Abstract
In humans, many diseases are associated with alterations in gut microbiota, namely increases or decreases in the abundance of specific bacterial groups. One example is the genus Faecalibacterium. Numerous studies have underscored that low levels of Faecalibacterium are correlated with inflammatory conditions, with inflammatory bowel disease (IBD) in the forefront. Its representation is also diminished in the case of several diseases, including colorectal cancer (CRC), dermatitis, and depression. Additionally, the relative presence of this genus is considered to reflect, at least in part, intestinal health status because Faecalibacterium is frequently present at reduced levels in individuals with gastrointestinal diseases or disorders. In this review, we first thoroughly describe updates to the taxonomy of Faecalibacterium, which has transformed a single-species taxon to a multispecies taxon over the last decade. We then explore the links discovered between Faecalibacterium abundance and various diseases since the first IBD-focused studies were published. Next, we examine current available strategies for modulating Faecalibacterium levels in the gut. Finally, we summarize the mechanisms underlying the beneficial effects that have been attributed to this genus. Together, epidemiological and experimental data strongly support the use of Faecalibacterium as a next-generation probiotic (NGP) or live biotherapeutic product (LBP).
Keywords: Faecalibacterium, keystone, next-generation probiotic, live biotherapeutic product, anaerobe, commensal, inflammation
The authors describe (i) updates to the taxonomy of Faecalibacterium, which has transformed a single-species taxon to a multispecies taxon over the last decade, (ii) the links that have been discovered between Faecalibacterium abundance and various diseases since the first IBD-focused studies were published, and (iii) current available strategies for modulating Faecalibacterium levels in the gut.
Introduction
Faecalibacterium is a genus of strictly anaerobic, extremely oxygen-sensitive (EOS), Gram-positive, rod-shaped, nonmotile, and nonspore-forming bacteria (Duncan et al. 2002a). Initially, it was thought to account for 5% of the human microbiota (Qin et al. 2010). Based on a metagenomic analysis of over 7900 human samples, a more recent study has suggested the mean and median may be 6.5% and 4.8%, respectively, in adults and could may even be as high as 75% (De Filippis et al. 2020). These results confirm the high relative abundance of this genus in the human gut. Moreover, Faecalibacterium is prevalent in human populations across the world—it was detected in 85% of gut samples (De Filippis et al. 2020)—and members of Faecalibacterium are considered to be ubiquitous in the gastrointestinal tracts (GITs) of healthy humans (Tap et al. 2009). Research also indicates that Faecalibacterium levels differ with age and potentially gender, where abundances are lower in women than in men (Aguirre de Carcer et al. 2011). Additionally, the prevalence of this genus is less pronounced in newborns, children, and the elderly (De Filippis et al. 2020). It is first detected around 6–7 months of age and persists at a low level until 2–3 years of age (Hopkins et al. 2005). This pattern in early infancy suggests that there must be a first wave of gut colonization for Faecalibacterium to become established. Indeed, the implantation of EOS bacteria depends on certain physicochemical conditions that are generated by other commensal bacteria (Tomas et al. 2013). Faecalibacterium abundance is higher in non-Westernized populations, highlighting its ancient synergistic relationship with humans that has only recently been disrupted by modern lifestyles (De Filippis et al. 2020). It has also been hypothesized that Faecalibacterium might act to stabilize the gut microbiota. Support for this idea was found in a study that monitored the microbiota of healthy human subjects over the course of a year: the abundance of this genus was negatively correlated with greater intraindividual variability in microbiota composition (Olsson et al. 2022), suggesting its potential role as a keystone taxon (Tudela et al. 2021). In this vein, Faecalibacterium is among the first genus of commensal bacteria to have been found to differ between individuals with and without inflammatory bowel disease (IBD), and its beneficial role appears to be associated with its anti-inflammatory properties (Sokol et al. 2008).
The genus Faecalibacterium
From a single species to multiple species
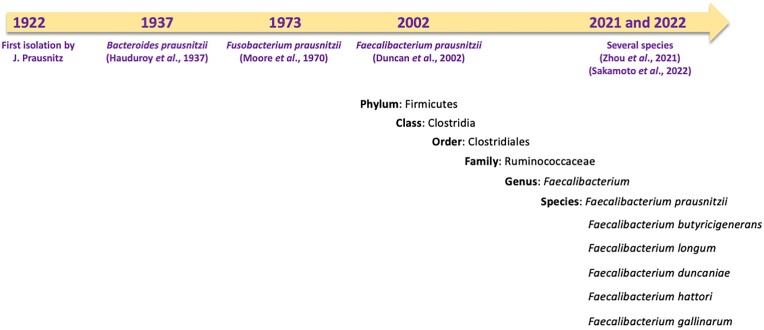
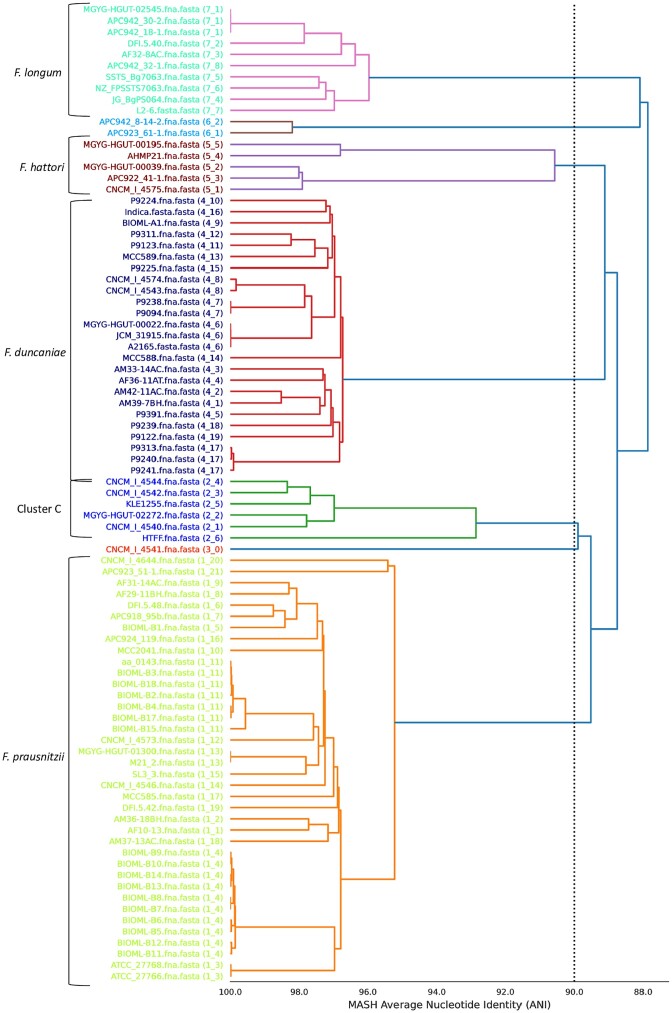
Faecalibacterium has undergone several taxonomic shifts since its discovery in 1922 by J. Prausnitz (Prausnitz 1922). Initially classified as Bacteroides prausnitzii in 1937 by Hauduroy et al. (1937), it was reclassified as Fusobacterium prausnitzii in 1974 (Cato et al.1974) and then as Faecalibacterium prausnitzii in 2002 (Duncan et al. 2002a) (Fig. 1). Initially, F. prausnitzii was the only member of the genus and was represented by two cultured strains—ATCC 27768 (the type strain) and ATCC 27766—as well as by two newly discovered strains (A2-165 and L2-6) (Duncan et al. 2002a). Despite its high prevalence and abundance, it has been challenging to isolate and culture this genus because its EOS status. In recent years, it has been possible to isolate Faecalibacterium strains using classical strategies, such as microbiological flow charts (Martin et al. 2017), or more complex strategies, including flow cytometry-assisted sorting under anaerobic conditions using anti-Faecalibacterium polyclonal antibodies (Bellais et al. 2022). Thanks to advances in anaerobic cultivation and next-generation sequencing techniques, the complexity of the genus has progressively been deciphered. First, a 16S rRNA gene-based approach divided F. prausnitzii into two phylotypes, which accounted for 97.9% of the amplified sequences found across several studies (Lopez-Siles et al. 2012). In 2017, an analysis of 17 Faecalibacterium isolates led to the identification of three phylotypes (A, B, and C) (Benevides et al. 2017). However, several strains did not belong to any of these phylotypes, setting the basis for the future identification of additional species. The number of phylogroups increased as a result of research comparing the average nucleotide identity (ANI) of 35 genomes (Fitzgerald et al. 2018). In 2020, more than 7900 human and 200 nonhuman primate metagenomes were analyzed leading to the identification of 22 Faecalibacterium-like metagenome-assembled genomes (MAGs) (De Filippis et al. 2020). A total of 12 were specific to humans, distributed across the globe, and displayed a degree of age-related variation. The others were specific to other particular niches (De Filippis et al. 2020). In 2021, following genome sequencing and phenotypic, chemotaxonomic, and phylogenetic characterization, two new species of Faecalibacterium were described: Faecalibacterium butyricigenerans (type strain: AF52-21T) and Faecalibacterium longum (type strain: CM04-06T) (Zou et al. 2021). Finally, in 2022, three novel species were proposed: Faecalibacterium duncaniae (type strain: A2-165 of human origin), Faecalibacterium hattorii (type strain: APC922/41–1T of human origin), and Faecalibacterium gallinarum (type strain: ic1379T of chicken/broiler origin) (Sakamoto et al. 2022). The type strain for F. prausnitziii remains ATCC 27768. The complexity of this genus is still being uncovered: recent phylogenetical analysis revealed that several strains do not seem to belong to any of the above species (Fig. 2) (Tanno et al. 2022). The discovery of new phylogroups and species has expanded possibilities for studying and understanding the general characteristics of this genus and the role it plays in host health. In a recent study, Tanno et al. (2023) found that 16S rRNA is not a suitable gene marker for quantifying Faecalibacterium due to sequence divergence within clusters/strains and even among copies of a single strain, which could lead to biased results (Tanno et al. 2022). The same group recently proposed that rpoA may serve as a better gene marker for identifying Faecalibacterium spp. (Tanno et al. 2023). These important results indicate that the knowledge gathered prior to the discovery of these new phylogroups and species is inaccurate because adequate tools were lacking. They also signal that new analyses must be performed that account for genus diversity and that utilize appropriate gene markers, notably in the case of studies that used the 16S gene (Tanno et al. 2022).
Figure 1.
Faecalibacterium taxonomy and evolutionary timeline.
Figure 2.
ANI-value-based hierarchical clustering of Faecalibacterium genomes. Complete or draft genome sequences of Faecalibacterium strains were obtained from the NCBI database. The program dRep (version 3.2.2) was run using the 87 available Faecalibacterium genomes as input (Olm et al. 2017). ANI values of 95% or higher indicate that a genome pair likely belongs to the same species, whereas ANI values below 95% indicate that a genome pair likely represents different species. Cluster C corresponds to phylogroup C, as described elsewhere (Benevides et al. 2017).
General metabolism and metabolic cross-feeding
Acetate, propionate, and butyrate are among the most abundant short-chain fatty acids (SCFAs) in the human gut (Martin-Gallausiaux et al. 2021). They are the primary end products resulting from bacterial fermentation of dietary fibers, including plant cell wall polysaccharides, resistant starches, soluble oligosaccharides, and endogenous products (e.g. mucin). Additionally, some bacterial species can ferment amino acids and proteins to produce SCFAs and branched fatty acids. Butyrate is the main energy source of colonocytes and plays a central role in host physiology (Martin-Gallausiaux et al. 2021). The major end products of glucose fermentation by Faecalibacterium strains are formate, small amounts of d-lactate (l-lactate is undetectable), and large amounts of butyrate (>10 mM in vitro) (Duncan et al. 2002a, Miquel et al. 2013).
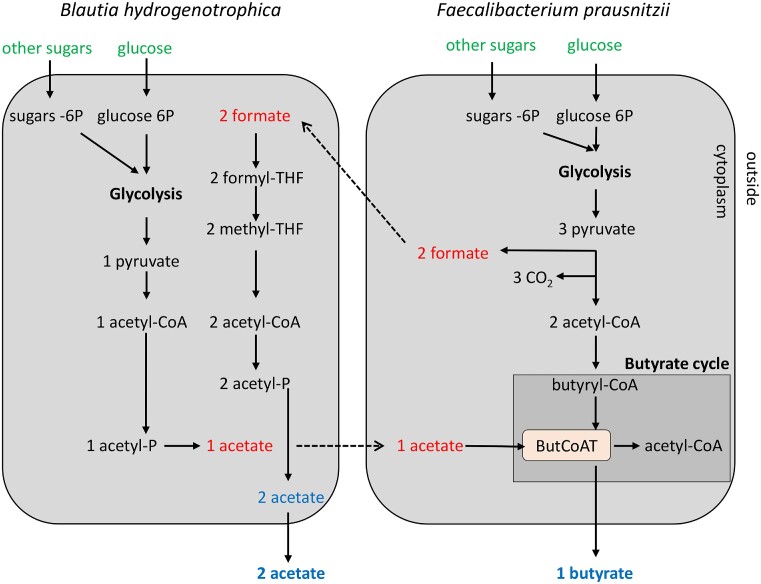
Cross-feeding occurs when a species metabolizes metabolites produced by another species (D’Souza et al. 2018). SCFAs in particular acetate, are among the most common cross-fed metabolites in the bacterial communities of the human gut (D’Souza et al. 2018). Acetate consumption is the major driver of butyrate production by members of Faecalibacterium genus (process known as acetate-cross feeding) in the healthy human gut (Miquel et al. 2013). The key enzyme in butyrate production is butyryl-CoA:acetate CoA-transferase (Fig. 3). It catalyzes the reaction whereby butyrate and acetyl-CoA are generated from extracellular acetate and intracellular butyryl-CoA, thus explaining the growth-promoting effects of acetate consumption (Duncan et al. 2002a,b, 2004, Charrier et al. 2006, Heinken et al. 2014). Indeed, supplementing the culture medium with acetate (33–50 mM) stimulates Faecalibacterium growth (Duncan et al. 2002a, Lopez-Siles et al. 2012, D’Hoe et al. 2018). In a coculture model, butyrate production by Faecalibacterium was increased via acetate cross-feeding with the acetate-producing bacteria Bifidobacteria adolescentis (Rios-Covian et al. 2015) and Blautia hydrogenotrophica (D’Hoe et al. 2018) (Fig. 3). In research using gnotobiotic rodents colonized by F. duncaniae and Bacteroidetes thetaiotaomicron, the phenomenon was observed in vivo by measuring butyrate and acetate levels in cecal samples (Wrzosek et al. 2013).
Figure 3.
Acetate and formate cross-feeding between B. hydrogenotrophica and F. prausnitzii. The metabolism of B. hydrogenotrophica centers on glycolysis and the acetate pathway. The metabolism of F. prausnitzii centers on glycolysis and the formate and butyrate pathways. The key enzyme in butyrate production is butyryl-CoA:acetate CoA-transferase (ButCoAT). In red are the SCFAs consumed (acetate, formate). In blue are the SCFAs produced (acetate, butyrate). In green are the sugars consumed.
Metabolic cross-feeding has been performed between F. duncaniae A2-165 and lactic acid bacteria (LAB) using F. duncaniae A2-165 cultured with cell-free supernatants (8% v/v) obtained from LAB grown in an acetate-rich, vitamin-poor medium (Lebas et al. 2020). A transcriptomics approach was used to characterize four LAB strains stimulating F. duncaniae A2-165 growth or delaying its lysis. These strains were Lactococcus lactis subsp. lactis CNCM I-1631, subsp. cremoris CNCMI-3558, Lactobacillus paracasei CNCM I-3689, and Streptococcus thermophilus CNCM I-3862. The results revealed that the supernatants had some shared effects, namely the upregulation of carbohydrate metabolism genes and cell wall-related genes as well as the downregulation of replication and mobilome genes. There were also LAB-specific effects. In particular, F. prausnitzii answer to the exposure of L. paracasei CNCM I-3689 supernatant, may stabilize cell wall formation, through the upregulation of some genes involved mainly in peptidoglycan formation, and the inhibition of cell wall degradation genes. This in vitro study suggests that LAB metabolites other than acetate may modify the metabolism of F. duncaniae A2-165.
While other members of the gut microbiome, such as B. thetaiotaomicron or Bifidobacterium, are known to be effective degraders and utilizers of polysaccharides, members of Faecalibacterium are less well-equipped to utilize complex carbon sources (Heinken et al. 2014). Indeed, the genome of F. duncaniae A2-165 (3.11 Mb) contains genes for just 31 glycoside hydrolases and one polysaccharide lyase. In contrast, the genome of B. thetaiotaomicron VPI 5482 contains genes for 255 glycoside hydrolases and 29 polysaccharide lyases (6.36 Mb). Moreover, glycolytic ability is likely strain dependent, as suggested by an in silico study (Blanco et al. 2019).
The second most relevant and best-documented compound involved in Faecalibacterium crossfeeding is fructose, which can result from the degradation of inulin, a dietary fiber and complex sugar (Ramirez-Farias et al. 2009). Even with limited carbohydrate-degradation machinery (Heinken et al. 2014), some Faecalibacterium members can metabolize inulin and pectin (Lopez-Siles et al. 2012, Rios-Covian et al. 2015, Martin et al. 2017). Research on inulin metabolism in Faecalibacterium was first explored in an interventional study where healthy volunteers took inulin supplements for 16 days. The result was an increase in populations of Faecalibacterium and B. adolescentis, with a concomitant increase in butyrate production (Ramirez-Farias et al. 2009). Under laboratory conditions, 9 of 11 strains were able to ferment inulin, but only two displayed efficient growth (Lopez-Siles et al. 2017). When Faecalibacterium metabolizes inulin, different products are released that can serve as substrates for other community members, thus contributing to broader-scale cross-feeding (Rios-Covian et al. 2015, Moens et al. 2016). Inulin degradation (glucosyl hydrolases) and uptake (ABC transporter) mechanisms were recently described in F. duncaniae A2-165 (Park et al. 2022). For this same strain, research has provided a molecular characterization of the phosphoenolpyruvate:carbohydrate phosphotransferase system transporter (PTS) involved in fructose uptake (EI,HPr and EIIABCFru, the general and specific PTS components, respectively) using the membranous protein extracts from cells grown on diverse carbohydrates and the purified PTS proteins (Kang et al. 2021). This result is consistent with the proteomic analysis of Park et al. (2022). Indeed, the fructose-specific PTS proteins were highly upregulated during growth in inulin (Park et al. 2022). Overall, the results suggest that teasing apart PTS functionality in Faecalibacterium could clarify how this taxon outcompetes other bacterial taxa in the human intestine.
Faecalibacterium can also take advantage of compounds generated by other members of the microbiome community, such as the products resulting from the degradation of ß-mannan complex polysaccharides, which are important components of hemicelluloses in plant cell walls. These products are released by metabolic processes in Bacteroides ovatus or Roseburia intestinalis, via a pathway encoded by specific, highly conserved genes with a worldwide distribution (Lindstad et al. 2021). Faecalibacterium is also able to metabolize the products of alginate metabolism released by Bacteroides species (Murakami et al. 2021).
Members of Faecalibacterium can also use other metabolites, such as N-acetylglucosamine, d-glucosamine, and d-glucuronic acid, allowing them to grow on substrates derived from their host, their host’s diet, and other bacteria (Lopez-Siles et al. 2012). Some Faecalibacterium can breakdown other, more unique compounds, such as avenanthramides—the phenolic alkaloids found in oats. Indeed, Faecalibacterium is the only known genus capable of transforming avenanthramides into dihydro-avenanthramides, bioactive compounds with anti-inflammatory and antioxidant properties in specific-pathogen free (SPF) and monocolonized mice (Wang et al. 2021).
Oxidative stress
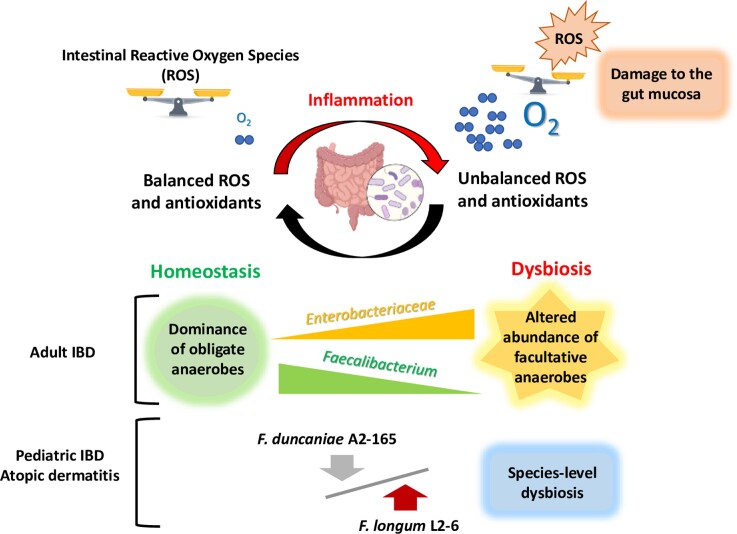
Although members of Faecalibacterium are considered to be EOS, F. duncaniae A2-165 can grow under low-oxygen conditions because it possesses an extracellular electron shuttle of flavins and thiols that transfers electrons to oxygen (O2) (Khan et al. 2012). This mechanism probably involves a flavin reductase, which might regenerate NAD+ from NADH and reduce O2 to H2O2 (Heinken et al. 2014). In addition, the health benefits of Faecalibacterium are directly related to the colon’s environmental conditions, given that O2 dynamics play a central role in intestinal homeostasis (Fig. 4). In the GIT of healthy individuals, O2 concentrations decrease longitudinally, from the stomach (∼5%) to the colon (0.1%–0.4%), and transversely, from the epithelial cells (5%) to the lumen (1%–2%) (Keeley and Mann 2019). Little is currently known about the relationship between variation in luminal O2 concentrations and intestinal diseases, including the subsequent effects on hosts. However, many gastrointestinal diseases are thought to be associated with high oxidative stress, which can influence luminal O2 levels and lead to dysbiosis (Singhal and Shah 2020). As chronic gut inflammation progresses, it can result in uncontrolled and persistent oxidative stress, where reactive oxygen species (ROS), such as H2O2, are overproduced and/or inadequately removed by antioxidant systems (Aviello and Knaus 2017, Burgueno et al. 2019). Thus, during gut inflammation, O2 and ROS levels may present a challenge for Faecalibacterium species. Indeed, one of the characteristics associated with IBD is a pronounced decrease in Faecalibacterium abundance in the gut (Sokol et al. 2009, Cao et al. 2014). Interestingly, compared to healthy individuals, certain patients with gut inflammation display relatively higher levels of F. longum L2-6 and relatively lower levels of F. duncaniae A2-165, suggesting that different Faecalibacterium species cope differently with oxidative stress (Song et al. 2016, Zhang et al. 2018).
Figure 4.
Graphic summary of relationships between Faecalibacterium representation within the intestinal microbiota and colon inflammation. The normal state of the microbiota greatly contrasts with the dysbiosis often encountered in digestive diseases such as IBD. Intestinal inflammation is accompanied by an overproduction of ROS as well as an increase in O2 levels, conditions that favour facultative anaerobes over strict aerobes such as Faecalibacterium. In some patient cohorts, abundance of F. duncaniae A2-165 decreases while that of F. longum L2-6 increases (Song et al. 2016, Zhang et al. 2018).
Little is known about how members of Faecalibacterium may remain unaffected by O2 and ROS except that, in vitro, F. duncaniae A2-165 has been found to have extracellular antioxidants, which transfer electrons to O2 in moderately oxygenated environments (Khan et al. 2014). Furthermore, when inorganic nitrate was used as an antioxidant, the Faecalibacterium population was maintained following total body irradiation in a mouse model (Wang et al. 2020). In addition, using a metabolic model exploring the effects of diet on O2 sensitivity, it was predicted that diets with a balanced carbohydrate and protein ratio could increase the O2 range over which Faecalibacterium could survive (Henson and Phalak 2017). Thus, the presence of antioxidants as well as relative dietary quantities of carbohydrates and proteins could impact the ability of Faecalibacterium species to tolerate oxidative stress during gut inflammation.
Faecalibacterium in relation to health and disease
When the normal microbial ecosystem is altered, various nonpredominant bacteria (mainly pathobionts) may thrive, a situation, i.e. sometimes associated with illness (Caballero-Flores et al. 2022). Such dynamics have been linked to many different types of diseases that may or may not be directly related to the GIT. Notably, the abundance of some Faecalibacterium species has been found to be altered across a wide range of diseases and disorders. It is essential to note that most of the results presented in this section are based on observational data, which do not allow to determine whether dysbiosis is the cause or consequence of disease. Therefore, these findings should be interpreted with caution. Indeed, the new taxonomic structure of the genus highlights the need for further research, particularly in the case of poorly studied diseases. Lastly, most of the intervention-focused or mechanistic studies described below have been performed in mice, which means that the relevance of their results in humans remains to be assessed.
Inflammatory bowel diseases (IBDs)
The two forms of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). Both are chronic conditions characterized by the heightened chronic activation of the gut mucosal immune system (Khor et al. 2011). This overactive immune response occurs in tandem with an imbalance in the gut microbiota: a decline in Faecalibacterium abundance has been observed in CD and UC (Sokol et al. 2008, Morgan et al. 2012). In addition, in IBD, there is a reduction in the richness of two mucosa-associated Faecalibacterium phylotypes (Lopez-Siles et al. 2015). The markers for IBD and its subtypes (with or without effects on the ileum) have revealed interesting patterns in a metaproteomics study that compared healthy individuals to individuals with the disease: the latter had reduced levels of Faecalibacterium (Henry et al. 2022). In CD patients with ileal involvement, the abundance of Faecalibacterium phylogroup II was lower (Lopez-Siles et al. 2016). In another study with the same target population, this decrease was associated with an increase in the levels of primary and secondary bile acids (Gonzalez et al. 2022). Such reductions in Faecalibacterium abundance have been linked to the decreased circulation of CCR6+CSCR6+DP8α regulatory T (Treg) leukocytes and increased values of disease activity metrics (Sarrabayrouse et al. 2014, Touch et al. 2022). Based on the above evidence, Faecalibacterium has been identified as playing a key role in inducing this subset of Treg cells, which have an important influence on microbiota–host cross-talk in cases of IBD. In individuals with active IBD, DP8α leucocytes from the peripheral blood and lamina propria do not respond to Faecalibacterium, whereas those from individuals who are healthy or in remission do respond (Sarrabayrouse et al. 2014). Moreover, DP8α T cells exhibit a regulatory phenotype in vitro (Sarrabayrouse et al. 2014, Godefroy et al. 2018, Alameddine et al. 2019) and dampen colitis severity in mice when combined with the administration of Faecalibacterium (Touch et al. 2022). IBD is also characterized by elevated levels of IgG antibodies in the mucus and serum. In a study where fecal samples from healthy individuals and IBD patients were incubated with autologous serum, the IgG-coated fractions from IBD patients displayed a lower-level response to Faecalibacterium. Thus, IBD patients displayed a higher relative level of immunological tolerance to this genus compared to other genera, including Streptococcus, Lactobacillus, and Lactococcus (Bourgonje et al. 2022).
Specific phages can infect Faecalibacterium and may play a role in several human diseases, including IBD (Cornuault et al. 2018). Faecalibacterium phages occur at higher levels in feces from humans with IBD than in feces from healthy controls (Cornuault et al. 2018). Such could partially explain the decline in bacterial abundance, along with redox imbalance during inflammation. In mice, treatments using Faecalibacterium or supernatant from Faecalibacterium cultures decreased inflammation severity in several murine models for colitis (Sokol et al. 2008, Carlsson et al. 2013, Qiu et al. 2013, Martin et al. 2014, Laval et al. 2015).
Irritable bowel syndrome
Irritable bowel syndrome (IBS) is a commonly occurring gastrointestinal disorder characterized by altered bowel habits, chronic abdominal pain, the absence of detectable structural abnormalities in the colon, and increased gut permeability (Marshall et al. 2004). Although less is known about the relationship between the microbiota and IBS than in the case of IBD, abnormalities in fecal microbiota have been found in a subgroup of IBS patients who experienced bloating, as well as altered intestinal motility or sensitivity (Collins et al. 2009). Furthermore, dysbiosis has been observed in at least some subsets of IBS (Bonfrate et al. 2013, Bennet et al. 2015, Sabo and Dumitrascu 2021, Chen et al. 2023). More specifically, individuals with alternating-type IBS (IBS-A) have significantly lower levels of Faecalibacterium. In contrast, Faecalibacterium abundance is unaltered in individuals with diarrhea-predominant IBS (IBS-D) and is either notably lower or unaltered in individuals with constipation-predominant IBS (IBS-C) (Rajilic-Stojanovic et al. 2011, Duboc et al. 2012, Rigsbee et al. 2012). In a recent study using multicenter amplicon sequencing data, Chen et al. (2023) found that, within a cohort of 708 individuals (354 IBS patients and 354 healthy controls), the genus Faecalibacterium was one of the depleted taxa in IBS patients. The same study showed that Faecalibacterium was among the top 10 hub taxa in all the IBS subtypes identified by co-occurrence network analysis and was also among the top three genera identified as potential microbial biomarkers for IBS using a random forest model (alongside Pseudoclostridium and Bifidobacterium) (Chen et al. 2023). Furthermore, a negative correlation has been observed between Faecalibacterium abundance and IBS severity in humans (Rajilic-Stojanovic et al. 2011). In murine models of visceral pain mimicking IBS, such as those using neonatal maternal separation (NMS) and partial restraint stress (PRS), hypersensitivity decreased in response to supplementation with F. duncaniae A2-165 bacteria (but not supernatant) (Miquel et al. 2016). Given that low-grade inflammation has been reported in a subset of human IBS patients (Collins 1992, Akiho et al. 2010), treatments using the A2-165 strain could possibly produce benefits because of the anti-inflammatory properties the bacterium displays. However, evidence for this hypothesis remains unclear given that, while both F. duncaniae A2-165 and its supernatant helped counteract impairment of the intestinal epithelial barrier in a murine model of chronic low-grade inflammation, only the bacterium was effective in murine models of NMS and PRS, indicating that other mechanisms likely explain the observed beneficial effects in mice (Martin et al. 2015).
Obesity, metabolic conditions, and liver disorders
Abnormal or excessive fat accumulation can present health risks. The Obesity Medicine Association defines obesity as ‘a chronic, relapsing, multifactorial, neurobehavioral disease, wherein an increase in body fat promotes adipose tissue dysfunction and abnormal fat mass physical forces, resulting in adverse metabolic, biomechanical, and psychosocial health consequences’. Obesity is associated with health concerns such as type 2 diabetes (T2D), hypertension, and cancer (Arroyo-Johnson and Mincey 2016).
Obese or T2D patients have lower microbiota diversity, and Faecalibacterium is one of the community members that has been found to differ (Le Chatellier et al. 2013). These results were corroborated by an Iranian study, which observed a negative correlation between Faecalibacterium levels and BMI (Navab-Moghadam et al. 2017). Chinese T2D patients displayed reduced abundances of Bacteroides, Akkermansia, and Faecalibacterium. Evidence of this shift was even detectable in prediabetic obese patients, reflecting the link between glucose intolerance and microbiota composition (Zhang et al. 2013). In other research, Faecalibacterium was enriched in T2D patients following weight loss (Hippe et al. 2016). Intriguingly, the same study found that samples from lean patients contained higher numbers of Faecalibacterium genes than did samples from obese and T2D patients; in contrast, lean patients had the lowest number of copies of the Faecalibacterium–associated butyryl-CoA:acetate CoA-transferase (BUT) gene. T2D patients had the highest butyrate levels. These findings could be interpreted as indicating that different Faecalibacterium species produce different levels of butyrate in vivo and that the taxon’s abundance varies between healthy and unhealthy individuals (Hippe et al. 2016). Consequently, although the exact mechanisms have yet to be determined, current research supports a relationship between BMI, blood glucose levels, and Faecalibacterium abundance. In mice, supplementation with F. duncaniae A2-165 bacteria holds promise for treating obesity and its related complications. Compared to control mice, HFD-fed mice given twice-weekly supplements of F. duncaniae A2-165 had smaller adipocytes as well as reduced hepatic inflammation, lipid accumulation in the liver, and inflammatory cell infiltration in adipose tissue (Munukka et al. 2017). While these results seem quite promising, studies on obesity in mice not always translate to humans and must, therefore, be carefully interpreted and validated by clinical intervention-based trials in humans.
In preliminary research, lower levels of Faecalibacterium were also observed in individuals with rare metabolic diseases such as propionic acidemia (Tims et al. 2022), phenylketonuria (Verduci et al. 2018), or glycogen storage disease type 1 (Ceccarani et al. 2020). Finally, patients with liver cirrhosis showed reduced amounts of certain Faecalibacterium phylogroups, especially those that are better butyrate producers (Chen et al. 2021). More research should be performed to clarify the significance of these observations.
Neurological conditions
The gut microbiota plays an important role in the complex crosstalk that takes place between the gut and the brain (Alonso-García et al. 2021). Thus, it is unsurprising that microbial dysbiosis has been observed in individuals with a range of neurological conditions, opening the door to research on the microbiota–gut–brain axis (Cryan et al. 2019). In addition, several neurological conditions have been associated with leaky gut syndrome and a proinflammatory shift in the colonic microbiota, which results from increased levels of Gram-negative bacteria containing immune-triggering lipopolysaccharides (LPSs) (Eicher and Mohajeri 2022). Correlations have also been found with lower abundances of Bifidobacterium, Coprococccus, Eubacterium, Lactobacillus, Prevotella, Roseburia, and Faecalibacterium (Eicher and Mohajeri 2022). The latter genus has been found to occur at lower levels in individuals with clinical depression (Ye et al. 2021). Exploratory studies have observed similar patterns in association with several additional psychiatric conditions, including bipolar disorder, anxiety, psychosis, and schizophrenia (Nikolova et al. 2021).
Alterations in gut microbiota have also been seen in proteinopathies such as Parkinson’s disease (PD) and Alzheimer’s disease (AD) (Nishiwaki et al. 2020, Xi et al. 2021). For example, Faecalibacterium levels were found to be lower in PD patients than in healthy controls (Nishiwaki et al. 2020). Recently, Faecalibacterium strains isolated from healthy human donors were tested in a murine model of AD; the administration of both living and dead bacteria improved cognitive impairment (Ueda et al. 2021). Furthermore, Faecalibacterium abundance was found to be higher in healthy individuals than in individuals with mild cognitive impairment (MCI) and AD patients, in whom it was positively correlated with cognitive performance (Ueda et al. 2021).
Recent preliminary research found that children on the autism spectrum had higher levels of Faecalibacterium (Xu et al. 2019, Iglesias-Vazquez et al. 2020). Unexpectedly, an intervention study discovered that autistic children who consumed gluten- and casein-restricted diets were better able to handle autism-related behavioral challenges and had higher Faecalibacterium levels than autistic children who consumed the control diet (Grimaldi et al. 2018). Members of this genus were found to be more abundant in individuals with epilepsy, although there were no links with severity status (Valles-Colomer et al. 2019, Cui et al. 2021).
Intriguing results aside, these results should be interpreted with caution because research relating Faecalibacterium abundance to neurological conditions is far more preliminary than that examining the links with diseases such as IBD.
Cancer
The gut microbiota also appears to have an impact on the efficacy of immune checkpoint inhibitors (Routy et al. 2018, Daillere et al. 2020, Effendi et al. 2022). Indeed, several studies have shown that a higher baseline level of Faecalibacterium, as well as that of other Firmicutes, positively correlates with responses to related treatments for various cancers, such as melanoma (Chaput et al. 2017, Gopalakrishnan et al. 2018, Coutzac et al. 2020, Limeta et al. 2020, Spencer et al. 2021), hepatocellular carcinoma (Li and Ye 2020), and nonsmall cell lung cancer (Newsome et al. 2022).
In a rat model where colorectal cancer (CRC) was induced via azoxymethane (AOM), treatment with F. prausnitzii reduced disease markers as well as lipid peroxidation (Dikeocha et al. 2022). Indeed, Kenyan patients with CRC were found to have intestinal mucosa-associated microbiota and microbial metabolic profiles different from those of healthy individuals (Obuya et al. 2022). In particular, Prevotella copri and Faecalibacterium species were less abundant, and alterations were observed in glutamate metabolism.
In another study, breast cancer patients showed reduced fecal levels of Faecalibacterium (Ma et al. 2020). Moreover, when Faecalibacterium and MCF-7 breast cancer cells were cocultured in vitro, proliferation of the latter was hampered through the inhibition of IL-6 production and the phosphorylation of JAK2/signal transducers and STAT3 activators (Ma et al. 2020). Interestingly, a recent study demonstrated that a mixture of four commensal Clostridiales strains (i.e. CC4: F. prausnitzii, R. intestinalis, Eubacterium hallii, and Anaerostipes caccae) displayed anticarcinogenic properties against different solid tumors (Montalban-Arques et al. 2021). The rationale behind CC4 composition was to combine different commensal strains capable of producing butyrate. In another preclinical rat model, treatment with F. duncaniae helped repair collateral damage in the colon that was caused by radiation therapy, a common treatment for cancer in the pelvic area. Mechanistically, this effect arose from reductions in hyperpermeability and neutrophil infiltration; the preservation of progenitor cells and colon cell morphology; and the upregulation of IL-18 expression (Lapiere et al. 2020).
Dermatitis and allergies
Atopic dermatitis is a condition in which the skin is chronically inflammed, resulting in redness, dryness, and irritation. It is characterized by a reduction in skin microbiota diversity, which facilitates colonization by pathogenic bacteria, such as Staphylococcus aureus, leading to increased disease severity (Paller et al. 2019). Gut dynamics in early childhood might have an important role to play. Indeed, metabolites production by some types of bacteria, from infancy to adulthood, results in heightened immune system priming and contributes to atopic dermatitis development, which means that early life experiences are likely key contributors to this condition (Gensollen and Blumberg 2017). Research has also shown that atopic dermatitis is associated with shifts in the gut microbiota. In a study conducted using 42 healthy individuals and 30 individuals with the condition, the latter had reduced butyrate and propionate levels in their feces; furthermore, they had lower abundances of Faecalibacterium subspecies related exclusively to F. duncaniae A2-165 (Song et al. 2016, Effendi et al. 2022). Faecalibacterium levels are reduced in individuals with psoriasis, but not in those with hidradenitis suppurativa, which can co-occur with IBD (Eppinga et al. 2016). Surprisingly, in a murine wound healing model, i.e. commonly employed to study healing dynamics and treatment efficacity, treatment with supernatant from Faecalibacterium cultures improved several types of skin lesions caused by intense, sustained inflammation, such as ulcers (Stefia et al. 2020). In NC/Nga mice in which atopic dermatitis had been induced using 2,5-dinitrochlorobencene (DNCB), the administration of Faecalibacterium improved symptoms by boosting the Th2 response, thus counterbalancing the excessive Th1 response elicited in the model (Lee et al. 2022). These promising results remain to be validated by clinical trials with humans.
Asthma is a chronic inflammatory condition that impacts lung airways. Research has suggested that shifts in the gut microbiome can impact the outcome of lung diseases. Indeed, the higher prevalence of lung diseases in individuals with gastrointestinal disorders supports the existence of a gut–lung axis, of which SCFAs may be major drivers (Correa et al. 2022). While findings for adults remain inconclusive (Wang et al. 2018b, Tikunov et al. 2021), children with allergic asthma had lower Faecalibacterium levels than did healthy children (Demirci et al. 2019). Finally, in a murine model, both living and dead F. duncaniae were able to alleviate dust mite-induced allergic asthma by modulating the microbiota and increasing butyrate production, an interesting finding that should be explored in future research (Hu et al. 2021).
Covid-19
Infection with SARS-CoV-2 causes the disease COVID-19, which mainly affects the respiratory system. Although COVID-19 commonly causes fever and respiratory tract symptoms, it may also lead to cardiac, gastrointestinal, hepatic, renal, neurological, olfactory, gustatory, ocular, cutaneous, and hematological symptoms (Zhang and Guo 2020). SARS-CoV-2 affects the gastrointestinal system because the virus enters cells via the angiotensin-converting enzyme 2 (ACE2) receptor, which is abundantly present on the glandular cells of the gastric, duodenal, and rectal epithelia as well as on the endothelial cells of the small intestine (Zhang and Guo 2020). Thus, the virus can also infect enterocytes, as revealed by the presence of viral RNA in feces (Trottein and Sokol 2020, Yeoh et al. 2021). Furthermore, several studies have described a pattern of infection-related dysbiosis, i.e. marked by a reduction in Faecalibacterium abundance, particularly in hospitalized patients with the severe phenotype (Yeoh et al. 2021, Hazan et al. 2022). Additionally, individuals suffering from postacute COVID-19 syndrome (PACS), otherwise known as Long COVID, have higher levels of Ruminococcus gnavus and Bacteroides vulgatus and lower levels of Faecalibacterium (Liu et al. 2022). Since Faecalibacterium holds promise for treating the intestinal issues caused by other gut-related disorders, its use has been proposed to help relieve COVID-19-related symptoms (He et al. 2021). Indeed, COVID-19 patients that received a fecal microbiota transfer (FMT) had higher levels of Faecalibacterium following the procedure than during the course of their infection (Liu et al. 2021). Moreover, in a preclinical model in hamsters, the oral administration of a Faecalibacterium strain to SARS-CoV-2 infected animals resulted in a 10-fold reduction in viral load (Monchatre-Leroy et al. 2021).
Host-Faecalibacterium cross-talk
Beneficial effects on the host
Several studies have sought to decipher how Faecalibacterium presence promotes GIT homeostasis using in vivo murine models or in vitro testing with human cell lines. Numerous types of evidence suggest that this genus plays an important role in immune system regulation, gut barrier protection, and microbiota modulation (Miquel et al. 2013), potentially via a range of mechanisms (Fig. 5). However, it is important to note that research on the interactions between Faecalibacterium and their hosts is in constant flux for two reasons. First, the taxonomy of the genus is undergoing shifts because of new discoveries. Second, it is difficult to separate out general effects, potentially attributable to multiple butyrate-producing bacteria, from effects specific to Faecalibacterium. Some studies have shown that the presence of specific species can modulate the host immune system, by upregulating IL-10 expression in dendritic cells and enhancing the proliferation of T cells, which suggests the presence of an effect specific to Faecalibacterium (Rossi et al. 2016). In support of this idea, in vitro tests revealed that the presence of different clinical isolates led to the upregulation of IL-10 in the human peripheral blood mononuclear cells (PBMCs) of healthy individuals; this result was seen especially, but not exclusively, in association with bacteria displaying higher levels of butyrate production (Martin et al. 2017). The presence of these clinical isolates could also block IL-8 production in human HT-29 cells that had been activated with TNF−α, although, in this model, there was no clear genus-specific effect tied to butyrate production (Martin et al. 2017). Additionally, treatment with F. duncaniae has been shown to increase levels of a specific subset of IL-10-secreting Treg cells (CD4CD8α) in the lamina propria of the colon; these cells are lacking in individuals with IBD. In a humanized murine model, the administration of DP8α cells and F. duncaniae protected against dextran sulfate sodium (DSS)-induced colitis induced (Touch et al. 2022). However, it is not yet known whether this effect is species-specific.
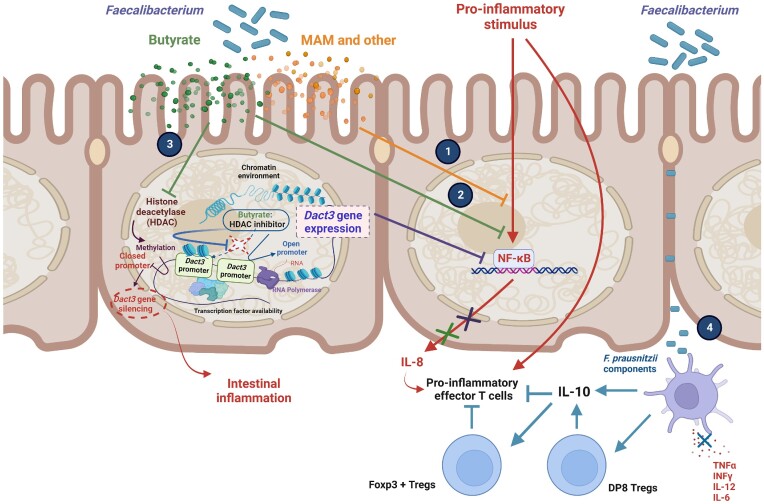
Figure 5.
Mechanisms of action associated with Faecalibacterium. (1) Supernatant from Faecalibacterium cultures blocks the activation of NF-κB induced by a proinflammatory stimulus. (2) Butyrate produced by Faecalibaterium inhibits NF-κB activation and blocks Il-8 production in TNF-α stimulated intestinal epithelial cells. (3) Butyrate inhibits HDAC, leading to the expression of Dact3, a gene encoding a negative regulator of the inflammatory Wnt/JNK signaling pathway, and the inhibition of IL-8 production. It is important to note that the silencing of Dact3 leads to a partial loss of the anti-inflammatory effects of Faecalibacterium supernatant in the intestinal epithelial cells. (4) Faecalibaterium components can induce the appearance of a specific subset of IL-10-secreting Treg cells—called DP8 cells—in the colonic lamina propria. Moreover, the presence of Faecalibacterium can boost IL-10 levels in antigen-presenting cells and DP8 cells, which may enhance the suppressive activity of Foxp3 + Treg cells and block proinflammatory effector T cells induced by various stimuli.
Faecalibacterium presence can also affect intestinal epithelial cells. In assays using human Caco-2 cells, IL-1β-induced NF-κB activity was abolished after the addition of supernatant from Faecalibacterium cultures but not after the addition of Faecalibacterium cells (Sokol et al. 2008). Furthermore, in a murine model where colitis was induced using dinitrobenzene sulfonic acid (DNBS), the butyrate produced by Faecalibacterium upregulated expression of the dishevelled binding antagonist of beta catenin 3 (Dact3) gene in TNFα−stimulated HT-29 cells (Lenoir et al. 2020). Dact3 is a negative regulator of the inflammatory Wnt/JNK signaling pathway and could be one of the host effectors that mediates the positive effects attributed to Faecalibacterium, given that its silencing leads to a partial loss of supernatant-mediated anti-inflammatory effects in intestinal epithelial cells. That said, the key role played by butyrate makes difficult to determine whether the presence of Faecalibacterium specifically contributes to activation (Lenoir et al. 2020).
The presence of Faecalibacterium also appears to have impacts on the intestinal barrier. Treatment with F. duncaniae A2-165 was found to improve gut permeability and function in a murine model of DNBS-induced intestinal barrier impairment (Sokol et al. 2008, Martin et al. 2014, Laval et al. 2015, Martin et al. 2015, Miquel et al. 2015a). Rossi et al. (2015, 2016) and Carlsson et al. (2013) also found that treatment helped restore intestinal permability in a model of DSS-induced colitis in mice. In all these cases, it is difficult to distinguish whether these beneficial effects arise from the anti-inflammatory properties of the bacteria or the physical effects of the bacteria on the barrier. However, in a noninflammatory model of gut alteration in mice, administration of F. duncaniae A2-165 led to the recovery of claudin-2 levels, which had been altered using NMS (Miquel et al. 2016). It also ameliorated the degree of radiation-induced colonic hyperpermeability by preserving the progenitor cells and colon cell morphology in mice (Lapiere et al. 2020). In a nonpathological model using germ-free rats colonized by B. thetaiotaomicron and F. duncaniae A2-165, the presence of the latter diminished the negative effects of Bacteroides on goblet cell differentiation and mucin glycosylation. This effect likely stems from the strain’s metabolic processes, in which acetate is consumed and butyrate is produced, and underscores how the presence of Faecalibacterium helps modulate the gut microbial community during host–microbiome interactions (Wrzosek et al. 2013).
Bacterial effectors
Metabolites
As mentioned previously, one of the main metabolites produced by Faecalibacterium is butyrate (Duncan et al. 2004), which has several effects on host physiology. For example, it can be used by colonocytes as a carbon source and bolster the gut barrier by strengthening the tight junctions (Liu et al. 2018). In continuous cocultures with primary epithelial cells, butyrate production by F. duncaniae A2-15 was correlated with the downregulation of TLR3 and TLR4 expression via the HDAC1 and NF-kB pathways (Zhang et al. 2021). The health effects of butyrate are reviewed elsewhere (Liu et al. 2018).
In a gnotobiotic model employing F. duncaniae A2-165 and E. coli, metabolomic analysis identified several metabolites—including salicylic acid, shikimic acid, and raffinose—that could play a role in the benefits attributed to the bacterium (Miquel et al. 2015b). Some studies have suggested that salicylic acid could help treat obesity, by inducing the browning of white adipocytes (Choi et al. 2022), or could serve as a preventive treatment for CRC (Imai et al. 2022). Furthermore, it is used as a coadjuvant in cancer treatments because it can boost the effects of antitumor immunotherapy (Sun et al. 2022). In the pharmaceutical industry, salicylic acid is used to produce the amine derivative 5-aminosalicylic acid (5-ASA or mesalamine), currently used to treat IBD (Messori et al. 1994). The effects of aspirin might be mediated via its transformation into salicylic acid, a process that can be carried out by members of the gut microbiota (Zhao et al. 2020). Through the achorismate synthase pathway, shikimic acid acts as a precursor for the synthesis of several aromatic compounds, including salicylic acid (Bochkov et al. 2012). Consequently, F. duncaniae may play a key role in the biosynthesis of salicylic acid, which is a precursor of 5-ASA. Both are anti-inflammatory molecules that could contribute to the anti-inflammatory effects observed in vivo in mice treated with Faecalibacterium. In addition, the oligosaccharide raffinose is a potential effector that could serve to address intestinal barrier dysfunction induced by Faecalibacterium presence during inflammation (Martin et al. 2018). It has yet to be determined whether Faecalibacterium produces these three metabolites, or if they are produced by host cells when Faecalibacterium is present.
Finally, we have also identified the butyrate as the F. prausnitzii effector responsible for Dact3 modulation (see the section ‘Beneficial effects on the host’). The elucidation of the impact of F. prausnitzii is Dact3 upregulation validated in vivo in both healthy and inflamed mice is currently in progress.
Extracellular vesicles
Extracellular vesicles (EVs) are spherical membranous vesicles used by prokaryotic and eukaryotic cells to release compounds into the extracellular space for functional reasons, such as responding to environmental changes or communicating with the host or other bacteria (Deatherage and Cookson 2012). Recent work has underscored that EVs could potentially serve as biomarkers or therapeutic tools, which has resulted in an increase in the number of EV-focused studies. EVs are released by all cellular organisms, including Gram-positive bacteria (Toyofuku et al. 2023), and, since they were first described, unexpected biophysical, biochemical, and functional heterogeneity has been observed (Buzas 2023). For F. duncaniae A2-165, EVs were first isolated and characterized by Jafari et al. in 2019. The EVs contain proteins ranging in size from 11 to 245 kDa (Jafari et al. 2019). In Caco-2 cells, a treatment utilizing EVs from A2-165 and A. municiphila augmented serotonin production and the expression of genes related to serotonin (Yaghoubfar et al. 2021). Similarly, the EVs alone strengthened the expression of genes encoding tight junction proteins [zonula occludens 1 (ZO1) and occludin (OCLN)] and ParR family compounds, results that confirm the role of EVs in maintaining gut barrier integrity (Moosavi et al. 2020). EVs from Faecalibacterium species are also involved in immune homeostasis: when used to treat Caco-2 cells, they significantly diminished levels of several proinflammatory cytokines (Rabiei et al. 2019). Similar results were obtained in a model utilizing Faecalibacterium EVs and the lung cancer cell line A549 (Jafari et al. 2019). Taken together, these results should encourage further research on the potential role played by Faecalibacterium EVs in hosts and drive additional, more conclusive research in this field.
Microbial anti-inflammatory molecule
The microbial anti-inflammatory molecule (MAM; ZP05614546.1) is 15 kDa in size and has a nonpolar residue prevalence of 53%. It was first described in a peptidomic analysis of supernatants from Faecalibacterium cultures, in which seven of its derivative peptides inhibited the NF-kB pathway in intestinal epithelial cells, displaying the potential for anti-inflammatory effects (Quevrain et al. 2016). Indeed, in DNBS and DSS models employing NF-κΒ−luciferase transgenic mice, a decrease in proinflammatory cytokines related to Th1 and Th17 responses was observed in mice gavaged with Lactococcus lactis strain delivering MAM cDNA (Breyner et al. 2017). Recent in vitro and in vivo studies have shown that MAM’s anti-inflammatory properties are diverse and species-specific. In murine models of DNBS-induced acute and chronic inflammation, the MAM from the F. prausnitzii strain M21-2 was more effective than the MAM from F. duncaniae A2-165 (Auger et al. 2022).
The effects of MAM were examined in db/db mice, which do not express leptin receptors and have lower levels of Faecalibacterium in their guts. Oral supplementation with E. coli-produced MAM suggest that the molecule interacts with ZO-1 and other tight junction proteins. Furthermore, the transfection of MAM into a cell line increased ZO-1 expression and restored epithelial barrier function (Xu et al. 2019). These results suggest that MAM has therapeutic potential in improving intestinal barrier function.
Extracellular polymeric matrix
The presence of a biofilm-forming strain, Faecalibacterium HTF-F, inhibited the production of proinflammatory signals in human dendritic cells that had been stimulated using L. plantarum. In the same study, the presence of the HTF-F strain was better than the presence of the A2-165 strain at improving symptoms of DSS-induced colitis (Rossi et al. 2015). The authors concluded that the greater anti-inflammatory effects associated with HTF-F could be related to the extracellular polymeric matrix (EPM) of the strain’s biofilm. However, given ongoing taxonomic shifts within Faecalibacterium, it is essential to carefully explore other differences between these two strains, which belong to different phylogenetic cluster.
Faecalibacterium as a therapeutic tool
Potential applications
In the last 15 years, the field of probiotics has exploded as a result of massive leaps in knowledge about the gut microbiota and its crucial role in human health. Probiotics are ‘live microorganisms that, when administered in adequate amounts, confer a health benefit to the host’ (Hill et al. 2014). They are usually microorganisms and most frequently a specific list of bacterial genera of proven safety with a long history of use in mainly healthy people (Martin and Langella 2019). However, this more classical definition of probiotics has evolved over time to include next-generation probiotics (NGPs) and live biotherapeutic products (LBPs). In contrast to conventional probiotics, these two new groups do not have an established history of safe use in humans. Instead, NGPs and LBPs are directly isolated from the human gut microbiome and then carefully chosen for their biological effects. Faecalibacterium holds great promise as an LBP because of its demonstrated role in host homeostasis and its many positive effects in preclinical murine models for various diseases (e.g. IBD, CRC, AD, and even radiation therapy). Its use is already being explored in livestock. For instance, research has characterized Faecalibacterum strains isolated from the stool of calves and piglets (Foditsch et al. 2014). Given the strong relationship between IBD in humans and lower levels of Faecalibacterium (Sokol et al. 2008, 2009, Cao et al. 2014), the genus is clearly a potential LBP candidate. It is being used for the first time in a clinical trial focused on maintaining both corticosteroid-induced clinical responses and remission in individuals with CD (MAINTAIN 2022). This phase 1 study has two parts: a open-label evaluation of safety using a small group of participants and a randomised, double-blind, placebo-controlled evaluation using a large group of participants (ClinicalTrials.gov Identifier: NCT05542355). Furthermore, a specific formulation has been proposed to keep these EOS bacteria alive under ambient conditions, which would be necessary to treat patients with intestinal dysbiosis-associated diseases (Khan et al. 2014).
Additionally, Faecalibacterium could serve as an effective disease biomarker and diagnostic support tool (Miquel et al. 2013). In 2008, a diagnostic test based on Faecalibacterium prevalence and leukocyte counts was developed to distinguish active CD from UC and displayed a high level of performance (79%–80% sensitivity and 98%–100% specificity) (Swidsinski et al. 2008). As-yet-undiscovered species and phylotypes could yield further insights and improve the accuracy of methods that utilize Faecalibacterium as a disease marker. Work addressing these issues is especially important since different ratios of strains and phylotypes have been seen in association with several health conditions, such as AD and obesity (Lopez-Siles et al. 2017).
Interventions to modulate faecalibacterium
Dietary interventions, including probiotics and prebiotics
Dietary interventions remain one of the most efficient ways for modifying the microbiota. For example, eating the fiber-rich Mediterranean diet can result in higher levels of Faecalibacterium (Godny et al. 2022). Several studies have investigated nutritional strategies for increasing the abundance of this genus in the gut. A 30-day diet of the alga Euglena gracilis was observed to increase Faecalibacterium levels (Nakashima et al. 2021). The same effect was achieved via a 6-day, lifestyle-based immersion program that included nutritional intervention: with 100% plant based meals and that incorporated whole foods (Ahrens et al. 2021). Similarly, levels of Faecalibacterium increased in children given a diet low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAPs), and the abundance of bacteria in this taxon correlated with symptom improvement (Chumpitazi et al. 2015). The FODMAP diet contains reduced quantities of fermentable carbohydrates and has been effective in reducing gastrointestinal symptoms in adults with IBD and IBS, even if it decreases Faecalibacterium levels (Hustoft et al. 2017, Cox et al. 2020). However, it remains unclear whether the relationship between this diet and Faecalibacterium abundance is consistent, an issue discussed in a recent systematic review (Vandeputte and Joossens 2020). In a 6-month controlled-feeding experiment, 217 healthy young adults were randomly assigned a low-, mid-, or high-fat diet; those consuming the low-fat diet showed a decrease in Faecalibacterium levels (Wan et al. 2019). In contrast, no such changes were seen during a 3-month single-center intervention where six patients with glucose transporter 1 deficiency syndrome were given a traditional ketogenic diet (i.e. high fat and low carbohydrate) (Tagliabue et al. 2017). Several studies in rodents have revealed the existence of a negative correlation between protein intake and Faecalibacterium abundance, as noted in a systematic review of how dietary proteins affect the gut microbiota (Wu et al. 2022). Research examining this relationship remains scarce, and few intervention studies have been conducted using healthy subjects. Finally, fasting has been found to increase Faecalibacterium levels in humans, confirming that dietary habits may shape Faecalibacterium occurrence in myriad ways (Remely et al. 2015).
Prebiotics are another tool that can be used to modulate the levels of Faecalibacterium. A prebiotic is a ‘non-digestible compound that, through its metabolization by microorganisms in the gut, modulates composition and/or activity of the gut microbiota, thus conferring a beneficial physiological effect on the host’ (Bindels et al. 2015). Faecalibacterium is a fibre fermenter, and several studies have found a positive correlation between levels of dietary fibre and the relative abundance of this genus (Benus et al. 2010). Historically, supplementation with inulin was one of the first strategies successfully used to increase the representation of Faecalibacterium within the microbial community of the human gut (Ramirez-Farias et al. 2009). A recent systematic review has highlighted the ability of prebiotics to boost Faecalibacterium abundance (Verhoog et al. 2019). Indeed, in germ-free mice conventionalized with human microbiota, the administration of a supplement containing resistant starch increased butyrate production and increased Faecalibacterium levels; the effect was more substantial when donors had lower initial abundances of the genus (Cherbuy et al. 2019). In a randomized placebo-controlled crossover study, the intake of polydextrose and soluble corn fiber boosted levels of Faecalibacterium (Hooda et al. 2012). In another crossover study, the consumption of chickpea and its main oligosaccharide, raffinose, had the same effect (Fernando et al. 2010).
It has been hypothesized that traditional probiotics can help treat disease-related bacterial imbalances in the gut microbiome. Some improve Faecalibacterium abundance, perhaps because traditional probiotics often produce acetate. Acetate promotes the growth of Faecalibacterium via cross-feeding (Ramirez-Farias et al. 2009). For instance, during a dietary intervention study that used L. plantarum PMO08, Faecalibacterium levels had significantly increased after 2 weeks of treatment (Oh et al. 2021). Host health improved and Faecalibacterium abundance increased in response to the administration of L. plantarum ZPL001, Enterococcus durans EP1, and L. paracasei N1115, among others (Carasi et al. 2017, Wang et al. 2018a).
Medications
It is well-established that antibiotics greatly impact the structural composition of the gut microbiota (Wang et al. 2023). For example, an 8-week amoxicillin treatment led to diminished levels of Faecalibaterium (Pallav et al. 2014), as did the administration of ciprofloxacin (Stewardson et al. 2015). In contrast, nitrofurantoin, a medication mainly used for urinary tract infections, increased the abundance of four genera of Firmicutes, including that of Faecalibacterium (Stewardson et al. 2015). In vaginal-delivered infants, Faecalibacterium levels were lower at the 1-year mark for babies that had received postnatal antibiotics at any point compared to babies that had not (Ainonen et al. 2022). Another study reported that antibiotic and antifungal treatments diminished Faecalibacterium abundance in children with leukemia (Dunn et al. 2022); the same result was seen in response to intraveinously administered antibiotics in individuals with multiple myeloma (D’Angelo et al. 2022).
Shifts in Faecalibacterium levels have also been observed following exposure to other drugs. For example, they have been seen to decline in individuals taking diabetes medications including metformine (Manor et al. 2020) or to display partial restoration in those taking vortioxetine for depression (Ye et al. 2021). In contrast, a cross-sectional study in humans (n = 8583 participants) found no significant correlations between Faecalibacterium abundance and levels of different drug metabolites in the blood (Dekkers et al. 2022).
Fecal microbiota transplants
Fecal microbiota transplants have been extensively used to deal with multirecurrent Clostridioides difficile infections (CDIs), and their use in other contexts is currently under exploration (Cammarota et al. 2017). Faecalibacterium levels have been found decreased in patients with CDIs and low levels of Faecalibacterium are proposed as predictive of CDIs recurrence (Han et al. 2020, Khanna et al. 2016, Lee et al. 2020, Milani et al. 2016, Stewart et al. 2019). Research indicates that fecal microbiota transplants could restore and maintain Faecalibacterium levels for 2–4 months (Bjorkqvist et al. 2021). Indeed, in a case study of coinfection with C. difficile and an extensively drug-resistant KPC-producing Klebsiella pneumoniae, the levels of Faecalibacterium increased after the fecal microbiota transplant (Bahl et al. 2020). In a murine model, transplants led to the recovery of Faecalibacterium levels that had diminished following treatment with antibiotics and chemotherapy (Le Bastard et al. 2018), confirming the promise of this strategy.
Technical and legislative challenges
Before EOS bacteria can be administered to human subjects, there is a 2-fold challenge to resolve: (1) bacteria need to be produced at large scales in the absence of animal compounds and under strictly anaerobic conditions and (2) sufficiently anoxic conditions must be maintained across all the postculture processing steps (e.g. centrifugation, filtration, or lyophilization). In addition, the bacteria should remain alive during storage, so as to reach the gut in an effective state. Faecalibacterium is not only EOS. It is also highly sensitive to slight increases (from 0.1% to 0.5%) in physiological concentrations of bile salts, and its optimal pH is between 5.7 and 6.7 (Lopez-Siles et al. 2012, Lopez-Siles et al. 2017). To solve this problem, Faecalibacterium could be encapsulated so that it could survive transit through the GIT (Raise et al. 2020). To further simplify this part of the process, nonviable bacteria could be employed. A recent study compared the preventive and therapeutic effects of administering live vs. inactivated F. prausnitzii ATCC27768 in murine models of DSS-induced colitis (Kawade et al. 2019). It was found that mice given live F. prausnitzii recovered better than did mice given inactivated F. prausnitzii (Kawade et al. 2019). That said, dead bacteria were effective in a murine model of AD (i.e. improved AD-related cognitive impairment) and in a murine model of allergic asthma (Hu et al. 2021, Ueda et al. 2021). To address this question more clearly, we need additional research that focuses on direct crosstalk between Faecalibacterium and the host. Furthermore, the efficacy of live versus dead bacteria could depend on design variables, including the strain or model employed, or various technical variables. To provide clarity, more research should be conducted on the mechanisms by which dead bacteria could have impacts.
As stated above, probiotic therapy often seeks to restore balance of the intestinal ecosystem. This objective could be achieved with naturally occurring commensal bacteria in the form of NGPs or LBPs (Miquel et al. 2015a). However, unlike traditional probiotic strains, these bacteria are not recognized under formal product safety regimes in either Europe [qualified presumption of safety (QPS)] or the USA [generally recognized as safe (GRAS)]. Furthermore, as they have a short track record of safe consumption—no safe use was documented in Europe prior to 1997—these NGPs must be treated as novel foods or drugs, and they will face more severe commercial requirements in Europe than their conventional counterparts did (Miquel et al. 2015b). Although Faecalibacterium is not on the QPS list as most of the commensal bacteria, it holds great promise as a safe treatment given that thousands of FMTs have been performed worldwide without any Faecalibacterium-induced side effects or infections occurring. Moreover, regulatory agencies in Europe and the USA have already given the green light for research on how LBPs, including Faecalibacterium, affect humans (Cani 2018, CAUSALITY 2021, MAINTAIN 2022).
Conclusions and future perspectives
Ever since low levels of Faecalibacterium were discovered to be associated with several diseases, researchers have sought to characterize the mechanisms underlying interactions between hosts and members of this genus. In addition, as scientists have gained greater clarity around the taxonomy of Faecalibacterium, studies are being conducted in which the effects of the bacteria can be assigned to specific strains or species. In addition, a robust literature exists documenting the anti-inflammatory properties of the genus, in which the effects of butyrate, EVs, and cell wall components are addressed. Taken together, these findings have piqued interest in utilizing the genus as a LBP.
However, conclusions based on this work should be made with caution. First, most research to date has focused on the single strain F. duncaniae A2-165. Second, the beneficial effects have largely been observed in preclinical murine models. Third, the relationship between Faecalibacterium occurrence and diverse health conditions has been described based on correlational data, making it difficult to evaluate the degree to which Faecalibacterium has a causal effect on disease reduction. Clinical studies are currently underway to address these concerns, and they will play an important part in supporting the growing knowledge base being generated by preclinical and in vitro models.
Acknowledgement
The authors would like to thank all the people involved in gathering the knowledge that served as the basis for this review. In particular, we are grateful to our past and present colleagues who have demonstrated great dedication to this work: S. Miquel, M. Thomas, C. Bridonneau, S. Hudault, V. Robert, S. Chadi, F. Chain, N. Breyner, M. Lenoir, P. González-Dávila, D. Seda, L. Hua, P. Ruffié, S. Touch, N. Rolhion, L. Wrzosek, C. Cherbuy, P. Kharrak, C. Aubry, C. Michon, S. Verstraeten, J.J. Gratadoux, and S. Blugeon. L.B.G.H. is the recipient of an ANR grant (ANR-21-CE18-0050-01), and S.K. was paid a salary from this grant.
Contributor Information
Rebeca Martín, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
David Rios-Covian, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Eugénie Huillet, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Sandrine Auger, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Sarah Khazaal, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Luis G Bermúdez-Humarán, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Harry Sokol, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France; Sorbonne Université, INSERM, Centre de Recherche Saint-Antoine, CRSA, AP-HP, Saint Antoine Hospital, Gastroenterology Department, F-75012 Paris, France; Paris Centre for Microbiome Medicine (PaCeMM) FHU, F-75012, Paris, France.
Jean-Marc Chatel, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Philippe Langella, Paris-Saclay University, INRAE, AgroParisTech, Micalis Institute, 78350, Jouy-en-Josas, France.
Conflict of interest statement
H.S. and P.L. are cofounders of the startup Exeliom Biosciences, whose objective is to generate an LBP based on F. prausnitzii that targets IBD. H.S. has received lecture fees, board membership compensation, or consultancy payments from Amgen, Fresenius, IPSEN, Actial, Astellas, Danone, THAC, Biose, BiomX, Eligo, Immusmol, Adare, Nestle, Ferring, MSD, Bledina, Pfizer, Biocodex, BMS, Bromatech, Gilead, Janssen, Mayoli, Roche, Sanofi, Servier, Takeda, or Abbvie and holds stocks issued by Enterome Biosciences. P.L. has received lecture fees, board membership compensation, or consultancy payments from Biose, Biostime, Boiron, Bonduelle, BMS, Bromatech, IPSEN, iTaK, Lallemand, Lesaffre, L’Oréal, Mayoli, Merck, Pilèje, Procter and Gamble, Second Genome, Therascience, and URGO.
References
- Aguirre de Carcer D, Cuiv PO, Wang Tet al. . Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J. 2011;5:801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens AP, Culpepper T, Saldivar Bet al. . A six-day, lifestyle-based immersion program mitigates cardiovascular risk factors and induces shifts in gut microbiota, specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A pilot study. Nutrients. 2021;13:3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainonen S, Tejesvi MV, Mahmud MRet al. . Antibiotics at birth and later antibiotic courses: effects on gut microbiota. Pediatr Res. 2022;91:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiho H, Ihara E, Nakamura K. Low-grade inflammation plays a pivotal role in gastrointestinal dysfunction in irritable bowel syndrome. World J Gastrointest Pathophysiol. 2010;1:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alameddine J, Godefroy E, Papargyris Let al. . Faecalibacterium prausnitzii skews Human DC to prime IL10-producing T cells through TLR2/6/JNK signaling and IL-10, IL-27, CD39, and IDO-1 induction. Front Immunol. 2019;10:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-García P, Martín R, Martínez-Pinilla E. Gut microbial imbalance and neurodegenerative proteinopathies: from molecular mechanisms to prospects of clinical applications. Explor Neuroprotective Ther. 2021;1:33–54. [Google Scholar]
- Arroyo-Johnson C, Mincey KD. Obesity epidemiology worldwide. Gastroenterol Clin North Am. 2016;45:571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger S, Kropp C, Borras-Nogues Eet al. . Intraspecific diversity of microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii. Int J Mol Sci. 2022;23:1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviello G, Knaus UG. ROS in gastrointestinal inflammation: rescue or sabotage?. Br J Pharmacol. 2017;174:1704–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl MI, Jorgensen SMD, Skriver AHet al. . Faecal microbiota transplantation for eradication of co-infection with Clostridioides difficile and extensively drug-resistant KPC-producing Klebsiella pneumoniae. Scand J Gastroenterol. 2020;55:626–30. [DOI] [PubMed] [Google Scholar]
- Bellais S, Nehlich M, Ania Met al. . Species-targeted sorting and cultivation of commensal bacteria from the gut microbiome using flow cytometry under anaerobic conditions. Microbiome. 2022;10:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevides L, Burman S, Martin Ret al. . New insights into the diversity of the genus Faecalibacterium. Front Microbiol. 2017;8:1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9:318–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benus RF, van der Werf TS, Welling GWet al. . Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr. 2010;104:693–700. [DOI] [PubMed] [Google Scholar]
- Bindels LB, Delzenne NM, Cani PDet al. . Towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol. 2015;12:303–10. [DOI] [PubMed] [Google Scholar]
- Bjorkqvist O, Rangel I, Serrander Let al. . Faecalibacterium prausnitzii increases following fecal microbiota transplantation in recurrent Clostridioides difficile infection. PLoS ONE. 2021;16:e0249861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco G, Sanchez B, Fdez-Riverola Fet al. . In silico approach for unveiling the glycoside hydrolase activities in Faecalibacterium prausnitzii through a systematic and integrative large-scale analysis. Front Microbiol. 2019;10:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov DV, Sysolyatin SV, Kalashnikov AIet al. . Shikimic acid: review of its analytical, isolation, and purification techniques from plant and microbial sources. J Chem Biol. 2012;5:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfrate L, Tack J, Grattagliano Iet al. . Microbiota in health and irritable bowel syndrome: current knowledge, perspectives and therapeutic options. Scand J Gastroenterol. 2013;48:995–1009. [DOI] [PubMed] [Google Scholar]
- Bourgonje AR, Roo-Brand G, Lisotto Pet al. . Patients with inflammatory bowel disease show IgG immune responses towards specific intestinal bacterial genera. Front Immunol. 2022;13:842911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyner NM, Michon C, de Sousa CSet al. . Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-kappaB pathway. Front Microbiol. 2017;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueno JF, Fritsch J, Santander AMet al. . Intestinal epithelial cells respond to chronic inflammation and dysbiosis by synthesizing H2O2. Front Physiol. 2019;10:1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas EI. The roles of extracellular vesicles in the immune system. Nat Rev Immunol. 2023;23:236–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Flores G, Pickard JM, Nunez G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat Rev Microbiol. 2022;21:347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G, Ianiro G, Tilg Het al. . European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD. Evaluation of the effects associated with the administration of Akkermansia muciniphila on parameters of metabolic syndrome related to obesity. Ottignies-Louvain-la-Neuve: Université Catholique de Louvain, 2018. [Google Scholar]
- Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract. 2014;2014:872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carasi P, Racedo SM, Jacquot Cet al. . Enterococcus durans EP1 a promising anti-inflammatory probiotic able to stimulate sIgA and to increase Faecalibacterium prausnitzii abundance. Front Immunol. 2017;8:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AH, Yakymenko O, Olivier Iet al. . Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand J Gastroenterol. 2013;48:1136–44. [DOI] [PubMed] [Google Scholar]
- Cato EP, Salmon CW, Moore WECY. Fusobacterium prausnitzii (Hauduroy et al.) Moore and Holdeman: emended description and designation of neotype strain. Int J Syst Evol Microbiol. 1974;24. 10.1099/00207713-24-2-225. [DOI] [Google Scholar]
- Ceccarani C, Bassanini G, Montanari Cet al. . Proteobacteria overgrowth and butyrate-producing taxa depletion in the gut microbiota of glycogen storage disease type 1 patients. Metabolites. 2020;10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput N, Lepage P, Coutzac Cet al. . Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with Ipilimumab. Ann Oncol. 2017;28:1368–79. [DOI] [PubMed] [Google Scholar]
- Charrier C, Duncan GJ, Reid MDet al. . A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology. 2006;152:179–85. [DOI] [PubMed] [Google Scholar]
- Chen H, Ou R, Tang Net al. . Alternation of the gut microbiota in irritable bowel syndrome: an integrated analysis based on multicenter amplicon sequencing data. J Transl Med. 2023;21:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu P, Liu Ret al. . Comprehensive strain-level analysis of the gut microbe Faecalibacterium prausnitzii in patients with liver cirrhosis. mSystems. 2021;6:e0077521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbuy C, Bellet D, Robert Vet al. . Modulation of the caecal gut microbiota of mice by dietary supplement containing resistant starch: impact is donor-dependent. Front Microbiol. 2019;10:1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HE, Jeon EJ, Kim DYet al. . Sodium salicylate induces browning of white adipocytes via M2 macrophage polarization by HO-1 upregulation. Eur J Pharmacol. 2022;928:175085. [DOI] [PubMed] [Google Scholar]
- Chumpitazi BP, Cope JL, Hollister EBet al. . Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther. 2015;42:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov . Xla1 Christensenella Minuta, phase I, randomized, partially placebo-controlled double-blind protocol, evaluating safety, tolerability and impact on the gut microbiota in healthy volunteers, overweight and obese adults (CAUSALITY). ClinicalTrials.gov Identifier: NCT04663139. Bordeaux: YSOPIA Bioscience, 2021. [Google Scholar]
- Collins SM, Denou E, Verdu EFet al. . The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig Liver Dis. 2009;41:850–3. [DOI] [PubMed] [Google Scholar]
- Collins SM. Is the irritable gut an inflamed gut?. Scand J Gastroenterol Suppl. 1992;192:102–5. [DOI] [PubMed] [Google Scholar]
- Cornuault JK, Petit MA, Mariadassou Met al. . Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa RO, Castro PR, Moser Ret al. . Butyrate: connecting the gut-lung axis to the management of pulmonary disorders. Front Nutr. 2022;9:1011732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutzac C, Jouniaux JM, Paci Aet al. . Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. 2020;11:2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SR, Lindsay JO, Fromentin Set al. . Effects of low FODMAP diet on symptoms, fecal microbiome, and markers of inflammation in patients with quiescent inflammatory bowel disease in a randomized trial. Gastroenterology. 2020;158:176–88. [DOI] [PubMed] [Google Scholar]
- Cryan JF, O'Riordan KJ, Cowan CSMet al. . The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. [DOI] [PubMed] [Google Scholar]
- Cui G, Liu S, Liu Zet al. . Gut microbiome distinguishes patients with epilepsy from healthy individuals. Front Microbiol. 2021;12:696632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo C, Sudakaran S, Asimakopoulos Fet al. . Perturbation of the gut microbiome and association with outcomes following autologous stem cell transplantation in patients with multiple myeloma. Leuk Lymphoma. 2022;64:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hoe K, Conterno L, Fava Fet al. . Prebiotic wheat bran fractions induce specific microbiota changes. Front Microbiol. 2018;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza G, Shitut S, Preussger Det al. . Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35:455–88. [DOI] [PubMed] [Google Scholar]
- Daillere R, Routy B, Goubet AGet al. . Elucidating the gut microbiota composition and the bioactivity of immunostimulatory commensals for the optimization of immune checkpoint inhibitors. Oncoimmunology. 2020;9:1794423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Pasolli E, Ercolini D. Newly explored Faecalibacterium diversity is connected to age, lifestyle, geography, and disease. Curr Biol. 2020;30:4932–43. [DOI] [PubMed] [Google Scholar]
- Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80:1948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers KF, Sayols-Baixeras S, Baldanzi Get al. . An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat Commun. 2022;13:5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci M, Tokman HB, Uysal HKet al. . Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol. 2019;47:365–71. [DOI] [PubMed] [Google Scholar]
- Dikeocha IJ, Al-Kabsi AM, Chiu HTet al. . Faecalibacterium prausnitzii ameliorates colorectal tumorigenesis and suppresses proliferation of HCT116 colorectal cancer cells. Biomedicines. 2022;10:1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H, Rainteau D, Rajca Set al. . Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:513–20. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Hold GL, Harmsen HJMet al. . Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002a;52:2141–6. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Barcenilla A, Stewart CSet al. . Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002b;68:5186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GEet al. . Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–23. [DOI] [PubMed] [Google Scholar]
- Dunn KA, MacDonald T, Rodrigues GJet al. . Antibiotic and antifungal use in pediatric leukemia and lymphoma patients are associated with increasing opportunistic pathogens and decreasing bacteria responsible for activities that enhance colonic defense. Front Cell Infect Microbiol. 2022;12:924707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi R, Anshory M, Kalim Het al. . Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-related diseases. Microorganisms. 2022;10:2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher TP, Mohajeri MH. Overlapping mechanisms of action of brain-active bacteria and bacterial metabolites in the pathogenesis of common brain diseases. Nutrients. 2022;14:2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinga H, Sperna Weiland CJ, Thio HBet al. . Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J Crohns Colitis. 2016;10:1067–75. [DOI] [PubMed] [Google Scholar]
- Fernando WM, Hill JE, Zello GAet al. . Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults. Benef Microbes. 2010;1:197–207. [DOI] [PubMed] [Google Scholar]
- Fitzgerald CB, Shkoporov AN, Sutton TDSet al. . Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genomics. 2018;19:931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foditsch C, Santos TM, Teixeira AGet al. . Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS ONE. 2014;9:e116465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensollen T, Blumberg RS. Correlation between early-life regulation of the immune system by microbiota and allergy development. J Allergy Clin Immunol. 2017;139:1084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godefroy E, Alameddine J, Montassier Eet al. . Expression of CCR6 and CXCR6 by gut-derived CD4(+)/CD8alpha(+) T-regulatory cells, which are decreased in blood samples from patients with inflammatory bowel diseases. Gastroenterology. 2018;155:1205–17. [DOI] [PubMed] [Google Scholar]
- Godny L, Reshef L, Sharar Fischler Tet al. . Increasing adherence to the Mediterranean diet and lifestyle is associated with reduced fecal calprotectin and intra-individual changes in microbial composition of healthy subjects. Gut Microbes. 2022;14:2120749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez CG, Mills RH, Zhu Qet al. . Location-specific signatures of Crohn’s disease at a multi-omics scale. Microbiome. 2022;10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi Let al. . Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi R, Gibson GR, Vulevic Jet al. . A prebiotic intervention study in children with autism spectrum disorders (ASDs). Microbiome. 2018;6:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Yi J, Kim JHet al. . Investigation of intestinal microbiota and fecal calprotectin in non-toxigenic and toxigenic Clostridioides difficile colonization and infection. Microorganisms. 2020;8:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauduroy P, Urbain GEG, Guillot Get al. . Dictionnaire Des Bactéries Pathogènes. Paris: Masson et Cie, 1937. [Google Scholar]
- Hazan S, Stollman N, Bozkurt HSet al. . Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022;9:e000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhao S, Li Y. Faecalibacterium prausnitzii: a next-generation probiotic in gut disease improvement. Can J Infect Dis Med Microbiol. 2021;2021:1–10. [Google Scholar]
- Heinken A, Khan MT, Paglia Get al. . Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol. 2014;196:3289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Bassignani A, Berland Met al. . Modern metaproteomics: a unique tool to characterize the active microbiome in health and diseases, and pave the road towards new biomarkers-example of Crohn’s Disease and ulcerative colitis flare-ups. Cells. 2022;11:1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson MA, Phalak P. Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC Syst Biol. 2017;11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid Get al. . Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–14. [DOI] [PubMed] [Google Scholar]
- Hippe B, Remely M, Aumueller Eet al. . Faecalibacterium prausnitzii phylotypes in type two diabetic, obese, and lean control subjects. Benef Microbes. 2016;7:511–7. [DOI] [PubMed] [Google Scholar]
- Hooda S, Boler BM, Serao MCet al. . 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr. 2012;142:1259–65. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT, Furrie Eet al. . Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol. 2005;54:77–85. [DOI] [PubMed] [Google Scholar]
- Hu W, Lu W, Li Let al. . Both living and dead Faecalibacterium prausnitzii alleviate house dust mite-induced allergic asthma through the modulation of gut microbiota and short-chain fatty acid production. J Sci Food Agric. 2021;101:5563–73. [DOI] [PubMed] [Google Scholar]
- Hustoft TN, Hausken T, Ystad SOet al. . Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol Motil. 2017;29:27747984. [DOI] [PubMed] [Google Scholar]
- Iglesias-Vazquez L, Van Ginkel Riba G, Arija Vet al. . Composition of gut microbiota in children with Autism spectrum disorder: a systematic review and meta-analysis. Nutrients. 2020;12:792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A, Horinaka M, Aono Yet al. . Salicylic acid directly binds to ribosomal protein S3 and suppresses CDK4 expression in colorectal cancer cells. Biochem Biophys Res Commun. 2022;628:110–5. [DOI] [PubMed] [Google Scholar]
- Jafari B, Ali Khavari Nejad R, Vaziri Fet al. . Evaluation of the effects of extracellular vesicles derived from Faecalibacterium prausnitzii on lung cancer cell line. Biologia. 2019;74:889–98. [Google Scholar]
- Kang D, Ham HI, Lee SHet al. . Functional dissection of the phosphotransferase system provides insight into the prevalence of Faecalibacterium prausnitzii in the host intestinal environment. Environ Microbiol. 2021;23:4726–40. [DOI] [PubMed] [Google Scholar]
- Kawade Y, Sakai M, Okamori Met al. . Administration of live, but not inactivated, Faecalibacterium prausnitzii has a preventive effect on dextran sodium sulfate–induced colitis in mice. Mol Med Rep. 2019;20:25–32. [DOI] [PubMed] [Google Scholar]
- Keeley TP, Mann GE. Defining physiological normoxia for improved translation of cell physiology to animal models and humans. Physiol Rev. 2019;99:161–234. [DOI] [PubMed] [Google Scholar]
- Khan MT, Duncan SH, Stams AJet al. . The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6:1578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, van Dijl JM, Harmsen HJ. Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium Faecalibacterium prausnitzii alive at ambient air. PLoS ONE. 2014;9:e96097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Montassier E, Schmidt Bet al. . Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment Pharmacol Ther. 2016;44:715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapiere A, Geiger M, Robert Vet al. . Prophylactic Faecalibacterium prausnitzii treatment prevents the acute breakdown of colonic epithelial barrier in a preclinical model of pelvic radiation disease. Gut Microbes. 2020;12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval L, Martin R, Natividad JNet al. . Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes. 2015;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bastard Q, Ward T, Sidiropoulos Det al. . Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci Rep. 2018;8:6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin Jet al. . Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. [DOI] [PubMed] [Google Scholar]
- Lebas M, Garault P, Carrillo Det al. . Metabolic response of Faecalibacterium prausnitzii to cell-free supernatants from lactic acid bacteria. Microorganisms. 2020;8:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AA, Rao K, Limsrivilai Jet al. . Temporal gut microbial changes predict recurrent Clostridiodes difficile infection in patients with and without ulcerative colitis. Inflamm Bowel Dis. 2020;26:1748–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Byeon HR, Jang SYet al. . Oral administration of Faecalibacterium prausnitzii and Akkermansia muciniphila strains from humans improves atopic dermatitis symptoms in DNCB induced NC/Nga mice. Sci Rep. 2022;12:7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Martin R, Torres-Maravilla Eet al. . Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes. 2020;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye J. Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: a Chinese population-based study. Medicine. 2020;99:e21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limeta A, Ji B, Levin Met al. . Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight. 2020;5:e140940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstad LJ, Lo G, Leivers Set al. . Human gut Faecalibacterium prausnitzii deploys a highly efficient conserved system to cross-feed on beta-mannan-derived oligosaccharides. Mbio. 2021;12:e0362820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ye S, Zhu Xet al. . Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang J, He Tet al. . Butyrate: a double-edged sword for health?. Adv Nutr. 2018;9:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kuang D, Li Det al. . Roles of the gut microbiota in severe SARS-CoV-2 infection. Cytokine Growth Factor Rev. 2022;63:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Duncan SH, Garcia-Gil LJet al. . Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Khan TM, Duncan SHet al. . Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012;78:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Abella Cet al. . Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015;81:7582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Suris-Valls Ret al. . Changes in the abundance of Faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis. 2016;22:28–41. [DOI] [PubMed] [Google Scholar]
- Ma J, Sun L, Liu Yet al. . Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020;20:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAINTAIN . EXL01 in the maintenance of steroid-induced clinical response/remission in participants with mild to moderate Crohn’s disease. 2022. https://clinicaltrials.gov/ct2/show/NCT05542355?term=faecalibacterium&draw=2&rank=5 (22 March 2023, date last accessed).
- Manor O, Dai CL, Kornilov SAet al. . Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JK, Thabane M, Garg AXet al. . Intestinal permeability in patients with irritable bowel syndrome after a waterborne outbreak of acute gastroenteritis in Walkerton, Ontario. Aliment Pharmacol Ther. 2004;20:1317–22. [DOI] [PubMed] [Google Scholar]
- Martin R, Bermudez-Humaran LG, Langella P. Searching for the bacterial effector: the example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Front Microbiol. 2018;9:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Chain F, Miquel Set al. . The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 2014;20:417–30. [DOI] [PubMed] [Google Scholar]
- Martin R, Langella P. Emerging health concepts in the probiotics field: streamlining the definitions. Front Microbiol. 2019;10:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Miquel S, Benevides Let al. . Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front Microbiol. 2017;8:1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Miquel S, Chain Fet al. . Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015;15:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gallausiaux C, Marinelli L, Blottiere HMet al. . SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc. 2021;80:37–49. [DOI] [PubMed] [Google Scholar]
- Messori A, Brignola C, Trallori Get al. . Effectiveness of 5-aminosalicylic acid for maintaining remission in patients with Crohn’s disease: a meta-analysis. Am J Gastroenterol. 1994;89:692–8. [PubMed] [Google Scholar]
- Milani C, Ticinesi A, Gerritsen Jet al. . Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Beaumont M, Martin Ret al. . A proposed framework for an appropriate evaluation scheme for microorganisms as novel foods with a health claim in Europe. Microb Cell Fact. 2015a;14:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Leclerc M, Martin Ret al. . Identification of metabolic signatures linked to anti-inflammatory effects of faecalibacterium prausnitzii. mBio. 2015b;6. 10.13140/RG.2.1.3628.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Lashermes Aet al. . Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci Rep. 2016;6:19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Martin R, Rossi Oet al. . Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–61. [DOI] [PubMed] [Google Scholar]
- Moens F, Weckx S, De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int J Food Microbiol. 2016;231:76–85. [DOI] [PubMed] [Google Scholar]
- Monchatre-Leroy Elodie BF, Picard-Meyer E, Servat Aet al. . Utilisation de Faecalibacterium prausnitzii pour traiter une infection par un virus respiratoire. FR2109498. 2021. [Google Scholar]
- Montalban-Arques A, Katkeviciute E, Busenhart Pet al. . Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. 2021;29:1573–88. [DOI] [PubMed] [Google Scholar]
- Moosavi SM, Akhavan Sepahi A, Mousavi SFet al. . The effect of Faecalibacterium prausnitzii and its extracellular vesicles on the permeability of intestinal epithelial cells and expression of ppars and ANGPTL4 in the Caco-2 cell culture model. J Diabetes Metab Disord. 2020;19:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol Het al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munukka E, Rintala A, Toivonen Ret al. . Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 2017;11:1667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Hashikura N, Yoshida Ket al. . Growth-promoting effect of alginate on Faecalibacterium prausnitzii through cross-feeding with Bacteroides. Food Res Int. 2021;144:110326. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Sasaki K, Sasaki Det al. . The alga Euglena gracilis stimulates faecalibacterium in the gut and contributes to increased defecation. Sci Rep. 2021;11:1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navab-Moghadam F, Sedighi M, Khamseh MEet al. . The association of type II diabetes with gut microbiota composition. Microb Pathog. 2017;110:630–6. [DOI] [PubMed] [Google Scholar]
- Newsome RC, Gharaibeh RZ, Pierce CMet al. . Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort. Genome Med. 2022;14:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova VL, Smith MRB, Hall LJet al. . Perturbations in gut microbiota composition in psychiatric disorders: a review and meta-analysis. JAMA Psych. 2021;78:1343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki H, Ito M, Ishida Tet al. . Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov Disord. 2020;35:1626–35. [DOI] [PubMed] [Google Scholar]
- Obuya S, Elkholy A, Avuthu Net al. . A signature of Prevotella copri and Faecalibacterium prausnitzii depletion, and a link with bacterial glutamate degradation in the Kenyan colorectal cancer patients. J Gastrointest Oncol. 2022;13:2282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YJ, Nam K, Kim Yet al. . Effect of a nutritionally balanced diet comprising whole grains and vegetables alone or in combination with probiotic supplementation on the gut microbiota. Prev Nutr Food Sci. 2021;26:121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olm RM, Brown CT, Brooks Bet al. . dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson LM, Boulund F, Nilsson Set al. . Dynamics of the normal gut microbiota: a longitudinal one-year population study in Sweden. Cell Host Microbe. 2022;30:726–39. [DOI] [PubMed] [Google Scholar]
- Pallav K, Dowd SE, Villafuerte Jet al. . Effects of polysaccharopeptide from Trametes versicolor and amoxicillin on the gut microbiome of healthy volunteers: a randomized clinical trial. Gut Microbes. 2014;5:458–67. [DOI] [PubMed] [Google Scholar]
- Paller AS, Kong HH, Seed Pet al. . The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Song WS, Lee Jet al. . An integrative multiomics approach to characterize prebiotic inulin effects on Faecalibacterium prausnitzii. Front Bioeng Biotechnol. 2022;10:825399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prausnitz C. Der Bacillus mucosus anaerobius. Zentralbl. Bakteriol Parasitenk Infektionskr Hyg Abt Orig. 1922;89:126–32. [Google Scholar]
- Qin J, Li R, Raes Jet al. . A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Zhang M, Yang Xet al. . Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J Crohns Colitis. 2013;7:e558–568. [DOI] [PubMed] [Google Scholar]
- Quevrain E, Maubert MA, Michon Cet al. . Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiei N, Ahmadi Badi S, Ettehad Marvasti Fet al. . Induction effects of Faecalibacterium prausnitzii and its extracellular vesicles on toll-like receptor signaling pathway gene expression and cytokine level in human intestinal epithelial cells. Cytokine. 2019;121:154718. [DOI] [PubMed] [Google Scholar]
- Raise ADS, Dupont S, Iaconelli Cet al. . Comparison of two encapsulation processes to protect the commensal gut probiotic bacterium Faecalibacterium prausnitzii from the digestive tract. J Drug Deliv Sci Technol. 2020;56:101608. [Google Scholar]
- Rajilic-Stojanovic M, Biagi E, Heilig HGet al. . Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–801. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Zet al. . Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–50. [DOI] [PubMed] [Google Scholar]
- Remely M, Hippe B, Geretschlaeger Iet al. . Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wien Klin Wochenschr. 2015;127:394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigsbee L, Agans R, Shankar Vet al. . Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol. 2012;107:1740–51. [DOI] [PubMed] [Google Scholar]
- Rios-Covian D, Gueimonde M, Duncan SHet al. . Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. 2015;362:fnv176. [DOI] [PubMed] [Google Scholar]
- Rossi O, Khan MT, Schwarzer Met al. . Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS ONE. 2015;10:e0123013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O, van Berkel LA, Chain Fet al. . Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6:18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa Let al. . Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- Sabo CM, Dumitrascu DL. Microbiota and the irritable bowel syndrome. Minerva Gastroenterol. 2021;67:377–84. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Sakurai N, Tanno Het al. . Genome-based, phenotypic and chemotaxonomic classification of Faecalibacterium strains: proposal of three novel species Faecalibacterium duncaniae sp. nov., Faecalibacterium hattorii sp. nov. and Faecalibacterium gallinarum sp. nov. Int J Syst Evol Microbiol. 2022;72:35416766. [DOI] [PubMed] [Google Scholar]
- Sarrabayrouse G, Bossard C, Chauvin JMet al. . CD4CD8αα lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PLoS Biol. 2014;12:e1001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R, Shah YM. Oxygen battle in the gut: hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J Biol Chem. 2020;295:10493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot Let al. . Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Nat Acad Sci USA. 2008;105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JPet al. . Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. [DOI] [PubMed] [Google Scholar]
- Song H, Yoo Y, Hwang Jet al. . Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137:852–60. [DOI] [PubMed] [Google Scholar]
- Spencer CN, McQuade JL, Gopalakrishnan Vet al. . Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefia LV, Lee J, Patel Jet al. . Secretome components from Faecalibacterium prausnitzii strains A2-165 and AHMP21 modulate cutaneous wound inflammation. J Invest Dermatol. 2020;140:2312–5. [DOI] [PubMed] [Google Scholar]
- Stewardson AJ, Gaia N, Francois Pet al. . Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin Microbiol Infect. 2015;21:344.e341–311. [DOI] [PubMed] [Google Scholar]
- Stewart DB Sr, Wright JR, Fowler Met al. . Integrated meta-omics reveals a fungus-associated bacteriome and distinct functional pathways in Clostridioides difficile infection. mSphere. 2019;4. 10.1128/mSphere.00454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Yu J, Hu Jet al. . Salicylic acid-based hypoxia-responsive chemodynamic nanomedicines boost antitumor immunotherapy by modulating immunosuppressive tumor microenvironment. Acta Biomater. 2022;148:230–43. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Bengmark Set al. . Bacterial biofilm suppression with antibiotics for ulcerative and indeterminate colitis: consequences of aggressive treatment. Arch Med Res. 2008;39:198–204. [DOI] [PubMed] [Google Scholar]
- Tagliabue A, Ferraris C, Uggeri Fet al. . Short-term impact of a classical ketogenic diet on gut microbiota in GLUT1 deficiency syndrome: a 3-month prospective observational study. Clin Nutr ESPEN. 2017;17:33–7. [DOI] [PubMed] [Google Scholar]
- Tanno H, Chatel JM, Martin Ret al. . New gene markers for classification and quantification of Faecalibacterium spp. In the human gut. FEMS Microbiol Ecol. 2023;99:fiad035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno H, Maeno S, Salminen Set al. . 16S rRNA gene sequence diversity in Faecalibacterium prausnitzii-complex taxa has marked impacts on quantitative analysis. FEMS Microbiol Ecol. 2022;98:fiac004. [DOI] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez Fet al. . Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–84. [DOI] [PubMed] [Google Scholar]
- Tikunov AY, Shvalov AN, Morozov VVet al. . Taxonomic composition and biodiversity of the gut microbiome from patients with irritable bowel syndrome, ulcerative colitis, and asthma. Vavilovskii Zhurnal Genet Selektsii. 2021;25:864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tims S, Marsaux C, Pinto Aet al. . Altered gut microbiome diversity and function in patients with propionic acidemia. Mol Genet Metab. 2022;137:308–22. [DOI] [PubMed] [Google Scholar]
- Tomas J, Wrzosek L, Bouznad Net al. . Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J. 2013;27:645–55. [DOI] [PubMed] [Google Scholar]
- Touch S, Godefroy E, Rolhion Net al. . Human CD4+CD8alpha+ tregs induced by Faecalibacterium prausnitzii protect against intestinal inflammation. JCI Insight. 2022;7:e154722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Schild S, Kaparakis-Liaskos Met al. . Composition and functions of bacterial membrane vesicles. Nat Rev Microbiol. 2023; 21:415–30. [DOI] [PubMed] [Google Scholar]
- Trottein F, Sokol H. Potential causes and consequences of gastrointestinal disorders during a SARS-CoV-2 infection. Cell Rep. 2020;32:107915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudela H, Claus SP, Saleh M. Next generation microbiome research: identification of keystone species in the metabolic regulation of host-gut microbiota interplay. Front Cell Dev Biol. 2021;9:719072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Shinkai S, Shiroma Het al. . Identification of Faecalibacterium prausnitzii strains for gut microbiome-based intervention in Alzheimer's-type dementia. Cell Rep Med. 2021;2:100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Colomer M, Falony G, Darzi Yet al. . The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4:623–32. [DOI] [PubMed] [Google Scholar]
- Vandeputte D, Joossens M. Effects of low and high FODMAP diets on human gastrointestinal microbiota composition in adults with intestinal diseases: a systematic review. Microorganisms. 2020;8:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduci E, Moretti F, Bassanini Get al. . Phenylketonuric diet negatively impacts on butyrate production. Nutr Metab Cardiovasc Dis. 2018;28:385–92. [DOI] [PubMed] [Google Scholar]
- Verhoog S, Taneri PE, Roa Diaz ZMet al. . Dietary factors and modulation of bacteria strains of Akkermansia muciniphila and Faecalibacterium prausnitzii: a systematic review. Nutrients. 2019;11:1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Wang F, Yuan Jet al. . Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut. 2019;68:1417–29. [DOI] [PubMed] [Google Scholar]
- Wang J, Ji H, Wang Set al. . Probiotic Lactobacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front Microbiol. 2018a;9:1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhang S, Yerke Aet al. . Avenanthramide metabotype from whole-grain oat intake is influenced by Faecalibacterium prausnitzii in healthy adults. J Nutr. 2021;151:1426–35. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li F, Liang Bet al. . A metagenome-wide association study of gut microbiota in asthma in UK adults. BMC Microbiol. 2018b;18:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu L, Chang Set al. . Total body irradiation-induced colon damage is prevented by nitrate-mediated suppression of oxidative stress and homeostasis of the gut microbiome. Nitric Oxide. 2020;102:1–11. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu B, Chen Het al. . Environmental factors and gut microbiota: toward better conservation of deer species. Front Microbiol. 2023;14:1136413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzosek L, Miquel S, Noordine MLet al. . Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Bhat ZF, Gounder RSet al. . Effect of dietary protein and processing on gut microbiota-A systematic review. Nutrients. 2022;14:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J, Ding D, Zhu Het al. . Disturbed microbial ecology in Alzheimer's disease: evidence from the gut microbiota and fecal metabolome. BMC Microbiol. 2021;21:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Xu X, Li Jet al. . Association between gut microbiota and Autism spectrum disorder: a systematic review and meta-analysis. Front Psychiatry. 2019;10:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubfar R, Behrouzi A, Zare Banadkoki Eet al. . Effect of Akkermansia muciniphila, Faecalibacterium prausnitzii, and their extracellular vesicles on the serotonin system in intestinal epithelial cells. Probiotics Antimicrob Proteins. 2021;13:1546–56. [DOI] [PubMed] [Google Scholar]
- Ye X, Wang D, Zhu Het al. . Gut microbiota changes in patients with major depressive disorder treated with vortioxetine. Front Psychiatry. 2021;12:641491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh YK, Zuo T, Lui GCet al. . Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang YJ, Yoon JYet al. . Primary human colonic mucosal barrier crosstalk with super oxygen-sensitive Faecalibacterium prausnitzii in continuous culture. Med. 2021;2:74–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv Biomark Sci Technol. 2020;2:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Deeke SA, Ning Zet al. . Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat Commun. 2018;9:2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shen D, Fang Zet al. . Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE. 2013;8:e71108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Coker OO, Wu Jet al. . Aspirin reduces colorectal tumor development in mice and gut microbes reduce its bioavailability and chemopreventive effects. Gastroenterology. 2020;159:969–83. [DOI] [PubMed] [Google Scholar]
- Zou Y, Lin X, Xue Wet al. . Characterization and description of Faecalibacterium butyricigenerans sp. nov. and F. longum sp. nov., isolated from human faeces. Sci Rep. 2021;11:11340. [DOI] [PMC free article] [PubMed] [Google Scholar]