Abstract
Very late factor 1 (VLF-1) of Autographa californica multicapsid nuclear polyhedrosis virus (AcMNPV) activates the transcription of two genes, polyhedrin (polh) and p10, during the final, occlusion-specific phase of infection. Using transient expression assays responsive to VLF-1, we identified linker scan mutations in the polh and p10 promoters which abolished or weakened the ability of the promoters to respond to stimulation by VLF-1. These mutations were located between the transcriptional and translational initiation sites, a region previously shown to be essential for the burst of expression during the very late phase. Addition of partially purified, epitope-tagged VLF-1 to DNA encompassing this “burst sequence” resulted in a shift in the gel electrophoretic mobility of the DNA, indicating that VLF-1 forms a complex with DNA. Addition of an antibody specific for the epitope tag of VLF-1 decreased the mobility of the DNA further, confirming the presence of VLF-1 in the complex. DNase I footprint assays revealed that VLF-1 partially purified from either insect cells or bacterial cells interacted with the burst sequences of both the polh and p10 very-late promoters. Linker scan mutations within the burst sequences severely impaired interaction between VLF-1 and the promoters. We propose that VLF-1 transactivates the polh and p10 promoters by interacting with the burst sequences.
The VLF-1 (very late factor 1) gene (vlf-1) of Autographa californica multicapsid nuclear polyhedrosis virus is required for expression of very late genes, e.g., polyhedrin (polh) and p10, during the final phase of infection involving the formation of occlusion bodies. vlf-1 was originally identified by characterization of an occlusion-defective mutant virus, tsB837, which produces only low levels of polh and p10 transcripts during the very late phase (11). In transient-expression assays, vlf-1 also stimulates expression from very late promoters but has no effect on expression from late promoters (22). Construction of recombinant viruses with altered vlf-1 expression revealed that polh expression is regulated by the timing of vlf-1 expression and/or the concentration of VLF-1 in the cell (27). vlf-1 itself is expressed as a late gene, and its product is an essential and limiting factor in polh expression (28).
Transcription of late and very-late baculovirus genes initiates from a TAAG sequence which is an essential element for both classes of promoters. The strength of expression from these promoters during the late phase depends on the context of the TAAG. The 18 bp encompassing the TAAG of the late vp39 promoter are the primary, if not the sole, determinants of expression levels from this promoter (12). In contrast, the strength of expression from the polh promoter during the very late phase of infection depends not only on the context of the TAAG but also on the nature of the sequence located between the TAAG and the translational initiation site, i.e., the sequence specifying the untranslated leader of very late mRNAs (10, 14, 17, 19, 25). This sequence is required for the burst of expression during the very late phase and is therefore referred to as the burst sequence. Mutations within the burst sequence reduce expression during the very late phase (e.g., 48 h postinfection) by 10- to 20-fold and lower both the steady-state levels of polh RNA and the rate of transcriptional initiation from the polh promoter (14). In contrast, mutations in sequences upstream of the TAAG sequence of the polh promoter have comparatively mild effects on the level of very late gene expression (14, 17, 19). Progressive deletions of the p10 promoter also suggest the presence of a burst sequence that is essential for strong expression during the very late phase (18, 24, 25).
The important roles of both VLF-1 and the burst sequence in hyperexpression of very late promoters suggest that they are two components of the same transactivation mechanism. We have examined the possibility that VLF-1 exerts its effect through interaction with very late promoters. We show that the burst sequence is important for a very late promoter to respond to stimulation by VLF-1 in transient-expression assays. Our data also suggest that VLF-1 interacts with the burst sequence in vitro.
MATERIALS AND METHODS
Cell line, plasmids, and recombinant viruses.
The Spodoptera frugiperda (fall armyworm) IPLB-SF-21 (SF-21) cell line (23) was grown at 27°C in TC-100 medium (GIBCO/BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (Intergen, Purchase, N.Y.) and 0.26% tryptose broth (15).
phcwt, phcLSXVII, phcLSVI, and phcLSVII contain a chloramphenicol acetyltransferase (CAT)-encoding gene driven by a wild-type (wt) or linker scan mutant polh promoter (14, 19). pCAPCAT contains a vp39 promoter-driven cat gene (21). pXA76.9 (28) and pXA76.9d (27) contain wt vlf-1 or frameshift mutant vlf-1 under the control of the p6.9 promoter.
p10hcBS, p10hc-81, p10hc-47, and p10hc-17 contain a cat gene driven by a wt or linker scan mutant p10 promoter (Fig. 1). p10hcBS was constructed by moving the KpnI-HindIII fragment of plasmid p10hc (22) containing the p10-promoted cat gene into pBluescriptII KS(+) (Stratagene, La Jolla, Calif.) between the SmaI and HindIII sites. Plasmids p10hc-81, p10hc-47, and p10hc-17 were constructed by site-directed mutagenesis (2) using p10hcBS as the template plasmid and oligomers P10hc-81 (5′-CTTATTTAACTATCCGGATCCGTGTTGGGTTG-3′), P10hc-47 (5′-GTATTTTAATTAATATGGCTCGAGATTGATAATAATTC-3′), and P10hc-17 (5′-GTAAATAAAATGTGCGGCCGCGTATAGTATTTTAA-3′), respectively, as mutagenic primers. pETvlf1 was constructed by inserting the intact vlf-1 open reading frame (ORF) between the ClaI and BamHI sites of pET-15b (Novagen, Madison, Wis.) so that a 6×His tag was fused to the N terminus of vlf-1. pETvlf1ΔSstIII is identical to pETvlf1, except that it lacks the sequence between the SstI and SstII sites in the vlf-1 ORF. Thus, pETvlf1ΔSstIII contains a 6×His-tagged vlf-1 truncation that encodes the first 151 residues of VLF-1.
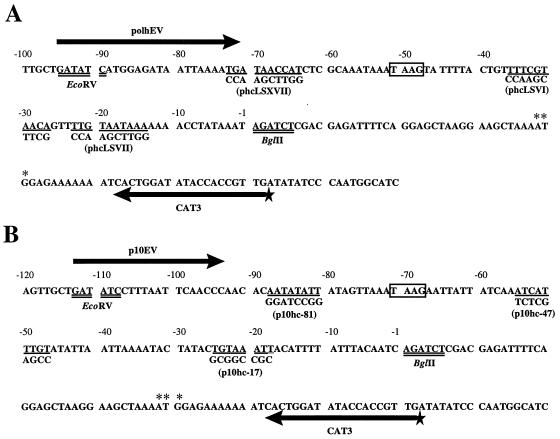
FIG. 1.
Sequences of the polh and p10 promoter regions within reporter plasmids. Plasmids phcwt and p10hcBC contain the cat gene driven by wt polh (A) and p10 (B) promoter sequences, respectively. Sequences modified by linker scan mutations are indicated by single underlining, and the mutant sequences and names of the corresponding reporter plasmids are shown below the mutated sequences. TAAG sequences are boxed. Restriction sites are doubly underlined. The numbering above the sequences is relative to the original translational initiation sites of the polh and p10 genes. In both cases, the ATG has been modified to a BglII site so that only the A (+1) remains. The translation initiation codon of the cat gene is marked by asterisks. Thick arrows indicate the positions of primers used to generate probes by PCR for DNase I footprint assays. The radioactively labeled 5′ end of the CAT3 primer is indicated by a star.
vcFgvlf1 is a recombinant of A. californica multicapsid nuclear polyhedrosis virus with a FLAG epitope tag inserted at the C terminus of the vlf-1 ORF (28).
Transient-expression assays.
Transient-expression assays used to assess the ability of mutant polh and p10 promoters to respond to stimulation by VLF-1 were similar to those described previously (22). In each transfection, 2 μg of the reporter plasmid and 0.5 μg of any additional plasmid were introduced into 2 × 106 SF-21 cells using Lipofectin (GIBCO/BRL) (15). CAT assays were performed 72 h posttransfection as described previously (4). A PhosphorImager 4000 (Molecular Dynamics, Sunnyvale, Calif.) was used for quantification.
Protein expression and purification.
SF-21 cells (40 × 106) were infected with recombinant virus vcFgvlf1 at a multiplicity of infection of 20 PFU per cell (15) and lysed 24 h postinfection with 5 ml of 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–1% Nonidet P-40. Cleared cell lysates were incubated with 200 μl of anti-FLAG M2 monoclonal antibody affinity gel (Kodak, New Haven, Conn.) with gentle agitation at 4°C for 4 h. The gel pellets were washed three times with 50 mM Tris-HCl (pH 8.0)–50 mM NaCl. Bound proteins were eluted with 100 μl of 200-μg/ml FLAG peptide (Kodak) in 10 mM Tris-HCl (pH 7.4)–150 mM NaCl. Glycerol was added to the eluates to a final concentration of 20%. Purified FLAG–VLF-1 was diluted with 10 mM Tris-HCl (pH 7.4)–150 mM NaCl–20% glycerol when necessary.
To express 6×His-tagged vlf-1 in Escherichia coli, strain BL2(DE3)pLysS (Novagen) was transformed with pETvlf1 or pETvlf1ΔSstIII. A single colony was picked to inoculate 2 ml of Luria-Bertani growth medium containing 50-μg/ml ampicillin and 34-μg/ml chloramphenicol and grown at 30°C overnight. The overnight cell culture was used to inoculate, at a 1:100 dilution, 50 ml of Luria-Bertani medium containing ampicillin and chloramphenicol. Cells were grown at 30°C until the optical density at 600 nm reached 0.5 to 0.6 and induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. Cells were harvested 3 h later and lysed by freezing and thawing in 5 ml of 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–1% Nonidet P-40. The supernatant of the cell lysate was incubated with 100 μl of Ni-nitrilotriacetic acid (NTA) resin (QIAGEN, Chatsworth, Calif.) at 4°C with gentle shaking for 4 h. The resin was washed three times with 50 mM Tris-HCl (pH 8.0)–50 mM NaCl before elution with 120 μl of 250 mM imidazole–10 mM Tris-HCl (pH 7.4)–150 mM NaCl. Glycerol was added to the eluate to 20%.
Immunoblot analysis.
Proteins of the FLAG–VLF-1 preparation were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and transferred to nylon membranes (Millipore). Blots were blocked in TBST buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Tween 20) in the presence of 5% nonfat dried milk, probed first with a 1:5,000 dilution anti-FLAG M2 monoclonal antibody and then a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G in TBST buffer, and visualized with an enhanced-chemiluminescence Western blot kit (Amersham).
Gel mobility shift assays.
To prepare radioactively labeled promoters, plasmids phcwt and p10hcBC were digested with EcoRV and BglII (Fig. 1). The BglII site was filled in with [α-32P]dATP by using T4 DNA nucleotide polymerase, and the DNA fragments containing the polh and p10 promoters were gel purified. A 1-μl sample of purified FLAG–VLF-1 was mixed with 10 μl of binding buffer (10 mM HEPES-NaOH [pH 7.9], 50 mM KCl, 0.1 mM EDTA, 10% glycerol) containing 50 ng of poly(dI-dC) and 20,000 cpm of the promoter-containing fragment (0.56 and 0.35 ng of the polh and p10 promoters, respectively). The mixtures were kept at room temperature for 20 min before being analyzed by electrophoresis in 0.5× Tris-borate-EDTA–6% polyacrylamide gels. For the supershift reactions, 1 μl of anti-FLAG M2 monoclonal antibody was added and the mixture was incubated for an additional 5 min before electrophoresis. For competition experiments, EcoRI-linearized pBluescript was used as a nonspecific competitor, while specific competitors were the same DNA fragments used as probes but lacked the radiolabel. Competition experiments were performed under the same conditions as described above with various amounts of competitors added simultaneously with probes.
DNase I protection assays.
Radioactively labeled polh, p10, and vp39 promoter fragments were made by PCR using plasmids phcwt, p10hcBC, and pCAPCAT as templates and primers polhEV, p10EV, and CAP104, respectively, plus a γ-32P-labeled CAT3 primer (Fig. 1). A footprint assay was initiated by adding 15 μl of the purified VLF-1 fusion (9.45 μg of FLAG–VLF-1 or 11.24 μg of 6×His–VLF-1) to 35 μl of 15 mM HEPES-NaOH (pH 7.9)–75 mM KCl–0.15 mM EDTA–15% glycerol containing 30,000 cpm of promoter fragment and allowing the binding reaction to proceed at room temperature for 20 min. A 50-μl volume of 10 mM MgCl2–5 mM CaCl2 was added to each reaction mixture, which was then cooled on ice for 5 min. The probe was digested with 1 μl of RQ1 RNase-free DNase I (Promega, Madison, Wis.) at 4°C for 5 min. The digestion was terminated by adding 90 μl of 200 mM NaCl–20 mM EDTA–1% sodium dodecyl sulfate–20-μg/ml single-stranded salmon sperm DNA. The DNA was purified by phenol-chloroform extraction and analyzed on a sequencing gel. A sequence ladder of the corresponding template plasmid was generated in parallel with the same radioactively labeled CAT3 primer for use as a marker.
RESULTS
Involvement of the burst sequence in the stimulation of expression from the polh and p10 promoters by vlf-1 in transient-expression assays.
To determine the region(s) of very late promoters involved in VLF-1 stimulation, we examined the effects of linker scan mutations on the ability of the polh and p10 promoters to respond to transcriptional stimulation by VLF-1 in transient-expression assays (22). Reporter plasmids containing a cat gene driven by a wt or mutant promoter were cotransfected into SF-21 cells with a set of genomic clones collectively containing all of the late expression factor genes (lef) required for late gene expression in transient-expression assays (9, 20) but lacking vlf-1. Either a truncated version of vlf-1 or wt vlf-1 was supplied in the transfections on a separate plasmid. The difference between CAT expression levels in the presence of wt or mutant vlf-1 was a reflection of the sensitivity of the promoter to VLF-1 stimulation. In these assays, both wt vlf-1 and mutant vlf-1 were under the control of the p6.9 promoter, which provided elevated expression of vlf-1 and greater stimulation of expression from very late reporter plasmids (27).
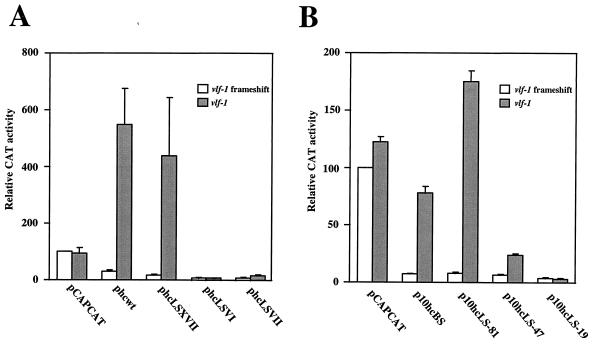
Mutant polh and p10 promoters with 10-bp linker mutations introduced at selected positions (Fig. 1) were examined and compared to wt promoters. While the late vp39 promoter (pCAPCAT) was not affected by addition of functional vlf-1 as expected, the wt polh promoter (phcwt) was stimulated approximately 20-fold in the presence of wt vlf-1 (Fig. 2 and Table 1). A linker scan mutation 13 nucleotides (nt) upstream of the polh TAAG (phcLSXVII) was also stimulated approximately 20-fold by VLF-1. Mutations 12 nt (phcLSVI) or 25 nt (phcLSVII) downstream of the TAAG reduced the level of expression approximately eightfold in the absence of wt vlf-1 but either abolished the response of the mutant polh promoter (phcLSVI) to VLF-1 stimulation or reduced the response to only twofold (phcLSVII) (Table 1). The p10 promoter series showed a similar pattern. The wt p10 promoter (p10hcBS) and the mutant p10 promoter with a linker 10 nt upstream of the TAAG (p10hc-81) were stimulated 10- and 22-fold by VLF-1. Mutations 13 nt (p10hc-47) or 43 nt (p10hc-17) downstream of the p10 TAAG were stimulated only three- to fourfold or not affected at all by VLF-1, respectively. The fact that the response of the polh and p10 promoters to stimulation by VLF-1 is severely impaired by mutations in their burst sequences suggests that VLF-1 exerts its effects through these sequences.
FIG. 2.
Response of mutated very-late promoters to stimulation by VLF-1 in transient-expression assays. All transfections included genomic clones BC5, HL8, HL5, ETL7, PstH4, PstH1, HC10, XmaB, HK5, IE15, and pSDEM2, which collectively supplied all of the lef genes required for late gene expression in transient-expression assays (22). A plasmid carrying p6.9 promoter-driven frameshift mutant vlf-1 (pXA76.9d) or wt vlf-1 (pXA76.9) was also transfected into the cells. The reporter plasmid used in each pair of transfections is shown at the bottom. The positions of their mutated sequences are shown in Fig. 1. pCAPCAT carries a late vp39 promoter-driven cat gene and served as a negative control. Cells were harvested 72 h after transfection, and levels of CAT activity were determined. The CAT activity of the transfection with pCAPCAT and frameshift mutant vlf-1 was arbitrarily set as 100%. Relative CAT activities of other transfections were calculated based on this standard. Standard errors were determined from results of triplicate experiments. (A) polh promoters. (B) p10 promoters.
TABLE 1.
Responses of promoters to VLF-1 stimulationa
| Expt no. and vlf-1 construct |
polh-promoted reporter plasmids
|
p10-promoted reporter plasmids
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pCAPCAT | phcwt | phcLSXVII | phcLSVI | phcLSVII | pCAPCAT | p10hcBS | p10LS-81 | p10LS-47 | p10LS-17 | |
| 1 | ||||||||||
| vlf-1 frameshift | 100 | 24.4 | 10.8 | 3.4 | 3.7 | 100 | 7.5 | 5.8 | 5.3 | 5.6 |
| vlf-1 | 132 | 441 | 128 | 4.9 | 10 | 114 | 66.7 | 159 | 21.4 | 4.8 |
| 2 | ||||||||||
| vlf-1 frameshift | 100 | 25.6 | 17.2 | 6.7 | 11.3 | 100 | 7.9 | 8.6 | 7.6 | 2.4 |
| vlf-1 | 76 | 803 | 828 | 6.4 | 19 | 125 | 85.5 | 191 | 26.7 | 1.9 |
| 3 | ||||||||||
| vlf-1 frameshift | 100 | 40.2 | 22.1 | 6.4 | 6.8 | 100 | 6.9 | 9.4 | 6.9 | 3.1 |
| vlf-1 | 74 | 402 | 361 | 5.6 | 18.1 | 129 | 81.8 | 176 | 23.6 | 2.4 |
For details, see the legend to Fig. 2. Each experiment was done three times, and percent CAT activities relative to that of the pCAPCAT control were determined.
Interaction between VLF-1 and the burst sequences of the polh and p10 promoters.
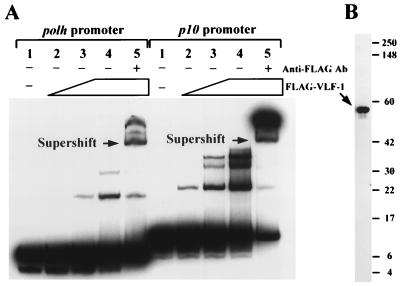
Based on these results, we explored the possibility that VLF-1 physically interacts with very late promoters. Gel electrophoresis mobility shift assays provided the first evidence of such interaction (Fig. 3). DNA fragments containing the polh promoter (from −1 to −92) and the p10 promoter (from −1 to −107) were radioactively labeled for use as probes. VLF-1 tagged with a FLAG epitope at the C terminus was purified from SF-21 cells infected with vcFgvlf1, a recombinant virus expressing the FLAG–vlf-1 fusion (28). As increasing amounts of purified FLAG–VLF-1 were added to the probes, two FLAG–VLF-1–polh promoter complexes and three FLAG–VLF-1–p10 promoter complexes were formed. The presence of FLAG–VLF-1 in these complexes was verified by the observations that they could be supershifted by the addition of a monoclonal antibody against the FLAG epitope (Fig. 3A, lanes 5) and that FLAG–VLF-1 was the only protein recognized by the anti-FLAG antibody in the preparation (Fig. 3B).
FIG. 3.
Gel mobility shift assays showing interactions between very late promoters and FLAG–VLF-1. (A) Complex formation of the polh and p10 promoters with FLAG–VLF-1. The polh and p10 promoters were cut from plasmids phcwt and p10hcBC, respectively, with EcoRV and BglII (Fig. 1), end labeled, and gel purified. Each probe was incubated with increasing amounts of partially purified FLAG–VLF-1: 0, 0.16, 0.32, 0.64, and 0.64 μg of total protein were used in lanes 1, 2, 3, 4, and 5 with the polh promoter, respectively, and 0, 0.04, 0.08, 0.16, and 0.16 μg of FLAG–VLF-1 were used in lanes 1, 2, 3, 4, and 5 with the p10 promoter, respectively. The resulting complexes were resolved by a Tris-borate-EDTA–6% polyacrylamide gel. Lanes 5 in each panel also included a monoclonal antibody (Ab) against the FLAG epitope to form antibody–FLAG–VLF-1 complexes and further altered the mobility of the DNA probes (supershifted DNA). (B) Western blot analysis showing cross-reactivity of the FLAG–VLF-1 preparation with the anti-FLAG monoclonal antibody. A 1.28-μg sample of total protein was analyzed. Molecular masses (in kilodaltons) of standard proteins are indicated on the right.
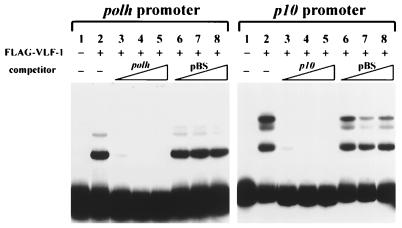
The specificity of interaction between FLAG–VLF-1 and very-late promoters was investigated by competition gel shift assays. Interaction of FLAG–VLF-1 with the probes was efficiently inhibited by addition of a 20- to 80-fold excess of the same DNA fragments as the probes (Fig. 4, lanes 3, 4, and 5). The same amounts of linearized pBluescript DNA had no significant effect on complex formation between FLAG–VLF-1 and the polh or p10 promoter (Fig. 4, lanes 6, 7, and 8), suggesting that interaction of FLAG–VLF-1 with very late promoters is specific.
FIG. 4.
Competition gel shift assays showing the specificity of interactions between very late promoters and FLAG–VLF-1. Lanes 3, 4, and 5 contained 20×, 40×, and 80× specific competitors (the polh or p10 promoter). Lanes 6, 7, and 8 contained 20×, 40×, and 80× nonspecific competitors (linearized pBluescript). Competitors were mixed with probes before addition to the reactions. Other conditions were as described in the legend to Fig. 3.
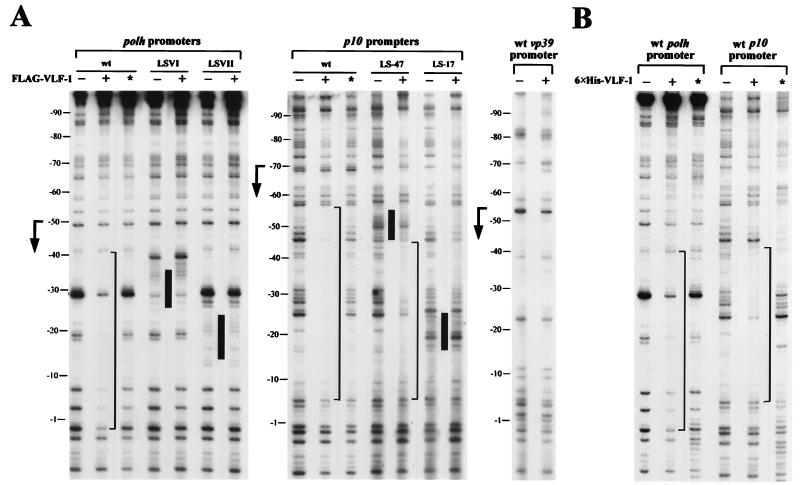
To confirm the interaction of FLAG–VLF-1 with very late promoters and to locate the regions within the polh and p10 promoters which were involved in complex formation, we performed DNase I protection assays. The polh and p10 promoters, labeled on the template strands, were incubated with partially purified C-terminally tagged VLF-1 and then subjected to DNase I digestion. As shown in Fig. 5A, the wt polh promoter was protected from −1 to −40 and the wt p10 promoter was protected from −5 to −56 in the presence of FLAG–VLF-1, which was produced in insect cells and purified with anti-FLAG M2 affinity gels. Neither the polh nor the p10 promoter was protected by equivalent extracts from wt virus-infected cell lysates lacking FLAG–VLF-1 (Fig. 5A, lanes with asterisks), indicating that the observed footprints were not due to proteins binding nonspecifically to the column. The vp39 promoter, a late promoter, was not protected by FLAG–VLF-1 (Fig. 5A).
FIG. 5.
Mapping of the binding site of VLF-1 by DNase I footprint assays. polh promoters, p10 promoters, and the vp39 promoter were amplified by PCR using primer polhEV (5′-GATATCATGGAGATAATTAAAATG-3′), p10EV (5′-GATATCCTTTAATTCAACCC-3′), or CAP104 (5′-GAATTTAAAATTTTATACAAC-3′) plus radioactively labeled primer CAT3 (5′-CAACGGTGGTATATCCAGTG-3′), respectively (Fig. 1). The wt, LSVI, and LSVII polh promoters were amplified from plasmids phcwt, phcLSVI, and phcLSVII (Fig. 1), respectively. The wt, −47, and −17 p10 promoters were amplified from plasmids p10hc, p10hc-47, and p10hc-17 (Fig. 1), respectively. Labeled promoters were digested with DNase I after incubation with purified FLAG–VLF-1 and analyzed on a sequencing gel. The markers on the left were determined by alignment with a sequence ladder generated with primer CAT3. Footprints are marked by brackets. Positions of mutations are indicated by black bars between lanes. (A) Protection of late and very late promoters by FLAG–VLF-1. vcFgvlf1, a recombinant virus expressing a FLAG-vlf-1 fusion, was used to infect SF-21 cells for production of FLAG–VLF-1, which was partially purified with an anti-FLAG M2 affinity gel. As a negative control, lysate from wt virus-infected cells was processed in parallel and used to protect the DNA probes (lanes marked with asterisks). (B) Protection of wt polh and p10 promoters by 6×His–VLF-1. Full-length 6×His-tagged VLF-1 and truncated 6×His–VLF-1 (lanes with asterisks) were partially purified from bacteria expressing plasmid-borne genes by using Ni-NTA resin and used to protect probes.
Similar footprints on very-late promoters were also revealed by protection with 6×His–VLF-1 that was produced in E. coli cells and purified with Ni-NTA resin (Fig. 5B). The footprint on the polh promoter is indistinguishable from that resulting from interaction with insect cell-derived FLAG–VLF-1 (Fig. 5A). The p10 promoter was protected by 6×His–VLF-1 from −5 to −45 (Fig. 5B), a region slightly shorter than that protected by FLAG–VLF-1. A 6×His–VLF-1 truncation containing the N-terminal two-fifths of VLF-1 was generated in parallel with 6×His–VLF-1 as a control and did not produce these footprints, although a small region (from −40 to −56) appeared to be protected. The basis for this other footprint is not clear. The fact that two preparations of full-length VLF-1 produced in completely different expression systems and purified by different affinity methods resulted in footprints at the same locations strongly suggests that VLF-1 is involved in the formation of these footprints.
Since the protected sites constitute the majority of the burst sequences, we examined interactions between FLAG–VLF-1 and mutant polh or p10 promoters containing linker scan mutations in their burst sequences. Three of the four examined promoters with mutated burst sequences were no longer protected by FLAG–VLF-1 under these conditions (Fig. 5A). Only a weaker and shorter footprint (from −10 to −44) was detected for the fourth one, the p10 (−47) promoter with a 10-bp mutation 13 nt downstream of the TAAG (Fig. 5A). These data indicate that the ability of VLF-1 to interact with burst sequences of very late promoters depends on the sequence of the burst sequence and correlates with VLF-1’s ability to transactivate expression from the promoters.
DISCUSSION
We have examined the ability of mutant very late promoters to respond to VLF-1. While the wt polh promoter and a mutant promoter with a linker mutation upstream of the TAAG sequence were stimulated approximately 20-fold by VLF-1 in transient-expression assays, the two mutant promoters with linker mutations in their burst sequences either failed to respond or responded only slightly to VLF-1 stimulation. Since VLF-1 has previously been shown to influence the level and timing of polh expression (11, 27), this observation indicates that the effect of VLF-1 is exerted through the burst sequence. Although the p10 promoter had not been mutationally analyzed as extensively as the polh promoter previously, our analysis of linker mutations in the p10 promoter confirmed the presence of a burst sequence between the TAAG and the translational initiation codon and further showed that VLF-1 stimulates expression from the p10 promoter through the burst sequence.
The connection between the burst sequence and VLF-1 transactivation is further supported by our observation that VLF-1 may physically interact with the polh and p10 promoters through their burst sequences. DNase I protection assays detected similar footprints within the burst sequences of both polh and p10 promoters when different VLF-1 fusion preparations generated in insect and E. coli cells were used. Thus, it appears that VLF-1 is able to bind to very-late promoters, although the possibility cannot be excluded that this interaction is facilitated by factors in infected insect cells. The observation that VLF-1 partially purified from insect cells resulted in a larger footprint on the p10 promoter than the VLF-1 partially purified from E. coli cells (Fig. 5) may be an indication that other factors copurifying with VLF-1 participated in the protection. Multiple VLF-1-containing complexes were detected in gel shift assays, and they may represent involvement of other factors or multimerization of VLF-1. The large size of the protected region (40 to 45 bp) suggests that VLF-1 binds as a multimer. Since VLF-1 is a relative of the λ phage integrase family (11, 13, 28), the members of which usually form tetramers upon association with DNA, it would not be surprising to find that VLF-1 binds to the burst sequence as a tetramer. Binding of VLF-1 to the burst sequence may be synergistic, since VLF-1 may form multimers and activation of the polh promoter requires a threshold level of VLF-1 (27). Although the burst sequences of both the polh and p10 promoters are capable of interacting with VLF-1, there is no obvious similarity between them, nor are there short consensus sequences reminiscent of the λ phage integrase binding sites. Both promoters are AT rich, which may facilitate VLF-1 complex formation. Interaction of VLF-1 with the p10 promoter is more readily detected than that with the polh promoter in both gel shift assays and DNase I protection assays, suggesting that VLF-1 has higher affinity to the burst sequence of the p10 promoter.
Mutational analyses show that complex formation of VLF-1 with the burst sequences of the polh and p10 promoters is closely correlated with transactivation of these promoters by VLF-1. In most cases, mutations in the burst sequences disrupted VLF-1 complex formation and also abrogated the responses of the corresponding very late promoters to stimulation by VLF-1. One of the p10 promoter mutants (−47) bound partially to VLF-1 and responded partially to VLF-1 stimulation. These data provide strong support for the view that the interaction of VLF-1 with the burst sequences of these promoters is essential for VLF-1 transactivation.
Both late and very late genes are transcribed by a novel virus-induced RNA polymerase (1, 3, 5, 7) whose major components are four viral gene products, LEF-8, LEF-9, LEF-4, and p47 (6). LEF-8 and LEF-9 have sequence motifs found in subunits of cellular RNA polymerases (8, 16). In vitro transcription studies suggest that the polymerase responsible for late gene transcription is biochemically distinguishable from the polymerase responsible for very late gene transcription, although the two activities may differ by only a subunit (26). Both late and very late promoters require a polymerase which can recognize and initiate at a TAAG sequence, while very late promoters apparently require an additional factor which can recognize the burst sequence. It is possible that interaction of VLF-1 with the burst sequence facilitates the recruitment of RNA polymerase to the promoter or stabilizes the transcription complex and thereby enhances transcription initiation. The interaction of VLF-1 with components of the very-late transcription complexes remains to be demonstrated.
Our current model is that very-late promoters have TAAG sequences which are in relatively poor contexts for polymerase initiation during the late phase, when VLF-1 levels are low. When VLF-1 accumulates above a threshold level (27), binding of VLF-1 to the burst sequence becomes sufficiently potent and facilitates polymerase interaction with and initiation from the TAAG sequence. Other very late factors may be involved in the interaction of VLF-1 with the burst sequence or with the RNA polymerase. However, no such additional factors have been identified in transient-expression assays (22), and regulation of polh expression is governed by the level of VLF-1 (27). Thus, VLF-1 seems to be the primary factor in the regulation of very late gene expression, and it appears to act by interaction with the burst sequence.
ACKNOWLEDGMENTS
We thank Yonghong Li, Jeanne McLachlin, Janet Hatt, Alex Harvey, Jeff Rapp, Domagoj Vucic, and Joyce Wilson for help and advice.
This work was supported in part by Public Health Service grant AI23719 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Beniya H, Funk C J, Rohrmann G F, Weaver R F. Purification of a virus-induced RNA polymerase from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells that accurately initiates late and very late transcription in vitro. Virology. 1996;216:12–19. doi: 10.1006/viro.1996.0029. [DOI] [PubMed] [Google Scholar]
- 2.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;48:641–646. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grula M A, Buller P L, Weaver R F. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J Virol. 1981;38:916–921. doi: 10.1128/jvi.38.3.916-921.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huh N E, Weaver R F. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1990;71:195–202. doi: 10.1099/0022-1317-71-1-195. [DOI] [PubMed] [Google Scholar]
- 8.Lu A, Miller L K. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J Virol. 1994;68:6710–6718. doi: 10.1128/jvi.68.10.6710-6718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mans R M, Knebel-Morsdorf D. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J Virol. 1998;72:2991–2998. doi: 10.1128/jvi.72.4.2991-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLachlin J R, Miller L K. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris T D, Miller L K. Mutational analysis of a baculovirus major late promoter. Gene. 1994;140:147–153. doi: 10.1016/0378-1119(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 13.Nunes-Duby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooi B G, Rankin C, Miller L K. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J Mol Biol. 1989;210:721–736. doi: 10.1016/0022-2836(89)90105-8. [DOI] [PubMed] [Google Scholar]
- 15.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 16.Passarelli A L, Todd J W, Miller L K. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J Virol. 1994;68:4673–4678. doi: 10.1128/jvi.68.7.4673-4678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Possee R D, Howard S C. Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1987;15:10233–10248. doi: 10.1093/nar/15.24.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, Liu A, Weaver R F. Studies on the control region of the p10 gene of the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1989;70:1273–1279. doi: 10.1099/0022-1317-70-5-1273. [DOI] [PubMed] [Google Scholar]
- 19.Rankin C, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene. 1988;70:39–50. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 20.Rapp J C, Wilson J A, Miller L K. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol. 1998;72:10197–10206. doi: 10.1128/jvi.72.12.10197-10206.1998. . . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiem S M, Miller L K. Differential gene expression mediated by late, very late, and hybrid baculovirus promoters. Gene. 1990;91:87–94. doi: 10.1016/0378-1119(90)90166-o. [DOI] [PubMed] [Google Scholar]
- 22.Todd J W, Passarelli A L, Lu A, Miller L K. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J Virol. 1996;70:2307–2317. doi: 10.1128/jvi.70.4.2307-2317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 24.Weyer U, Possee R D. Analysis of the promoter of the Autographa californica nuclear polyhedrosis virus p10 gene. J Gen Virol. 1989;70:203–208. doi: 10.1099/0022-1317-70-1-203. [DOI] [PubMed] [Google Scholar]
- 25.Weyer U, Possee R D. Functional analysis of the p10 gene 5′ leader sequence of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1988;16:3635–3654. doi: 10.1093/nar/16.9.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu B, Guarino L A. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J Virol. 1995;69:2912–2917. doi: 10.1128/jvi.69.5.2912-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Miller L K. Control of baculovirus polyhedrin gene expression by very late factor 1 gene. Virology. 1998;248:131–138. doi: 10.1006/viro.1998.9272. [DOI] [PubMed] [Google Scholar]
- 28.Yang S, Miller L K. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology. 1998;245:99–109. doi: 10.1006/viro.1998.9152. [DOI] [PubMed] [Google Scholar]