Significance
Light-inducible proteins have had tremendous impact for the precise control of cells in space and time. We found a new type of light-induced behavior in the BcLOV4 photoreceptor, which simultaneously clusters and translocates to the membrane in mammalian cells. This unique combination opens the door to a new class of optogenetic tools. We also find that clustering and membrane translocation are linked such that enhancing clustering enhances membrane translocation. This allows us to rapidly generate more potent and sensitive variants of BcLOV-based optogenetic probes. Because BcLOV4 works in cells from diverse organisms including yeast, flies, zebrafish, ciona, and humans, our findings can be applied broadly.
Keywords: optogenetics, clustering, cell signaling, intrinsically disordered regions
Abstract
Optogenetic tools respond to light through one of a small number of behaviors including allosteric changes, dimerization, clustering, or membrane translocation. Here, we describe a new class of optogenetic actuator that simultaneously clusters and translocates to the plasma membrane in response to blue light. We demonstrate that dual translocation and clustering of the BcLOV4 photoreceptor can be harnessed for novel single-component optogenetic tools, including for control of the entire family of epidermal growth factor receptor (ErbB1-4) tyrosine kinases. We further find that clustering and membrane translocation are mechanistically linked. Stronger clustering increased the magnitude of translocation and downstream signaling, increased sensitivity to light by ~threefold-to-fourfold, and decreased the expression levels needed for strong signal activation. Thus light-induced clustering of BcLOV4 provides a strategy to generate a new class of optogenetic tools and to enhance existing ones.
Optogenetics enables optical control of proteins by coupling them to naturally evolved photoreceptor switches. These switches can undergo one of a handful of inducible behaviors, including conformational changes (1, 2), homo-/hetero-dimerization (3–8), clustering (9, 10), and membrane translocation (11, 12), each of which has been leveraged to control numerous aspects of cell physiology. In some cases, a photoreceptor can possess multiple such functions. One example is Arabidopsis Cry2, which, in addition to heterodimerizing with CIB1, also forms light-induced clusters (8–10, 13).
BcLOV4 is a photoreceptor that translocates from the cytosol to the plasma membrane under blue light (11). Although the molecular details of this process are not fully understood, BcLOV4 contains a canonical LOV domain that associates with a flavin mononucleotide cofactor (FMN) (11). Blue light absorption by the FMN triggers a conformational change that propagates through the LOV domain and, in the case of BcLOV4, exposes an amphipathic helix. The exposed helix increases the protein’s affinity for anionic phospholipids, resulting in translocation to the plasma membrane. This mechanism has now allowed light-induced translocation of BcLOV4 in cells from a broad range of organisms including yeast, flies, zebrafish, ciona, and humans (11, 14–17).
BcLOV4 translocation has been leveraged for multiple probes of cell signaling, including of Rho GTPases, Ras, and PI3K signaling. While live-cell microscopy showed that stimulated BcLOV4 translocates to the plasma membrane, experiments with purified protein found that BcLOV4 can also undergo light-induced aggregation (11). However, clustering was not observed in the presence of lipid membranes. Within water-in-oil emulsions, light-stimulated BcLOV4 localized to the membrane and did not visibly aggregate, although aggregation was still observed in the center of large emulsion droplets where the diffusive distance to the membrane was largest (11). These results suggested that BcLOV4 clustering and membrane association may be mutually antagonistic. However, whether BcLOV4 forms clusters at the membrane, and whether clustering plays a role in BcLOV4 translocation, has not been formally tested.
The inability to observe BcLOV4 clustering in cells could be explained if membrane-associated clusters were sufficiently small. Small clusters will not appear punctate under conventional fluorescence imaging due to measurement limitations including the diffraction limit of light and a low signal-to-noise of the aggregated fluorophore against the fluorescence background (18). A similar effect can be observed with the bona fide clustering module Cry2, which clusters in response to blue light, but whose clusters can only be observed above an expression threshold (9, 18–21). Recently our group developed the CluMPS reporter to indicate the presence of protein clusters as small as trimers (18). When applied to Cry2, CluMPS revealed the presence of small light-induced Cry2 clusters at all expression levels, including at low expression levels where clusters could not be otherwise observed (18).
Clarifying the existence of BcLOV4 clustering in cells would be impactful for several reasons. Optogenetic clustering of Cry2 has been a powerful approach, for example, in studies of cell signaling (9, 22), stem cell differentiation (23, 24), neurodegenerative aggregation (25), and protein phase separation (26) However, Cry2 remains the only photoreceptor whose light-induced clustering has been used for optogenetic control. Additional such methods would expand the applications toward which optogenetic clustering could be applied. Further, understanding the molecular details of BcLOV4 activation may yield insights to understand and mitigate the unique temperature-sensitivity of BcLOV4, which spontaneously self-inactivates within ~ 1 h of strong light stimulation above ~30 °C (16).
In this work, we find that BcLOV4 is a multifunctional protein that simultaneously clusters and translocates to the membrane under light stimulation in cells. We leverage this multifunctionality to generate new tools for the activation of the epidermal growth factor receptor (EGFR) kinase, which could not be activated by membrane translocation alone. We further apply this same strategy to regulate the entire ErbB receptor family in a modular manner and find receptor-specific signal dynamics. Surprisingly, in contrast to previous evidence that clustering and translocation are antagonistic, we find that clustering potentiates BcLOV4 membrane translocation, sensitizes stimulation to lower levels of light, and diminishes temperature-dependent inactivation. Our work thus uncovers new features of BcLOV4 stimulation and provides a platform to engineer a unique class of optogenetic tools and enhance existing ones.
Results
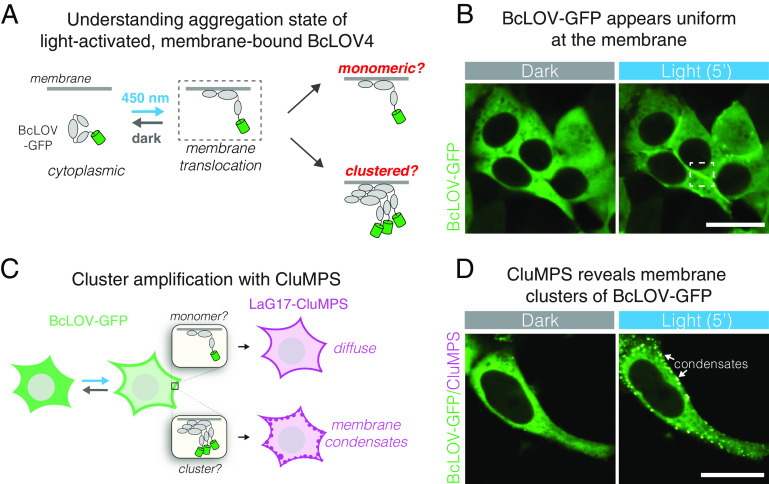
To examine whether BcLOV4 formed light-induced clusters at the membrane, we transiently transfected BcLOV-GFP in HEK 293T cells and observed its distribution after blue light stimulation (Fig. 1A). As reported before (11, 14–16, 27), fluorescence appeared mostly uniform at the membrane (Fig. 1B). However, we noticed that in cells with high expression, light-induced fluorescent puncta could be observed at the membrane (SI Appendix, Fig. S1A). Since cluster size depends on concentration, we reasoned that smaller clusters may be forming in low-expressing cells as well, but at a submicroscopic scale. To test this possibility, we repeated our imaging experiment in the presence of a CluMPS reporter. CluMPS reporters amplify small protein clusters through multivalent interactions that generate large fluorescent condensates in the presence of a clustered target (18) (Fig. 1C). In cells that coexpressed BcLOV-GFP with a CluMPS reporter of GFP clustering (LaG17-CluMPS), light stimulation rapidly triggered the formation of condensates at the membrane, regardless of BcLOV4 expression level (Fig. 1D, SI Appendix, Fig. S1B, and Movie S1). Notably, CluMPS did not produce reporter condensates in response to membrane recruitment of GFP through optogenetic heterodimerization [iLid and sspB-GFP (5)], suggesting that clustering and CluMPS activation was not a general property of membrane translocation (SI Appendix, Fig. S1C).
Fig. 1.
BcLOV4 forms light-induced clusters at the membrane. (A) BcLOV-GFP translocates to the plasma membrane when stimulated with blue light. However, it is unclear whether it forms clusters at the membrane. (B) Representative image of membrane recruitment of BcLOV-GFP upon blue-light stimulation in HEK 293T cells. Dashed box shows membrane localization as depicted in (A). (Scale bar, 20 μm.) (C) The CluMPS reporter for GFP clustering (LaG17-CluMPS) was coexpressed with BcLOV to amplify and visualize potential submicroscopic membrane-associated clusters of BcLOV-GFP. (D) Representative images of membrane recruitment of BcLOV-GFP in the presence of LaG17-CluMPS. CluMPS amplifies and visualizes membrane-associated BcLOV condensates in the light. [Scale bar, 20 μm (Movie S1).] See SI Appendix, Fig. S1 for additional controls and SI Appendix, Table S1 for details of optogenetic illumination parameters.
The unique ability of BcLOV4 to both translocate and cluster in response to blue light carries the potential for new types of optogenetic tools. The optogenetic clustering protein Cry2 has been applied to cluster and activate receptor tyrosine kinases (RTKs) (22, 28–31). However, these tools require either constitutive anchoring to the plasma membrane, which could raise basal signaling levels (28–30, 32), or require a separate interaction partner anchored at the membrane, which can necessitate stoichiometric tuning between the two components for optimal function (32, 33). We reasoned that BcLOV4 could implement a simpler, single-component variant of such tools, without the need for membrane anchoring.
We first sought to stimulate EGFR (ErbB1), a receptor important for cell growth and survival that is commonly misregulated in human cancers. We fused BcLOV-mCh to the N terminus of the EGFR intracellular domain (BcLOV-EGFR, Fig. 2A). To assay activation, we observed activity of the downstream Erk kinase using the ErkKTR reporter, a fluorescent probe that translocates from the nucleus to the cytoplasm upon Erk activation (Fig. 2B) (34). Within seconds after light stimulation, BcLOV-EGFR translocated to the membrane, and within minutes, ErkKTR-miRFP moved from the nucleus to the cytoplasm, indicating Erk activation (Fig. 2C and Movie S2). BcLOV-EGFR signaling could be stimulated and inactivated over multiple cycles (Fig. 2D). Immunofluorescence staining for phospho-Erk (ppErk) confirmed light-induced Erk activation and also showed a lack of basal pathway activation in transfected but unstimulated (dark state) cells (SI Appendix, Fig. S2 A–E). BcLOV-EGFR stimulated signaling in multiple cell types, including in NIH 3T3s, which do not express endogenous EGFR (SI Appendix, Fig. S2 F–J), demonstrating that BcLOV-EGFR does not require the endogenous receptor for its activity.
Fig. 2.
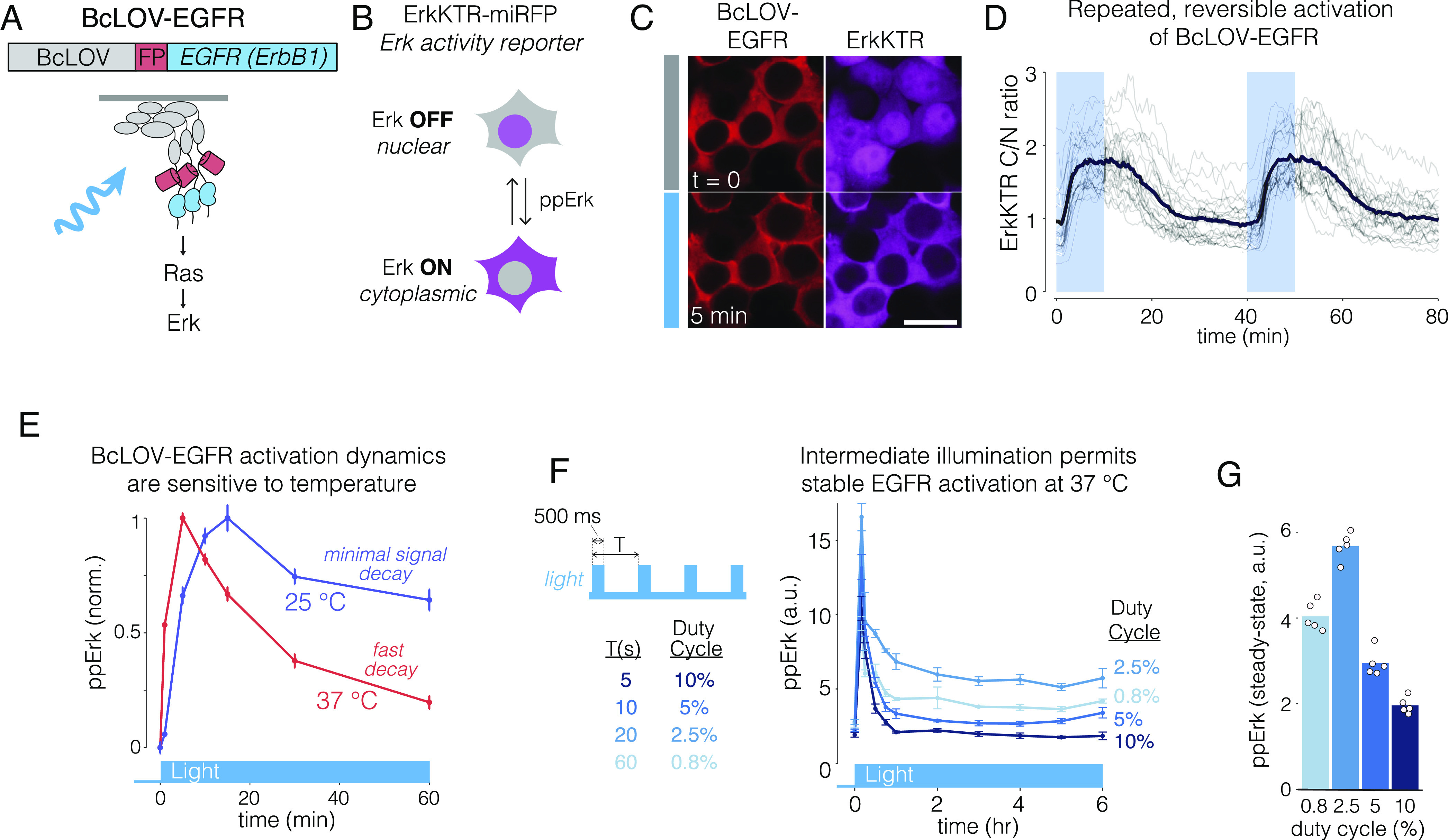
Membrane translocation and clustering allows activation of EGFR. (A) The intracellular domain of EGFR was fused to BcLOV-mCherry to generate BcLOV-EGFR. Light-induced BcLOV-EGFR activity was assessed by measuring activity of downstream Ras-Erk signaling. (B) ErkKTR is a fluorescent biosensor of Erk activity. ErkKTR is nuclear when Erk is off and is cytoplasmic when Erk is on. (C) HEK 293T cells that coexpress BcLOV-EGFR and ErkKTR-miRFP show Erk activation upon light stimulation. [Scale bar, 20 μm (Movie S2).] (D) Erk activity could be stimulated reversibly over multiple cycles. Gray traces represent mean ErkKTR C/N ratios of individual cells (n = 25). Black trace represents mean of these traces. (E) BcLOV-EGFR signal kinetics depend on temperature. Erk signal can be stably maintained with light at 25 °C but decays more rapidly 37 °C. Data represent mean ± SEM of four replicates, with each replicate representing the mean of ~1,000 to 4,000 cells. (F) BcLOV-EGFR activity can be stably maintained at 37 °C at intermediate light doses. Cells were stimulated with pulse trains of light of variable duty cycles. (G) Maximal steady-state Erk levels were achieved at 2.5% duty cycle (500 ms ON every 20 s). Data in (G) represents the mean ± SEM of four replicates, with each replicate representing the mean of ~300 to 1,700 cells. Datapoints in (G) are the mean steady-state (from 2 h to 6 h) ppErk levels shown in (F). See SI Appendix, Table S1 for details of optogenetic stimulation for all experiments. a.u., arbitrary units.
Notably, clustering was required for EGFR activation, since membrane recruitment of EGFR through 1:1 heterodimerization of the iLid/sspB system was insufficient to stimulate ErkKTR or to elevate ppErk (SI Appendix, Fig. S3 A–E). Moreover, single-cell analysis revealed that EGFR activation was observed at even the lowest detectable levels of BcLOV-EGFR expression, confirming that clustering occurs even at these low levels (SI Appendix, Fig. S3F). Collectively, our data show that 1) both membrane translocation and clustering are required for activation of the EGFR intracellular domain, and 2) BcLOV4 clustering at the membrane can be leveraged for novel optogenetic signaling tools.
BcLOV4 membrane translocation dynamics depend on both light and temperature (16). Although BcLOV4 binds the membrane at all temperatures, it then spontaneously dissociates within ~1 h at a rate that depends on both light exposure and temperature above ~30 °C (16). We observed similar temperature-dependent behavior for BcLOV-EGFR: Erk phosphorylation could be stably maintained at 25 °C but decayed within ~1 h of strong light stimulation at 37 °C (Fig. 2E). However, intermediate but sustained levels of pathway activation (>6 h) could be achieved at mammalian temperatures by using intermediate doses of stimulating light (Fig. 2 F and G). Such intermediate doses sustain signaling presumably because they stimulate a sufficient amount of BcLOV for EGFR activation but also a small enough amount such that only a small fraction of BcLOV undergoes inactivation, leaving a large reservoir of activatable protein to maintain signal activity under sustained pulsatile stimulation.
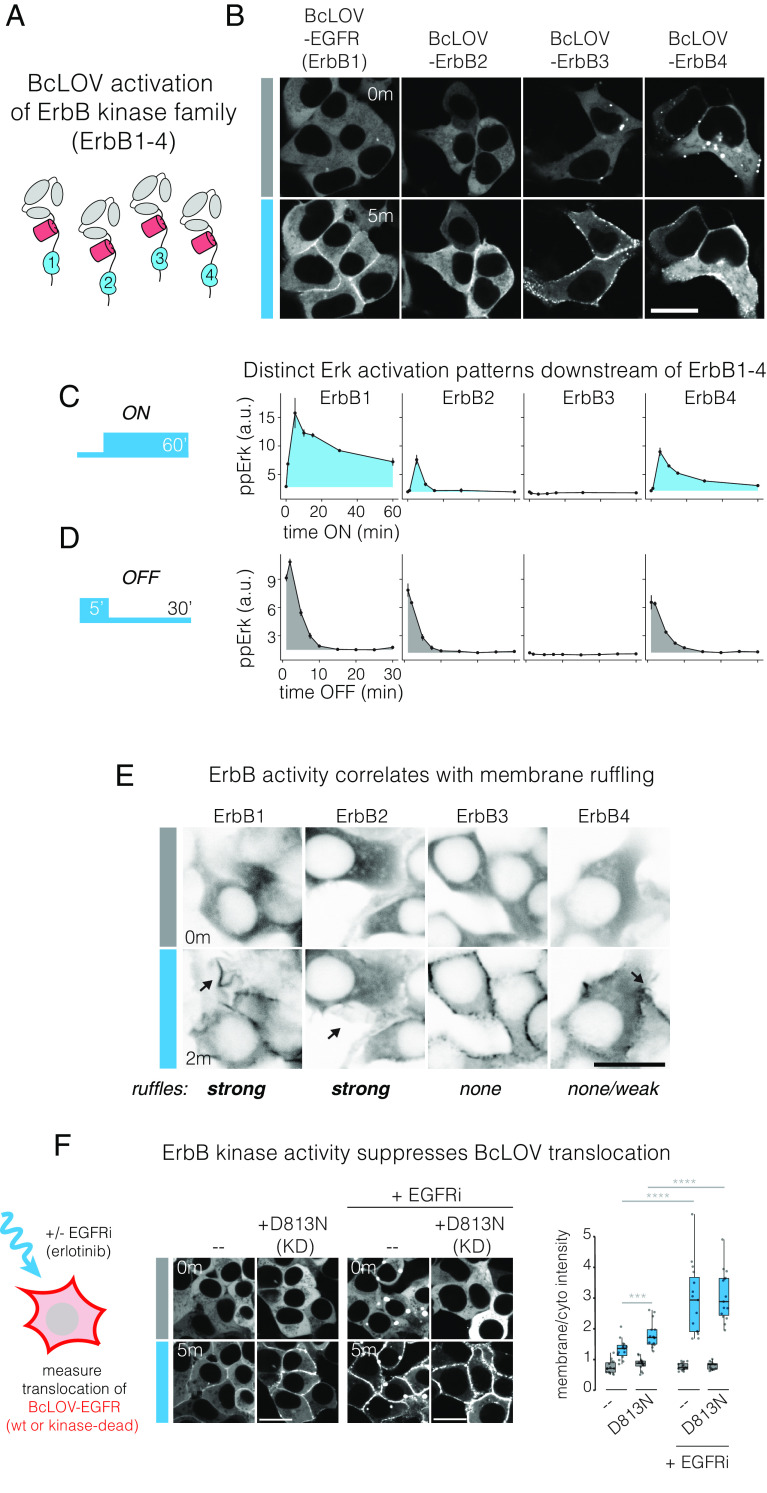
EGFR is a member of the ErbB receptor family, whose members (ErbB1-4) play important roles in development as well as cancer (35). However, it is currently challenging to study the specific activity of each ErbB family member in isolation for several reasons. First, receptor ligands can activate multiple family members. Second, ErbB2 has no known ligand. Third, the ErbB family members can heterodimerize with each other upon ligand activation. Chemical and optical probes have been developed to overcome this challenge for EGFR and ErbB2 (31, 36, 37), although a single method that can stimulate each member of the ErbB receptor family individually has not been reported. We asked whether BcLOV clustering could be used to stimulate ErbB2-4 in the same manner as for EGFR. We fused the intracellular domains of each ErbB receptor to the C-terminus of BcLOV-mCh and observed membrane translocation under light stimulation (Fig. 3A). Each fusion rapidly localized to the plasma membrane after stimulation with blue light (Fig. 3B). Intriguingly, the magnitude of translocation differed between fusions. ErbB1(EGFR) and ErbB2(Her2) showed weak-to-moderate translocation, with apparent uniformity of fluorescence at the membrane. In striking contrast, ErbB3 showed the strongest translocation, even stronger than the original BcLOV-mCh fusion, and showed obvious clusters at the membrane. ErbB4 showed strong membrane translocation and moderate membrane clustering, lower than ErbB3 but more than ErbB1/2. Notably, ErbB3 and ErbB4 fusions on occasion formed cytoplasmic condensates in the dark, and these condensates would dissolve in favor of membrane translocation after light stimulation (Fig. 3B and Movie S3).
Fig. 3.
BcLOV4 clustering at the membrane allows for modular activation of the entire ErbB receptor family. (A) The intracellular domains of ErbB1-4 were fused to the C-terminus of BcLOV-mCherry. (B) Membrane translocation of BcLOV-ErbB1-4 in response to blue light. (Scale bar, 20 μm.) See also Movie S3. (C and D) Erk activation dynamics downstream of BcLOV-ErbB1-4 in response to ON and OFF steps of blue light. See also SI Appendix, Fig. S4. Data represent mean ± SEM of two replicates, with each replicate representing the mean of ~500 to 2,400 cells. (E) Membrane ruffling (black arrows) downstream of stimulation of BcLOV-ErbB1-4, indicative of RTK stimulation. Ruffling is strongest for ErbB1 and ErbB2, less for ErbB4, and absent for ErbB3 activation. (Scale bar, 20 μm.) See also Movie S4. (F) Kinase activity suppresses BcLOV-EGFR membrane translocation. Translocation was observed under light stimulation of BcLOV-EGFR harboring a kinase-inactivating D813N mutation, in either the presence or absence of 1 µM EGFR inhibitor erlotinib (EGFRi). (Scale bar, 20 μm.) Quantification (Right) shows ratios of mean membrane and cytoplasmic fluorescence of 15 cells per condition. Significance was tested by one-way t tests between individual groups and was assessed by comparing P values to Bonferroni-corrected significance level of α/3. ****P < 0.00001, ***P < 0.0001.
We next asked whether translocation of the BcLOV-ErbB fusions activated downstream signaling by measuring downstream Erk phosphorylation. Each receptor fusion elicited distinct Erk activation dynamics (Fig. 3C). In response to sustained stimulation, ppErk activation was strongest for ErbB1, whereas ErbB2 and ErbB4 showed weaker signaling despite equivalent expression levels (SI Appendix, Fig. S4). ErbB1 and ErbB4 also showed sustained signal above baseline, whereas ErbB2 signaled with a transient pulse and rapidly returned to baseline (Fig. 3C and SI Appendix, Fig. S4). ErbB3, by contrast, showed no Erk phosphorylation, in line with the fact that ErbB3 is a pseudokinase and lacks enzymatic activity (38–40). We found no major differences in the OFF-kinetics between each tool, as measured after 5 min of light stimulation and subsequent light removal (Fig. 3D). The half-life of signal decay was ~5 min for Erb1, 2, and 4, with complete loss of signal by 15 min. We further confirmed signal activation using the ErkKTR reporter (SI Appendix, Fig. S5) and through the observation of membrane ruffling, indicative of PI3K/Rac1 activation downstream of receptor activation (Fig. 3E and Movie S4). Collectively, our results show that BcLOV4 can be applied in a modular fashion, with no further optimization, to generate optogenetic tools for each of the ErbB family members.
Successful control of the ErbB receptor family led us to ask whether the modularity of BcLOV4 would extend to other families of RTK signals. We generated fusions of BcLOV4 to the intracellular domain of two other RTKs, fibroblast growth factor 1 (FGFR1) and platelet derived growth factor (PDGFRβ). Both constructs could stimulate the ErkKTR reporter in HEK 293T cells (SI Appendix, Fig. S6). However, for BcLOV-FGFR1, high basal ppErk and Erk activity were observed and were strongly correlated with expression levels of the fusion, such that optimal switching of Erk (OFF in dark, ON in light) could only be achieved in low-expressing cells (SI Appendix, Fig. S6A). FGFR1 stimulation was substantially weaker when recruited to the membrane through iLid/sspB heterodimerization, although Erk signaling could still be stimulated in a fraction of cells (SI Appendix, Fig. S6B). BcLOV-PDGFR showed no elevation of basal ppErk in low/medium-expressing cells, and ppErk induction required medium-high expression (SI Appendix, Fig. S6C). Clustering was required for PDGFR activation, as membrane recruitment with iLid/sspB heterodimerization did not increase ppErk levels (SI Appendix, Fig. S6D). In summary, we find that BcLOV4 can regulate diverse RTKs in a modular manner, although receptor-specific expression, host-cell dependencies, and molecular context will dictate optimization for each individual RTK, as observed previously (29, 30, 36, 37).
Among the BcLOV-ErbB probes, BcLOV-ErbB3 showed the strongest translocation to the membrane despite a lack of downstream signaling (Fig. 3B). Because ErbB3 is the only pseudokinase among the ErbB family, we asked whether kinase activity might suppress membrane translocation. We compared translocation of BcLOV-EGFR to the same construct with a kinase-inactivating D813N mutation, which blocks EGFR kinase activity but does not perturb its multimerization state (41). BcLOV-EGFR(D813N) exhibited stronger membrane translocation than BcLOV-EGFR(wt), suggesting that kinase activity indeed may suppress BcLOV4 translocation (Fig. 3F). As expected, translocation of D813N did not activate the Ras-Erk pathway due to the inactive EGFR kinase (SI Appendix, Fig. S7A). As a further test, we also measured translocation of BcLOV-EGFR in the presence of EGFR inhibitor erlotinib (1 µM, EGFRi). EGFRi treatment yielded a strong increase in translocation compared to untreated cells (Fig. 3F). Notably, EGFRi further enhanced translocation relative to levels seen with the D813N kinase-inactivating mutation. Thus, kinase inhibition may only partly explain the boost in translocation from erlotinib treatment, which could also be explained by drug-induced increase in EGFR dimerization (42, 43). Interestingly, EGFRi also enhanced EGFR endodomain recruitment through the heterodimeric iLid/sspB system (SI Appendix, Fig. S7 B–D). Collectively, our results show that EGFR kinase activity indeed can suppress BcLOV4 translocation, potentially through direct phosphorylation of the membrane-binding domain of BcLOV4, or through recruitment of downstream adapters that sterically interfere with membrane binding. However, enhanced multimerization potential (e.g., due to drug treatment or differences between ErbB family members) could also contribute to increases in translocation.
Previous in vitro experiments suggested that BcLOV4 clustering and membrane association may be mutually antagonistic (11). However, throughout our study, we observed a direct correlation between increased clustering and stronger membrane binding (Figs. 1D and 3 B and F). We thus directly tested the role of clustering on membrane association by strengthening the clustering potential of BcLOV4 with intrinsically disordered regions (IDRs), which have previously been used to potentiate the clustering strength of Cry2 (26) (Fig. 4A). We tested two IDRs: the FUS low complexity domain [FUS(LC)] and the RGG domain from LAF-1, two well-characterized domains that have been used to engineer protein phase separation (26, 44, 45). Both IDR fusions dramatically enhanced optogenetic membrane translocation of BcLOV-mCh, supporting a positive role for clustering on translocation (Fig. 4B and Movie S5). Notably, both IDR fusions retained clear membrane localization of BcLOV4 even after two hours of stimulation at 37 °C, whereas wt BcLOV4 translocation decayed back to unstimulated levels, as observed previously (16) (Fig. 4C). Thus, the modulation of cluster properties can tune the amplitude and temperature-dependent dynamics of BcLOV4 stimulation (Fig. 4D).
Fig. 4.
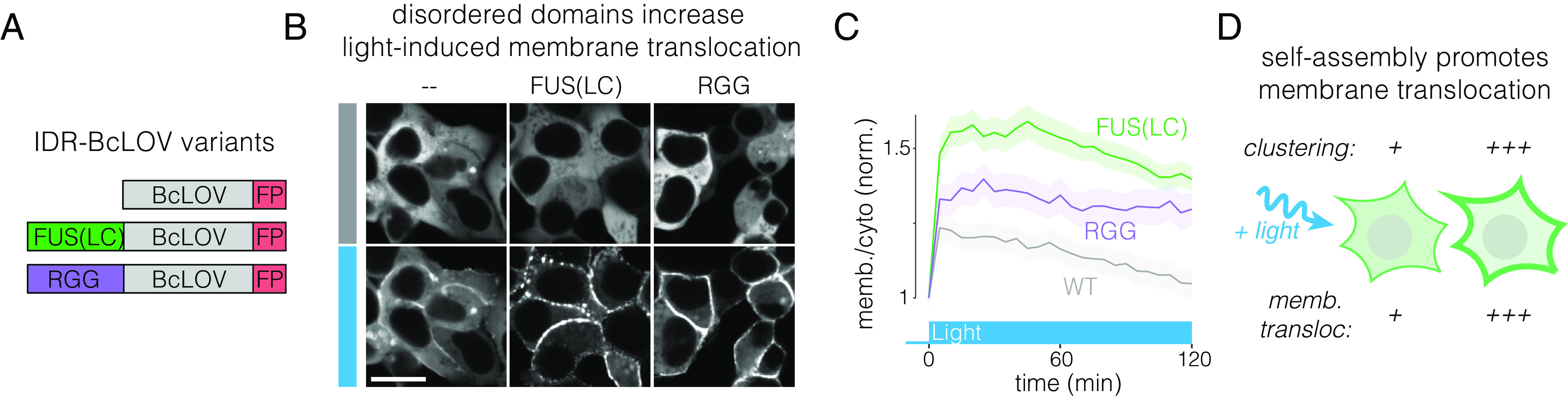
Enhanced clustering strengthens light-induced membrane binding of BcLOV4. (A) The IDRs FUS(LC) and RGG were fused to BcLOV-mCh to test whether increasing BcLOV4 clustering strength could tune the magnitude of membrane binding. (B) Both IDR-fused variants of BcLOV-mCh showed dramatic enhancement in membrane translocation. (Scale bar, 20 μm.) See also Movie S5. (C) IDR-BcLOV fusions maintained strong membrane localization even after 2 h of stimulation at 37 °C, whereas wt BcLOV-mCh decays back to unstimulated levels. Data represent mean BcLOV4 membrane/cytoplasmic ratios of ~350 to 750 cells. Ribbons = 95% CI (see Methods section for quantification details). (D) The clustering strength of BcLOV4 can tune its ability to translocate to the membrane in response to light stimulation.
The ability of IDRs to enhance membrane association suggested that IDRs might also increase the sensitivity of BcLOV-based tools. To test this, we first measured the effects of IDRs on activation of BcLOV-EGFR (Fig. 5A). Both wt and FUS- or RGG-fused BcLOV-EGFR expressed well and stimulated the ErkKTR reporter in HEK 293T cells (Fig. 5B). To quantify the effects of the IDRs, we performed a dose–response experiment to examine ppErk levels in response to a range of blue light intensities. FUS-BcLOV-EGFR drove a 30% higher maximum ppErk activation relative to wt BcLOV-EGFR and showed a two-fold increased sensitivity to light (1.2 vs. 2.5 mW/cm2 to reach half-max amplitude of wt, Fig. 5C). Interestingly, RGG-fused BcLOV-EGFR showed no benefit over the wt variant (Fig. 5C). The divergent effects of FUS and RGG IDRs demonstrate that distinct IDRs can have distinct effects on the activity of BcLOV-based tools, potentially through differential abilities to enhance either membrane localization or clustering. Nevertheless, the increased sensitivity and responsiveness of FUS-BcLOV-EGFR permitted higher levels of ppErk during both short- and long-term stimulation as compared to the wt BcLOV-EGFR probe (Fig. 5D). As before, maximal steady-state ppErk stimulation was observed at low-intermediate light patterns (2.5% duty cycle of 160 mW/cm2 blue light) (Fig. 5E). Importantly, FUS also increased the efficiency of the probe, where equivalent signal strength could be achieved at lower expression levels compared to unmodified BcLOV-EGFR. (Fig. 5F). Thus, amplification of BcLOV4 clustering and membrane translocation can generate optogenetic probes with higher sensitivity and signal strength, stronger sustained signaling, and lower requirements for probe expression levels.
Fig. 5.
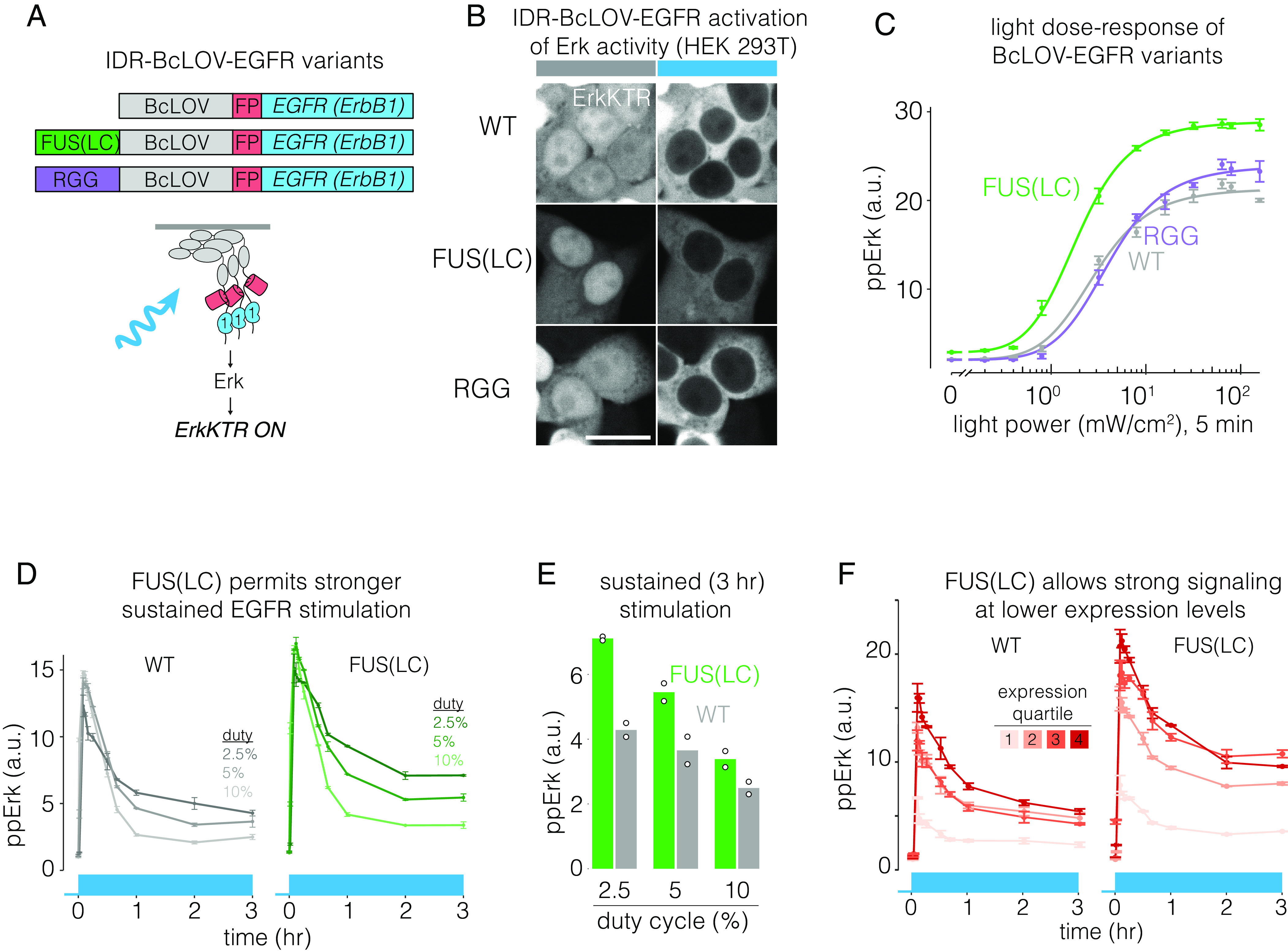
IDRs enhance sensitivity and strength of BcLOV-EGFR. (A) IDR-fused variants of BcLOV-EGFR. (B) IDR-BcLOV-EGFR variants stimulated ErkKTR-miRFP under blue light in HEK 293T cells. (Scale bar, 20 μm.) (C) Dose-response of light intensity on ppErk after 5 min of constant illumination at the indicated light dosages. Data represent the mean ± SEM of four replicates, each representing the mean from ~2,000 to 4,000 cells. (D) Comparison of sustained stimulation of wt or FUS-fused BcLOV-EGFR at variable duty cycles of stimulation. See SI Appendix, Table S1 for details of optogenetic stimulation parameters. (E) Steady-state levels of ppErk after 3 h of stimulation. FUS-BcLOV-mCh allowed stronger steady state levels of ppErk at all duty cycles tested. For (D), data represent mean ± SEM of two replicates, each representing the mean of ~2,000 to 5,000 cells. Datapoints in (E) are the mean steady-state ppErk levels at 3 h shown in (E). (F) FUS(LC) decreases the concentration of the BcLOV probe required to achieve a given signaling level. Data points represent mean ± SEM of two replicates, with each replicate representing the mean of 400 to 1,500 cells per expression quartile.
To determine whether benefits of increased clustering potential would extend to other BcLOV-based tools, we tested the effects of IDRs on BcLOV-SOScat, which stimulates Ras-Erk signaling, and which also self-inactivates at 37 °C (16) (Fig. 6A). Both IDR-BcLOV-SOScat variants stimulated ErkKTR in NIH 3T3 cells (Fig. 6B). While Erk activity began to decay shortly after its rapid activation by wt BcLOV-SOScat, activity was more sustained and showed slower decay when driven by either IDR-BcLOV-SOScat variant (Fig. 6C). Dose–response experiments showed that while all variants reached equivalent maximal ppErk levels, both IDR variants were 3-4X more sensitive to light than wt BcLOV-SOScat (intensity for half-max activation: wt: 20 mW/cm2, FUS: 5.1 mW/cm2, RGG: 6.7 mW/cm2, Fig. 6D). When illuminated at light levels that gave equivalent max ppErk response, the IDR variants yielded more sustained and higher integrated ppErk signals over 1 h of constant stimulation (Fig. 6E). We also compared signaling in response to a strong but pulsatile light input, a commonly used stimulation pattern that minimizes phototoxicity (46) (Fig. 6F). Here, both IDR variants achieved >two-fold higher maximal signal and more sustained activity compared to wt BcLOV-SOScat.
Fig. 6.
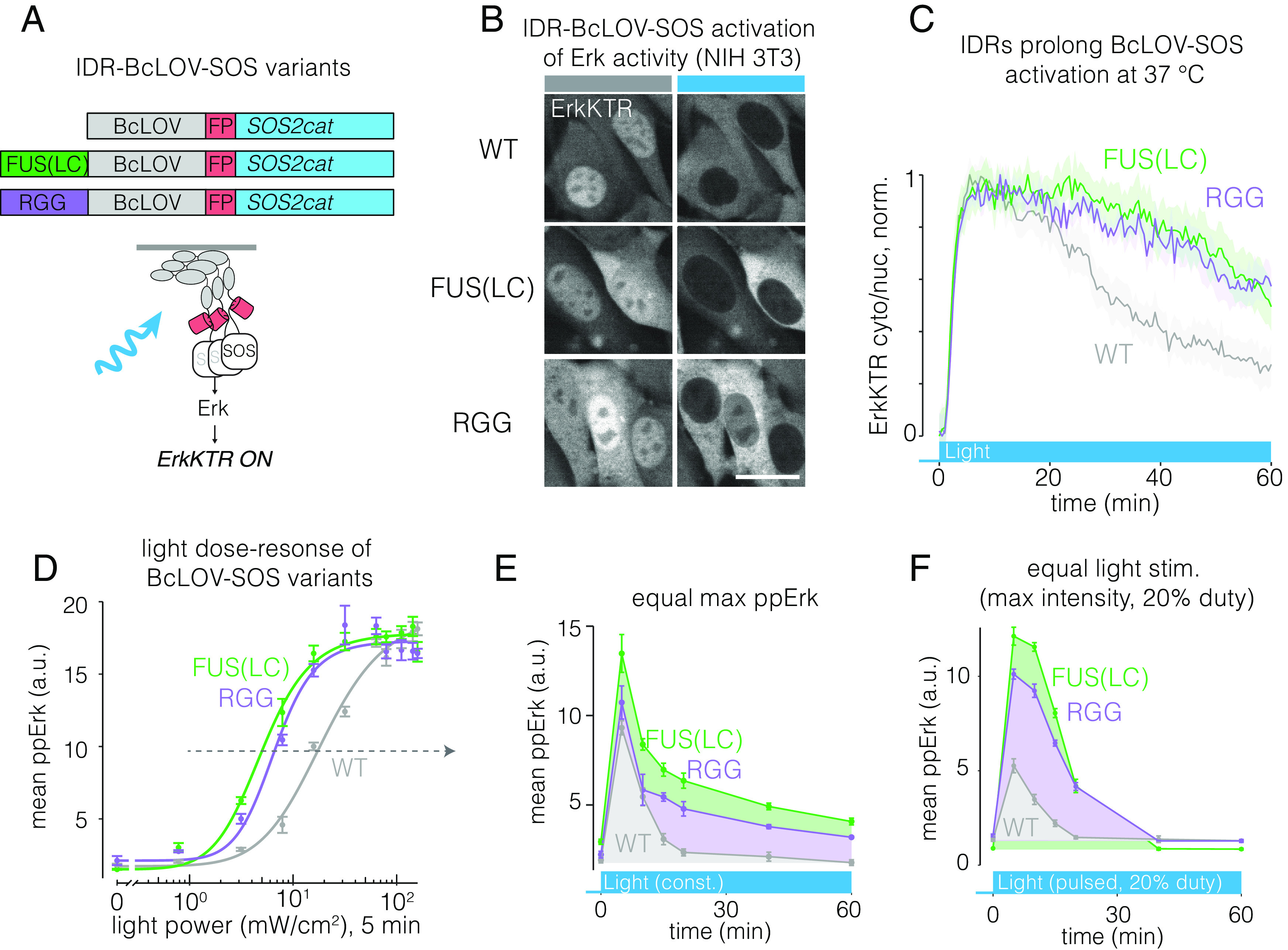
IDRs enhance sensitivity and strength of BcLOV-SOScat. (A) IDR-fused variants of BcLOV-SOScat. (B) The Ras/Erk pathway was activated in cells by IDR-BcLOV-SOScat variants in NIH 3T3 fibroblasts, as measured by the ErkKTR reporter. (Scale bar, 20 μm.) (C) Quantification of ErkKTR activity during 1 h of stimulation by BcLOV-SOScat variants. IDR variants show slower pathway decay. See SI Appendix, Table S1 for details of optogenetic illumination parameters. (D) Dose–response of light intensity on ppErk after 5 min of constant illumination at the indicated light dose. Data represent mean ± SEM of four replicates, each representing the mean signal from ~200 to 1,000 cells. (E) Comparison of ppErk activation dynamics by BcLOV-SOScat variants, each stimulated at a constant light intensity that produced equivalent max ppErk, as determined in (D) (dotted arrow). IDR variants showed higher sustained and integrated signaling over 1 h of stimulation. Data represent the mean ± SEM of four replicates, each representing the mean of ~50 to 400 single cells. (F) Comparison of ppErk activation dynamics by BcLOV-SOScat variants in response to pulsatile (20% duty cycle) maximum intensity light. IDR variants achieved >twofold higher maximal signal and more sustained and integrated activity compared to wt BcLOV-SOScat. Data represent mean ± SEM of four replicates, each representing the mean of ~100 to 600 cells.
Taken together, our results for both BcLOV-EGFR and BcLOV-SOScat show that cluster strength serves as a tuning knob to that can offer stronger, more stable, and less perturbative stimulation of BcLOV-based optogenetic tools.
Discussion
BcLOV4 is a photoreceptor protein that, upon light stimulation, both clusters and translocates to the plasma membrane. This dual translocation and clustering can be leveraged for new optogenetic signaling probes, including of the entire ErbB RTK family. Potentiation of clustering with IDRs allowed for a higher amplitude of signal induction, increased sensitivity to light, extended durations of signaling, and a higher efficiency of signaling (signal per unit of BcLOV probe expression). BcLOV4 represents, to our knowledge, the second described photosensor whose light-induced clustering can be co-opted for optogenetic control. The first, Arabidopsis Cry2 (9), has found widespread use across diverse systems of study (9, 22, 24–26, 28, 29, 47, 48). As the importance of protein condensation continues to emerge (49), we expect that BcLOV4 clustering will find extensive use cases, particularly for classes of condensation that occur at the membrane. Furthermore, the availability of multiple optogenetic clustering systems now provides more options for optogenetic control with distinct clustering properties (e.g., sensitivity, size, subcellular localization) and could further allow for multiplexed control of distinct clustering phenomena using the same blue light input.
The present work resolves the apparent paradox whereby BcLOV4 was observed to cluster in vitro but not in the presence of a lipid membrane (in vitro or in cellulo). In previous studies, clustering was not observed because the membrane-associated clusters were likely too small to resolve due to detection limitations including the diffraction limit and high fluorescence background. Our work overcomes these limitations to provide clear evidence of clustering by 1) co-expressing BcLOV-GFP with a CluMPS reporter (18) that amplified the size of small GFP clusters (Fig. 1), and 2) coupling clustering to a biochemical readout (EGFR/Erk activation) and measuring Erk activation in all cells with detectable BcLOV-EGFR expression, even when clustering was not visible (Fig. 2 and SI Appendix, Fig. S3). Further experiments with BcLOV-ErbB3, FUS-BcLOV, and BcLOV-EGFR with erlotinib treatment (Figs. 3 E and F and 4B) showed that in certain molecular contexts, clusters of stimulated BcLOV fusions can be large enough to see by fluorescence microscopy alone.
We leveraged the dual translocation and clustering of BcLOV4 to regulate RTK signaling, with a focus on EGFR and the entire ErbB receptor family. These studies further demonstrate the remarkable modularity of BcLOV4 as an optogenetic actuator, building on its previous application to control GTPases, guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs), and phosphatidyl inositol-3 kinase (PI3K) (14–16). We also found that by simply exchanging the intracellular domain of EGFR for the analogous domain of other ErbB family members, we could generate probes to control those receptors with no further optimization. Although this strategy also allowed stimulation of other families of RTKs including FGFR and PDGFR, we did observe RTK-family-specific effects including high basal signaling and limited activation strength. These results confirm the unique character of distinct RTK families that demands further optimization for their optimal activation, as has been observed previously (29, 30, 36, 37). These previous studies, as well as studies that optimized BcLOV4 for other signaling applications (14, 15, 27), provide a roadmap for future engineering of BcLOV-based RTK stimulation.
As with any synthetic strategy for stimulating intracellular signals, the events observed downstream of BcLOV-RTKs may not fully recapitulate the events downstream of endogenous receptors. Differences could occur, for example, because of differences in the oligomeric state of ligand-stimulated endogenous RTKs versus light-stimulated BcLOV4, due to differences in expression levels of endogenous versus exogenous receptor, or due to idiosyncratic interactions of the signaling domain with BcLOV4 (e.g., differential translocation of ErbB family members, Fig. 3B). Such potential effects must be considered to best contextualize experimental results.
Our ability to enhance the strength and sensitivity of BcLOV4 translocation through addition of IDRs has important practical implications. Two complications of optogenetic approaches are 1) toxicity from extensive blue light stimulation, and 2) elevated basal levels of signaling from expression of the optogenetic probe. Potentiation of BcLOV4 membrane translocation (here using IDRs) addresses both of these concerns, allowing comparable signal induction with ~four-fold less light (Fig. 6D) or with lower expression levels of the probe (Fig. 5F). In addition, and specifically for BcLOV4, the increased sensitivity allows one to slow the spontaneous signal decay observed at mammalian temperatures (37 °C) in two ways. First, because decay depends on both light and temperature, lower light levels lead to slower decay (16). Second, because IDRs can boost the amplitude of signal, the signal will remain above a given threshold for longer than for probes that lack the IDR.
While we found that IDR-mediated enhancements in BcLOV4 translocation generally translated to signal activation of optogenetic probes, we also found important exceptions. For example, despite increased membrane translocation arising from both the FUS(LC) and RGG IDRs (Fig. 4 B and C), only FUS(LC) potentiated signaling of BcLOV-EGFR (Fig. 5D). By contrast, both IDRs potentiated signaling of BcLOV-SOScat (Fig. 6D). Furthermore, despite strong membrane localization over 2 h with IDR-fused variants of BcLOV-mCh (Fig. 4C), BcLOV-SOScat activity could only be extended, but not sustained indefinitely (Fig. 6F). Future work will define the molecular details of BcLOV4 thermal sensitivity and provide additional strategies by which to mitigate or eliminate its effects. Collectively, these results highlight that a probe’s activation dynamics can be influenced by many factors, including the molecular nature of the signaling event, probe expression level, the host cell environment, and even the enzymatic activity of the probe itself (Fig. 3F and SI Appendix, Fig. S7 D and E).
In summary, BcLOV4 is a multifunctional photoreceptor that uniquely both clusters and translocates to the membrane in mammalian cells. BcLOV clustering can be leveraged both for new types of single-component optogenetic tools and to enhance existing ones.
Methods
Cell Culture.
All cell lines were maintained at 37 °C and 5% CO2 in a cell culture incubator. Lenti-X HEK 293T cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). NIH 3T3 fibroblast cells were cultured in DMEM containing 10% calf serum (CS) and 1% P/S.
Plasmid Design and Assembly.
Constructs were assembled using Gibson assembly. DNA fragments for the inserts and backbone were generated via PCR with primers obtained from Genewiz (Azenta Life Sciences) and inserted into the backbone using HiFi cloning mix (New England Biolabs). All constructs were verified with Sanger sequencing. The DNA sequence encoding BcLOV4 was described previously (11). GFP-binding nanobody LaG17 was obtained from Michael P. Rout (50). LaG17-CluMPS was previously described (18). ErkKTR-miRFP670 was previously described (16). EGFR/ErbB1 was sourced from Opto-hEGFR, which was a kind gift from Harold Janovjak (36). ErbB2 was sourced from MSCV-human Erbb2-IRES-GFP, which was a gift from Martine Roussel (Addgene plasmid # 91888; http://n2t.net/addgene:91888; RRID:Addgene_91888). ErbB3 was sourced from pDONR223-ERBB3, which was a gift from William Hahn & David Root (Addgene plasmid # 23874; http://n2t.net/addgene:23874; RRID:Addgene_23874). ErbB4 was sourced from pDONR223-ERBB4, which was a gift from William Hahn & David Root (Addgene plasmid # 23875; http://n2t.net/addgene:23875; RRID:Addgene_23875). iLID, sspB, and SOScat were sourced from previously described constructs (16). FUS(LC) (1 to 163) and LAF-1 RGG were kindly provided by Matthew Good. A list of constructs used in this study is provided in SI Appendix, Table S2.
Plasmid Transfection.
Lenti-X HEK 293T cells were transfected using Lipofectamine™ 3000 transfection reagent (ThermoFisher Scientific) following the manufacturer’s protocol. The transfection mixture contained 100 ng/µL DNA, 2% Lipofectamine™ reagent, and 2% P3000 reagent and was brought up to a final volume of 10 µL with Opti-MEM™ (ThermoFisher Scientific). The transfection mixture was incubated for 15 min at room temperature and was then added to the cells. For cells seeded in 96-well plates, 10 µL of transfection mixture was added per well. Cells seeded in 384-well plates received 2.5 µL of transfection mixture per well.
Lentiviral Packaging and Cell Line Generation.
Lentivirus was packaged by contransfecting the pHR transfer vector, pCMV-dR8.91 (Addgene, Catalog #12263), and pMD2.G (Addgene, Catalog #12259) into Lenti-X HEK 293T cells. Cells were seeded 1 d prior to transfections at a density of 700,000 cells/mL in a six-well plate. Cells were transfected using the calcium phosphate transfection method: for 300 µL of transfection mix, 1.5 µg of transfer vector, 1.33 µg of pCMV-dR8.91, 0.17 µg of pMD2.G, 150 µL of 2X HEPES-buffered saline (HeBS), and H2O up to 132 µL were mixed. Then, 18 µL of 2.5 mM CaCl2 was then added, the mixture was incubated for 1 min 45 s at room temperature, and then the mixture was added dropwise to the cells. One day post-transfection, media was removed from the plate and was replaced with fresh media. Two days posttransfection, media containing virus was collected and centrifuged at 800 × g for 3 min. The supernatant from centrifuged media was then collected and filtered through a 0.45-µm filter. Then, 500 µL of filtered virus was added to 100,000 cells (Lenti-X HEK293T or NIH 3T3) seeded in a six-well plate. Cells were observed for transduction by checking for fluorescence ~1 to 2 d postinfection. Cells were expanded over multiple passages. Successfully transduced cells were enriched through cell sorting using a BD FACSAria Fusion.
Preparation of Cells for Plate-Based Experiments.
For experiments, cells were seeded in 96- or 384-well plates (Cellvis 96-well plate with glass-like polymer bottom, catalog number P96-1.5P; Greiner Bio-One CELLSTAR 384-well, Cell Culture-Treated, Flat-Bottom Microplate, catalog number 781091). First, wells were coated with 30 µL (for 96-well plate) or 12 µL (for 384-well plate) of 10 µg/mL of MilliporeSigma™ 597 Chemicon™ Human Plasma Fibronectin Purified Protein in 1X PBS for 15 min at 37 °C. For 96-well plate experiments, 25,000 Lenti X HEK 293T or 12,000 NIH 3T3 cells were seeded in 150 µL of P/S-free cell-culture medium (DMEM + 10% FBS or 10% CS) in each well. For 384-well plate experiments, 3500 Lenti-X HEK 293T or 2500 NIH 3T3’s were seeded. Following the seeding step, the plates were spun down at 20 × g for 1 min to promote uniform distribution of cells throughout the well. For experiments with stable cell lines, cells were starved after 24 h by performing seven 80% washes (for 384-well plate) or four 75% washes (for 96-well plate) with starvation media (DMEM +1% P/S) using an automated plate washer (BioTEK ELx405). Experiments were performed 3 to 4 h post-starvation. For experiments with transiently transfected cells, cells were starved 6 h post-transfection to remove lipofectamine reagent from cells, and experiments were performed after overnight starvation.
Optogenetic Stimulation.
For live-cell imaging experiments, the 488-nm laser was used to stimulate BcLOV4 tools for membrane translocation. For fixed-cell experiments, cells were stimulated with a single-color blue LED optoPlate-96 (46). LED intensities were calibrated using a Thorlabs power meter (catalog number PM16-140). Briefly, each well of the optoPlate was turned on to maximum intensity. The power meter was used to scan the well, and the maximum intensity reading from that well was recorded. This process was repeated for all wells. The ratio of each LED intensity to the dimmest LED intensity found was then calculated, and this value was used as a “scaling factor”, such that each LED was scaled down to emit at the same intensity as the weakest LED. In this way, all LEDs were set to the same power output. For stimulation experiments, the light intensity was configured to stimulate the wells with a range of light intensities spanning from 0 to 160 mW/cm2. The Arduino IDE (v1.8) was used to program the Arduino Micro present on the optoPlate-96. A 20-mm tall black adaptor was used for even light diffusion across each of the wells on the 384-well plate. For time course experiments, time points were assigned to individual wells, and stimulations were run in a sequential manner to allow simultaneous fixing of cells at the end of the experiment. The apparatus was arranged inside a standard cell culture incubator set at 37 °C and 5% CO2, and the experiments were run under dark conditions to avoid unwanted light exposure. Prior to experiments, optoPlate stimulation protocols were tested to ensure that no sample heating occurred due to heat generation from the device. Sample temperatures were measured using a custom-built immersion temperature sensor.
Immunofluorescence Staining.
Immediately following completion of a stimulation protocol,16% paraformaldehyde (Paraformaldehyde Aqueous Solution, Electron Microscopy Sciences, catalog number 15710) was added to each well to a final concentration of 4%, and cells were incubated for 10 min in the dark. Cells were then permeabilized 1X PBS + 0.1% Triton X-100 for 10 min at room temperature (RT). Cells were further permeabilized with ice-cold 100% methanol at −20 °C for 10 min. After permeabilization, the cells were blocked with 1% BSA in 1X PBS for 30 min at RT. The cells were then incubated in primary antibody diluted in 1X PBS + 0.1% BSA (phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), Cell Signaling, catalog number 4370L, 1:400 dilution) at 4 °C overnight. After overnight incubation, the primary antibody was removed and the plate was washed 5 times in PBS + 0.1% Tween-20 (PBS-T). Cells were then incubated with secondary antibody (Jackson Immunoresearch Alexa Fluor 488 AffiniPure goat anti-rabbit IgG (H+L), 1:500) and DAPI (ThermoFisher Scientific, catalog number D1306, 300 nM) in 1X PBS + 0.1% BSA for 1 h at RT. The secondary antibody was removed, and the plate was washed 5 times in PBS-T.
Immunoblotting.
Cells were lysed with RIPA containing protease inhibitors. The lysates were electrophoretically separated on an SDS-PAGE gel. Proteins were then transferred onto a nitrocellulose membrane and probed for endogenous EGFR [EGF Receptor (D38B1) XP® Rabbit mAb, Cell Signaling, catalog number 4267, 1:1,000 dilution] with α-Tubulin [α-Tubulin (DM1A) Mouse mAb, Cell Signaling, catalog number 3873, 1:1,000 dilution] as a loading control.
Imaging.
Live-cell imaging.
Live-cell imaging was done using a Nikon Ti2E microscope equipped with a Yokagawa CSU-W1 spinning disk, 405/488/561/640 nm laser lines, an sCMOS camera (Photometrics), a motorized stage, and an environmental chamber (Okolabs). HEK 293T and NIH 3T3 cells with desired constructs were plated in 96- or 384-well plates and imaged with a 40X oil immersion objective at 37 °C and 5% CO2. For the EGFR inhibition experiments, cells were treated with 1 µM of erlotinib 30 min prior to imaging. Erlotinib was kindly provided by Arjun Raj.
High-content imaging.
For fixed-cell experiments, samples were imaged using a Nikon Ti2E epifluorescence microscope equipped with DAPI/FITC/Texas Red/Cy5 filter cubes, a SOLA SEII 365 LED light source and motorized stage. High-content imaging was performed using the Nikon Elements AR software. Image focus was ensured using image-based focusing in the DAPI channel.
Image Processing and Analysis.
Live-cell ErkKTR quantification.
To determine the cytoplasmic/nuclear (C/N) fluorescence ratios of ErkKTR reporter from the live-cell imaging experiments for Fig. 2D, ImageJ (51) was used to manually compare the pixel intensities of the mean cytoplasmic and nuclear intensities for 25 cells in the same field of view. The obtained values were exported into R (version 4.2.2) for data analysis using the dplyr (52) and ggplot2 (53) packages.
Immunofluorescence quantification.
Cell Profiler (54) was used to quantify ppErk levels in the fixed-cell experiments. Cells were segmented using the DAPI channel and the cytoplasm was identified by expanding a 5-pixel ring from the nuclei. The obtained cytoplasmic and nuclear fluorescence values were exported into R for further analysis.
Membrane recruitment.
Membrane recruitment of BcLOV4 in Fig. 4C was quantified using the MorphoLibJ Plugin for ImageJ (55). All experiments were performed in cells stably expressing a fluorescent membrane marker (GFP-CAAX). Images of the membrane marker were used to automatically segment single cells using the “Morphological Segmentation” feature of the MophoLibJ with a threshold of 150. Segmentation of each membrane marker image was exported as a separate tiff image. Segmentation images were imported to CellProfiler along with the corresponding images of BcLOV-mCherry variants. Membrane values of mCh were then determined by designating a 1-pixel-wide perimeter of each cell’s membrane. The membrane intensity and total cell intensity of BcLOV4 was then measured and recorded for each cell. R was used to process these values, normalizing membrane BcLOV4 intensity of each cell by the whole-cell intensity and averaging these single-cell values for each time point.
Curve fitting.
ppErk levels for the dose-response curves of IDR-fused variants of BcLOV-EGFR and BcLOV-SOScat (Figs. 5D and 6D) were fit to a Hill function of the form
where X is the power of light used, a is the maximal amount of Erk activation, b is the parameter defining steepness of the curve, and c is the power of light needed to achieve half-maximal activation of Erk. A MATLAB function was written to determine the parameters, and the curves were fitted on RStudio.
Statistics.
Statistics were calculated using R version 4.2.2 using the “stats” package.
Supplementary Material
Appendix 01 (PDF)
CluMPS amplifies and visualizes membrane-associated BcLOV4 condensates. Confocal imaging of BcLOV-GFP translocation with or without CluMPS in HEK 293T cells upon stimulation with 488 nm light. Time is in minutes:seconds. Blue square indicates light stimulation. Scale bar = 10 µm.
The Ras-Erk pathway was activated in cells by BcLOV-EGFR. Confocal imaging of BcLOV-EGFR translocation and activation of ErkKTR-miRFP in HEK 293T cells upon stimulation with 488 nm light. Blue square indicates light stimulation.
Magnitude of membrane translocation differs between fusions of ErbB family members with BcLOV4. Confocal imaging of BcLOV-mCh-ErbB1-4 translocation to the membrane upon blue light stimulation with 488 nm light. Blue square indicates light stimulation.
ErbB activity correlates with membrane ruffling. Confocal imaging of BcLOV-mCh-ErbB1-4 translocation and induced membrane ruffling upon stimulation with 488 nm light. Blue square indicates light stimulation.
Disordered domains increase light-induced membrane translocation of BcLOV4. Confocal imaging of IDR-fused variants of BcLOV-mCh in HEK 293T cells upon stimulation with 488 nm light. Blue square indicates light stimulation.
Acknowledgments
We thank Erin Berlew for helpful discussions on this work. This work was supported by the NIH (R35GM138211 for L.J.B) and the NSF (Graduate Research Fellowship Program to W.B., CAREER 2145699 to L.J.B., and CAREER MCB1652003 for B.Y.C). Cell sorting was performed on a BD FACSAria Fusion that was obtained through NIH S10 1S10OD026986 and is operated through the Penn Cytomics and Cell Sorting Resource Laboratory.
Author contributions
A.A.P., W.B., Z.H., B.Y.C., and L.J.B. designed research; A.A.P., W.B., and Z.H. performed research; W.B. and T.R.M. contributed new reagents/analytic tools; A.A.P., W.B., T.R.M., and L.J.B. analyzed data; and A.A.P. and L.J.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Fluorescence imaging data underlying each figure can be found at the following link: bit.ly/palbenman.
Supporting Information
References
- 1.Harper S. M., Neil L. C., Gardner K. H., Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Wu Y. I., et al. , A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu-Sato S., Huq E., Tepperman J. M., Quail P. H., A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Levskaya A., Weiner O. D., Lim W. A., Voigt C. A., Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guntas G., et al. , Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 112–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwerdtfeger C., Linden H., VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22, 4846–4855 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strickland D., et al. , TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy M. J., et al. , Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S., Schaffer D. V., Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Más P., Devlin P. F., Panda S., Kay S. A., Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Glantz S. T., et al. , Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Proc. Natl. Acad. Sci. U.S.A. 115, E7720–E7727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L., et al. , Optical control of membrane tethering and interorganellar communication at nanoscales. Chem. Sci. 8, 5275–5281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H., et al. , Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Berlew E. E., Kuznetsov I. A., Yamada K., Bugaj L. J., Chow B. Y., Optogenetic Rac1 engineered from membrane lipid-binding RGS-LOV for inducible lamellipodia formation. Photochem. Photobiol. Sci. 19, 353–361 (2020), 10.1039/c9pp00434c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berlew E. E., et al. , Single-component optogenetic tools for inducible RhoA GTPase signaling. Adv. Biol. 5, 2100810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benman W., et al. , Temperature-responsive optogenetic probes of cell signaling. Nat. Chem. Biol. 18, 152–160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao J., Peng H., Dong B., Development and application of an optogenetic manipulation system to suppress actomyosin activity in Ciona epidermis. Int. J. Mol. Sci. 24, 5707 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumford T. R., et al. , Visual detection of submicroscopic protein clusters with a phase-separation-based fluorescent reporter. bioRxiv [Preprint] (2022). 10.1101/2022.07.13.499962 (Accessed 1 December 2022). [DOI]
- 19.Lee S., et al. , Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods 11, 633–636 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Taslimi A., et al. , An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park H., et al. , Optogenetic protein clustering through fluorescent protein tagging and extension of CRY2. Nat. Commun. 8, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bugaj L. J., et al. , Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat. Commun. 6, 6898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Repina N. A., McClave T., Bao X., Kane R. S., Schaffer D. V., Engineered illumination devices for optogenetic control of cellular signaling dynamics. Cell Rep. 31, 107737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenbloom A. B., et al. , β-catenin signaling dynamics regulate cell fate in differentiating neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 117, 28828–28837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., et al. , Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, e39578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin Y., et al. , Spatiotemporal control of intracellular phase transitions using light-activated optodroplets. Cell 168, 159–171.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berlew E. E., et al. , Designing single-component optogenetic membrane recruitment systems: The Rho-family GTPase signaling toolbox. ACS Synth. Biol. 11, 515–521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N., et al. , Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem. Biol. 21, 903–912 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Chang K.-Y., et al. , Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun. 5, 4057 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Duan L., et al. , Optical activation of TrkA signaling. ACS Synth. Biol. 7, 1685–1693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farahani P. E., et al. , Substratum stiffness regulates Erk signaling dynamics through receptor-level control. Cell Rep. 37, 110181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnamurthy V. V., et al. , A generalizable optogenetic strategy to regulate receptor tyrosine kinases during vertebrate embryonic development. J. Mol. Biol. 432, 3149–3158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dine E., Reed E. H., Toettcher J. E., Positive feedback between the T cell kinase Zap70 and its substrate LAT acts as a clustering-dependent signaling switch. Cell Rep. 35, 109280 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regot S., Hughey J. J., Bajar B. T., Carrasco S., Covert M. W., High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Citri A., Yarden Y., EGF-ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Grusch M., et al. , Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leopold A. V., Thankachan S., Yang C., Gerashchenko D., Verkhusha V. V., A general approach for engineering RTKs optically controlled with far-red light. Nat. Methods 19, 871–880 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Kim H. H., Vijapurkar U., Hellyer N. J., Bravo D., Koland J. G., Signal transduction by epidermal growth factor and heregulin via the kinase-deficient ErbB3 protein. Biochem. J. 334, 189–195 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy P. M., Platko J. V., Cantley L. C., Cerione R. A., Carraway K. L. III, Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc. Natl. Acad. Sci. U.S.A. 91, 8132–8136 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jura N., Shan Y., Cao X., Shaw D. E., Kuriyan J., Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y., et al. , Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife 5, e14107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bublil E. M., et al. , Kinase-mediated quasi-dimers of EGFR. FASEB J. 24, 4744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haubrich J., Zwier J. M., Charrier-Savournin F., Prézeau L., Pin J.-P., Different conformations of EGF-induced receptor dimers involved in signaling and internalization. bioRxiv [Preprint] (2022). 10.1101/2022.04.19.488777 (Accessed 1 May 2023). [DOI] [PubMed] [Google Scholar]
- 44.Schuster B. S., et al. , Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Bugaj L. J., Lim W. A., High-throughput multicolor optogenetics in microwell plates. Nat. Protoc. 14, 2205–2228 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Repina N. A., et al. , Optogenetic control of Wnt signaling for modeling early embryogenic patterning with human pluripotent stem cells. bioRxiv [Preprint] (2019). 10.1101/665695 (Accessed 1 December 2022). [DOI]
- 48.Gropp M. H. M., Klaips C. L., Hartl F. U., Formation of toxic oligomers of polyQ-expanded Huntingtin by prion-mediated cross-seeding. Mol. Cell 82, 4290–4306.e11 (2022), 10.1016/j.molcel.2022.09.031. [DOI] [PubMed] [Google Scholar]
- 49.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fridy P. C., et al. , A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham H., François R., Henry L., Müller K., dplyr: A grammar of data manipulation (R package version 0.8. 0.1, 2019). https://dplyr.tidyverse.org/. Retrieved 13 January 2020.
- 53.Wickham H., ggplot2: ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011). [Google Scholar]
- 54.Lamprecht M. R., Sabatini D. M., Carpenter A. E., CellProfilerTM: Free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Legland D., Arganda-Carreras I., Andrey P., MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
CluMPS amplifies and visualizes membrane-associated BcLOV4 condensates. Confocal imaging of BcLOV-GFP translocation with or without CluMPS in HEK 293T cells upon stimulation with 488 nm light. Time is in minutes:seconds. Blue square indicates light stimulation. Scale bar = 10 µm.
The Ras-Erk pathway was activated in cells by BcLOV-EGFR. Confocal imaging of BcLOV-EGFR translocation and activation of ErkKTR-miRFP in HEK 293T cells upon stimulation with 488 nm light. Blue square indicates light stimulation.
Magnitude of membrane translocation differs between fusions of ErbB family members with BcLOV4. Confocal imaging of BcLOV-mCh-ErbB1-4 translocation to the membrane upon blue light stimulation with 488 nm light. Blue square indicates light stimulation.
ErbB activity correlates with membrane ruffling. Confocal imaging of BcLOV-mCh-ErbB1-4 translocation and induced membrane ruffling upon stimulation with 488 nm light. Blue square indicates light stimulation.
Disordered domains increase light-induced membrane translocation of BcLOV4. Confocal imaging of IDR-fused variants of BcLOV-mCh in HEK 293T cells upon stimulation with 488 nm light. Blue square indicates light stimulation.
Data Availability Statement
Fluorescence imaging data underlying each figure can be found at the following link: bit.ly/palbenman.