Abstract
The herpes simplex virus type 1 γ34.5 gene product and the cellular GADD34 protein both contain similar domains that can regulate the activity of eukaryotic initiation factor 2 (eIF2), a critical translation initiation factor. Viral mutants that lack the GADD34-related function grow poorly on a variety of malignant human cells, as activation of the cellular PKR kinase leads to the accumulation of inactive, phosphorylated eIF2 at late times postinfection. Termination of translation prior to the completion of the viral reproductive cycle leads to impaired growth. Extragenic suppressors that regain the ability to synthesize proteins efficiently in the absence of the viral GADD34-related function have been isolated. These suppressor alleles are dominant in trans and affect the steady-state accumulation of several viral mRNA species. We demonstrate that deregulated expression of Us11, a virus-encoded RNA-binding, ribosome-associated protein is necessary and sufficient to confer a growth advantage upon viral mutants that lack a GADD34-related function. Ectopic expression of Us11 reduces the accumulation of the activated cellular PKR kinase and allows for sustained protein synthesis. Thus, an RNA-binding, ribosome-associated protein (Us11) and a GADD34-related protein (γ34.5) both function in a signal pathway that regulates translation by modulating eIF2 phosphorylation.
Phosphorylation of the cellular translation initiation factor eukaryotic initiation factor 2 (eIF2) in response to a variety of stresses, such as heat shock, viral infection, and growth factor withdrawal, results in the inhibition of translation (13, 21, 22, 24, 32, 36). Thus, agents that modulate eIF2 phosphorylation are poised to globally regulate polypeptide synthesis (for a review, see reference 9). While kinases that phosphorylate eIF2 in vitro have been identified, the detailed series of events that culminate in the accumulation of phosphorylated eIF2 remain to be elucidated.
Much of our knowledge concerning how eIF2 phosphorylation controls protein synthesis stems from analysis of both Saccharomyces cerevisiae and viruses (for reviews, see references 19, 27, 32, and 36). As viruses intervene and usurp important cellular regulatory pathways, they can serve as probes for understanding vital functions in mammalian cells. Herpesviruses are particularly valuable in this regard, as they establish stable latent infections in their host and periodically reemerge from this latent state to cause productive infections. These viruses are responsible for a variety of pathological conditions ranging from benign sores to malignancies. Herpes simplex virus type 1 (HSV-1), for example, remains latent in neurons but upon reactivation can grow productively at epithelial sites, causing blisters, or in the central nervous system, resulting in encephalitis (for a review, see reference 41). Lytic replication of these viruses requires acute changes in host cell metabolism to produce predominately viral polypeptides. In addition, alphaherpesviruses, exemplified by HSV-1, have evolved mechanisms to override normal regulatory processes important for host defense or stress response (7). To sustain protein synthesis throughout the infection, HSV has parasitized a functional domain from the cellular GADD34 protein that prevents the accumulation of phosphorylated eIF2.
The carboxy terminus of the γ34.5 gene encoded by HSV-1 shares substantial similarity to a region within the cellular GADD34 gene (6, 33, 54). The GADD designation signifies a set of genes coordinately expressed upon exposure of cells to agents that induce growth arrest, DNA damage, and differentiation (15, 20). Mutants that affect the HSV-1 GADD-like gene fail to grow productively in neurons of the central nervous system and are thus nonneurovirulent (1, 4, 31). Additionally, infection of a variety of human neoplastic cells with γ34.5 mutants results in premature cessation of protein synthesis due to the accumulation of phosphorylated eIF2 (5, 7). This inhibition of protein synthesis is accompanied by the activation of the cellular PKR kinase (7). The cellular GADD34 gene complements viral mutants for growth on nonpermissive cultured cells, thus demonstrating that one function of GADD34 is to preclude the accumulation of phosphorylated eIF2 (17). The results of a recent study suggest that the γ34.5 gene product functions in a complex that contains the cellular protein phosphatase 1α, thus maintaining steady-state pools of unphosphorylated eIF2 by fostering its dephosphorylation (16).
Recently, HSV-1 γ34.5 mutants that regain the ability to grow on neoplastic cells have been described. As these variants have all rearranged a specific viral DNA element and lack the coding sequences for the viral γ34.5 gene, they are second-site suppressors (35). The viral locus defined by these rearrangements has been termed the SUP locus. Remarkably, discrete 583-bp deletions which define the SUP locus enable γ34.5 mutants to sustain protein synthesis on otherwise nonpermissive cells and may completely preclude the accumulation of phosphorylated eIF2 (3, 35). This report demonstrates that the suppressor mutations alter expression of a viral RNA-binding, ribosome-associated protein, and this compensates for the loss of a GADD34-related function to regulate viral growth and protein synthesis. It further suggests a role for RNA-binding, ribosome-associated proteins in GADD34-mediated translational control.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (African green monkey kidney cells) were from the American Type Culture Collection (ATCC), adapted for growth in calf serum, and propagated in Dulbecco modified Eagle medium (DMEM) plus 5% calf serum. U373 human glioblastoma cells were from the ATCC and propagated in DMEM supplemented with 5% fetal bovine serum and 5% calf serum. 143tk− cells were from the ATCC and propagated in DMEM plus 10% fetal calf serum. The HSV-1 Patton strain was used exclusively in the work described here. The SUP1 virus used in these studies was constructed at the New York University School of Medicine.
Isolation of tk− recombinant viruses.
Δ34.5 mutant viral DNA was either transfected alone or cotransfected with rescue plasmid 27P, 11S, 11f.s., 11AS, or 5-3 into permissive Vero cells (seeded at 106 per 60-mm-diameter dish the previous day). After the appearance of cytopathic effect (CPE), a lysate was prepared by freeze-thawing. Dilutions of this lysate were prepared and used to infect 143tk− cells. These cells were maintained in DMEM supplemented with 1% fetal bovine serum and 100 μg of bromodeoxyuridine (BUdR) per ml. Once plaques were visible, the monolayers were overlaid with agarose, and plaques were picked. Isolates were subjected to two rounds of plaque purification on 143tk− cells in the presence of BUdR, and stocks were then prepared on Vero cells. Southern analysis of viral DNA (isolated as described in reference 35) verified that they contained the correct thymidine kinase (tk−) DNA insertion and that the BamHI Z fragment was intact and unrearranged. Lysates from the 5-3 transfection served as a control to monitor the BUdR selection, as this plasmid does not target the viral tk locus and cannot create a tk− recombinant above spontaneous background levels.
Marker rescue.
Δ34.5 mutant viral DNA was either transfected alone or cotransfected with a specific rescue plasmid into permissive Vero cells. The rescue plasmids were all wild type (WT) except for the different internal deletions each contained. After the appearance of CPE, a cell-free lysate was prepared by freeze-thawing, and dilutions from this lysate were used to infect nonpermissive U373 human glioblastoma cells. After a single pass of the transfection lysate on U373 cells, the viral stock was diluted and used to infect (i) fresh 60-mm-diameter dishes of U373 cells which are then fixed and stained with crystal violet and (ii) Vero cells for the analysis of viral DNA. Viral DNA was isolated from Vero cells as described previously (35). A second passage through U373 cells was included when tk targeting vectors were tested, as the resulting multimutated rescuants were highly crippled. In this instance, the 5-3 control transfection was also passaged twice on U373 cells.
Analysis of total viral protein synthesis.
Infections with viruses with high multiplicity of infection (MOI) and labeling with [35S]methionine and [35S]cysteine were performed as described previously (35).
Construction of SUP targeting vectors.
Throughout this work, (i) nucleotide numbers (nt) refer to the sequenced HSV-1 strain 17 (GenBank accession no. X14112), (ii) HSV-1 Patton strain viral DNA was used exclusively, and (iii) all PCR products were sequenced (Amersham catalog no. US70990) to confirm that no mutations were introduced by the polymerase. The plasmid pSXZY contains HSV-1 sequences from the SalI site at nt 143481 in the BamHI-X fragment to the BstEII site at nt 147040 in the BamHI-Y fragment inserted into pGEM9zf−. Restriction sites were introduced into pSXZY via PCR with mutagenic oligonucleotides. pSXZY12F is an isogenic variant containing an engineered HindIII site at nt 145581 that destroys the Us12 ATG. pSXZY12F5-3 is derived from pSXZY12F and contains an XbaI site at nt 145415. pSXZY12F2-9 is also derived from pSXZY12F and contains an XbaI site introduced immediately before the natural ApaLI site. Deletions were introduced into 12F5-3 and 12F2-9 by standard molecular biological methods.
Construction of tk targeting vectors.
The minimal α27 promoter (48) from the BamHI site at nt 113324 to the HinfI site at nt 113646 was amplified with cloned Pfu polymerase (Stratagene catalog no. 600153-81) and 5 ng of pEcoRI-B, a plasmid with the Patton strain EcoRI-B genomic fragment cloned into the EcoRI site of vector pACYC184 (a gift from Thomas Jones). The primers (Genelink) used were 5′-GCCACGTGTAGCCTGGATCCCAAC-3′, corresponding to nt 113308-113331, and 5′-CGGAATTCGGTAACCGGGGAGAGGCACCGA AG-3′, whose 3′ end corresponds to nt 113629 to 113646 immediately followed by BstEII and EcoRI sites on the 5′ end. Following digestion with BamHI and EcoRI, the amplified product was ligated into BamHI-EcoRI-cut pBluescript II SK(+) to create pBS/Bam/Hinf. The plasmid p7H1-7-12 (a gift from Thomas Jones) which contains the Patton strain BglII-I genomic fragment cloned into the BglII site of vector pKC7 was cut with SalI, filled in with Klenow fragment, and subsequently digested with BglII. To generate p5′TK-α27, the 2,400-bp fragment corresponding to nt 50255 to 47855 was purified and ligated into pBS/Bam/Hinf that was digested with SpeI, filled in with Klenow fragment, and subsequently cleaved with BamHI. The following plasmids were assembled with p5′TK-α27.
(i) p5′TK-α27-senseUs11.
The plasmid pSXZY12F was cleaved with SalI, filled in with Klenow fragment, and cut with BstEII. The 1,835-bp fragment corresponding to nt 143481 to 145316 that contained the Us11 open reading frame (ORF) was purified and ligated into p5′TK-α27 that had been digested with HindIII, filled in with Klenow fragment, and cleaved with BstEII.
(ii) p5′TK-α27-antisenseUs11.
pSXZY-12F was cut with PflM1, resected with T4 DNA polymerase, and cut with HindIII. The 867-bp fragment corresponding to nt 144714 to 145581 that contained the Us11 ORF was ligated into p5′TK-α27 that had been digested with BstEII, filled in with Klenow fragment, and subsequently cleaved with HindIII.
To complete the tk targeting constructs 11AS and 27P, a cassette that contained 3′ tk sequences was inserted next. The plasmid p7H1-6-38 (gift from Thomas Jones) contains the Patton strain BglII-M genomic fragment cloned into the BglII site of vector pKC7. To prepare a fragment that contains the 3′ tk region, p7H1-6-38 was cut with SacI, resected with T4 DNA polymerase, cut with BamHI, and filled in with Klenow fragment. The 2,303-bp fragment corresponding to nt 47358 to 45055 was purified and ligated into p5′TK-α27-antisenseUs11 and p5′TK-α27 digested with BstEII and SalI and filled in with Klenow fragment to generate 11AS and 27P targeting plasmids, respectively. Clones with the correct orientation of the 3′ tk cassette, namely, those that had the SacI site of the 2,303-bp fragment fused to the BstEII site of p5′TK-α27-antisenseUs11 and p5′TK-α27, were identified by restriction digestion analysis.
To complete the targeting constructs 11S and 11f.s., an alternate cloning strategy that facilitated the assembly of the frameshift variant 11f.s. was employed. The pBluescript II SK(+) derivative pBSΔSac/2800Bam-Bgl/Kan contains the following features: (i) a kanamycin resistance cassette; (ii) a destroyed SacI polylinker site; (iii) the 2,800-bp BamHI-BglII fragment from p7H1-6-38 corresponding to HSV-1 nt 45055 to 47855 (3′ tk) ligated into the polylinker BamHI site such that the viral BglII site is proximal to the polylinker SalI site; and (iv) a unique SacI site within the HSV-1 3′ tk sequences. To generate p5′TK-α27-senseUs11-3′TK/Kan, p5′TK-α27-senseUs11 was digested with PflM1, resected with T4 DNA polymerase, and cut with SalI. The 3,329-bp fragment corresponding to the 5′TK-α27-Us11 sequences was purified and ligated into pBSΔSac/2800Bam-Bgl/Kan digested with SacI, resected with T4 DNA polymerase, and cut with SalI. To release the insert, p5′TK-α27-senseUs11-3′TK/Kan was cleaved with SalI, filled in with Klenow fragment, and digested with XbaI. The 5,632-bp insert was purified and ligated into pBluescript II SK(+) cleaved with KpnI, resected with T4 DNA polymerase, and digested with XbaI to generate the targeting plasmid, 11S. The only two XhoI sites in this construct are in the Us11 ORF, thus enabling the exchange of WT and mutant XhoI fragments.
A PCR product that has a cytosine insertion between nt 145239 and 145240 was generated by PCR. This shifts the Us11 reading frame at codon 3. This product was digested with XhoI and ligated into XhoI-cut 11S to generate the targeting plasmid, 11f.s. Isolates containing a single copy of the mutant insert in the correct orientation were verified by DNA sequencing.
Prior to transfection, the targeting constructs were digested with the following enzymes to release the HSV-1 insert from the vector sequences: for 11S and 11f.s., XbaI plus SalI; for 11AS and 27P, SalI; and for XN1 and 5-3, PvuII.
Nucleic acid blotting and hybridization.
For Northern analysis, total RNA was isolated with BIOTEC-X reagent (Houston, Tex.) as directed by the manufacturer. Cycloheximide (CHX) (100 μg/ml) and phosphonoacetate (PAA) (300 μg/ml) were used at the indicated concentrations. Poly(A)+ RNA was isolated by using the Promega polyATract system according to the manufacturer’s instructions. Ten micrograms of total RNA or poly(A)+ RNA derived from an equivalent of 10 μg of total RNA was run on a 1.2% agarose–6% formaldehyde gel and transferred to a nylon membrane (Schleicher & Schuell catalog no. 77407). Membranes were hybridized in a solution containing 0.25 M Na2HPO4, 7% sodium dodecyl sulfate (SDS), 1% bovine serum albumin fraction V, and 1 mM EDTA overnight at 85°C with approximately 2.5 × 107 cpm of riboprobe. Riboprobe was prepared as described previously (39a). Membranes were washed once for 30 min at 85°C in a solution containing 20 mM Na2HPO4, 5% SDS, and 1 mM EDTA and once for 1 h at 85°C in a solution containing 20 mM Na2HPO4, 1% SDS, and 1 mM EDTA.
DNA for Southern analysis was transferred to nylon membranes as described elsewhere (44a). Membranes were hybridized as described above for Northern analysis, namely, overnight at 68°C with approximately 2.5 × 107 cpm of labeled probe and 100 μg of sonicated salmon sperm DNA per ml. Membranes were washed twice for 30 min each time at 68°C in a solution containing 20 mM Na2HPO4, 5% SDS, and 1 mM EDTA.
Preparation of S10 cellular protein extracts.
Sets of three confluent 10-cm-diameter dishes of U373 cells were infected at an MOI of 3 with either of the following tk− recombinants: 27P (27P-B-5d), 11fs (11fs-B-7a), 11AS (11AS-A-4a), and 11S (11S-A-3d [fs]). Infections with Δ34.5, SUP-1, and WT HSV-1 (Patton) were performed in parallel. At 16 h postinfection, the plates were scraped, and the cells were washed first with 1 ml of cold phosphate-buffered saline followed by 1 ml of cold buffer 1 (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM magnesium acetate [MgOAc]). After centrifugation for 4 min at 1,000 × g, the final cell pellet was resuspended in a volume of cold buffer 1 (approximately 100 to 150 μl) equivalent to the volume of packed cells, and vortexed extensively. One pellet volume of cold lysis buffer (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgOAc, 0.5% Nonidet P-40) was added, and the extract was mixed by gentle inversion. An amount of cold buffer E (100 mM HEPES-KOH [pH 7.6], 550 mM KCl, 50% glycerol, 2.5 mM spermidine, 3.5 mM β-mercaptoethanol) equivalent to 0.4 times the volume of packed cells was added, followed by the addition of phenylmethylsulfonyl fluoride to a final concentration of 200 μM. After mixing by gentle inversion, samples were incubated for 5 min on ice, and centrifuged at 10,000 × g for 4 min at 4°C. The S10 supernatant was recovered, and aliquots were quick-frozen and stored at −80°C. Typically, this procedure yields S10 extracts with protein concentrations of 3 to 4 mg/ml.
PKR kinase assay.
Kinase reactions were prepared with 42 μl of the appropriate S10 protein extract, 30 μCi of [γ-32P]ATP (6,000 Ci/mmol; NEN Life Science), 30 μM ATP, 5 mM MgOAc, 20 mM HEPES-KOH (pH 7.4), and 100 μM phenylmethylsulfonyl fluoride. The volume of the reaction mixture was brought to 50 μl with MilliQ H2O and incubated at 30°C for 30 min. Fifty microliters of a 10% protein A Sepharose (Pharmacia catalog no. 17-0780-01) slurry equilibrated in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris-HCl [pH 7.5]) was added, and the reaction mixtures were incubated on ice for 25 min. After the beads were spun down, the supernatant was transferred to a new tube, 1 μg of rabbit anti-PKR polyclonal antibody (Santa Cruz Biotechnology sc-707) was added, and the samples were incubated for 1 h on ice. One hundred microliters of the 10% protein A Sepharose slurry was added, and the samples were rocked at 4°C for 50 min. The beads were then washed three times with 1 ml of cold RIPA buffer, and the final pellet was resuspended in 40 μl of 1× Laemmli buffer. Aliquots (20 μl) of each sample were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography. To quantitate the amount of 32P incorporated into p68 PKR, bands were excised from the gel and counted in liquid scintillant.
RESULTS
Mapping the SUP locus by internal deletions.
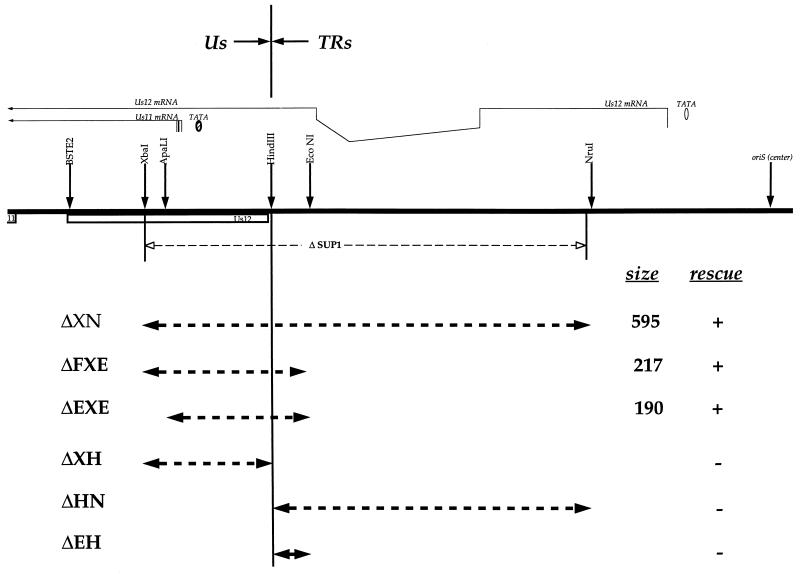
To further define the elements within SUP that must be rearranged to produce the suppressor phenotype, more-defined mutations within the SUP locus were created. New restriction sites were introduced into plasmid DNA clones to facilitate this analysis. The boundary between Us and TRs was denoted by a HindIII site that also destroyed the ATG for the Us12 ORF (Fig. 1). In addition, an XbaI site was introduced at the Us endpoint of the SUP1 deletion (plasmid 12F5-3) or immediately before the natural ApaLI site (plasmid 12F2-9) (Fig. 1). These two plasmids were then modified to generate a series of internal deletions. The new mutant plasmids were tested for their ability to restore growth of Δ34.5 mutants on nonpermissive human glioblastoma (U373) cells in a marker rescue protocol (Fig. 2A) (35). Only plasmids that contain rearrangements in the SUP locus generate CPE on U373 cells in this assay. While the parent plasmids (12F5-3 and 12F2-9) and a related construct that destroys the Us11 ATG were unable to generate the suppressor phenotype, the ΔXN deletion did confer the SUP phenotype on Δ34.5 mutant viral DNA (Fig. 1 and 2B). This deletion establishes a positive point of reference from which all other results can be evaluated and further demonstrates that the completely synthetic ΔXN deletion is indistinguishable from the natural deletion specified by the original SUP1 virus. A larger BstE2-NruI deletion also results in a suppressor virus (not shown).
FIG. 1.
Structure of the HSV-1 SUP locus and summary of mapping data. Deletion plasmids and the extent of the deletions appear below the map. The map represents an enlargement of the Us-TRs junction segment contained in the HSV-1 BamHI Z fragment. Characterized ORFs are represented as open boxes. The Us11 ATG is on the extreme left, while oriS lies on the right. The spliced Us12 transcript and the Us11 transcript (note heterogeneous start sites) appear above the map. Restriction sites are indicated by the arrows over the map. The deletion contained in the SUP1 isolate described by Mohr and Gluzman (35) is shown immediately below the map. The rescue column summarizes data presented in Fig. 2A and B (+, rescues; −, does not rescue).
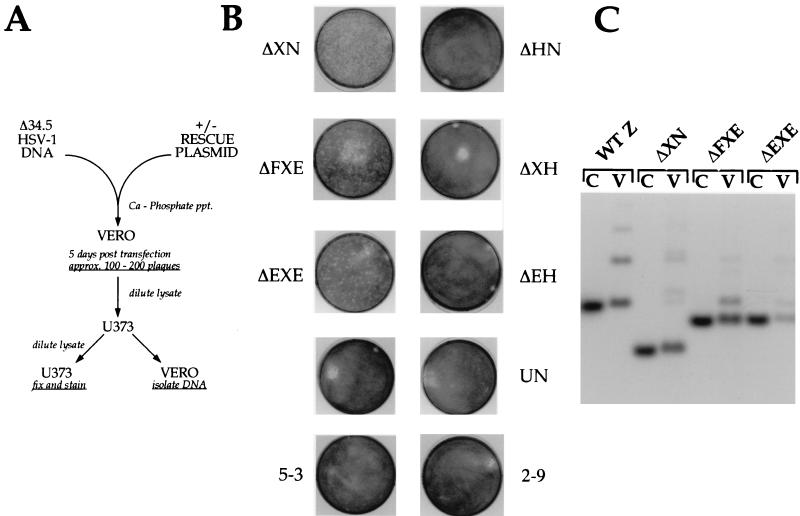
FIG. 2.
Small deletions which cross the boundary between Us and TRs can generate the SUP phenotype. (A) Outline of the marker rescue protocol used in this study. Δ34.5 mutant viral DNA was either transfected alone or cotransfected (+) with a specific rescue plasmid into permissive Vero cells. The rescue plasmids were all WT except for the different internal SUP deletions each contained. After the appearance of CPE, a cell-free lysate was prepared by freeze-thawing, and this lysate was used to infect nonpermissive U373 cells. After passage of the transfection lysate on U373 cells, the viral stock was used to infect freshly confluent 60-mm-diameter dishes of U373 cells and Vero cells. The U373 dishes were fixed, stained, and photographed, while the infected Vero cells were used to isolate viral DNA. ppt., precipitation. (B) After a single passage of the transfection lysates on U373 cells, a cell-free lysate was prepared by freeze-thawing and used to infect a fresh set of confluent U373 cells on 60-mm-diameter dishes. Photographs of these plates, after fixing and staining with crystal violet, are shown. The transfected rescue plasmid appears next to the image of the stained plate. UN, not transfected. (C) Analysis of viral genomes. Lysates from duplicate U373 plates were used to prepare viral DNA in Vero cells. Rescue plasmid DNA clones (C lanes) harboring various deletions within the SUP locus and the corresponding population of rescued viral DNA isolated from Vero cells (V lanes) were digested with BamHI, fractionated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled BamHI-BstE2 DNA fragment that contains the unique portion of the BamHI Z fragment (shown partially in Fig. 1). The filter was washed, and the autoradiogram is shown. As Δ34.5 mutant viruses fail to replicate efficiently on U373 cells, the viral population is enriched with rare recombinants that have acquired a SUP mutation from the targeting plasmid. This Southern analysis demonstrates that viral populations displaying the suppressor phenotype consist of predominantly recombinant viruses which have acquired the genotype specified by the rescue plasmid used in the transfection. The WT BamHI Z fragment of Δ34.5 HSV-1 mutant viruses (WT Z) comigrates with the WT HSV-1 BamHI Z fragment. Δ34.5 viral DNA digested was prepared from stocks propagated only on Vero cells. Additional slow-migrating bands in viral DNA samples (for example, the WT Z fragment in the Δ34.5 mutant) represent expansions within the repetitive TRs regions.
Deletions smaller than ΔXN were subsequently constructed around the HindIII site and examined in this assay. As Us and TRs join at this point, we may discern whether elements in Us, TRs, or both are responsible for the suppressor phenotype. Figure 2B shows that deletions spanning the HindIII site rescue the growth of Δ34.5 mutants in this assay, while those terminating at the HindIII site do not. For example, ΔXH, ΔHN, and ΔEH individually do not rescue, but the contiguous deletions ΔXN, ΔEXE, and ΔFXE all do (Fig. 1 and 2B and C). The smaller deletions in ΔFXE and ΔEXE were all produced synthetically and were not found in any of the SUP viruses isolated initially (35). They are, however, completely contained within the boundaries of the larger SUP1 deletion. Southern analysis of this selected population of viruses formed in the marker rescue experiment reveals them to be homogeneous and stable (Fig. 2C). It further demonstrates that viruses which display the suppressor phenotype acquire the genotype specified by the rescue plasmid used in the transfection (Fig. 2C).
Thus, deletion of both a 140-bp Us and 51-bp TRs component within the SUP locus is required to confer the suppressor phenotype upon γ34.5 mutant viruses. As the only characterized genes in this region are confined to the Us segment, the deletions defining the SUP locus appear to delineate a novel genetic element that contains both Us and TRs components. Furthermore, the nature of these deletions does not support models that simply invoke loss of functions encoded in the Us region.
The SUP locus mutants are dominant in trans.
To determine whether the suppressor phenotype results from a gain or loss of function, we performed complementation analysis. Complementation is assessed by the ability of a coinfecting virus to provide a function that enables a γ34.5 mutant virus to sustain protein synthesis on normally nonpermissive cells. As such studies require both viruses to coinhabit the same host cell, these infections must be performed at a MOI of approximately 5 to 10. The conditions for coinfection were established by using WT virus to complement the Δ34.5 mutant, as it has been demonstrated that the γ34.5 gene encodes a polypeptide which acts in trans (8). Replicate cultures of human U373 cells, which are nonpermissive for the growth of γ34.5 mutants, were each infected with a mixture composed of either WT virus and Δ34.5, WT virus and SUP1 (SUP1 is a suppressor virus and therefore of the genotype Δ34.5ΔSUP), or Δ34.5 and SUP1. The MOI for each individual virus present in the mixture was 5, yielding a total MOI of 10 for the coinfections. Cultures infected with each of the individual viruses in parallel at an MOI of 5 served as controls. After pulse-labeling with 35S-labeled amino acids at late times postinfection, total protein synthesis was examined by SDS-PAGE followed by autoradiography (Fig. 3). Under these conditions, cells infected with a mixture composed of the WT and Δ34.5 viruses were able to direct late protein synthesis. These two viruses are isogenic except for differences at the γ34.5 loci. The WT γ34.5 gene product is able to complement the null allele in the Δ34.5 virus. The allele in the Δ34.5 virus is a recessive genetic element and thus behaves as a loss of function. This control recapitulates the trans complementation observed by others (8) and serves to validate our experimental system.
FIG. 3.
The SUP1 deletion behaves as a trans-dominant mutant allele. U373 cells were infected either with each individual virus at an MOI of 5 or infected with a mixture composed of two viruses where each virus was present at an MOI of 5 (the total MOI was 10). Labeled proteins synthesized at late times postinfection were fractionated on SDS-polyacrylamide gels and visualized by autoradiography. MW, molecular weight markers.
Cells coinfected with the Δ34.5 and SUP1 viruses do not display a shutoff of protein synthesis similar to the one observed in cells infected with Δ34.5 alone (Fig. 3). The SUP rearrangements thus do not behave as recessive elements and are not likely to represent a loss of function. The WT SUP locus present in the Δ34.5 virus does not mediate the premature cessation of translation and does not behave as a trans-dominant allele. Instead, cells infected with a mixture composed of Δ34.5 virus and SUP1 viruses are able to sustain protein synthesis at late times postinfection (Fig. 3). These viruses are isogenic except for differences that occur at the SUP locus. Thus, in the complete absence of the γ34.5 gene product, the mutant SUP allele (in the Δ34.5ΔSUP1 virus) is able to override signals that would, if left unchecked, lead to the cessation of late protein synthesis. This is consistent with the proposals that SUP locus rearrangements (i) affect the synthesis of a specific product (s) and (ii) behave as either dominant, gain-of-function, or dominant-negative mutant alleles.
To confirm that both the SUP1 and Δ34.5 viruses do indeed coinhabit the same infected cell, we call attention to the rate of Us11 protein synthesis. Us11 is normally regulated as a true-late (γ2) gene. Rearrangements of the SUP locus that allow γ34.5 mutants to replicate on nonpermissive cells remove most of the Us12 coding sequences and sever the Us11 ORF from the cis elements that govern its transcription (3, 35). Although Us11 accumulates to similar steady-state levels in SUP1-infected cells, the kinetics of its production differ markedly, as it is now produced as an immediate-early protein (18). Deletion of upstream ATGs in the Us12 ORF allows the Us11 protein to be produced from an immediate-early transcript (Fig. 4). This most likely initiates from the Us12 immediate-early promoter. The rate of Us11 synthesis at late times postinfection relative to the intensities of other viral protein bands within a given lane is enhanced in cells coinfected with the SUP1 and Δ34.5 viruses compared to cultures singly infected with the SUP1 virus (Fig. 3). As the SUP1 virus does not produce significant amounts of Us11 as a late protein, the most likely source of a late mRNA encoding Us11 is the Δ34.5 viral chromosome. Late mRNAs, such as the Us11 mRNA, are produced in cells singly infected by Δ34.5 viruses but are not translated due to the premature cessation of protein synthesis (5). Thus, a late mRNA derived from the Δ34.5 chromosome is translated in cells coinfected with the SUP1 deletion. This is consistent with the presence of both Δ34.5 and SUP1 viruses within the same infected cell.
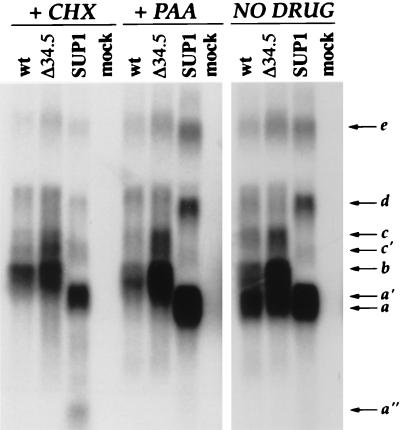
FIG. 4.
Alterations to the SUP locus affects the steady-state accumulation of multiple RNA species. U373 cells were either infected with each individual virus at high MOI or mock infected in the presence (+) or absence of drug (CHX or PAA). At 6 h postinfection, total RNA was harvested, fractionated by electrophoresis through formaldehyde-agarose gels, and transferred to nylon membranes. To detect transcripts that span the Us-TRs junction, strand-specific RNA probes were prepared from the BstE2-XbaI region (see map in Fig. 1). RNAs were detected only with probes antisense to the previously characterized Us11 and Us12 ORFs; furthermore, identical RNAs were observed in WT virus- and Δ34.5-infected cells when an antisense probe within the SUP1 deletion (ApaLI-EcoNI) was employed (not shown). Approximate sizes of the RNA species (shown to the right of the gel) are as follows: a (Us11), 1.4 kb; b (Us12), 1.9 kb; c, 2.6 kb; d, 3.4 kb; e, 7.9 kb.
SUP locus alterations affect the steady-state accumulation of multiple RNA species.
To characterize the dominant SUP product, the RNA products derived from the Us-TRs junction were analyzed in detail. U373 cells were either infected with each individual virus at high MOI or mock infected in the presence or absence of drug (CHX or PAA). Cycloheximide allows only for the synthesis of immediate-early transcripts, thus reducing the complexity of the RNA population substantially. The DNA polymerase inhibitor PAA, on the other hand, blocks cells at the early-to-late transition just prior to the initiation of viral DNA replication. Immediate-early, early, as well as leaky late RNAs (γ1) are present in PAA-treated cells, but γ2 or true-late RNAs are absent. Both pharmacological blocks were effective, as evidenced by the lack of the Us11 γ2 or true-late mRNA from both the WT and Δ34.5 CHX- and PAA-treated lanes (Fig. 4).
Northern analysis demonstrates that the steady-state accumulation of multiple RNA species can be detected with a probe, antisense to the Us11 and Us12 transcripts, that hybridizes to transcripts emerging from the SUP locus in cells infected with WT HSV-1 (Fig. 4). The two most abundant transcripts correspond to the late Us11 (RNA a) and immediate-early Us12 (RNA b) mRNAs. However, several uncharacterized transcripts of lesser abundance are also detected (RNA c, d, and e); furthermore, these novel transcripts are polyadenylated (not shown). In cells infected with the SUP1 mutant, the following alterations to the steady-state levels of multiple transcripts are observed. (i) The Us12 mRNA (RNA b) is not produced, as most of the US12 ORF is deleted. (ii) Us11 is regulated as an abundant immediate-early mRNA (RNA a′) that accumulates in CHX-treated cells (Fig. 4). This RNA is slightly longer than the bona fide Us11 mRNA (RNA a), as the SUP1 deletion fuses RNAs initiating from the Us12 promoter to the Us11 ORF. Accordingly, an oligonucleotide probe immediately upstream of the Us12 immediate-early promoter fails to detect RNA a′ but hybridizes to transcript c (not shown). (iii) Novel RNA d is considerably more abundant, while transcript c is absent, perhaps replaced by RNA c′ (Fig. 4). In addition, these novel SUP products (RNA a′, c′, and d) are produced with either immediate-early, or early or leaky late kinetics, and are all temporally poised to preclude the cessation of translation.
Assessing the role of immediate-early Us11 expression in generating the suppressor phenotype.
The suppressor mutants synthesize the Us11 protein from an abundant immediate-early mRNA. Us11 is normally a γ2, or true-late gene whose expression is strictly regulated by viral DNA replication. Although it is normally produced late in infection, Us11 is assembled into the tegument, an ill-defined proteinaceous structure between the nucleocapsid and the envelope, and is thus a component of infectious virions. As a tegument polypeptide, Us11 is thus delivered into the cytosol of infected cells at times which precede the expression of viral immediate-early genes. While the role of Us11 remains to be clarified, it is known to be an RNA-binding protein that can associate with at least one HSV mRNA (encoding the UL34 gene product), and it reportedly contains a transactivator activity similar to ones encoded by complex retroviruses (11, 43). Furthermore, Us11 is found stably associated with ribosomes following its introduction into the host cytosol (44). Thus, the mere presence of Us11 protein at early times may not be sufficient to overcome the block to protein synthesis that occurs in nonpermissive cells infected with Δ34.5. It is possible, however, that the suppressor phenotype requires the continued expression of Us11, beginning at immediate-early times. Recent studies indicate Us11 expression in cultured cells enhances survival following heat shock. Although heat shock normally leads to the accumulation of phosphorylated eIF2 and the inhibition of translation, the Us11 protein fostered the resumption of protein synthesis (12). The contribution immediate-early expression of Us11 makes towards the suppressor phenotype was therefore investigated.
To avoid potentially introducing a deletion that spans the Us-TRs region into the endogenous SUP locus via gene conversion, we chose to ectopically express Us11 from a heterologous immediate-early promoter located at a distinct site in the viral chromosome. To express Us11, we made use of the well-characterized minimal α27 immediate-early promoter and its ability to direct immediate-early transcription from within the viral tk locus (48). Thus, the Δ34.5 recombinant that produces Us11 with immediate-early kinetics will have a WT endogenous SUP locus, a dominant mutant SUP allele within the tk locus, a deletion that inactivates the tk gene (UL23), and a mutation in the neighboring UL24 gene.
Several targeting plasmids were constructed (Fig. 5A) such that they would integrate into the viral tk locus and create tk− viruses that: (i) express an RNA capable of producing Us11 as an immediate-early protein (11S); (ii) express an RNA antisense to the Us11 RNA (11AS); (iii) express an RNA that introduces a frameshift mutation into the US11 ORF at codon 3 (11f.s.); and (iv) insert the α27 promoter cassette into the tk locus in the absence of a US11-related transcription unit (27P). These tk targeting constructs were transfected onto permissive Vero cells in the presence of Δ34.5 viral DNA, and cell-free lysates were prepared after plaques were visible. Lysates were passed through U373 cells according to the marker rescue protocol. Only transfection mixtures that contained 11S targeting vector yielded lysates capable of generating extensive CPE on U373 cells (Fig. 5B). An isogenic targeting construct that differs only by the insertion of a single nucleotide at codon 3 of the Us11 ORF does not rescue in this assay. 11AS and 27P constructs also do not rescue. As an additional control, a BamHI Z targeting vector that does not rescue in this assay (5-3; Fig. 2B) was transfected along with Δ34.5 DNA in parallel. Figure 5B illustrates the profound growth defect of Δ34.5 tk+ viruses under these conditions, relative to the extensive CPE caused by the Δ34.5 tk− suppressor represented by 11S.
FIG. 5.
Expression of the Us11 gene product is necessary and sufficient to confer a growth advantage upon Δ34.5 mutants in nonpermissive cells. (A) Schematic of the US11 expression constructs designed to integrate into the HSV-1 tk locus and create tk− recombinants. (B) Fixed plates, stained with crystal violet resulting from a marker rescue experiment. The targeting plasmid used in the transfection appears to the left of the plate. The columns designated A and B refer to two transfections handled independently. (C) Genome analysis of the rescued viral population. A lysate from a duplicate set of U373 plates identical to those shown in panel B was used to infect Vero cells, and viral DNA was isolated. Following digestion with EcoRI, DNA was fractionated by agarose gel electrophoresis, blotted onto a nylon membrane, and probed with a fragment from the 3′ tk region. Note that almost all of the DNA in the population, in samples derived from two independent transfections, contains the 11S:tk insertion specified by the 11S targeting construct.
To analyze the genotype of the resulting rescued population of viruses, a lysate from a parallel set of U373 plates was used to infect Vero cells and viral DNA was isolated. Following digestion with EcoRI, DNA was fractionated by agarose gel electrophoresis, blotted onto a nylon membrane, and probed with a fragment from the 3′ tk region. Note that almost all of the DNA in the population, in samples derived from two independent transfections, contains the 11S:tk insertion specified by the 11S targeting construct (Fig. 5C, compare 11S construct lane with 11S - A and 11S - B lanes). This dramatic enrichment for tk− alleles in the complete absence of a tk− selection regimen demonstrates that immediate-early expression of the Us11 polypeptide confers a growth advantage upon Δ34.5 mutant viruses in cells nonpermissive for the growth of Δ34.5 mutants. The faint band in the 11S - A and 11S - B lane comigrates with WT tk fragment in the Δ34.5 parent virus. This reflects: (i) the slower growth of the tk− suppressors on confluent U373 monolayers (note trace amounts of WT BamHI Z fragments are not observed in populations of tk+ suppressors [Fig. 2C]) and (ii) the dominance of the suppressor mutants in trans, which will allow the persistence of small amounts of Δ34.5 tk+ genotype within the overall population. Southern analysis revealed that the endogenous SUP locus at the Us-TRs junction in the rescued population was structurally intact and not rearranged (not shown).
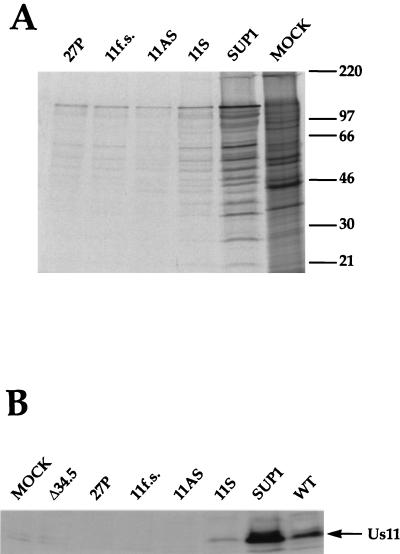
Individual tk− viruses were obtained by selecting and plaque purifying isolates from the Vero transfection lysate on 143tk− cells in the presence of BUdR. Southern analysis of these homogeneous isolates verified that the tk locus contained the correct insertion and that the BamHI Z locus was structurally intact and not rearranged (not shown). These isolates were then screened for their ability to sustain protein synthesis on nonpermissive U373 cells at late times postinfection. The SUP1 positive control is tk+, while all of the remaining recombinant viruses are tk−. Figure 6 demonstrates that the 11S virus is capable of enhanced protein synthesis at late times postinfection relative to 11f.s., 27P, and 11AS. As the 11f.s. virus differs only by virtue of a single nucleotide insertion at codon 3 of the Us11 ORF, expression of the Us11 protein product is responsible for this enhanced translation.
FIG. 6.
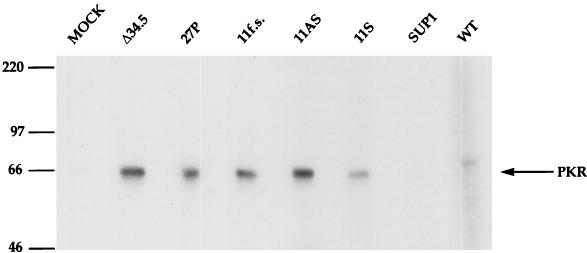
(A) Individual plaque-purified tk−:Δ34.5 isolates that express the Us11 polypeptide are capable of enhanced protein synthesis. Nonpermissive U373 cells were infected with the individual viral isolates at high MOI. Proteins labeled in a 1-h pulse with [35S]methionine at 13 h postinfection were fractionated by SDS-PAGE and visualized by autoradiography. The positions of molecular size markers (in kilodaltons) are indicated to the right of the gel. (B) Analysis of steady-state Us11 levels in SUP1 versus Δ34.5 tk−:11S. S10 extracts were prepared from infected U373 cells as described in Materials and Methods. Aliquots were fractionated by SDS-PAGE, electroblotted to Immobilon, and probed with a monoclonal antibody against HSV-1 Us11 (44). Identical results were obtained with extracts prepared directly in 1× Laemmli buffer (not shown).
The SUP1 virus synthesizes more total protein than the tk−:11S recombinant (Fig. 6A). Western analysis revealed that Us11 accumulates to greater steady-state levels in cells infected with the tk+:SUP1 mutant than in cells infected with the tk−:11S recombinant (Fig. 6B). This raises the possibility that the penetrance of the suppressor phenotype correlates with overall levels of Us11 polypeptide in the infected cell. The differences in levels may reflect differences in promoter strength between the minimal α27 promoter used in the tk− insertions compared to the Us12 (α47) promoter used in the bona fide SUP1 mutant. The loss of the TRs component from the suppressor mutants shown in Fig. 1 may reflect the loss of an element that further downregulates the Us12 (α47) immediate-early promoter. Alternatively, reduced Us11 levels could also reflect the decreased overall replication levels of the multimutated tk−:Us11 recombinant relative to the tk+:SUP1 virus.
Us11 expression reduces the accumulation of the activated cellular PKR protein kinase.
Previous studies demonstrate that the shutoff of protein synthesis that occurs in nonpermissive cells infected with γ34.5 mutants is accompanied by the activation of the cellular PKR protein kinase (7). The effect of expressing Us11 as an immediate-early protein on the activation of PKR was therefore investigated. S10 lysates were prepared from U373 cells infected with either the 11S, 11f.s., 11AS, or 27P virus (unpublished data). Figure 7 demonstrates that PKR is activated, as assessed by autophosphorylation, in lysates prepared from cells infected with either 27P, 11S, or 11f.s. Expression of Us11 as an immediate-early protein effectively reduces the accumulation of activated PKR by 50%. This correlates with the enhanced overall levels of protein synthesis observed in tk−:11S-infected cells (Fig. 6B). Furthermore, the considerably greater levels of Us11 in SUP1-infected cells results in higher levels of protein synthesis and even less activated PKR than that observed in cells infected with tk−:11S recombinant (Fig. 6 and 7). Finally, lysates derived from 11A.S.-infected cells incorporate 20% more 32P into p68 PKR than 11f.s. extracts. This modest hyperactivation is reproducible, perhaps reflecting the formation of a double-stranded RNA (dsRNA) species between the endogenous Us11 sense mRNA and the ectopically expressed 11AS RNA. Endogenous Us11 mRNA, although present, is not translated due to the efficient shutoff of protein synthesis that ensues following the onset of DNA replication in cells infected with γ34.5 mutants (5).
FIG. 7.
Expression of the Us11 polypeptide at immediate-early times precludes the hyperactivation of the cellular PKR kinase. S10 extracts prepared from infected U373 cells were incubated for 30 min at 30°C in the presence of 30 μCi of [γ-32P]ATP. PKR was then immunoprecipitated, and the resulting immune complexes were fractionated by SDS-PAGE and visualized by autoradiography. Bands containing PKR were excised from the gel and counted in liquid scintillant. The positions of molecular size markers (in kilodaltons) are indicated to the left of the gel.
DISCUSSION
HSV-1 γ34.5 mutants that lack the GADD34-related viral function grow poorly on a variety of malignant human cells due to the accumulation of phosphorylated eIF2 at late times postinfection (7). Recently, we isolated γ34.5 deletion variants that regained the ability to grow efficiently and sustain protein synthesis on nonpermissive cells (35). All of these variants suffered rearrangements in the HSV-1 genome where the Us component joins the TRs. These mutations are both necessary and sufficient to create the suppressor phenotype. In this report, we demonstrate that these dominant suppressor alleles compensate for the absence of the GADD34 function by overproducing a virus-encoded RNA-binding, ribosome-associated protein (Us11) at immediate-early times postinfection. As a consequence of expressing Us11 at immediate-early times, the accumulation of active PKR kinase is reduced (Fig. 8). This raises the intriguing possibility that GADD34 signal pathways, in response to agents that promote growth arrest, DNA damage, and differentiation may utilize components that have the intrinsic ability to bind RNA, associate with ribosomes, and prevent the dsRNA-mediated activation of PKR. In addition, it demonstrates that HSV encodes at least two distinct functions designed to preclude the accumulation of phosphorylated eIF2: a GADD34 homolog (γ34.5) and a ribosome-associated, RNA-binding protein (Us11). Although other families of large DNA viruses, such as poxviruses, encode multiple functions that regulate eIF2 phosphorylation, GADD34 homologs are not involved (10). African swine fever virus, an unrelated, unclassified large DNA virus, infects cells of the monocyte-macrophage lineage and apparently encodes a GADD34-related function (49). It would certainly be of interest to ascertain if African swine fever virus also directly targets a ribosomal function. The peculiarities different viruses have adopted to prevent the accumulation of phosphorylated eIF2 may reflect important aspects of the differentiated host cells they infect.
FIG. 8.
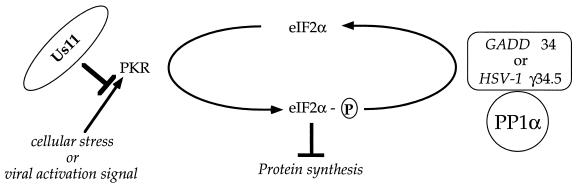
Us11 inhibits PKR activation and thus allows for sustained translation in the absence of a GADD34-related function. Suppressor viruses display enhanced growth on nonpermissive cells due to their ability to overcome a protein synthesis checkpoint guarded by the cellular PKR kinase. As Us11 is involved in mediating the suppressor phenotype, it could prevent activation of the PKR kinase either by intercepting the activator or inhibiting the activation process (as assessed by autophosphorylation) at a step subsequent to dsRNA binding. P, phosphate; PP1α, protein phosphatase 1α.
Although Us11 is not essential for growth of HSV-1 in cultured cells and is not required for neurovirulence in animals (2, 30, 38, 52), the loss of Us11 function is tolerated only in the presence of a GADD34-related function. Normally, approximately 600 molecules of Us11 are delivered into the cytosol of the host cell by the entering virion, whereupon it is found associated with host polysomes (44). While this very early role of Us11 can be eliminated in the presence of a viral GADD34-related function, continuous synthesis of Us11, beginning at immediate-early times, is required to preclude the activation of PKR in the absence of a GADD34-related function (Fig. 8). Both viral functions and their respective homologous cellular counterparts may contribute to whether a given cell type is permissive for the growth of γ34.5 mutants.
Us11 and PKR can each associate with ribosomes, and this interaction is mediated through their respective RNA-binding domains (42, 55). As Us11 appears to block PKR activation, ribosomal proteins and ribosomes may be involved in regulating PKR. Regulated exposure of structured dsRNA regions within ribosomes may be a component of the cellular stress response, creating the potential for structured rRNA segments to modulate PKR activity. The ribosome association of Us11 could be a means of precluding the dsRNA activator from interfacing with the dsRNA-binding domains of PKR. Alternatively, Us11 could inhibit PKR autophosphorylation at a step subsequent to ligand binding. Different structured rRNA segments could conceivably have different effects on PKR, with some structures activating and others inhibiting the enzyme (40). Such a model does not necessarily exclude a role for other cytosolic dsRNA species in modulating PKR activation.
Us11 is a ribosome-associated protein encoded by HSV-1. Cellular ribosomal proteins may execute functions similar to those performed by Us11 upon induction of GADD34 signal pathways in uninfected cells. Ribosome-associated polypeptides may participate, either enzymatically or as end points, in a variety of signaling pathways that affect rates of protein synthesis in response to stimuli that induce growth arrest, DNA damage, and differentiation. They can thus exert global effects on the growth and development of multicellular organisms (14, 28, 53). At the cellular level, ribosomal proteins and ribosomes affect a variety of processes fundamentally important to cell biology, such as the regulation of cell proliferation and the cellular stress response (23, 25, 26, 34, 37, 46).
Finally, the intimate association herpesviruses exhibit with a variety of differentiated cells may necessitate discrete modification of the host cell ribosome. Epstein-Barr virus, a herpesvirus that colonizes and immortalizes B lymphocytes, encodes small RNAs (EBERs) that bind L22 (51). Although the EBER RNAs are not essential for the growth or transforming functions of Epstein-Barr virus in cultured cells, they have been shown to modulate PKR activity in vitro (47, 50). L22 is a common target in herpesvirus-infected cells, as the HSV-1 α4 transcriptional regulatory protein binds L22 and leads to its accumulation in the nucleoplasm (29). While the L22 gene is involved in a common chromosomal translocation seen in leukemias (39), the significance of its relocalization in herpesvirus-infected cells remains a mystery.
Further studies on ribosome-associated proteins are necessary to understand how they participate in such a wide variety of cellular processes. In particular, studies on the herpesvirus Us11 protein may help illuminate how these proteins operate in pathways that involve dsRNA and GADD34 signaling.
ACKNOWLEDGMENTS
We are especially grateful to Winship Herr for critical review of the manuscript, Rich Roller for providing the anti-Us11 monoclonal antibody, Joel Baines for expert advice on generating tk− recombinants, and Tom Jones for Patton strain plasmids. We also thank John Blaho, Rich Roller, Kol Zarember, and David Frendewey for discussions.
ADDENDUM IN PROOF
After this article was submitted, Cassady et al. (K. A. Cassady, M. Gross, and B. Roizman, J. Virol. 72:8620–8626) reported that a GST-US11 fusion protein inhibits PKR activation and subsequent eIF2 phosphorylation in a cell-free system.
Footnotes
I.M. dedicates this work to the everlasting memories of his father, Donald Mohr, and Yasha Gluzman, a close friend, colleague, and mentor.
REFERENCES
- 1.Bolovan C A, Sawtell N M, Thompson R L. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J Virol. 1994;68:48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown S M, Harland J. Three mutants of herpes simplex virus type 2: one lacking the genes US10, US11, and US12 and two in which Rs has been extended by 6 Kb to 0.91 map units with loss of Us sequences between 0.94 and the Us/TRs junction. J Gen Virol. 1987;68:1–18. doi: 10.1099/0022-1317-68-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Cassady K, Roizman B. The second-site mutation in the herpes simplex virus recombinants lacking the γ34.5 genes precludes the shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J Virol. 1998;72:7005–7011. doi: 10.1128/jvi.72.9.7005-7011.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus 1 neurovirulence to γ34.5, a gene nonessential for growth in cell culture. Science. 1990;252:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 5.Chou J, Roizman B. The γ34.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou J, Roizman B. Herpes simplex virus 1 γ34.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Chen J J, Gross M, Roizman B. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF2α and premature shutoff of protein synthesis after infection with γ34.5− mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou J, Poon A P W, Johnson J, Roizman B. Differential response of human cells to deletions and stop codons in the γ134.5 gene of herpes simplex virus. J Virol. 1994;68:8304–8311. doi: 10.1128/jvi.68.12.8304-8311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 607–630. [Google Scholar]
- 10.Davies M V, Chang H W, Jacobs B L, Kaufman R J. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz J J, Dodon M D, Schaerer-Uthurralt N, Simonin D, Kindbeiter K, Gazzolo L, Madjar J J. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature (London) 1996;379:273–277. doi: 10.1038/379273a0. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Latoud C, Diaz J J, Fabre-Jonca N, Kindbeiter K, Madjar J J, Arrigo A P. Herpes simplex virus Us11 protein enhances recovery of protein synthesis and survival in heat shock treated HeLa cells. Cell Stress Chaperones. 1997;2:119–131. doi: 10.1379/1466-1268(1997)002<0119:hsvupe>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan R, Hershey J W B. Heat-shock induced translational alterations in HeLa cells: initiation factor modifications and the inhibition of translation. J Biol Chem. 1984;259:11882–11889. [PubMed] [Google Scholar]
- 14.Fisher E M, Beer-Romero P, Brown L G, Ridley A, McNeil J A, Lawrence J B, Willard H F, Bieber F R, Page D C. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 15.Fornace A J., Jr Mammalian genes induced by radiation: activation of genes associated with growth control. Annu Rev Genet. 1992;26:507–526. doi: 10.1146/annurev.ge.26.120192.002451. [DOI] [PubMed] [Google Scholar]
- 16.He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J Virol. 1997;71:6049–6054. doi: 10.1128/jvi.71.8.6049-6054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He B, Chou J, Liebermann D A, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by the double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch A G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman B, Liebermann D A. Molecular controls of apoptosis: differentiation/growth arrest primary response genes, proto-oncogenes, and tumor suppressor genes as positive and negative modulators. Oncogene. 1994;9:1807–1812. [PubMed] [Google Scholar]
- 21.Hu B R, Wieloch T A. Stress induced inhibition of protein synthesis initiation: modulation of initiation factor 2 and guanine nucleotide exchange factor activities follwing transient cerebral ischemia in the rat. J Neurosci. 1993;13:1830–1838. doi: 10.1523/JNEUROSCI.13-05-01830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B R, Yang Y-B O, Wieloch T. Heat shock inhibits protein synthesis and eIF-2 activity in cultured cortical neurons. Neurochem Res. 1993;18:1003–1007. doi: 10.1007/BF00966760. [DOI] [PubMed] [Google Scholar]
- 23.Iordanov M S, Pribnow D, Magun J L, Dinh T H, Pearson J A, Magun B E. Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J Biol Chem. 1998;273:15794–15803. doi: 10.1074/jbc.273.25.15794. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Jagus R, May W S. Interleukin 3 stimulates protein synthesis by regulating double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:7455–7459. doi: 10.1073/pnas.91.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferies H B J, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 389–410. [Google Scholar]
- 26.Jiang H, Lin J J, Tao J, Fisher P B. Suppression of human ribosomal protein L23A expression during cell growth inhibition by interferon-beta. Oncogene. 1997;14:473–480. doi: 10.1038/sj.onc.1200858. [DOI] [PubMed] [Google Scholar]
- 27.Katze M G. Translational control in cells infected with influenza virus and reovirus. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 607–630. [Google Scholar]
- 28.Kongsuwan K, Yu Q, Vincent A, Frisardi M C, Rosbash M, Lengyel J A, Merriam J. A Drosophila Minute gene encodes a ribosomal protein. Nature (London) 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- 29.Leopardi R, Roizman B. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc Natl Acad Sci USA. 1996;93:4572–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longnecker R, Roizman B. Generation of an inverting herpes simplex virus type 1 mutant lacking the L-S junction α sequences, an origin of DNA synthesis, and several genes including those specifying glycoproteins E and α47. J Virol. 1986;58:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLean A R, Ul-Fareed M, Robertson L, Harland J, Brown S M. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the ‘a’ sequence. J Gen Virol. 1991;72:631. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- 32.Mathews M B. Virus cell interactions. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- 33.McGeoch D J, Barnett B C. Neurovirulence factor. Nature (London) 1991;353:609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- 34.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNA’s in eukaryotes. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 35.Mohr I, Gluzman Y. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 1996;15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 36.Montine K S, Henshaw E C. Serum growth factors cause rapid stimulation of protein synthesis and dephosphorylation of eIF2 in serum depleted Ehrlich cells. Biochim Biophys Acta. 1989;1014:282–288. doi: 10.1016/0167-4889(89)90224-3. [DOI] [PubMed] [Google Scholar]
- 37.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The US9,10,11,12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 39.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER1. Proc Natl Acad Sci USA. 1993;90:7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Promega. Protocols and applications guide. Madison, Wis: Promega; 1996. [Google Scholar]
- 40.Robertson H, Manche L, Mathews M. Paradoxical interactions between human delta hepatitis agent RNA and the cellular protein kinase PKR. J Virol. 1996;70:5611–5617. doi: 10.1128/jvi.70.8.5611-5617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roizman B R, Sears A E. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 11–68. [Google Scholar]
- 42.Roller R J, Monk L L, Stuart D, Roizman B. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J Virol. 1996;70:2842–2851. doi: 10.1128/jvi.70.5.2842-2851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roller R J, Roizman B. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J Virol. 1991;65:5873–5879. doi: 10.1128/jvi.65.11.5873-5879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roller R J, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Schneider R J. Adenovirus and vaccinia virus translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 575–606. [Google Scholar]
- 46.Seshadri T, Uzman J A, Oshima J, Campisi J. Identification of a transcript that is down-regulated in senescent human fibroblasts. Cloning, sequence analysis, and regulation of the human L7 ribosomal protein gene. J Biol Chem. 1993;268:18474–18480. [PubMed] [Google Scholar]
- 47.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNA’s EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spector D, Purves F, Roizman B. Mutational analysis of the promoter region of the alpha 27 gene of herpes simplex virus 1 within the context of the viral genome. Proc Natl Acad Sci USA. 1990;87:5268–5272. doi: 10.1073/pnas.87.14.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sussman M D, Lu Z, Kutish G, Alfonso C L, Roberts P, Rock D L. Identification of an African swine fever virus gene with similarity to a myeloid differentiation primary response gene and a neurovirulence-associated gene of herpes simplex virus. J Virol. 1992;66:5586–5589. doi: 10.1128/jvi.66.9.5586-5589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toczyski D P, Matera A G, Ward D C, Steitz J A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umene K. Conversion of a fraction of the unique sequence to part of the inverted repeats in the S component of the herpes simplex virus type 1 genome. J Gen Virol. 1986;67:1035–1048. doi: 10.1099/0022-1317-67-6-1035. [DOI] [PubMed] [Google Scholar]
- 53.Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhan Q, Lord K A, Almo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Lieberman D A, Fornace A J., Jr The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu S, Romano P R, Wek R C. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J Biol Chem. 1997;272:14434–14441. doi: 10.1074/jbc.272.22.14434. [DOI] [PubMed] [Google Scholar]