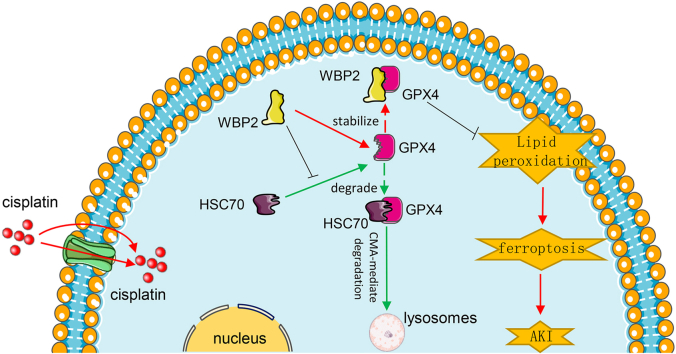
Abstract
Cisplatin is one of the major causes of acute kidney injury (AKI) in clinical practice, and ferroptosis is an essential form of cell death in cisplatin-induced AKI (CP-AKI). WW domain binding protein-2 (WBP2), a molecular chaperon, is involved in the progression of various malignancies, but its role in renal injuries has not been investigated. Our present study employed bioinformatics analysis to identify WBP2 as a potential modulator of AKI and ferroptosis. Preliminary laboratory investigations showed that WBP2, highly expressed in renal proximal tubular cells, was downregulated in CP-AKI. Further studies demonstrated that WBP2 decelerated ferroptosis to alleviate CP-AKI. Mechanistically, WBP2 interacted with glutathione peroxidase 4 (GPX4, a key detoxicating enzyme for ferroptosis) via its PPXY1 motif to inhibit ferroptosis. Furthermore, the in-depth investigations revealed that WBP2 competed with heat shock cognate protein 70 (HSC70) for the binding with the KEFRQ-like motifs of GPX4, leading to the deceleration of chaperon-mediated autophagy of GPX4. All in all, this study indicated the beneficial effect of WBP2 in CP-AKI and its relevance with ferroptosis, thus providing a novel insight into the modulation of ferroptosis in cisplatin-related nephropathy.
Keywords: WBP2, GPX4, Ferroptosis, Cisplatin, Acute kidney injury
Graphical abstract
Highlights
-
•
WBP2, ferroptosis, GPX4, chaperon-mediated autophagy.
1. Introduction
Cisplatin is a widely used chemotherapeutic drug for various solid malignancies, such as prostate cancer, testicular cancer, and ovarian cancer [1]. However, the clinical administration of cisplatin is largely hindered by its commonly observed nephrotoxicity. It is estimated that about 30% of cisplatin (high dose)-treated patients presented different manifestations of renal injuries, and its nephrotoxic effects can be accumulated after repeated use [2]. The major target of cisplatin-induced AKI (CP-AKI) is renal proximal tubules, where most cisplatin in the glomerular ultrafiltrate is reabsorbed [3]. Therefore, renal proximal tubular cells were constantly used in the laboratory investigations of CP-AKI. Previous publications revealed that various cell death pathways were incorporated in the progression of CP-AKI [4], but their exact regulatory machinery has not been fully elucidated; thus, no effective treatments were available in the clinical practice.
Ferroptosis is a type of regulated cell death identified in 2012, and it was initially used to describe erastin-induced cell death [5]. Later on, it was discovered that ferroptosis was operative in various pathophysiological scenarios, including ischemia/reperfusion injury, degenerative disorder, and chemotherapy of malignancies [[6], [7], [8]]. Unlike other types of regulated cell death, ferroptosis is characterized by decreased crista and condensed inner membrane in mitochondrial morphology, iron-catalyzed excessive lipid peroxidation, and the independency of caspase [9]. When cells were under overwhelming ferroptotic pressure, the key terminator of ferroptosis GPX4, together with glutathione (GSH), was involved to catalyze lethal lipid hydroperoxides to non-toxic lipid alcohols, leading to the inhibition of ferroptosis [10]. The role of ferroptosis in renal injuries has been investigated in multiple acute or chronic pathological states [11,12], and our previous investigation presented the initial evidence that ferroptosis was an integral process in CP-AKI [13]. However, the modulation of ferroptosis was poorly investigated in it.
WW domain binding protein-2 (WBP2) was initially identified as a chaperon of Yes-associated Protein (YAP), and their interaction led to the activation of estrogen and progesterone receptors [14,15]. Recent studies have revealed multiple binding partners of WBP2 and its pivotal role in various aspects of signaling transduction, such as steroid signaling pathway, Wnt signaling pathway, and Hippo signaling pathway [[15], [16], [17], [18]]. Notably, three PPXY motifs are present in the C-terminal domain of WBP2, which dominates the interaction with its targeted proteins [19]. The oncogenic property of WBP2 has been well described in various malignancies, and recent publications showed that WBP2 was also incorporated in the pathogenesis of hearing loss, fertility disorders, and hepatic steatosis [[20], [21], [22]]. The role of WBP2 in renal diseases has not been investigated. This present work used bioinformatics analysis to identify WBP2 as a latent regulator of AKI-related ferroptosis. Further investigations showed that WBP2 interacted with and stabilized GPX4 to inhibit ferroptosis in CP-AKI, suggesting that WBP2 might be a novel therapeutic target for it.
2. Methods
Bioinformatics. Gene expression profiles of AKI were filtrated through the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). Inclusion criteria were as follows: [1] Availability of kidneys from AKI patients in the dataset; [2] five or more kidney samples in the dataset; [3] Samples of kidney transplant patients undergoing acute rejection. Three eligible datasets were selected, including GSE1563 and GSE30718, and GSE61739.
All microarray data were submitted to the GEO database (http://www.ncbi.nih.gov/geo). The raw data were downloaded as MINiML files. It contains the data for all platforms, samples, and GSE records. The extracted data were normalized by log2 transformation. The microarray data were normalized by the normalized quantiles function of the preprocess Core package in R software (version 4.4.2). Probes were converted to gene symbols according to the annotation information of the normalized data in the platform. Probes matching multiple genes were removed from these datasets. As in different datasets or the same dataset but in different platforms, extracting multiple data sets with common gene symbols, marking different datasets or platforms as different batches, used the remove Batch Effect function of the limma package in the R software to remove batch effects [23]. The result of the data preprocessing was assessed by Density plot, and the UMAP plot was drawn to illustrate the samples before and after batch effect.
The weighted gene co-expression network analysis (WGCNA) was performed using the R package WGCNA (version 1.70) [24]. Before analysis, the hierarchical clustering analysis was performed using the Hclust function in R language to exclude the outlier samples. Subsequently, the appropriate soft powers β (ranged from 1 to 20) was selected using the function of “pickSoftThreshold” in the WGCNA package according to the standard of scale-free network. Next, the soft power value β and gene correlations matrix among all gene pairs calculated by Pearson analysis were used to build adjacency matrix, which was calculated by the formula: aij = |Sij|β (aij: adjacency matrix between gene i and gene j, Sij: similarity matrix which is composed of Pearson correlation coefficients of all gene pairs, β: soft power value). After choosing the power of 6, the adjacency was transformed into a topological overlap matrix (TOM), which could measure the network connectivity of a Gene defined as the sum of its adjacency with all other Genes for network Gene ration, and the corresponding dissimilarity (1-TOM) was calculated. To classify Genes with similar expression profiles into Gene modules, average linkage hierarchical clustering was conducted according to the TOM-based dissimilarity measure with a minimum size (Gene group) of 100 for the Genes dendrogram. To further analyze the module, we calculated the dissimilarity of module eigen Genes, chose a cut line of 0.5 for module dendrogram and merged some module. Finally, we obtained 8 co-expression modules. In this study, the soft threshold β was 2 in the WGCNA analysis of AKI. The other parameters were the following: networkType = “unsigned”, minModuleSize = 100, mergeCutHeight = 0.50 and deepSplit = 2.
For network visualization, the Cytoscape.js library was used as previously reported [25]. In order to uncover the biological function related to the network nodes, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using clusterprofiler R package [26].
Cell culture and treatment. BUMPT cells, mouse renal proximal tubular cell line, were initially obtained from Drs. William Lieberthal and John Shwartz at Boston University. Cell culture media, containing DMEM, 10% FBS, and antibiotics, were used to culture the cells in a humidified environment. The cell culture incubator had 5% CO2 and 37 °C in temperature. Lentivirus was used to manipulate the expression of WBP2 in BUMPT cells. HK-2 cells (human proximal tubular cell line) and HMC cells (human mesangial cell line) were also used in our study. HK-2 cell and HMC cells were maintained in DMEM/F12 cell culture media and the identical culture environment was used for them. The incorporated lentiviruses in this study were: WBP2 overexpression lentivirus (Lv-WBP2), WBP2 knockdown lentivirus (Lv-shWBP2), WBP2 overexpression lentivirus for HK-2 cells (Lv-WBP2-HK-2), WBP2 knockdown lentivirus for HK-2 cells (Lv-shWBP2-HK-2), and their empty vector viruses (Lv-NC and Lv-shNC). Cells were seeded into different plates and treated with 20 μM cisplatin (Sigma-Aldrich, #P4394) for 12–24 h. Cisplatin used in vitro studies was initially dissolved in dimethylformamide (DMF) before adding into the culture media. For several biochemistry experiments, 293t cells were used to increase the transfection rate. The same culture media were used for 293t cells. Other treatments used in this study were: Fer-1(Sigma-Aldrich, #SML0583, 0.4 μM), myoglobin (Sigma-Aldrich, #M1882, 10 mg/ml), Chloroquine (MCE, #HY-17589A,20 mM), NH4Cl (Sigma-Aldrich, #213330,20 μM), and MG132 (MCE, #HY-13259, 15 μM). For hypoxia/reoxygenation treatment, cells were maintained in a hypoxia chamber for 12 h and they were then cultured in a normal environment for 6 h.
Animals. 8–12 weeks old C57BL/6J male mice were used in vivo studies, and the mice were divided into multiple groups accordingly (each group had 8-10 mice). For the induction of CP-AKI, cisplatin was dissolved in PBS solution at the concentration of 1 mg/ml, and cisplatin solution was intraperitoneally injected into the mice at the dose of 25 mg/kg. Mice were sacrificed two days later, and the kidneys and blood samples were harvested. For the inhibition of ferroptosis, Fer-1 (5 mg/kg) was administrated intraperitoneally 45 min before the injection of cisplatin. Intraparenchymal injection of adenovirus solution in the renal cortex was used to manipulate the expression of WBP2 in mice. Briefly, the mouse was anesthetized, and the left kidney was exposed. The left renal pedicle was clamped, and three sites were selected for adenovirus injection with a 31-gauge needle (30 μL adenovirus solution in each site). The needle was maintained for 15 s after each injection to reduce the volume of leakage. Besides, the clamp was maintained for another 5 min after the removal of the needle to ensure the permeation of adenovirus. The duration of renal artery occlusion was about 8 min for each kidney. The adenovirus used in this study was at the concentration of 1.5–2 ✕ 1012 particles/ml. The incorporated adenoviruses in this study were: WBP2 overexpression adenovirus (Ad-WBP2), WBP2 knockdown adenovirus (Ad-shWBP2), and their empty vector adenoviruses (Ad-NC and Ad-shNC).
Preparation of human samples: Patients pathologically diagnosed with acute tubular necrosis (ATN) were incorporated into the AKI group, and their renal biopsy sections were used for further analysis. Patients undergoing radical nephrectomy were classified into the control group. Normal renal tissues (at least 4 cm away from the border of the tumor) were embedded into the paraffin, and the renal sections were obtained.
For the preparation of human primary tubular cells (HPT cells), normal renal cortex (at least 4 cm away from the border of the tumor) was immediately obtained and stored in cold Hanks solution after the surgery. The renal samples were cut with a scissor, and they were then transferred into a 15 ml tube filled with pre-heated Hanks solution. The renal samples in the tube were then subjected to centrifuge-mediated purification twice (1000 rpm, 5 min). The supernatants were removed, and red ball cells were erased. Collagenase solution (1 mg/ml) was used to digest the samples for 1 h at 37 °C, and intermittent shanking was used to aid in the digestion. The mixture was then transferred into a 50 ml tube, and Hanks solution was used to resuspend the mixture. Subsequently, the mixture was purified with a 70 μm-filter in the centrifuge (1000 rpm, 5 min). The supernatant was removed, and the cells were seeded into the collagen-coated plates. Cells were maintained in a humidified environment for 48 h to assure attachment. When cell fusion was observed (usually 5–7 days), HPT cells were identified by CK18 staining, and cisplatin treatment (20 μM, 20 h) was applied.
Morphological studies. For in vitro studies, cells were immediately scanned by light microscopy after the treatment. For in vivo studies, H & E staining was used. 4-μm paraffin sections were deparaffinized and rehydrated with routing protocols. The sections with stained with hematoxylin for 3 min and eosin for 30 s. After the rinse by tap water, the sections were mounted and evaluated by light microscopy.
CCK8 assay. CCK8 assay was conducted to evaluate the cell survival rate. About 5000 cells were seeded into each well of the 96-well plate, and cells were maintained overnight for attachment. Cells were treated with cisplatin or RSL3 for an appropriate time before the removal of cell culture media. 200 μL CCK solution (TargetMol, #C0005) was added in each well, and the plate was maintained at 37 °C for 30 min to complete the reaction. The plate was subjected to 5 min’ shank, and OD 450 nm reading was obtained from the microplate reader.
Renal function analysis. For the evaluation of renal function, serum creatinine and blood urea nitrogen (BUN) levels were detected. A Serum Creatinine Colorimetric Assay Kit (Jiancheng, China, #C011-2-1) was used to detect serum creatinine levels as requested by the instruction. Briefly, 6 μL double distilled water (used as the blank group), serum samples, or standards were added in the wells containing 180 μL Enzyme Solution A. The 96-well plate was then maintained at 37 °C for 5 min, and OD 546 nm readings were obtained. Subsequently, 60 μL Enzyme Solution B was added into each well, and the mixture was harbored in the environment of 37 °C for 5 min. Then, OD 546 nm readings were obtained for another time. The concentration of serum creatinine was then calculated accordingly. For the evaluation of serum BUN levels, BUN assay kits (Jiancheng, China, #C013-2-1) was used according to its instruction. Briefly, 20 μL double distilled water (used as the blank group), serum samples or standards were mixed with 250 μL urease solution, and the reaction was maintained at 37 °C for 10 min. After that, 1 ml phenol solution and 1 ml sodium hypochlorite solution were added, and the mixtures were maintained in 37 °C water bath for 10 min. Finally, OD 640 nm readings were obtained, and the data was calculated accordingly.
Evaluation of ROS generation. DHE staining was used to evaluate the overall ROS generation. BUMPT cells and frozen sections were stained with 10 μM DHE solution at 37 °C for 30 min, and the stained sections were examined by fluorescence microscopy. For lipid ROS evaluation in vitro, C11 BODIPY 581/591 kit (ThermoFisher, #C10445) was used to stain the cells accordingly. Briefly, cells were stained with 10 μM Image-iT® Lipid Peroxidation Sensor (Component A) for 30 min at 37 °C after the treatment. After that, the media were removed, and the cells were washed with PBS three times. Finally, the cells were subjected to fluorescence microscopic analysis and quantification. For lipid ROS evaluation in vivo, immunofluorescence staining of 4-HNE was applied to the renal sections, and quantification was applied accordingly.
Western blot studies. Renal cortex or cells were lysed by RIPA buffer containing proteinase inhibitors. BCA assay (Takara, #T9300A) was used to evaluate the protein concentration, and equal amounts of protein were loaded into the SDS-PAGE gel. Electrophoresis was performed to separate the proteins, which were then transferred onto the PVDF membranes. The membranes were blocked by 10% milk for 1 h at room temperature, and they were then incubated with primary antibody dilutions overnight at 4 °C. The membranes were washed and incubated with secondary antibody dilutions for 1–2 h at room temperature. After three times of washing, the membranes were finally evaluated by ECL chemiluminescence system. The incorporated antibodies were: WBP2(Proteintech, #12030-1-AP and Santa Cruz, #sc-514247, 1:1000), NCOA4 (Bethyl Laboratories, #A302-272A, 1: 1000), GPX4 (Abcam, #ab125066, 1: 1000), FTH1 (Abcam, #ab183781, 1: 1000, and Cell Signaling Technology, #4393,1:1000), KIM-1 (R&D Systems, #AF1817, 0.25 μg/mL), NGAL (R&D Systems, #AF1857, 0.25 μg/mL), Lamp1 (Abmart, #TD7033S, 1: 1000), Flag (Sigma-Aldrich, #F1804, 1: 5000), HA (Santa Cruz, #sc-7392, 1: 1000), Myc (Proteintech, #16286-1-AP, 1: 5000), HSC70 (Proteintech, #10654-1-AP, 1: 5000), Lamp2a(Proteintech, #66301-1-AP, 1: 1000), GFP (Proteintech, #66002-1-Ig, 1: 5000), β-Actin (Proteintech, #66009-1-Ig, 1: 5000), secondary antibody anti-Mouse (Proteintech, #SA00001-1, 1:5000), secondary antibody anti-Rabbit (Proteintech, #SA00001-2, 1:5000), and secondary antibody anti-Goat (Proteintech, #SA00001-4, 1:5000),
qRT-PCR analysis. Cells were homogenized by TRIzol Reagent (Invitrogen, catalog 15-596-026), and the routing protocols were used to exact mRNA from the cells. The remaining gDNA was erased, and the cDNA was obtained from the mRNA by using a TAKARA kit (TaKaRa, #RR037A). Equal amounts of cDNA were subjected to quantitative PCR reaction by using a Vazyme kit (Vazyme, #Q111-02). The Cq values of each sample were obtained and calculated accordingly. β-actin was used as the internal control, and ΔΔCq values were used for the statistical analysis. The primers used in our studies were indicated in the Supplemental Table 1.
Immunohistochemistry staining. 4-μm paraffin sections were obtained after harvesting the renal samples. The sections were heated and deparaffinized before the rehydration. After washing, heat-induced antigen retrieval was applied to the sections, and endogenous peroxidase was suppressed. Goat serum was used to incubate the sections for 1 h at room temperature to block the background. Subsequentially, the sections were incubated with primary antibody dilutions overnight at 4oC. After three times of washing, the sections were incubated with secondary antibody dilutions for 1 h at room temperature. PBS was used to wash the sections three times, and DAB solution was applied to the sections for 30–60 s accordingly. Hematoxylin was used to stain the nuclei for 30 s. Finally, the sections were dehydrated and mounted before the light microscopic examination. The incorporated antibodies were: WBP2(Proteintech, #12030-1-AP, 1:200), NCOA4 (Bethyl Laboratories, #A302-272A, 1: 200), GPX4 (Abcam, #ab125066, 1: 200), FTH1 (Abcam, #ab183781, 1: 200), Anti-Mouse IgG-HRP (Abcam, #ab6789, 1: 500), and Anti-Rabbit IgG-HRP (Abcam, #ab97051, 1: 500).
Immunofluorescence staining. BUMPT cells were seeded on the coverslips, and cisplatin was used to treat the cells. After the treatment, cells were fixed with 4% paraformaldehyde before the permeation. 4-μm paraffin sections of renal samples were deparaffinized, rehydrated, and permeated before the antigen retrieval. All sections were subjection to 5% BSA solution incubation (1 h, room temperature) for blocking. The sections were then incubated with primary antibody dilution overnight at 4oC. After three times washing, secondary antibody dilutions were used to incubate the sections for 1–2 h at room temperature. The sections were rinsed, and the nuclei were stained with DAPI solution. Finally, the sections were subjected to fluorescence microscopic evaluation. The incorporated antibodies were: WBP2(Proteintech, #12030-1-AP, 1:200), Megalin (Abcam, # ab184676; 1:500), GPX4(Santa Cruz, #sc-166570, 1: 200), Lamp1 (Abmart, #TD7033S, 1: 100), CK18 (AMSBIO, #A01357-1, 1:100), 4-HNE (R&D system, MAB3249, 1:100), GFP (Proteintech, #66002-1-Ig, 1: 200), Anti-Rabbit IgG-Alexa Fluor 488 (Abcam, #ab150077, 1: 500), Anti-Mouse IgG-Alexa Fluor 488 (Abcam, #150113, 1: 500), Anti-Rabbit IgG-Alexa Fluor 594 (Abcam, #ab150080, 1: 500), and Anti-Mouse IgG-Alexa Fluor 594 (Abcam, #150116, 1: 500).
Evaluation of protein-protein interaction. The potential interaction between WBP2 and GPX4 was initially explored by String analysis, and its website is: STRING: functional protein association networks (string-db.org). After that, co-immunoprecipitation (CO-IP) was used to validate their interaction. Cells were lysed by CO-IP lysis buffer with proteinase inhibitors. The protein concentration was initially detected by BCA assay, and about 750 μg protein (about 250 μl in volume) was incubated with 1–2 μg primary antibody overnight at 4oC. 20 μl protein A/G beading solution (Santa-cruz, #sc-2003) was added into the mixture, and they were shanked for 4 h at room temperature to precipitate the targeted proteins. The mixtures were centrifuged, and the supernatants were removed. The beads were washed six times before boiling with 2✕loading buffer. After cooling down, the supernatants were loaded into the SDS-PAGE gel, and a routing western blot procedure was applied to develop the bands. The incorporated antibodies were: Flag (Sigma-Aldrich, #F1804), HA (Proteintech, #51064-2-AP), Myc (Proteintech, #16286-1-AP), GPX4(Abcam, #ab125066), Rabbit IgG (Proteintech, #B900610), and Mouse IgG (Proteintech, #B900620).
Evaluation of GPX4 activity. Homogenization buffer (pH 7.4) was initially prepared, which contains 0.1 M KH2PO4/K2HPO4, 0.15 M KCl, 0.05% [wt/vol] CHAPS, 5 mM β-mercaptoethanol, and protease inhibitors. About 2✕106 cells or 200 μg renal cortex were homogenized in 200 μl homogenization buffer. The homogenates were centrifuged (10000 rpm, 15 min) at 4oC, and the supernatants were harvested. Bradford assay was used to measure the protein concentration of the supernatants, and their concentrations were normalized with homogenization buffer. Subsequently, GPX4 assay buffer (pH 7.8) was made, which contains 5 mM EDTA, 5 mM GSH (MCE,#HY-D0187), 0.1% [vol/vol] Triton X-100, 180 IU/mL glutathione reductase (Beyotime, #P2372S), and 160 mM NADPH/H+ (Beyotime, #ST360). 50 μL supernatants were mixed with 1 ml GPX4 assay buffer, and the mixtures were maintained at 22 °C for 5 min. After that, 5ul of 30 mM cumene hydroperoxide (Aladdin, #C109598) was added into the mixture to ignite the reaction. OD340nm readings were obtained every 10 s until the reading was stabilized. The rate of changes in OD340 readings was calculated as GPX4 activity.
TUNEL staining. The state of DNA damage was evaluated by TUNEL staining kit (Roche, #11684795910). Briefly, cells were washed with PBS three times after the treatment, and they were then fixed with 4% paraformaldehyde for 1 h at room temperature. Permeation was applied to the cells at 4 °C with 2 min’ incubation of 0.1% Triton X-100 (dissolved in 0.1% sodium citrate). TUNEL regent was used to stain the cells for 60 min at 37 °C. After two times of PBS washing, the sections were mounted and evaluated by fluorescence microscopy.
Preparation of lysosomal and lysosome-free fractionations: The isolation of lysosomes was performed as previously indicated in the kit (Solarbio, #EX1230). Cells were harvested and washed with PBS solution twice. 1 ml Solution A was used to resuspend the cells, and the cells were shaken for 10 min at 4 °C. Dounce homogenizer was used to facilitate the process. The mixture was sequentially centrifuged at 1000×g for 5 min and 1000×g for 10 min, and the supernatant was collected. Subsequently, the supernatant was centrifuged at 20000×g for 20 min, and the pellet was harvested. 500 μL solution B was added and resuspended, and the mixture was centrifuged at 20000×g for 20 min, and the pellet was collected, which was dissolved by 100 μL solution C. After 30 min’ shaking, the mixture was centrifuged at 12000×g for 15 min, and the supernatant with abundant lysosomal proteins was used for further studies.
Evaluation of labile iron levels. FerroOrange (DOJINDO, #F374) staining was used to detect the labile iron in BUMPT cells. Briefly, DMSO was used to dissolve FerroOrange to make the stock solution (1 mM). Working solution of FerroOrange (2 μM) was made by adding stocking solution into cell culture media (1:500). Cells were stained with working solution for 30 min at 37oC. Fluorescence microscopy was used to examine the cells immediately after the staining.
Plasmid construction. The cDNA templates of WBP2 and GPX4 were acquired from Miaolingbio (Wuhan, China). They were truncated with routing protocols. The full-length and truncated forms of WBP2 and GPX4 were subcloned in the backbone plasmid, including PCDH-3✕Myc, PCDH-3✕Flag, and PCDH-3✕HA. For point mutation, mutated sites were designed in the primers, and the cDNA sequence was divided into two parts by the PCR procedure. Subsequently, fusion PCR was performed to obtain the mutated full-length cDNA sequence. For the expression of GPX4, a selenoprotein-expressing lentivirus plasmid, i.e. seleno-GFP-3✕flag-PCDH, was generated. The details of the synthesis were indicated in the supplemental files.
Statistics. For bioinformatics analysis, statistical procedures were performed by the R statistical software tool version 3.6.1 (www.r-project.org). For experimental data, Graphpad Prism 9.0 was used to calculate the differences between each group. Mean ± SD was used to indicate the values of results. 1-way ANOVA with Dunn's multiple comparisons was used for statistics.
Study approval. Animal procedures used in this study were approved by the Animal Care and Use Committee of the Second Xiangya Hospital of Central South University, China (20220515). Human studies were approved by the Second Xiangya Hospital of Central South University (LYF2022146). Written informed consent from participants or their guardians was obtained.
3. Results
3.1. WBP2, a novel modulator of AKI and ferroptosis, participated in the occurrence of CP-AKI
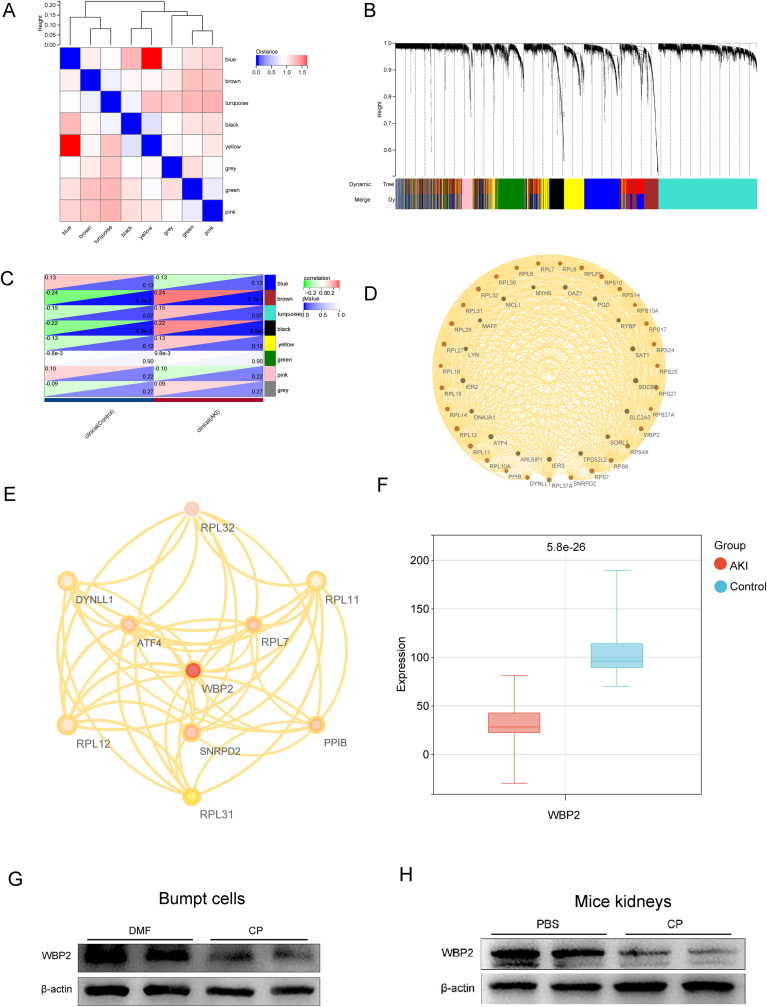
In order to identify novel modulators of AKI & ferroptosis, a series of bioinformatics analyses was performed. After quality control and removal of the batch effect between batches (Supplemental Figs. 1A–1E, see “Methods”), a total of 83 AKI patients and 68 controls from 3 datasets (GSE1563, GSE30718, and GSE61739) were enrolled in this study (Supplemental Table 2). Subsequently, WGCNA analysis was used to investigate correlation patterns among genes across microarray renal samples, which identified 8 modules (Fig. 1A & Supplemental Table 3). Notably, the eight modules corresponded to 6840 genes correlated with AKI (Fig. 1B). Among these, Black and Brown modules showed significance in both comparisons, both showing upregulation of the eigenvectors in the AKI samples (Fig. 1C and Supplemental Figs. 1F–1G). The core-related genes (hub genes) from the Brown and Black modules were analyzed, and their interactions were demonstrated by a network map (Fig. 1D). In order to explore the physiological processes captured by the network, we evaluated the cellular and molecular progress significantly enriched in all the network nodes (Supplemental Fig. 1H - 1O). Importantly, the majority of them belong to canonical pathways involved mainly in apoptosis and inflammation, indicative of their essential role in the progression of AKI.
Fig. 1.
WBP2, a novel modulator of AKI and ferroptosis, participated in the occurrence of CP-AKI. A The identification of WGCNA co-expression modules; B The determination of the clinical features of AKI; C The correlation analysis of the genes in all modules and the clinical features of AKI; D The network map of Brown and black modules; E The merging of WGCNA network and ferroptosis-related gene network, and the identification of WBP2 as the hub gene; F The downregulation of WBP2 in kidneys of AKI patients; G-H Western blot studies revealed that the expression of WBP2 was decreased in cisplatin-treated BUMPT cells and kidneys. DMF: dimethylformamide, CP: cisplatin. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Since the functional analysis of the core modules in the WGCNA network indicated the involvement of cell death, especially ferroptosis, we speculated that the ferroptosis is related to renal pathologic changes in AKI. In this regard, the ferroptosis-related gene network was generated using 265 ferroptosis-related genes (FRGs) from the Ferroptosis Database (FerrDb). The interaction network based on FRGs was obtained from the integrated interactions database (IID), including 1360 genes filtered by 2 or more studies of experimental validation in kidneys (Supplemental Table 4). In order to visualize the consistency between the WGCNA network and the ferroptosis-related gene network, Cytoscape was used to merge the two networks. Finally, WBP2 was identified as a hub gene after applying 12 algorithms in the plug-in cyto-Hubba (Fig. 1E & Supplemental Table 5). According to a previously integrated transcriptome dataset, WBP2 was significantly under-expressed in AKI patients (Fig. 1F).
The expression of WBP2 was also investigated by various laboratory experiments. Immunofluorescence staining showed that WBP2 (green) was extensively expressed in renal proximal tubular cells (marked by megalin, red) (Supplemental Fig. 2A). Subsequently, CP-AKI was incorporated for further investigations. Western blot studies demonstrated that WBP2 was conceivably decreased in cisplatin-treated kidney and BUMPT cells (mouse renal proximal tubular cell line), similar to the observations in bioinformatics analysis (Fig. 1G–H). The downregulation of WBP2 in CP-AKI was further confirmed by the immunofluorescence staining of BMUPT cells and immunohistochemistry staining of renal sections (Supplemental Figs. 2B and 2C). All in all, our work indicated that WBP2, initially identified as a modulator of AKI & ferroptosis by bioinformatics, was involved in the pathogenesis of CP-AKI.
3.2. WBP2 alleviated CP-AKI
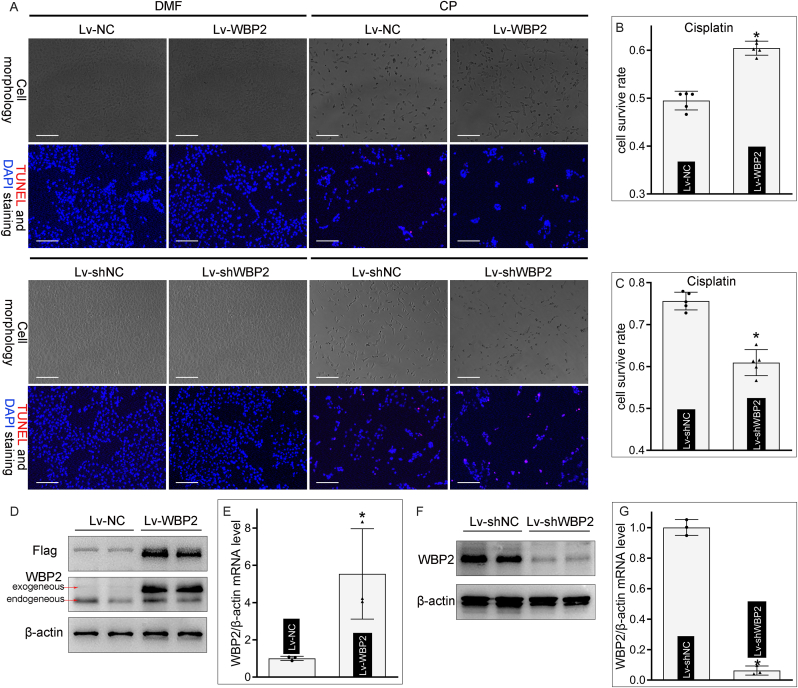
For in vitro studies, lentivirus was applied to BUMPT cells to intervene in the expression profile of WBP2, and the state of WBP2 overexpression or knockdown was validated by western blot studies (WBP2 & Flag) and qRT-PCR analysis (WBP2) (Fig. 2D–G). Light microscopic analysis revealed that cisplatin treatment led to morphological shrinkage and a numerical decrease in BUMPT cells (Fig. 2A). Interestingly, those aberrant alterations were attenuated by WBP2 overexpression but accentuated by its gene disruption (Fig. 2A). TUNEL staining showed that cisplatin-induced DNA damage was modulated by the expression profile of WBP2 in BUMPT cells (Fig. 2A). Besides, the cell survival rate was evaluated by CCK8 assay. It was noted that conceivable cell death was induced in cisplatin-treated BUMPT cells, which was alleviated by WBP2 overexpression but aggravated by WBP2 knockdown (Fig. 2BandC). Taken together, these data suggested that WBP2 ameliorated cisplatin-induced BUMPT cell injuries.
Fig. 2.
WBP2 alleviated BUMPT cell injuries induced by cisplatin. A Light microscopic analysis showed that WBP2 overexpression alleviated, while its gene disruption accentuated, cellular morphological changes in cisplatin-treated BUMPT cells; TUNEL staining showed that cisplatin-induced DNA damage in BUMPT cells was modulated by the expression profile of WBP2; B - C CCK8 assay revealed that WBP2 overexpression improved cellular survival rate in cisplatin-treated BUMPT cells, while WBP2 knockdown had the opposite effect (n = 5; *P < 0.0001 compared with Lv-NC or Lv-shNC group, Two-tailed student’ t-test); D - G Western blot studies (Flag and WBP2) and qRT-PCR analysis confirmed the overexpression and downregulation of WBP2 in lentivirus-transfected BUMPT cells (n = 3; *P = 0.0318 compared with Lv-NC group,*P < 0.0001 compared with Lv-shNC group, Two-tailed student’ t-test). DMF: dimethylformamide, CP: cisplatin, Lv-NC: empty vector lentivirus for WBP2 overexpression, Lv-WBP2: lentivirus-mediated WBP2 overexpression, Lv-shNC: empty vector lentivirus for WBP2 knockdown, Lv-shWBP2: lentivirus-mediated WBP2 knockdown. Data are presented as mean ± SD. Scale bars: 100 μm.
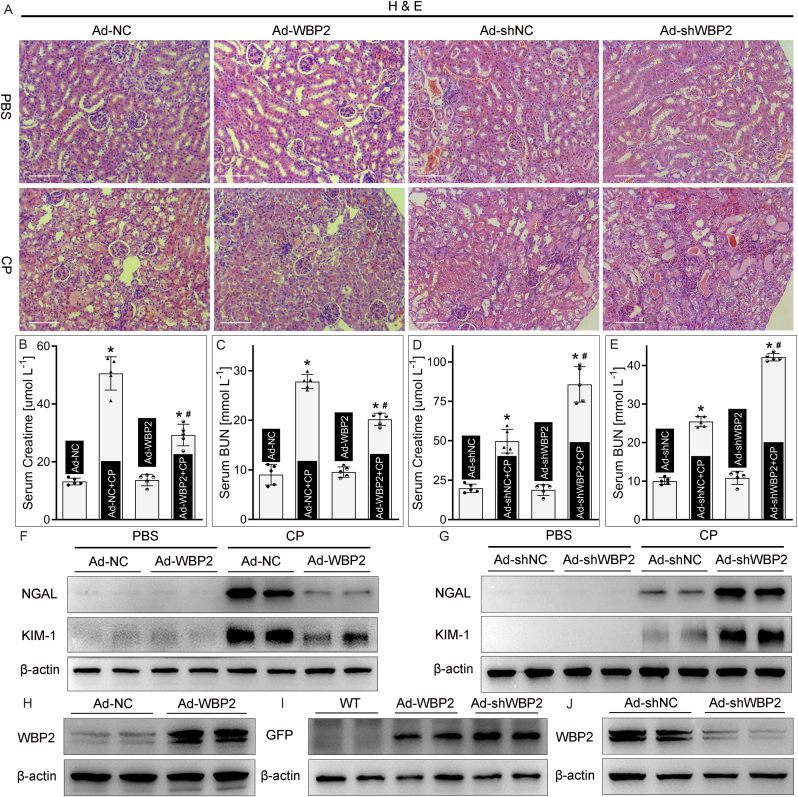
For in vivo studies, intraparenchymal injection of adenovirus was applied to wide-type mice to manipulate the expression profile of WBP2 in renal tissues. The successful delivery of adenovirus and gene intervention were confirmed by multiple procedures, including western blot studies (WBP2 and GFP) and immunofluorescence staining (GFP) (Fig. 3H–J and Supplemental Fig. 3). HE staining showed that severe tubular damage, such as tubular enlargement, cast formation, and interstitial edema, was observed in cisplatin-treated kidneys (Fig. 3A). Noteworthy, the renal morphological changes induced by cisplatin were attenuated by WBP2 overexpression but accentuated by WBP2 knockdown (Fig. 3A). Renal function was evaluated by serum creatinine levels and blood urea nitrogen (BUN) levels. Our data showed that cisplatin-induced deterioration in renal functions was regulated by the expression profile of WBP2 (Fig. 3B–E). Besides, the extent of tubular damage was evaluated by the expression of kidney injury molecule-1 (KIM-1) and neutrophil gelatinase-associated lipocalin (NGAL). Western blot studies showed that WBP2 overexpression attenuated, while its gene disruption accentuated, cisplatin-induced upregulation of KIM-1 and NGAL in kidneys (Fig. 3FandG). Collectively, these data indicated that WBP2 protected against cisplatin-induced tubular injuries in vivo.
Fig. 3.
WBP2 attenuated cisplatin-induced renal injuries. A H & E staining showed that cisplatin-induced renal morphological changes were attenuated by WBP2 overexpression but accentuated by WBP2 knockdown; B - E Renal functional analysis showed that WBP2 overexpression alleviated, while its gene disruption aggravated, cisplatin-induced increase in serum creatinine levels and BUN levels (n = 5; *P < 0.0001 compared with Ad-NC or Ad-shNC group, #P < 0.0001 compared with Ad-NC + CP or Ad-shNC + CP group, 1-way ANOVA with Dunn's multiple comparisons); F - G Western blot studies revealed that WBP2 overexpression attenuated cisplatin-induced upregulation of NGAL and KIM-1, while WBP2 knockdown presented the opposite effect; H - J Western blot studies showed that WBP2 was manipulated by the intraparenchymal injection of adenovirus, and GPF was also expressed in adenovirus-injected kidneys. CP: cisplatin, Ad-NC: empty vector adenovirus for WBP2 overexpression, Ad-WBP2: adenovirus-mediated WBP2 overexpression, Ad-shNC: empty vector adenovirus for WBP2 knockdown, Ad-shWBP2: adenovirus-mediated WBP2 knockdown. Data are presented as mean ± SD. Scale bars: 100 μm.
3.3. WBP2 inhibited ferroptosis in CP-AKI
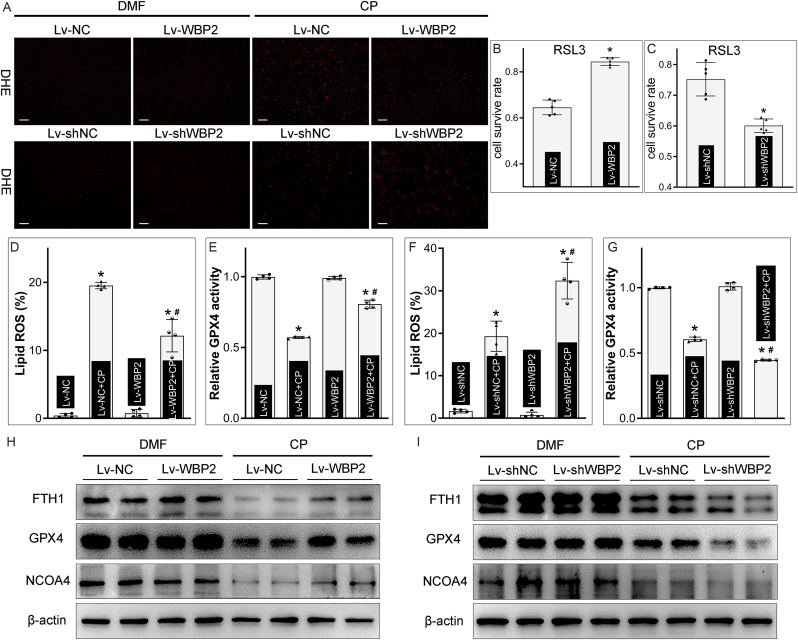
Our initial bioinformatics analysis revealed that WBP2 was a potential ferroptosis modulator in AKI, therefore, its relevance with ferroptosis was investigated. RSL3, an inhibitor of GPX4, was used to treat BUMPT cells to induce ferroptosis-specific cell death. CCK8 assay revealed that RSL3-induced ferroptosis in BUMPT cells was attenuated by WBP2 overexpression but accentuated by WBP2 knockdown (Fig. 4BandC). ROS generation, an essential process for ferroptosis, was explored by DHE staining. Our data showed that cisplatin-induced increased ROS generation in BUMPT cells was regulated by the expression profile of WBP2 (Fig. 4A). Lipid ROS generation, as evaluated by C11-BODIPY 591/581 staining, was also increased in cisplatin-treated BUMPT cells, and the state of lipid peroxidation was attenuated by WBP2 overexpression but accentuated by WBP2 knockdown (Fig. 4D & F). Furthermore, GPX4 activity was decreased in cisplatin-primed BUMPT cells, which was accentuated by WBP2 knockdown but partially restored by WBP2 overexpression (Fig. 4E & G). The expression of GPX4 was also decreased in cisplatin-treated BUMPT cells, and its decreased expression was modulated by the expression of WBP2 (Fig. 4HandI). The progression of ferroptosis is usually accompanied by ferritinophagy, a process where nuclear receptor coactivator 4 (NCOA4) mediated ferritin into the lysosomes for degradation [27]. Therefore, the simultaneous degradation of NCOA4 and FTH1 (heavy chain of ferritin) indicated the occurrence of ferroptosis. Our western blot studies demonstrated that NCOA4 and FTH1 were depleted in cisplatin-treated BUMPT cells (Fig. 4HandI). Interestingly, their degradation was accelerated by WBP2 knockdown but decelerated by WBP2 overexpression (Fig. 4HandI). Besides, cisplatin-induced increase in labile iron levels was also modulated by the expression profile of WBP2 (Supplemental Fig. 4). The modulation of ferroptosis by WBP2 was also observed in cisplatin-treated HK-2 cells (human renal proximal tubular cell line) (Supplemental Fig. 5). Hypoxia/reoxygenation treatment or heme treatment was also used in our study, since ferroptosis was operative in renal ischemia/reperfusion- or rhabdomyolysis-induced AKI. Within expectation, our data showed WBP2 decelerated ferroptosis in hypoxia/reoxygenation- or heme-treated BUMPT cell injuries (Supplemental Fig. 6). All in all, our work showed that WBP2 inhibits cisplatin-induced ferroptosis in BUMPT cells.
Fig. 4.
WBP2 inhibited ferroptosis in cisplatin-treated BUMPT cells. A DHE staining showed that WBP2 overexpression attenuated, while WBP2 knockdown accentuated, cisplatin-induced ROS generation in cisplatin-treated BUMPT cells; B - C CCK8 assay revealed that RSL3-induced ferroptosis in BUMPT cells was alleviated by WBP2 overexpression but aggravated by WBP2 knockdown (n = 5; *P < 0.0001 compared with Lv-NC group,*P = 0.0004 compared with Lv-shNC group, Two-tailed student’ t-test); D & F WBP2 overexpression mitigated, while its gene disruption accelerated, cisplatin-induced lipid peroxidation in BUMPT cells, as evaluated by C11-BODIPY 591/581 staining (n = 4; *P < 0.0001 compared with Lv-NC or Lv-shNC group, #P < 0.0001 compared with Lv-NC + CP or Lv-shNC + CP group, 1-way ANOVA with Dunn's multiple comparisons); E & G Cisplatin-induced decrease in GPX4 activity was modulated by the expression profile of WBP2 in BUMPT cells (n = 4; *P < 0.0001 compared with Lv-NC or Lv-shNC group, #P < 0.0001 compared with Lv-NC + CP or Lv-shNC + CP group, 1-way ANOVA with Dunn's multiple comparisons); H - I Cisplatin-induced depletion of FTH1, GPX4 and NCOA4 was attenuated by WBP2 overexpression but enhanced by WBP2 knockdown in BUMPT cells, as validated by western blot studies. DMF: dimethylformamide, CP: cisplatin, Lv-NC: empty vector lentivirus for WBP2 overexpression, Lv-WBP2: lentivirus-mediated WBP2 overexpression, Lv-shNC: empty vector lentivirus for WBP2 knockdown, Lv-shWBP2: lentivirus-mediated WBP2 knockdown. Data are presented as mean ± SD. Scale bars: 100 μm.
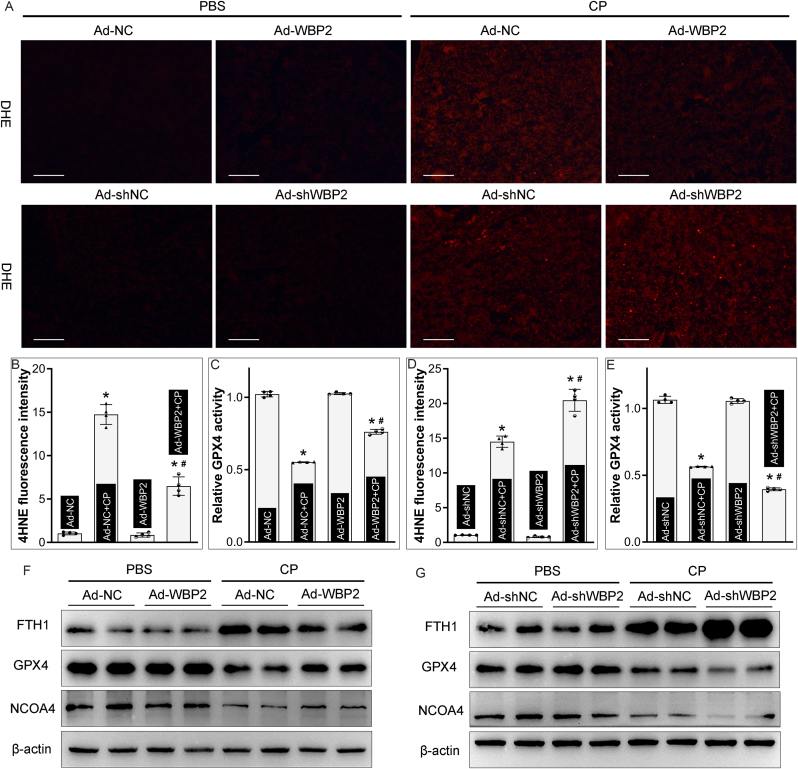
The role of WBP2 in ferroptosis was further confirmed in vivo studies. The state of ROS generation in renal tissues was explored by DHE staining, which showed increased fluorescence intensity following cisplatin treatment (Fig. 5A). Interestingly, the increased ROS generation was attenuated by WBP2 overexpression but accentuated by WBP2 knockdown (Fig. 5A). The extent of lipid peroxidation, as evaluated by 4-Hydroxynonenal (4-HNE) staining, was also increased in cisplatin-treated kidneys, which was regulated by the expression of WBP2 (Fig. 5B & D). Moreover, GPX4 activity was also explored, and our data revealed that WBP2 overexpression mitigated, while its gene disruption aggravated, cisplatin-induced decrease in GPX4 activity in cisplatin-treated kidneys (Fig. 5C & E). The protein levels of NCOA4 and GPX4 were conceivably decreased in cisplatin-treated kidneys, and their decrease was regulated by the expression profile of WBP2, as detected by western blot studies (Fig. 5FandG). Noteworthily, FTH1 was strongly upregulated in cisplatin-treated kidneys (Fig. 5FandG), a protective and feedback mechanism in the state with high iron levels as previously reported [13,28]. Cisplatin-induced upregulation of FTH1 was also modulated by the expression of WBP2 (Fig. 5FandG). Taken together, these data suggested that WBP2 decelerated ferroptosis to alleviate CP-AKI.
Fig. 5.
WBP2 decelerated ferroptosis in CP-AKI in vivo. A DHE staining revealed that WBP2 overexpression alleviated, while WBP2 knockdown aggravated, cisplatin-induced oxidative stress in kidneys; B & D WBP2 overexpression attenuated, while its gene disruption accentuated, cisplatin-induced increase in 4-HNE levels in the renal cortex, as validated by the immunofluorescence staining (n = 4; *P < 0.0001 compared with Ad-NC or Ad-shNC group, #P < 0.0001 compared with Ad-NC + CP or Ad-shNC + CP group, 1-way ANOVA with Dunn's multiple comparisons); C & E Cisplatin-induced decreased in GPX4 activity was modulated by the expression profile of WBP2 in kidneys (n = 4; *P < 0.0001 compared with Ad-NC or Ad-shNC group, #P < 0.0001 compared with Ad-NC + CP or Ad-shNC + CP group, 1-way ANOVA with Dunn's multiple comparisons); F-G Cisplatin-induced upregulation of FTH1 and downregulation of GPX4 and NCOA4 in kidneys were mitigated by WBP2 overexpression but aggravated by WBP2 knockdown, as detected by western blot studies. CP: cisplatin, Ad-NC: empty vector adenovirus for WBP2 overexpression, Ad-WBP2: adenovirus-mediated WBP2 overexpression, Ad-shNC: empty vector adenovirus for WBP2 knockdown, Ad-shWBP2: adenovirus-mediated WBP2 knockdown. Data are presented as mean ± SD. Scale bars: 100 μm.
3.4. WBP2 and ferroptosis-related markers were depleted in the kidneys of AKI patients
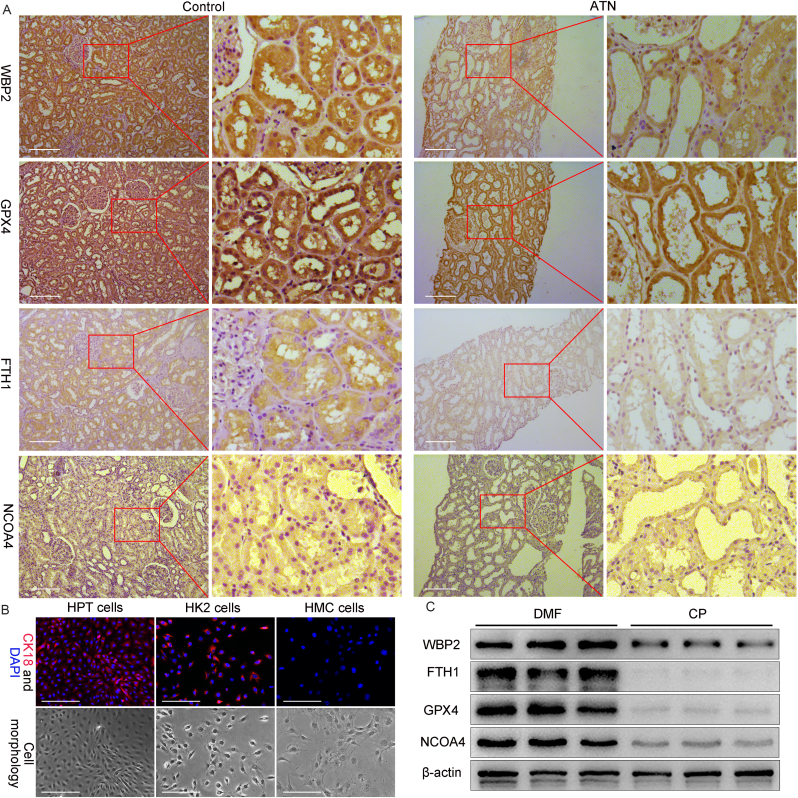
The expression of WBP2 and ferroptosis-related markers (NCOA4, FTH1, and GPX4) was detected in human samples. Immunohistochemistry staining revealed that WBP2, NCOA4, FTH1, and GPX4 were abundantly expressed in renal tubules, the major target of CP-AKI (Fig. 6A). Interestingly, their expression was simultaneously decreased in patients with acute tubular necrosis (ATN) (Fig. 6A). Due to the lack of renal biopsy tissue from patients of CP-AKI, human primary tubular cells (HPT cells) were obtained from patients undergoing radical nephrectomy. The identification of HPT cells was achieved by immunofluorescence staining of CK18, and positive control (HK-2 cells) and negative control (HMC cells) were used to assure the authenticity (Fig. 6B). Cisplatin was used to treat HPT cells, and the expression of WBP2, NCOA4, FTH1, and GPX4 was detected by western blot studies. Interestingly, the above-mentioned proteins were conceivably downregulated in cisplatin-treated cells (Fig. 6C). All in all, WBP2 and ferroptosis-related markers were downregulated in renal tubules of AKI patients.
Fig. 6.
WBP2 and ferroptosis-related markers were depleted in the kidneys of AKI patients. A Immunohistochemistry staining showed that the expression of WBP2, GPX4, FTH1, and NCOA4 was decreased in the renal samples of ATN patients, as compared with the controls; B CK18 staining showed conceivable fluorescence in human primary tubular cells (HPT cells) and HK-2 cells, but no fluorescence was observed in HMC cells; C Cisplatin treatment led to decreased expression of WBP2, GPX4, FTH1, and NCOA4 in HPT cells. ATN: acute tubular necrosis, DMF: dimethylformamide, CP: cisplatin. Scale bars: 100 μm.
3.5. Fer-1 alleviated WBP2 knockdown-accelerated ferroptosis in CP-AKI
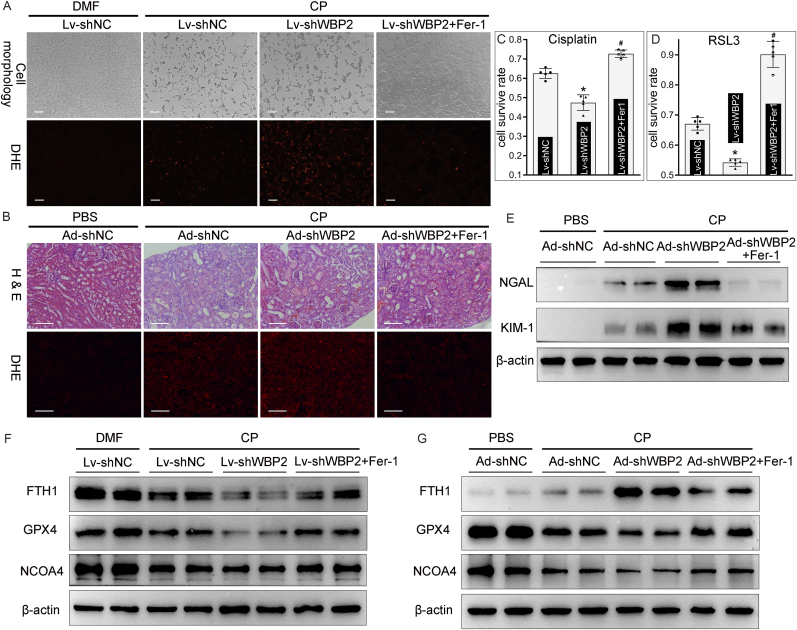
In order to confirm the dependence of ferroptosis in WBP2 knockdown-mediated renal injuries in CP-AKI, Fer-1 (a potent ferroptosis inhibitor) was used in vitro and in vivo studies. Light microscopic analysis demonstrated that cisplatin-induced morphological shrinkage in BUMPT cells was accentuated by WBP2 knockdown, and this aberrant change was conceivably attenuated by Fer-1 treatment (Fig. 7A). Similarly, WBP2 knockdown-aggravated renal morphological disruptions induced by cisplatin were also alleviated by the administration of Fer-1, as evaluated by H & E staining (Fig. 7B). DHE staining showed that WBP2 knockdown accentuated cisplatin-induced increased ROS generation in BUMPT cells and renal tissues, which was attenuated by Fer-1 treatment (Fig. 7AandB). Besides, WBP2 knockdown-accelerated cell death induced by cisplatin or RSL3 was also mitigated by the usage of Fer-1 (Fig. 7CandD). The extent of tubular injuries, as evaluated by the protein levels of NGAL and KIM-1, revealed that cisplatin-induced injuries were accentuated by WBP2 knockdown, which was conceivably alleviated by Fer-1 (Fig. 7E). Finally, the changes of ferroptosis-related markers (NCOA4, FTH1, and GPX4) were explored by western blot studies. Our data showed that WBP2 knockdown accentuated the aberrant changes of ferroptosis-related markers in CP-AKI, and those changes were ameliorated by the administration of Fer-1 (Fig. 7FandG). Taken together, we speculated that WBP2 knockdown-enhanced ferroptosis in CP-AKI was alleviated by Fer-1.
Fig. 7.
Fer-1 alleviated WBP2 knockdown-accelerated ferroptosis in CP-AKI. A Light microscopical analysis showed Fer-1 alleviated WBP2 knockdown-aggravated morphological shrinkage in cisplatin-treated BUMPT cells, DHE staining revealed that WBP2 knockdown-promoted ROS generation was attenuated by Fer-1; B H & E staining demonstrated that WBP2 knockdown-accentuated renal morphological disruption was partially restored by Fer-1 in cisplatin-treated kidneys, DHE staining revealed that Fer-1 mitigated WBP2 knockdown-accelerated ROS generation in cisplatin-treated kidneys; C - D CCK8 assay showed that RSL3 or cisplatin-induced cell death was accelerated by WBP2 knockdown in BUMPT cells, which was attenuated by Fer-1 treatment (n = 5; *P < 0.0001 compared with Lv-shNC group, #P < 0.0001 compared with Lv-shWBP2 group, 1-way ANOVA with Dunn's multiple comparisons); E Western blot studies demonstrated that WBP2-accelerated increase in NGAL and KIM-1 levels was mitigated by Fer-1 treatment in cisplatin-treated kidneys; F-G Western blot studies revealed that cisplatin-induced disruptions in ferroptosis-related markers (FTH1, GPX4 and NCOA4) was accentuated by WBP2 knockdown in BUMPT cells and kidneys, and these changes were attenuated by Fer-1 treatment. DMF: dimethylformamide, CP: cisplatin, Lv-shNC: empty vector lentivirus for WBP2 knockdown, Lv-shWBP2: lentivirus-mediated WBP2 knockdown; Ad-shNC: empty vector adenovirus for WBP2 knockdown, Ad-shWBP2: adenovirus-mediated WBP2 knockdown. Data are presented as mean ± SD. Scale bars: 100 μm.
3.6. WBP2 interacted with GPX4 to inhibit its lysosomal degradation
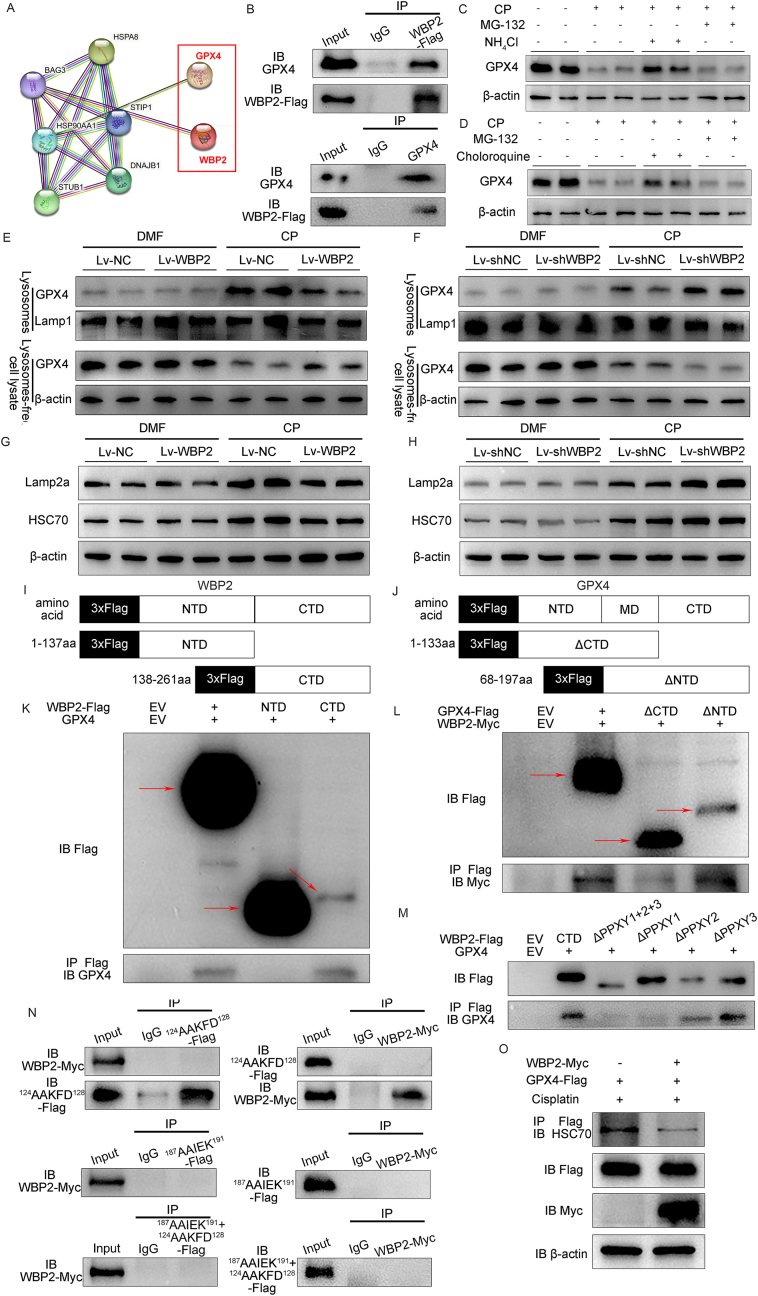
In order to explore the molecular mechanism of WBP2 in the modulation of ferroptosis, string analysis was performed. Our data showed that WBP2 might interact with GPX4 in BUMPT cells, which was further confirmed by CO-IP analysis (Fig. 8AandB). In regard to the modulation of GPX4 expression by WBP2 in CP-AKI and their interaction in BUMPT cells, we speculated that WBP2 might participate in the modulation of GPX4 stability in CP-AKI. Previous publications have indicated two types of degradation routes for GPX4, including chaperon-mediated autophagy (CMA)-induced degradation [29] or proteasomal degradation [30]. Our study found that cisplatin-induced GPX4 depletion was attenuated by NH4Cl (an inhibitor of lysosomes) and chloroquine (an inhibitor of autophagy), not by MG132 (an inhibitor of proteasomes), suggesting the involvement of the CMA pathway (Fig. 8C–D). Besides, CMA-related markers, including HSC70 and Lamp2a, were considerably upregulated in cisplatin-treated BUMPT cells, which was modulated by the expression of WBP2 (Fig. 8G–H). In this regard, lysosomal fractionation and lysosome-free fractionation were isolated in BUMPT cells, and GPX4 expression in those types of fractionations was evaluated. Western blot studies showed that the expression of GPX4 was increased in lysosomes but decreased in lysosome-free fractionation following cisplatin treatment, and those changes were modulated by the expression of WBP2 (Fig. 8E–F), indicating the modulation of CMA-mediated GPX4 degradation by WBP2. Similar results were observed in the colocalization between GPX4 and Lamp1 (Lysosomal-associated membrane protein 1) (Supplemental Fig. 7).
Fig. 8.
WBP2 interacted with GPX4 to inhibit its lysosomal degradation. A String analysis showed that WBP2 might interact with GPX4; B CO-IP analysis confirmed the interaction between WBP2 and GPX4; C - D Western blot studies demonstrated that cisplatin-induced GPX4 degradation was attenuated by chloroquine (an inhibitor of autophagy) and NH4Cl (an inhibitor of lysosomes), but not MG132 (an inhibitor of proteasome); E – F Western blot studies showed that the expression of GPX4 was increased in lysosomes but decreased in lysosome-free fractionation following cisplatin treatment, and those changes were modulated by the expression of WBP2; G – H Cisplatin-induced upregulation of Lamp2a and HSC70 in BUMPT cells was mitigated by WBP2 overexpression but aggravated by WBP2 knockdown, as detected by western blot studies; I - J The diagram for the truncation of WBP2 and GPX4; K - L Western blot studies showed that the truncated forms of WBP2 (Flag-tag) and GPX4 (Flag-tag) could be expressed in 293t cells, CO-IP analysis revealed that CTD of WBP2 (flag-tag) co-precipitated with GPX4, and ΔNTD of GPX4 co-precipitated with WBP2 (Myc-tag); M PPXY motifs of WBP2 (Flag-tag) were genetical deleted, and CO-IP studies showed that mutated WBP2 with ΔPPXY1 and ΔPPXY1+2 + 3 failed to bind with GPX4; N The two KEFRQ-like motifs 124NVKFD128 and 187QVIEK191 were mutated into 124AAKFD128 and 187AAIEK191 in GPX4 (Flag-tag), and CO-IP studies showed that WBP2 (Myc-tag) failed to co-precipitate with GPX4 (Flag-tag) when the two motifs were alternatively or simultaneously mutated; O In cisplatin-treated BUMPT cells, WBP2 overexpression disrupted the interaction between GPX4 (Flag-tag) and HSC70, as validated by CO-IP studies. DMF: dimethylformamide, CP: cisplatin, Lv-NC: empty vector lentivirus for WBP2 overexpression, Lv-WBP2: lentivirus-mediated WBP2 overexpression, Lv-shNC: empty vector lentivirus for WBP2 knockdown, Lv-shWBP2: lentivirus-mediated WBP2 knockdown, Ad-NC: empty vector adenovirus for WBP2 overexpression, Ad-WBP2: adenovirus-mediated WBP2 overexpression, Ad-shNC: empty vector adenovirus for WBP2 knockdown, Ad-shWBP2: adenovirus-mediated WBP2 knockdown. EV: empty vector, NTD: N-terminal domain, CTD: C-terminal domain, MD: middle domain, ΔCTD: a truncated protein containing MD domain and NTD domain, ΔNTD: a truncated protein containing MD domain and CTD domain, ΔPPXY1: a mutated protein with PPXY1 motif deletion, ΔPPXY2: a mutated protein with PPXY2 motif deletion, ΔPPXY3: a mutated protein with PPXY3 motif deletion, ΔPPXY1+2 + 3: a mutated protein with PPXY1, PPXY2, and PPXY3 motifs deletion.Scale bars: 100 μm.
In order to clarify the interaction pattern between WBP2 and GPX4, their truncated mutations were generated as indicated in Fig. 8I–J. However, GPX4, a selenoprotein, cannot be expressed in commonly-used plasmids. The synthesis of GPX4 incorporates a key process where selenocysteine is recognized by UGA (a stop codon in the normal state) inside its mRNA sequence [31]. In this regard, a new plasmid termed “seleno-GFP-3✕flag-PCDH plasmid” was generated to express GPX4 and its truncated forms (Supplemental Figs. 8–9). CO-IP analysis with truncated proteins revealed that C-terminal domain of WBP2 (CTD-WBP2) was involved in its interaction with GPX4 (Fig. 8K). Previous publications revealed that WBP2 interacted with its targeted proteins via the three PPXY motifs in CTD-WBP2. In this regard, mutated WBP2 plasmids with genetic deletion of PPXY motifs were used. Our studies revealed that GPX4 bands were not observed in ΔPPXY1 and ΔPPXY1+2 + 3 immunoprecipitates, suggesting that PPXY1 motif was involved in their interaction (Fig. 8M).
GPX4 was also truncated, and the CO-IP data revealed that C-terminal domain of GPX4 was involved in its interaction with WBP2 (Fig. 8L). As mentioned above, WBP2 inhibited CMA-induced degradation of GPX4, a process where HSC70 interacted with the two KEFRQ-like motifs (located in the C-terminal domain) of GPX4 to drive its lysosomal degradation. Therefore, we speculated that the two KEFRQ-like motifs of GPX4 might be involved in the interaction between WBP2 and GPX4. In this regard, the two motifs (124NVKFD128 and 187QVIEK191) were mutated (124AAKFD128 and 187AAIEK191) in GPX4 as previously reported [29]. Our CO-IP data demonstrated that the binding between WBP2 and GPX4 was not observed when the two KEFRQ-like motifs were alternatively or simultaneously mutated (Fig. 8N), indicative of their essential role in the interaction. Besides, no interaction between WBP2 and HSC70 was observed in BUMPT cells (Supplemental Fig. 10), thus ruling out the possibility that HSC70 acted as a linker between WBP2 and GPX4. Further investigations showed that WBP2 overexpression partially disrupted the interaction between GPX4 and HSC70 in cisplatin-treated BUMPT cells (Fig. 8O), indicating the competition between WBP2 and HSC70 in binding with GPX4. Collectively, these data demonstrated that WBP2 (PPXY1 motif) competed with HSC70 for binding with GPX4 (the two KEFRQ-like motifs), leading to the inhibition of CMA-mediated GPX4 degradation.
4. Discussion
Cisplatin-induced AKI (CP-AKI) is commonly observed in the clinic, and it is also widely used in laboratory investigations to explore the pathogenesis of renal injuries. The molecular machinery of AKI is quite complicated, previous publications have revealed various aspects of mechanisms in CP-AKI, such as DNA damage, mitochondrial disruption, vascular disorder, and ferroptosis [4,[32], [33], [34]]. Despite decades of intensive investigation, the exact mechanism of CP-AKI still remains blurry. In order to explore novel modulators for AKI, our present study employed bio-informatics methods to identify WBP2 as a potential regulator for AKI & ferroptosis (Fig. 1). WBP2 was initially regarded as a transcription coactivator with oncogenic properties [35]. Previous investigations revealed that WBP2 interacted with estrogen receptor and progesterone receptor to increase their transcriptional activity, thus promoting the progression of breast cancer [36]. A similar pattern was observed in its interaction with β-catenin, YAP and tafazzin, which led to the progression of multiple malignancies [15]. Recently, the role of WBP2 in non-oncological pathological states was also recognized. It has been demonstrated that WBP2 was involved in the maintenance of cochlea function [21] and oocyte activation during fertilization [20]. Besides, one recent study by Zheng et al. showed that WBP2 interacted with AMPKβ1 to activate the AMPK pathway, leading to the attenuation of diet-induced liver steatosis [22]. Herein, our work presented the initial evidence that WBP2 was highly expressed in renal proximal tubular cells, and its expression was conceivably decreased in CP-AKI (Fig. 1 & Supplemental Fig. 2). In addition, WBP2 overexpression ameliorated, while its gene disruption accentuated, cisplatin-induced proximal tubular injuries (Fig. 2, Fig. 3). Taken together, our data demonstrated that WBP2 was involved in the pathogenesis of CP-AKI.
Since WBP2 was initially identified as a potential modulator of AKI & ferroptosis by bioinformatics, and ferroptosis is an integral process for CP-AKI, it is conceivable that WBP2 might inhibit ferroptosis to alleviate cisplatin-induced proximal tubular injuries. Ferroptosis, an iron-catalyzed non-apoptotic regulated cell death, is intimately related to toxic lipid peroxidation, ferrous iron-catalyzed Fenton reaction, and disrupted amino acid metabolism [37,38]. In the past decade, ferroptosis has been intensively investigated in various physiopathological contexts, including the progression of malignancies, cardiac injuries, and hepatic damage [[39], [40], [41]]. In the realms of AKI, previous studies demonstrated that pharmaceutical inhibition of ferroptosis by Fer-1 attenuated the renal injuries induced by ischemia/reperfusion, folic acid, and rhabdomyolysis [[42], [43], [44]]. Our previous publication revealed that ferroptosis was an essential form of cell death in CP-AKI [13], which was later on validated by three groups of investigators [[45], [46], [47]]. Our present data showed that WBP2 alleviated RSL3-induced ferroptosis in BUMPT cells (Fig. 4), indicating the direct relationship between ferroptosis and WBP2. The detection of ferroptosis is somewhat difficult due to the lack of specific markers. The therapeutic effect of ferroptosis inhibitors and increased generation of lipid peroxide were usually used to evaluate the occurrence of ferroptosis. In this regard, 4-HNE (a marker for lipid peroxide), free iron levels, ferroptosis-associated proteins (GPX4, NCOA4, and FTH1), and Fer-1 (a ferroptosis inhibitor) were used to determine the progression of ferroptosis in this study. Our data showed that WBP2 attenuated the aberrant alterations in ferroptosis-related parameters in CP-AKI (Fig. 4, Fig. 5). Moreover, the detrimental effect of WBP2 knockdown in CP-AKI was conceivably ameliorated by the administration of Fer-1 (Fig. 7). Noteworthily, unlike the in vitro observations, the expression of FTH1 was increased in cisplatin-treated kidneys, which might be induced by a feedback mechanism. This unique phenomenon was initially presented by Gao et al., whose work showed that endogenous FTH1 was upregulated by increased free iron-induced transcription, while exogenous FTH1 was constantly degraded, in erastin (a ferroptosis inducer)-treated human fibrosarcoma cells [28]. Besides, our previous work also demonstrated the involvement of this unique feedback mechanism in CP-AKI(13). Interestingly, the expression profile of WBP2 and the ferroptosis markers was conceivably deceased in renal sections of ATN patients and cisplatin-treated human primary tubular cells (HPT cells) (Fig. 6). However, no renal samples of CP-AKI patients were used since those patients were not subjected to kidney biopsy in the clinical practice, which restricted the clinical relevance of our study. All in all, our data demonstrated that WBP2 decelerated ferroptosis to alleviate CP-AKI.
Next, we proceed to explore the exact mechanism involved in the modulation of ferroptosis by WBP2. The preliminary string analysis and CO-IP experiments revealed that WBP2 interacted with GPX4, a key enzyme for the inhibition of ferroptosis, in BUMPT cells (Fig. 8). The pathogenesis of ferroptosis is intimately related to the excessive generation of lipid peroxides, which can be decomposed into toxic derivatives to execute the cells [9]. Notably, GPX4, together with GSH, catalyzes the lethal lipid peroxides into non-toxic lipid alcohol, leading to the termination of ferroptotic process [48]. It has been reported that tamoxifen-induced GPX4 gene deletion in mice led to severe lipid-oxidation-related renal injuries [49], and considerable downregulation of GPX4 was observed in CP-AKI [45]. In fact, the inhibition of GPX4 or GSH synthesis was constantly used to induce ferroptosis in laboratory investigations. Previous data suggested that the expression of GPX4 can be regulated in transcriptional [45] or post-translational manners [29,30]. Due to the interaction between WBP2 and GPX4, we speculated that post-translational events might be involved in our study. Two types of post-translational modulatory mechanisms, i.e. proteasomal degradation and chaperon-mediated autophagy (CMA)-induced degradation, have been identified in the regulation of GPX4 stability [29,30,50]. Our data showed that the inhibition of lysosomes (NH4Cl) or autophagy (chloroquine), rather than the inhibition of proteasome (MG132), attenuated the degradation of GPX4 in cisplatin-treated BUMPT cells, indicating the involvement of CMA-related mechanism (Fig. 8). Similar observations were presented by Chen et al., who reported that CMA-induced GPX4 degradation was operative in erastin-treated primarily cultured mouse renal tubular cells [50]. CMA-induced GPX4 degradation refers to a unique type of selective autophagy guided by HSC70, which drives the lysosomal phagocytosis of GPX4 for degradation [29,50]. Our present studies showed that the expression profile of WBP2 modulated cisplatin-induced co-localization of GPX4 and lysosomes in renal proximal tubules, suggesting that CMA-mediated GPX4 degradation was regulated by WBP2 (Fig. 8 & Supplemental Fig. 5). Taken together, our studies demonstrated that WBP2 interacted with GPX4 to inhibit its lysosomal degradation in CP-AKI, leading to the deceleration of ferroptosis.
Finally, various biochemical experiments were performed to investigate the molecular machinery for the interaction between WBP2 and GPX4. Previous publications revealed that WBP2 conceives three PPXY motifs in its C-terminal domain, among which, PPXY2 and PPXY3 motifs usually dominate the interaction with its targeted proteins [15]. However, our study demonstrated that PPXY1 motif of WBP2 interacted with GPX4, a quite rare binding pattern (Fig. 8). One possible explanation for this might be the different amino acid sequences of the three PPXY motifs. PPXY2 and PPXY3 motifs of WBP2 share the same sequence of PPPY, while its PPXY1 motif has a different sequence of PPGY [15]. The binding sites in GPX4 were also explored. Since CMA-induced GPX4 degradation was modulated by WBP2, it is conceivable that the KEFRQ-like motifs might be involved [29]. Previous data suggested that two KEFRQ-like motifs of GPX4, i.e. 124NVKFD128 and 187QVIEK191, were involved in the interaction with HSC70, a vital process for CMA-mediated degradation of GPX4 [29]. Our CO-IP data showed that GPX4 with mutations in the two KEFRQ-like motifs failed to bind with WBP2, and WBP2 overexpression disrupted the interaction between GPX4 and HSC70 in cisplatin-treated BUMPT cells (Fig. 8). Since no interaction was observed between WBP2 and HSC70 (Supplemental Fig. 8), we speculated that WBP2 competed with HSC70 in binding with the KEFRQ-like motifs of GPX4. All in all, these data suggested that the PPXY1 motif of WBP2 interacted with the two KEFRQ-like motifs of GPX4.
In conclusion, this present study revealed the beneficial role of WBP2 in the pathogenesis of CP-AKI, and its protective effect was related to the preservation of GPX4 and the deceleration of ferroptosis, thus providing novel insights into the treatment of CP-AKI. However, it is noteworthy that various forms of cell death have been reported in the progression of CP-AKI(4, 47). Herein, only ferroptosis was investigated since the initial bioinformatic analysis targeted AKI & ferroptosis-related genes. Further investigations are still needed to elucidate the relevance between WBP2 and other forms of cell death in renal pathological states. Besides, Due to resource limitation, WBP2 knockout mice were not available in our present study, which should be incorporated in WBP2-related studies in the future.
Funding
This work is supported by the Innovative Platform and Talents Project of Hunan Province (2021RC2039), China Postdoctoral Science Foundation (2021M693568), National Natural Science Foundation of China (82100733), Hunan Province Natural Science Foundation (2021JJ40827), the Excellent Youth Foundation of Hunan Provincial Natural Science Foundation Committee (2023JJ20083), the Scientific Research Launch Project for new employees of the Second Xiangya Hospital of Central South University, and Guangzhou Science and Technology Project (2023A03J0863 & 2023A04J1211).
Ethics approval
Animal procedures used in this study were approved by the Animal Care and Use Committee of the Second Xiangya Hospital of Central South University, China (20220515). Human studies were approved by the Second Xiangya Hospital of Central South University (LYF2022146). Written informed consent from participants or their guardians was obtained.
Declaration of competing interest
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102826.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg. Chem. 2019;88 doi: 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- 2.Sears S., Siskind L. Potential therapeutic targets for cisplatin-induced kidney injury: lessons from other models of AKI and fibrosis. J. Am. Soc. Nephrol. 2021;32(7):1559–1567. doi: 10.1681/ASN.2020101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamano H., et al. Diphenhydramine may be a preventive medicine against cisplatin-induced kidney toxicity. Kidney Int. 2021;99(4):885–899. doi: 10.1016/j.kint.2020.10.041. [DOI] [PubMed] [Google Scholar]
- 4.Deng F., et al. Regulated cell death in cisplatin-induced AKI: relevance of myo-inositol metabolism. Am. J. Physiol. Ren. Physiol. 2021;320(4):F578–F595. doi: 10.1152/ajprenal.00016.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon S.J., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., et al. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019;26(11):2284–2299. doi: 10.1038/s41418-019-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan H., et al. Ferroptosis and its potential as a therapeutic target. Biochem. Pharmacol. 2021;186 doi: 10.1016/j.bcp.2021.114486. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., et al. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol. Cancer. 2022;21(1):47. doi: 10.1186/s12943-022-01530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockwell B.R., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat. Commun. 2021;12(1):1589. doi: 10.1038/s41467-021-21841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z., et al. Emerging role of ferroptosis in acute kidney injury. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/8010614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S., et al. Characterization of ferroptosis in kidney tubular cell death under diabetic conditions. Cell Death Dis. 2021;12(2):160. doi: 10.1038/s41419-021-03452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng F., et al. Myo-inositol oxygenase expression profile modulates pathogenic ferroptosis in the renal proximal tubule. J. Clin. Invest. 2019;129(11):5033–5049. doi: 10.1172/JCI129903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H.I., Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U. S. A. 1995;92(17):7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabatabaeian H., et al. The emerging roles of WBP2 oncogene in human cancers. Oncogene. 2020;39(24):4621–4635. doi: 10.1038/s41388-020-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., et al. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011;18(8):1346–1355. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J., et al. microRNA-485-5p inhibits the progression of hepatocellular carcinoma through blocking the WBP2/Wnt signaling pathway. Cell. Signal. 2020;66 doi: 10.1016/j.cellsig.2019.109466. [DOI] [PubMed] [Google Scholar]
- 18.Hum M., et al. WBP2 promotes gastric cancer cell migration via novel targeting of LATS2 kinase in the Hippo tumor suppressor pathway. Faseb. J. 2021;35(2) doi: 10.1096/fj.202000393R. [DOI] [PubMed] [Google Scholar]
- 19.Chen S., et al. WW domain-binding protein 2: an adaptor protein closely linked to the development of breast cancer. Mol. Cancer. 2017;16(1):128. doi: 10.1186/s12943-017-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buniello A., et al. Wbp2 is required for normal glutamatergic synapses in the cochlea and is crucial for hearing. EMBO Mol. Med. 2016;8(3):191–207. doi: 10.15252/emmm.201505523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton L.E., et al. WBP2 shares a common location in mouse spermatozoa with WBP2NL/PAWP and like its descendent is a candidate mouse oocyte-activating factor. Biol. Reprod. 2018;99(6):1171–1183. doi: 10.1093/biolre/ioy156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z., et al. WW domain-binding protein 2 overexpression prevents diet-induced liver steatosis and insulin resistance through AMPKbeta1. Cell Death Dis. 2021;12(3):228. doi: 10.1038/s41419-021-03536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie M.E., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancias J.D., et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M., et al. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26(9):1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z., et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2019;116(8):2996–3005. doi: 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Y., et al. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J. Hematol. Oncol. 2021;14(1):19. doi: 10.1186/s13045-020-01016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novoselov S.V., et al. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104(19):7857–7862. doi: 10.1073/pnas.0610683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hama T., et al. DNA damage is overcome by TRIP13 overexpression during cisplatin nephrotoxicity. JCI Insight. 2021;6(22) doi: 10.1172/jci.insight.139092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maekawa H., et al. Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Rep. 2019;29(5):1261–12673 e6. doi: 10.1016/j.celrep.2019.09.050. [DOI] [PubMed] [Google Scholar]
- 34.Tanimura S., et al. Renal tubular injury exacerbated by vasohibin-1 deficiency in a murine cisplatin-induced acute kidney injury model. Am. J. Physiol. Ren. Physiol. 2019;317(2):F264–F274. doi: 10.1152/ajprenal.00045.2019. [DOI] [PubMed] [Google Scholar]
- 35.Lim Y.X., et al. WBP2 promotes BTRC mRNA stability to drive migration and invasion in triple-negative breast cancer via NF-kappaB activation. Mol. Oncol. 2022;16(2):422–446. doi: 10.1002/1878-0261.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim S.K., et al. Hippo/MST blocks breast cancer by downregulating WBP2 oncogene expression via miRNA processor Dicer. Cell Death Dis. 2020;11(8):669. doi: 10.1038/s41419-020-02901-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mou Y., et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 2019;12(1):34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X., et al. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei G., et al. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer. 2022;22(7):381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., et al. PRMT4 promotes ferroptosis to aggravate doxorubicin-induced cardiomyopathy via inhibition of the Nrf2/GPX4 pathway. Cell Death Differ. 2022 doi: 10.1038/s41418-022-00990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., et al. Ubiquitin ligase E3 HUWE1/MULE targets transferrin receptor for degradation and suppresses ferroptosis in acute liver injury. Cell Death Differ. 2022 doi: 10.1038/s41418-022-00957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linkermann A., et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2014;111(47):16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Sanchez D., et al. Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid-induced AKI. J. Am. Soc. Nephrol. 2017;28(1):218–229. doi: 10.1681/ASN.2015121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerrero-Hue M., et al. Curcumin reduces renal damage associated with rhabdomyolysis by decreasing ferroptosis-mediated cell death. Faseb. J. 2019;33(8):8961–8975. doi: 10.1096/fj.201900077R. [DOI] [PubMed] [Google Scholar]
- 45.Hu Z., et al. VDR activation attenuate cisplatin induced AKI by inhibiting ferroptosis. Cell Death Dis. 2020;11(1):73. doi: 10.1038/s41419-020-2256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishima E., et al. Drugs repurposed as antiferroptosis agents suppress organ damage, including AKI, by functioning as lipid peroxyl radical scavengers. J. Am. Soc. Nephrol. 2020;31(2):280–296. doi: 10.1681/ASN.2019060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda Y., et al. Role of ferroptosis in cisplatin-induced acute nephrotoxicity in mice. J. Trace Elem. Med. Biol. 2021;67 doi: 10.1016/j.jtemb.2021.126798. [DOI] [PubMed] [Google Scholar]
- 48.Yao Y., et al. Selenium-GPX4 axis protects follicular helper T cells from ferroptosis. Nat. Immunol. 2021;22(9):1127–1139. doi: 10.1038/s41590-021-00996-0. [DOI] [PubMed] [Google Scholar]
- 49.Friedmann Angeli J.P., et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., et al. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis. 2021;12(1):65. doi: 10.1038/s41419-020-03362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.