucleotide and amino acid changes in genes and encoded proteins of influenza viruses
| Virus host and/or geographic locationa | Gene | Analyzed coding region | Mean no. of nucleotide changes ± SE | Mean no. of amino acid changes ± SE | Mean % of AAC/ NCb ± SE |
|---|---|---|---|---|---|
| Chicken, Hong Kongc | HA1 | 77–1042 | 6.8 ± 3.6 | 0.6 ± 0.5 | 5.9 ± 5.2 |
| Human, Hong Kongd | 77–1042 | 9.5 ± 2.9 | 5.0 ± 2.4 | 50.5 ± 18.7 | |
| Chicken, Mexicoe | 77–1042 | 40.8 ± 19.1 | 20.9 ± 10.1 | 57.3 ± 16.7 | |
| Chicken, Pennsylvaniaf | 77–1042 | 5.0 | 3.0 | 60.0 | |
| Duckg | 77–1042 | 41.5 ± 12.5 | 5.6 ± 0.8 | 15.1 ± 3.4 | |
| Chicken, Hong Kongc | NA | 64–1381 | 11.6 ± 5.9 | 4.2 ± 2.1 | 36.6 ± 6.4 |
| Chicken, Mexicoe | 64–1381 | 12.0 ± 9.3 | 3.6 ± 2.7 | 37.9 ± 25.9 | |
| Chicken, Hong Kongc | PB2 | 34–2304 | 16.5 ± 9.6 | 7.0 ± 3.6 | 46.9 ± 14.8 |
| Chicken, Mexicoe | 34–2304 | 22.2 ± 18.1 | 5.4 ± 4.4 | 24.3 ± 0 | |
| Avianh | 34–2304 | 294.7 ± 58.2 | 12.3 ± 4.1 | 4.9 ± 2.3 | |
| Chicken, Hong Kongc | PB1 | 34–2238 | 17.4 ± 11.9 | 4.2 ± 2.9 | 25.3 ± 10.9 |
| Chicken, Mexicoe | 34–2238 | 25.8 ± 21.1 | 3.6 ± 2.8 | 14.0 ± 0 | |
| Aviani | 34–2238 | 96.3 ± 7.4 | 2.0 ± 0 | 2.1 ± 0.1 | |
| Chicken, Hong Kongc | PA | 25–2172 | 13.1 ± 9.6 | 4.5 ± 2.7 | 51.3 ± 30.7 |
| Chicken, Mexicoe | 25–2172 | 24.4 ± 19.2 | 10.6 ± 8.5 | 41.3 ± 6.7 | |
| Avianj | 25–2172 | 71.0 ± 20.8 | 10.0 ± 2.5 | 10.6 ± 4.6 | |
| Chicken, Hong Kongc | NP | 46–1542 | 9.7 ± 5.7 | 2.7 ± 1.2 | 35.5 ± 21.8 |
| Chicken, Mexicoe | 46–1542 | 12.2 ± 9.6 | 0.6 ± 0.4 | 5.0 ± 0 | |
| Aviank | 46–1542 | 72.0 ± 9.0 | 3.0 ± 1.2 | 4.3 ± 2.2 |
With the exception of those viruses classified as avian, all viruses compared were of the H5 subtype.
AAC/NC, amino acid changes per nucleotide changes.
CHK258-97, DHK46-97, CHK915-97, CHK283-97, CHK786-97, and GHK355-97.
HK156, A/Hong Kong/481/97 (H5N1), A/Hong Kong/482/97 (H5N1), and A/Hong Kong/483/97 (H5N1).
CMEX94, CHID94, CPUE1-94, CPUE2-94, and CQUE95.
A/Chicken/Pennsylvania/1/83 (H5N2) and A/Chicken/Pennsylvania/1370/83 (H5N2).
DHK205-77, Dk/HK/342/78, DHK698-79, and DP84.
A/Budgerigar/Hokkaido/1/77 (H4N6), A/Ruddy Turnstone/New Jersey/47/85 (H4N6), A/Turkey/Minnesota/833/80 (H4N2), and A/Mallard/New York/6750/78 (H2N2).
A/Turkey/Minnesota/833/80 (H4N2), A/Mallard/New York/6750/78 (H2N2), and A/Gull/Maryland/704/77 (H13N6).
A/Pintail/Alberta/119/79 (H4N6), A/Turkey/Minnesota/833/80 (H4N2), A/Ruddy Turnstone/New Jersey/47/85 (H4N6), and A/Duck/Hokkaido/8/80 (H3N8).
DHK205-77, DHK698-79, A/Duck/Hong Kong/7/75 (H3N2), and A/Duck/Beijing/1/78 (H3N6).
RESULTS
Phylogenetic analysis of H5N1 isolates from poultry in Hong Kong.
Preliminary studies of the index human virus from Hong Kong and of the initial chicken isolate showed two distinct groups based on phylogenetic analysis of the HA (33). We now establish that the eight genes of the H5N1 viruses isolated from poultry were closely related but comprised two phylogenetic sublineages.
Genetic homology among the H5N1 isolates varied from 97.9 to 100% (Table 2). The viruses that were most homologous with the individual gene segments of H5N1 viruses from Hong Kong were from the Eurasian lineage. Despite the limited number of gene sequences available for the polymerase (PB2, PB1, and PA) genes in GenBank, the most homologous viruses were from Asia. The PB1, NA, M, and NS genes were most related to influenza viruses from China. The largest quantity of information available is for the NP gene, and the virus with greatest NP homology was A/Mallard/Astrakhan/244/82 (H14N6), a virus isolated from Central Asia. For the NA gene, the closest related virus was one isolated from an unidentified migrating aquatic bird in Hong Kong (ABHK603-98). Sequence information for additional Eurasian viruses is needed before any projection can be made as to the origins of other gene segments.
TABLE 2.
Genetic homology of the H5N1 Hong Kong avian influenza viruses
| Gene segment | Region sequenced | % Homology among H5N1 virusesa | Virus with greatest homology to CHK915-97b | % Homology with CHK915-97 |
|---|---|---|---|---|
| PB2 | 40–2306 | 98.4–100.0 | A/Budgerigar/Hokkaido/1/77 (H4N6) | 90.1 |
| PB1 | 31–2258 | 98.6–99.9 | A/Duck/Nanchang/266/98 (H4N6) | 90.6 |
| PA | 11–2165 | 98.0–99.8 | A/Swine/Hong Kong/81/78 (H3N2) | 91.0 |
| HA | 21–1746 | 98.4–99.9 | A/Turkey/England/91 (H5N1) | 93.5 |
| NP | 29–1543 | 98.5–99.8 | A/Mallard/Astrakhan/244/82 (H14N6) | 93.6 |
| NA | 64–1381 | 98.6–99.9 | A/Aquatic bird/Hong Kong/m603/98 (H11N1) | 91.1 |
| M | 28–1000 | 98.6–100.0 | A/Duck/Hong Kong/698/79 (H5N3) | 98.5 |
| NS | 21–867 | 97.9–100.0 | A/Duck/Nanchang/1944/93 (H7N4) | 93.7 |
HK156-97, CHK258-97, DHK46-97, GHK355-97, CHK915-97, CHK728-97, and CHK786-97 were used to analyze the homology among the H5N1 Hong Kong viruses.
CHK915-97 was chosen as the representative virus and was used to search the sequences in GenBank and our local database.
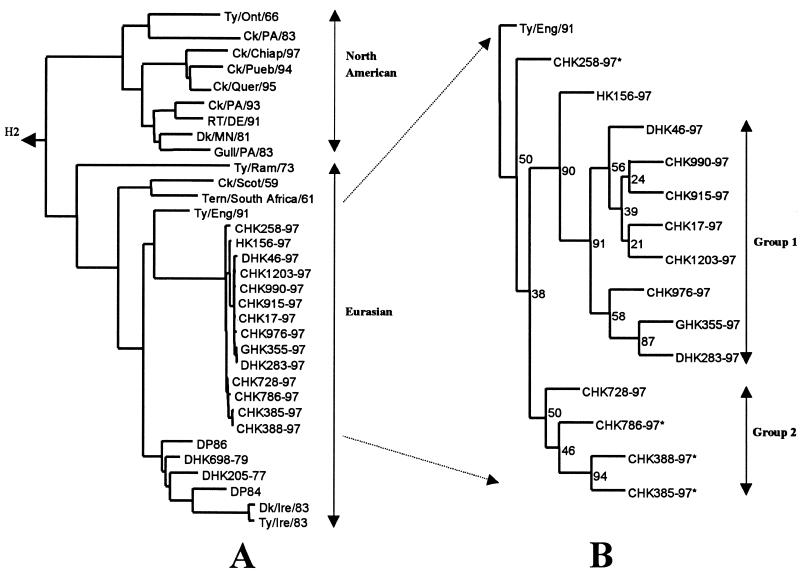
Despite the high degree of homology of the Hong Kong H5N1 viruses, phylogenetic analysis showed that there were two subgroups of each gene. Figure 1 shows the phylogenetic subgroups based on HA1. In one group, most viruses possessed a carbohydrate side chain at residue 154 of HA1; the other group, with one exception, lacked the carbohydrate. The viruses of both subgroups possessed a series of basic amino acids (RERRRKKR) at the connecting peptide between HA1 and HA2 and were highly pathogenic in chickens (Table 1). The sequences encoding the receptor binding sites were identical in the two groups and indicated binding to terminal SAα2,3-galactose determinants, as was described for the index human virus (10, 35).
FIG. 1.
Phylogenetic trees of the HA1 genes of H5 influenza A viruses. (A) Evolutionary relationship of the H5N1 Hong Kong viruses to various H5 viruses isolated previously in North America and Eurasia. The tree was generated by using the neighbor-joining algorithm in PHYLIP version 3.57c software (14) and is rooted to A/Pintail/Praimoric/625/76 (H2N2). Horizontal lines are proportional to the numbers of nucleotide substitutions between branch points. (B) One hundred-replication bootstrap resampling of the H5 Hong Kong viruses from panel A; the tree is rooted to A/Turkey/England/50-92/91. The number at each branch point indicates the percent probability of the accuracy of the resulting topology. Asterisks indicate the presence of a potential glycosylation site at amino acid 154 of HA. Abbreviations: Ty/Ont/66, A/Turkey/Ontario/7732/66 (H5N9); Ck/PA/83, A/Chicken/Pennsylvania/1/83 (H5N2); Ck/Chiap/97, A/Chicken/Chiapis/15224/97 (H5N2); Ck/Pue/94, CPUE1-94; Ck/Que/95, CQUE95; Ck/PA/93, A/Chicken/Pennsylvania/13609/93 (H5N2); RT/DEL/93, A/Ruddy turnstone/Delaware/244/93 (H5N2); Dk/MN/81, DMN81; Gull/PA/83, GPA83; Ty/Ram/73, TRAM73; Ck/Scot/59, A/Chicken/Scotland/59 (H5N1); Tern/South Africa/61, A/Tern/South Africa/61 (H5N1); Ty/Eng/91, A/Turkey/England/50-92/91 (H5N1); Dk/Ire/83, A/Duck/Ireland/113/83 (H5N8); Ty/Ire/83, A/Turkey/Ireland/1378/83 (H5N8).
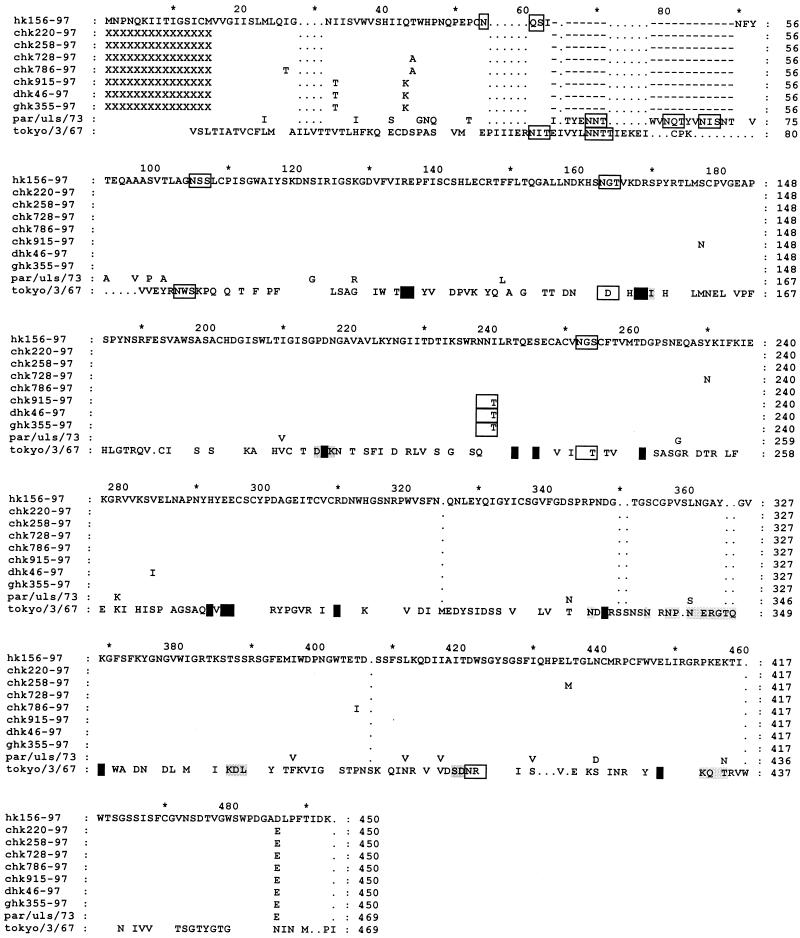
The NAs of the H5N1 Hong Kong influenza viruses isolated from poultry all had a 19-amino-acid deletion in the stalk (Fig. 2). Three glycosylation sites in the N1 protein are removed by this deletion. Partial sequencing of NA genes from 13 additional poultry H5N1 viruses confirmed that the deletion is present in all of the H5N1 viruses. This deletion consists of 4% of the NA molecule and was detectable by electrophoresis of PCR products of the NA gene. To check the difference in the sialic acid binding site and antigenic sites between the NA of HK156-97 and the poultry H5N1 viruses, the NA sequences of the Hong Kong viruses were aligned with the N2 protein of A/Tokyo/3/67, which has been defined by structural analysis (11). The alignment was made as described by Colman et al. (12) (Fig. 2). According to the alignment, all amino acids in the sialic acid binding site of N2 are conserved in the H5N1 Hong Kong viruses. No difference in antigenic determinants was observed for HK156-97 and the poultry H5N1 viruses. There are three potential glycosylation sites conserved in the head region of NA of A/Tokyo/3/67 (N2), A/Parrot/Ulster/77 (N1), and the H5N1 Hong Kong viruses (N1). The H5N1 viruses belonging to group 1 possessed an additional potential glycosylation site at residue I204T in the head region.
FIG. 2.
Alignment of NA amino acid sequences of H5N1 influenza viruses from Hong Kong. Amino acids in the open boxes are potential glycosylation sites, those in the dark background are enzyme activity sites, and those in the light background are antigenic sites. Dashes represent deletions; dots represent the gaps resulting from aligning the N1 proteins with N2. par/uls/73, A/Parrot/Ulster/73 (H7N1); Tokyo/3/67, A/Tokyo/3/67 (H2N2).
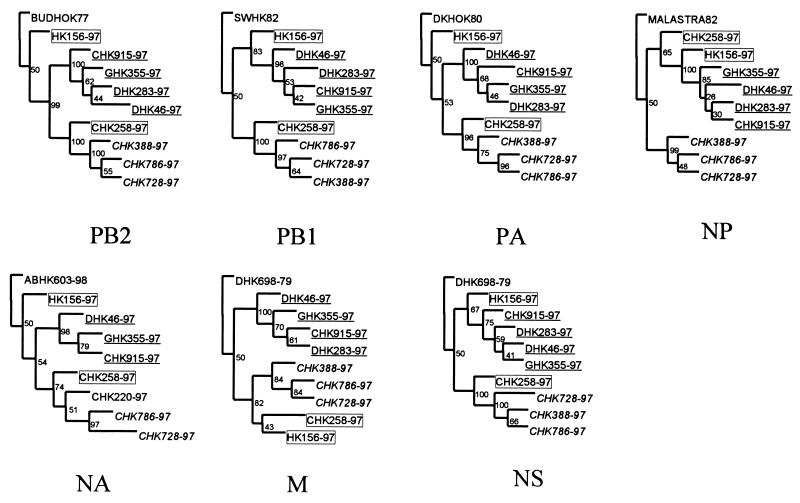
Phylogenetic analysis of the other seven gene segments of the H5N1 isolates from poultry confirmed that they each belong to the Eurasian avian lineage. For conservation of space, only the terminal branches of the phylogenetic trees of the NA and internal genes are shown in Fig. 3. Each of the genes of H5N1 poultry viruses from Hong Kong can be divided into the two subgroups given above for the HA. However, the human index virus, HK156-97, and the early chicken isolate CHK258-97 are intermediate between the two groups. Viruses from both subgroups caused severe disease in humans (6, 33, 41).
FIG. 3.
Terminal branches of the phylogenetic trees of the NA and internal genes of H5N1 influenza viruses from poultry in Hong Kong. The phylogenetic trees were generated as described in the legend to Fig. 1B. The viruses underlined belong to group 1, the viruses italicized belong to group 2, and the viruses in the boxes are intermediates. BUDHOK, A/Budgerigar/Hokkaido/1/77 (H4N6); SWHK82, A/Swine/Hong Kong/126/82 (H3N2); DKHOK80, A/Duck/Hokkaido/8/80 (H3N8); MALASTRA82, A/Mallard/Astrakhan/244/82 (H14N6).
The rate of evolution of the HA gene is disproportionately low.
When an influenza virus is first introduced into a new host, it usually undergoes a period of rapid evolutionary change (18, 27). Nucleotide changes that result in amino acid changes are usually most numerous in the HA, due to immunological selection, and are less numerous in the internal genes. Table 3 shows the number of nucleotide changes, the number of amino acid changes, and the percentage of changes in six of the gene sequences of H5N1 viruses. Where possible, we compared sequence information with that for H5N2 influenza viruses that had apparently emerged from wild aquatic birds and then spread through the chicken population of Mexico and become highly pathogenic (16). We also used the available but limited sequence information on influenza viruses from aquatic birds for comparison. In the HA gene of H5N1 poultry viruses, only 5.9% of the nucleotide changes produced amino acid changes, which was an order of magnitude less than the percentage of similar changes in the PA gene (Table 3). The NA gene and the genes comprising the replication complex (the genes for PB2, PB1, PA, and NP) all had a higher percentage of nonsilent nucleotide substitutions than did the HA gene.
In H5N1 viruses isolated from poultry in Hong Kong, the percentage of coding changes in the HA gene (5.9%) was an order of magnitude less than that found in H5N2 virus from chickens in Mexico (57.3%). This finding indicates a surprisingly low rate of evolution of the HA gene. Only the HA gene of influenza viruses from aquatic birds had a comparably low proportion of coding changes (H5 from duck, 15.1%). After transmission to humans the H5 HA gene evolved rapidly, which is expected in a new host. The rates of coding changes in the NA and other genes (those for PB2, PB1, PA, and NP) in H5 viruses from Hong Kong and Mexico were higher than those found in influenza viruses from aquatic birds. This rapid rate of change is typical of a virus in a new host.
These studies indicate that the HA gene of the H5N1 influenza viruses isolated from poultry in Hong Kong is optimally adapted, while the other gene segments are evolving rapidly. After transmission to humans the HA genes shows the expected high rate of change.
Correlation between host range and amino acids in PA, PB2, NP, and M2 gene products.
Previous studies have indicated that the NP, PB2, and M2 gene products are important in determining the host range of influenza viruses (8, 28, 34). We therefore compared the amino acid sequences of these proteins in the Hong Kong poultry H5N1 isolates with those of other viruses. We also compared the sequences of the PA proteins.
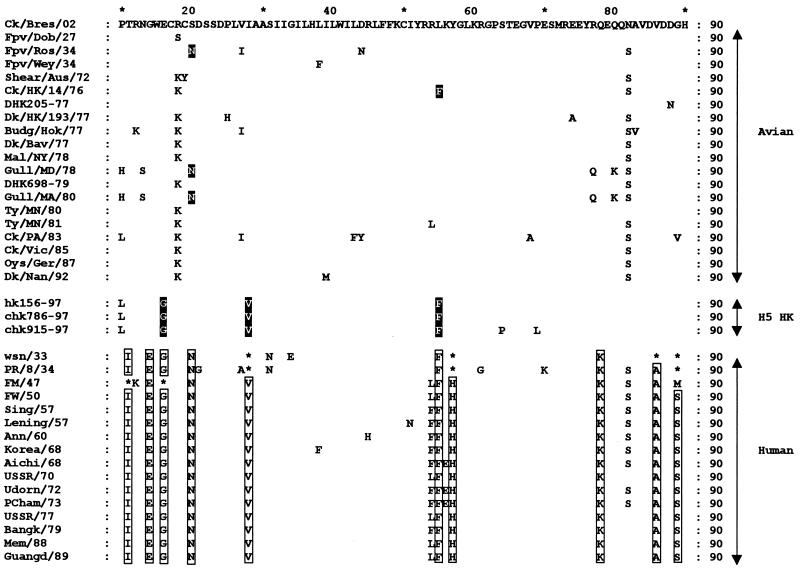
When available sequence information for the avian and human influenza virus proteins was compared, certain amino acids appeared to be specific to avian or human viruses. In the case of M2 proteins, 10 amino acids have previously been found almost exclusively in human influenza viruses; we found 3 of these amino acids in the M2 protein of H5N1 viruses from poultry in Hong Kong (Fig. 4). In each of the poultry isolates, the amino acids at M2 residues 16, 28, and 55 were G, V, and F, respectively, while in 20 other avian influenza viruses these amino acids were E, I, and L (A/Chicken/Hong Kong/14/76 also had an F at position 55). Thus, three amino acids in M2 of the chicken H5N1 influenza viruses are highly related to the sequence in human influenza viruses. Among the other internal gene products (PB2, PB1, PA, NP, M1, and NS) of the H5N1 viruses from Hong Kong, “human-like” amino acids were detected in PB2 (S199, T661, and R702), PA (N409), and NP (M136) (Table 4). No human-like substitutions were detected in the PB1, M1, or NS proteins. Thus, the H5N1 viruses isolated from poultry in Hong Kong possess several amino residues characteristically found in human strains.
FIG. 4.
Alignment of amino acids of the M2 proteins of human and avian influenza A viruses. The open boxes indicate amino acids found almost exclusively in human influenza viruses. The solid boxes indicate human-like amino acids that are present in avian viruses. Asterisks indicate avian-like amino acids that are present in human viruses. Abbreviations for avian viruses: Ck/Bres/02, A/Chicken/Brescia/19/02 (H7N7); Fpv/Dob/27, A/FPV/Dobson/27 (H7N7); Fpv/Ros/34, A/FPV/Rostock/34 (H7N1); Fpv/Wey/34, A/FPV/Weybridge (H7N7); Shear/Aus/72, A/Shearwater/Australia/1/72 (H6N5); Ck/HK/14/76, A/Chicken/Hong Kong/14/76 (H1N1); Dk/HK/193/77, A/Duck/Hong Kong/193/77 (H1N2); Budg/Hok/77, A/Budgerigar/Hokkaido/1/77 (H4N6); Dk/Bav/77, A/Duck/Bavaria/2/77 (H1N1); Mal/NY/78, A/Mallard/New York/6750/78 (H2N2); Gull/MD/78, A/Gull/Maryland/1824/78 (H13N6); Gull/MA/80, A/Gull/Massachusetts/26/80 (H13N6); Ty/MN/80, A/Turkey/Minnesota/833/80 (H4N2); Ty/MN/81, A/Turkey/Minnesota/166/81 (H1N1); Ck/PA/83, A/Chicken/Pennsylvania/1370/83 (H5N2); Ck/Vic/85, A/Chicken/Victoria/1/85 (H7N7); Oys/Ger/87, A/Oyster catcher/Germany/87 (H1N1); Dk/Nan/92, A/Duck/Nanchang/1749/92 (H11N2). Abbreviations for human viruses: wsn/33, A/WSN/33 (H1N1); PR/8/34, A/Puerto Rico/8/34 (H1N1); FM/47, A/Fort Monmouth/1/47 (H1N1); FW/50, A/Fort Warren/1/50 (H1N1); Sing/57, A/Singapore/1/57 (H2N2); Lening/57, A/Leningrad/134/57 (H2N2); Ann/60, A/Ann Arbor/6/60 (H2N2); Korea/68, A/Korea/426/68 (H2N2); Aichi/68, A/Aichi/2/68 (H3N2); USSR/70, A/USSR/90/70 (H1N1); Udorn/72, A/Udorn/72 (H3N2); PCham/73, A/Port Chalmers/1/73 (H3N2); USSR/77, A/USSR/90/77 (H1N1); Bangk/79, A/Bangkok/1/79 (H3N2); Mem/88, A/Memphis/8/88 (H3N2); Guangd/89, A/Guangdong/39/89 (H3N2).
TABLE 4.
Human-like amino acids found in H5N1 Hong Kong avian influenza virusesa
| Host | Amino acid at position in internal protein
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PB2
|
PA
|
NP
|
M2
|
|||||
| 199 | 661 | 702 | 409 | 136 | 16 | 28 | 55 | |
| Avian | A | A | K | S | L | E | I | L |
| Chicken (Hong Kong H5N1) | S/A | T | K/R | N/S | M | G | V | F |
| Human | S | T | R | N | M | G | V | F |
Numbers of strains used in this analysis: PB2, 8 avian, 12 human, and 6 chicken (Hong Kong); PA, 6 avian, 8 human, and 6 chicken (Hong Kong); NP, 30 avian, 21 human, and 6 chicken (Hong Kong); and M2, 20 avian, 16 human, and 6 chicken (Hong Kong).
DISCUSSION
Sequence analysis of each of the gene segments of H5N1 viruses isolated from poultry in Hong Kong established that of two distinguishable groups of the viruses cocirculated in domestic poultry. The origin of these two groups is unresolved. They may have originated through geographical separation of the original reassortant H5N1 possessing a chicken-adapted H5 gene and seven genes from a still-unidentified Eurasian avian influenza virus. Separation may have occurred in different parts of Asia providing poultry to Hong Kong or in different avian species in Hong Kong. Since the original chicken H5N1 isolate (CHK258-97) is intermediate between the two lineages (Fig. 1 and 3), the two lineages may be a recent evolutionary development that occurred in different markets in Hong Kong. Each gene segment of the two groups of viruses encoded amino acids that distinguish the two groups (results not shown). However, the facts that both groups infected humans and caused similar disease patterns (6, 33, 41) suggest that these amino acids are not directly involved in host range transfer.
The receptor specificity of influenza virus HA usually depends on the host species from which the virus is isolated. However, it was found that the H5N1 isolate from a human (HK156-97) has the same sequence in the receptor binding site of HA as found in poultry H5N1 viruses (10, 19, 33). This finding indicates that an avian virus can be transmitted to humans without a change in its receptor binding properties. However, this does not mean that avian viruses can be transmitted to humans as effectively as human virus. The difference in host cell receptor specificity may have played a role in restricting transmission of these H5N1 viruses among humans. Although a large human population in Hong Kong may have had contact with the infected chickens, only 18 cases were found to be positive by virus isolation. Serological studies with humans are still in progress, but the available evidence indicates little if any human-to-human transmission.
When influenza virus is introduced into a new host, the proportion of nucleotide changes that result in amino acid changes is usually highest in the HA gene. In the H5N1 influenza viruses isolated from Hong Kong poultry markets, this was not the case, the highest percentage of coding changes that altered amino acids was approximately six times higher in the PA gene than in the HA gene. The emergence of highly pathogenic H5 influenza viruses in chickens in Mexico represents the most comparable case for which information is available. Those isolates showed a large proportion of amino acid changes in the HA1 protein (57.3%) and a lower proportion in the products of the internal genes. This reversal of the pattern of coding changes suggests that the HA gene is better adapted to chickens than are the other genes. The Hong Kong H5N1 viruses may be reassortants that acquired the HA gene from an H5 virus that is well adapted to domestic poultry, while the other seven genes may have come from another source, such as wild aquatic birds. The HA could possibly have come from A/Goose/Guangdong/1/96 (H5N1), a virus that was highly lethal in geese (40). Guangdong Province lies adjacent to Hong Kong and provides much of Hong Kong’s domestic poultry. The other seven genes are of Eurasian avian origin, but their source is undetermined. Additional sequence information from Eurasian influenza viruses should elucidate the origin of these gene segments.
When H2 was introduced into humans in 1957 the molecule did change rapidly. Schafer et al. (27) reported that the earliest stages in the evolution of the human lineage appear to have been under greater selective pressures than the later branches as judged by their ratios of coding to noncoding changes. Initially, 1.6 nucleotide changes resulted in amino acid changes; later, 3.7 nucleotide changes were required per amino acid change. This was also seen in the case of European swine virus. When the H1N1 avian-like viruses appeared in swine in Europe in 1979, the NP genes of these new viruses evolved at a higher rate than NP genes in human and classic swine viruses over the period of 1930 to 1988. We do not see more rapid evolutionary changes in H3 for 1968 because we do not know when the avian HA was transmitted to humans. Some serological studies suggest that H3 was introduced into humans several years ahead of isolation of the virus in humans in 1968 (21).
The large proportion of amino acid changes in the NAs and internal gene products of the Hong Kong H5N1 isolates indicates that these viruses are evolving rapidly and therefore that chickens are a new host. These findings support the hypothesis that chickens represent a host population distinct from aquatic birds. As such, they may play a role in the evolution of the virus and may be an intermediate host in zoonotic transmission (19). Surveillance in poultry in Hong Kong between 1975 and 1985 detected H5 influenza viruses in ducks and geese but not in chickens (31), again suggesting that this virus was introduced into chickens relatively recently.
Specific amino acids in the M2 and PB2 proteins that have been associated with host range variants (8, 22, 28, 34) were not implicated by these studies. However, it must be noted that the host range of influenza viruses is a polygenic trait that can vary from virus to virus. The functional domains of PB2, PB1, PA, and NP that are associated with RNA synthesis, chain extension, and nuclear localization are being resolved (7, 23, 37). We found that the H5N1 Hong Kong viruses contain two human-like amino acids (T661 and R702) that are located in the functional domain (C-terminal 124 amino acids) of PB2 responsible for interaction with other polymerase components (26). The human-like amino acid (M136) in the NP of H5N1 viruses is located in the RNA binding domain of the protein (1), and the human-like amino acid (V28) in M2 of the H5N1 viruses is located in the domain serving as an ion channel (15). Although we do not know the function of these molecules in determining host range, we speculate that these amino acids are involved in the proliferation of H5N1 viruses in chickens and in humans. Identification of these host range-specific amino acids will provide a starting point for studies aimed at identifying the functional sites that mediate host range.
Our analysis of available sequence information from GenBank reveals that a number of influenza viruses isolated from humans contain avian-specific amino acids. These amino acids were present in the older human strains (A/WSN/33 [H1N1] and A/Puerto Rico/8/34 [H1N1]) identified as descendants of the virus causing the 1918 Spanish influenza pandemic and may be remnants of avian influenza viruses that were transmitted to humans. It is noteworthy that the partial sequences of the NP and M protein of the 1918 virus (36) contain more avian-like amino acids than in A/WSN/33. This indicates that the avian-like amino acids in A/WSN/33 were probably not derived from passage in chicken eggs.
The information that is now available underscores the wisdom of the slaughter of 1.6 million domestic fowl in Hong Kong in 1997. The H5N1 virus was evolving rapidly in this host. It had acquired a number of amino acids that correlate with replication in humans. Eradication of the chicken population in Hong Kong eliminated the immediate opportunity for the H5N1 viruses to be transmitted to humans.
ACKNOWLEDGMENTS
These studies were supported by Public Health Research Grant AI29680 and Cancer Center Support (CORE) grant CA-21765 from the National Institutes of Health and by the American Lebanese Syrian Associated Charities, the World Health Organization, the Department of Health of the Hong Kong government, and the Committee on Research and Conference Grants and the Office of the Vice-Chancellor, The University of Hong Kong.
We thank Enid M. Bedford and Kimberly Hampton for preparation of the manuscript and Sharon Naron for editorial assistance.
REFERENCES
- 1.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond J W. A single gene determines the host range of influenza virus. Nature. 1977;270:617–618. doi: 10.1038/270617a0. [DOI] [PubMed] [Google Scholar]
- 3.Banks J, Speidel E, Alexander D J. Characterization of an avian influenza A virus isolated from humans. Is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. doi: 10.1007/s007050050329. [DOI] [PubMed] [Google Scholar]
- 4.Bean W J, Kawaoka Y, Wood J M, Pearson J E, Webster R G. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J Virol. 1985;54:151–160. doi: 10.1128/jvi.54.1.151-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 6.Bender C, Huang J, Hall H, Klimov A, Cox N, Subbarao K. American Society for Virology 17th Annual Meeting. 1998. Molecular analysis of the surface protein genes of human (H5N1) influenza viruses, abstr. W29-7; p. 110. [Google Scholar]
- 7.Biswas S K, Nayak D P. Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J Virol. 1996;70:6716–6722. doi: 10.1128/jvi.70.10.6716-6722.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckler-White A J, Murphy B R. Nucleotide sequence analysis of the nucleoprotein gene of an avian and a human influenza virus strain identifies two classes of nucleoproteins. Virology. 1986;155:345–355. doi: 10.1016/0042-6822(86)90198-4. [DOI] [PubMed] [Google Scholar]
- 9.Campbell C H, Webster R G, Breese S S., Jr Fowl plague virus from man. J Infect Dis. 1970;122:513–516. doi: 10.1093/infdis/122.6.513. [DOI] [PubMed] [Google Scholar]
- 10.Claas E C J, Osterhaus A D, Van Beek R, de Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A (H5N1) virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 11.Colman P M, Varghese J N, Laver W G. Structure of catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 12.Colman P M, Hoyne P A, Lawrence M C. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol. 1993;67:2972–2980. doi: 10.1128/jvi.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jong J C, Claas E C, Osterhaus A D, Webster R G, Lim W L A. Pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP (phylogenetic I reference package) version 3.5. Seattle: Department of Genetics, University of Washington; 1993. . Distributed by the author. [Google Scholar]
- 15.Holsinger L J, Nichani D, Pinto L H, Lamb R A. Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster R G. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology. 1995;213:223–230. doi: 10.1006/viro.1995.1562. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Couceiro J N, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig S, Stitz L, Planz O, Van H, Fitch W M, Scholtissek C. European swine virus as a possible source for the next pandemic? Virology. 1995;212:555–561. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich M N, Gambaryan A S, Teneberg S, Piskarev V E, Yamnikova S S, Lvov D K, Robertson J S, Karlsson K A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 21.Monto A S, Maassab H F. Serologic responses to nonprevalent influenza A viruses during intercyclic period. Am J Epidemiol. 1981;113:236–244. doi: 10.1093/oxfordjournals.aje.a113092. [DOI] [PubMed] [Google Scholar]
- 22.Murphy B R, Buckler-White A J, London W T, Snyder M H. Characterization of the M protein and nucleoprotein genes of an avian influenza A virus which are involved in host range restriction in monkeys. Vaccine. 1989;7:557–561. doi: 10.1016/0264-410x(89)90283-1. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa Y, Kimura N, Toyoda T, Mizumoto K, Ishihama A, Oda K, Nakada S. The RNA polymerase PB2 subunit is not required for replication of the influenza virus genome but is involved in capped mRNA synthesis. J Virol. 1995;69:728–733. doi: 10.1128/jvi.69.2.728-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas K B, Nicholas H B., Jr . Gene Doc: a tool for editing and annotating multiple sequence alignments. 1997. Distributed by the author. [Google Scholar]
- 25.Paulson J C. Interactions of animal viruses with cell surface receptors. In: Conn P M, editor. The receptors. Vol. 2. Orlando, Fla: Academic Press; 1985. pp. 131–219. [Google Scholar]
- 26.Perales B, De La Luna S, Palacios I, Ortin J. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer J R, Kawaoka Y, Bean W J, Suss J, Senne D, Webster R G. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 28.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 29.Scholtissek C, Schultz U, Ludwig S, Fitch W M. The role of swine in the origin of pandemic influenza. In: Hannoun C, Kendal A P, Klenk H-D, Ruben F L, editors. Options for the control of influenza II. Amsterdam, The Netherlands: Elsevier Science Publisher B. V.; 1993. pp. 193–201. [Google Scholar]
- 30.Shortridge K F, Butterfield W K, Webster R G, Campbell C H. Isolation and characterization of influenza A viruses from avian species in Hong Kong. Bull W H O. 1977;55:15–20. [PMC free article] [PubMed] [Google Scholar]
- 31.Shortridge K F. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- 32.Shu L L, Bean W J, Webster R G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subbarao E K, London W, Murphy B R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 36.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 37.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster R G, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med. 1981;304:911. doi: 10.1056/NEJM198104093041515. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 39.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, Cox N, Guo Y. American Society for Virology 17th Annual Meeting. 1998. Emergence of influenza A (H5N1) viruses in geese in China, abstr. W29-5; p. 110. [Google Scholar]
- 41.Yuen K Y, Chan P K S, Peiris M, Tsang D N C, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T F, Sung R, Cheng A F B Members of the H5N1 Study Group. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 42.Zhou N, He S, Zhang T, Zou W, Shu L, Sharp G B, Webster R G. Influenza infection in humans and pigs in southeastern China. Arch Virol. 1996;141:649–661. doi: 10.1007/BF01718323. [DOI] [PubMed] [Google Scholar]