Significance
Segmentation of a visual scene is one of the most important steps in visual processing as it allows one to distinguish between regions of the scene that belong to different visual objects. Previous studies reported cells encoding different aspects of segmentation in random locations of visual areas, but a systematic characterization of segmentation tuning properties across visual areas is missing. Here, we combined fMRI and electrophysiology to identify “segmentation patches” that preferentially encode segmentation features. Our findings suggest visual cortex may be organized into modules not only for specific features such as orientation but also for a specific computation, segmentation. This advances our understanding of visual cortical organization and makes the question of how the brain computes segmentation more tractable.
Keywords: segmentation, border ownership, modularity, figure-ground
Abstract
Segmentation, the computation of object boundaries, is one of the most important steps in intermediate visual processing. Previous studies have reported cells across visual cortex that are modulated by segmentation features, but the functional role of these cells remains unclear. First, it is unclear whether these cells encode segmentation consistently since most studies used only a limited variety of stimulus types. Second, it is unclear whether these cells are organized into specialized modules or instead randomly scattered across the visual cortex: the former would lend credence to a functional role for putative segmentation cells. Here, we used fMRI-guided electrophysiology to systematically characterize the consistency and spatial organization of segmentation-encoding cells across the visual cortex. Using fMRI, we identified a set of patches in V2, V3, V3A, V4, and V4A that were more active for stimuli containing figures compared to ground, regardless of whether figures were defined by texture, motion, luminance, or disparity. We targeted these patches for single-unit recordings and found that cells inside segmentation patches were tuned to both figure-ground and borders more consistently across types of stimuli than cells in the visual cortex outside the patches. Remarkably, we found clusters of cells inside segmentation patches that showed the same border-ownership preference across all stimulus types. Finally, using a population decoding approach, we found that segmentation could be decoded with higher accuracy from segmentation patches than from either color-selective or control regions. Overall, our results suggest that segmentation signals are preferentially encoded in spatially discrete patches.
Segmentation is believed to be a crucial step in visual processing that allows a visual system to distinguish which regions of a visual scene belong to the background, which regions belong to different objects, and where the borders are that constrain the regions of these objects. Previous studies suggest that the brain uses a diversity of cues, including luminance, texture, disparity, and motion, to segregate objects from the background (1–4).
Several major candidates for segmentation signals have been identified in visual cortex, including figure-ground encoding cells that respond more strongly to a figure in the receptive field than to background (3–5), cells that encode curvature (6, 7), cells that encode kinetic boundaries for motion-defined figures (8–10), and so-called border-ownership cells that respond more strongly if a border in the receptive field belongs to an object on one specific side of it (2). To date, these various types of segmentation-encoding cells have been recorded in random locations of macaque visual areas V1, V2, V3, and V4. However, a recent study suggests that at least border-ownership cells may be clustered within V4 (11). The general functional organization of segmentation-encoding cells is thus a question of great interest. Moreover, since in the studies above only a limited number of stimulus types were presented, it is unclear whether cells exist that encode figure-ground or border ownership consistently, invariant to whether the segmentation is defined by luminance, texture, motion, disparity, or higher-level cues. Indeed, one study found that putative border-ownership cells recorded from random locations in V2 and V3 do not encode border ownership consistently when presented with a larger battery of artificial and natural stimuli (12). However, this result leaves open the possibility that consistent cells can be found when recording from appropriate functional modules.
Computational studies have shown that neural network models for object recognition can benefit from separating representations of object appearance and object contours (13–16). These findings hint at the possibility that visual cortex may exploit this modular organization, with some parts of visual cortex specialized for extracting texture, and other parts specialized for computing the segmentation of a visual scene. A vast amount of literature has reported modularity in retinotopic cortex for different simple features, such as color in thin stripes and motion/disparity in thick stripes (17). In contrast, here, we investigate modularity for a higher-level computational function, segmentation of a visual scene, rather than low-level features.
We used functional magnetic resonance imaging (fMRI)-guided electrophysiology to investigate whether visual cortex has a functional organization for segmentation, such that certain regions harbor a higher density of segmentation-encoding cells. Using fMRI, we found specific patches within V2, V3, V3A, V4, and V4A (18) that were preferentially activated by figures versus background; moreover, activation was consistent across four different cues (texture, luminance, motion, and disparity). When we targeted a subset of these activations for electrophysiology, we found a higher percentage of cells modulated by figure vs. ground than in control regions. Moreover, figure/ground and border ownership could be decoded with significantly higher accuracy from populations of neurons in these regions than in control regions. Finally, we found clustering of border-ownership cells that were consistent across all modalities within a segmentation patch in area V3.
The findings described in the present study suggest that visual cortex possesses discrete modules spanning mid-level visual areas V2, V3, V3A, V4, and V4A for segmenting visual scenes. These segmentation modules are functionally distinct from both color-selective regions and other neighboring regions in that they harbor cells that encode border-ownership invariant to cue and allow accurate decoding of the segmentation of a visual scene. The existence of an architecture for segmentation spanning multiple areas across retinotopic cortex may make future studies on mechanisms of segmentation more tractable.
Results
To search for segmentation-selective modules, we compared fMRI activation to stimuli containing figures on a background with activation to corresponding control stimuli containing only background. The figures and their respective backgrounds were created using four modalities (texture, luminance, motion, or disparity) (Fig. 1). This resulted in eight types of stimuli (four modalities × figure/background). For texture, stimuli consisted of lines of a given orientation at random positions; lines within the figure had a different orientation than in the background. For luminance, figure and background were defined by simple black-and-white contrast. For motion, dots at random positions moved in one direction for the figures and in the opposite direction for the background. For disparity, random dots were displaced in the two eyes to evoke the perception of disparity-defined figures on a background, compared to backgrounds with uniform disparity (19).
Fig. 1.
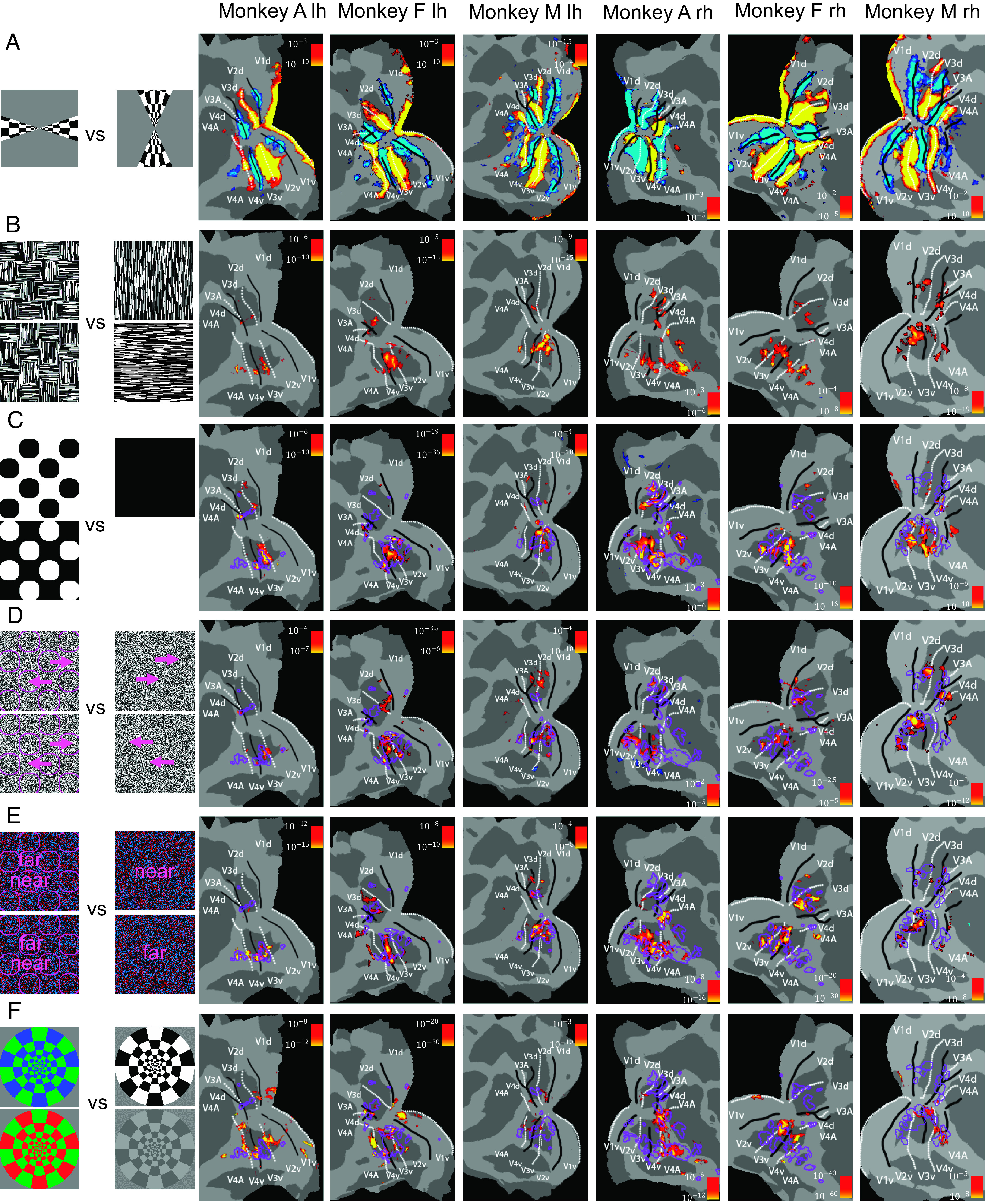
Patches of segmentation-related signals in visual cortex. A series of fMRI experiments were performed in three monkeys and activations were overlaid on patches of the left and right hemispheres of flattened brain surfaces. Rows correspond, from top to bottom, to (A) retinotopy, (B) texture figures versus background, (C) luminance figures versus background, (D) motion figures versus background, (E) disparity figures versus background, (F) color versus black and white. Color bars indicate P values of contrasts; lh: left hemisphere, rh: right hemisphere.
We performed fMRI in three monkeys (monkey A, monkey F, and monkey M), while each monkey passively viewed the stimuli described above, with each of the eight stimulus types presented in a separate block. For each of the four modalities, we compared activations when stimuli contained figures on a background versus only background (Fig. 1). To map area boundaries of V1, V2, V3, V4, and V4A, we used a standard retinotopy localizer comparing activations to vertical versus horizontal checkerboard wedges (Fig. 1A).
Comparison of activation to texture-defined figures versus background revealed multiple hotspots in areas V2, V3, V3A, V4, and V4A (Fig. 1B). When we compared these hotspots with those defined by activations to figure versus background for the other modalities (Fig. 1 C–E; regions of interest (ROIs) for significant texture-defined figure contrasts indicated by purple outlines), we found that activations for texture-, luminance-, motion-, and disparity-defined figure versus background all overlapped to a considerable degree. Importantly, this consistent activation of the same patches of visual cortex was not just due to a higher signal-to-noise ratio in these regions: as a control, we compared activations to color vs. grayscale stimuli and found activations in mostly nonoverlapping regions, suggesting that modules specialized in color processing may be separate from segmentation-related regions (Fig. 1F, see also Fig. 2C for quantification of overlap). For the remainder of this report, we will refer to these figure-ground activated hotspots as segmentation patches.
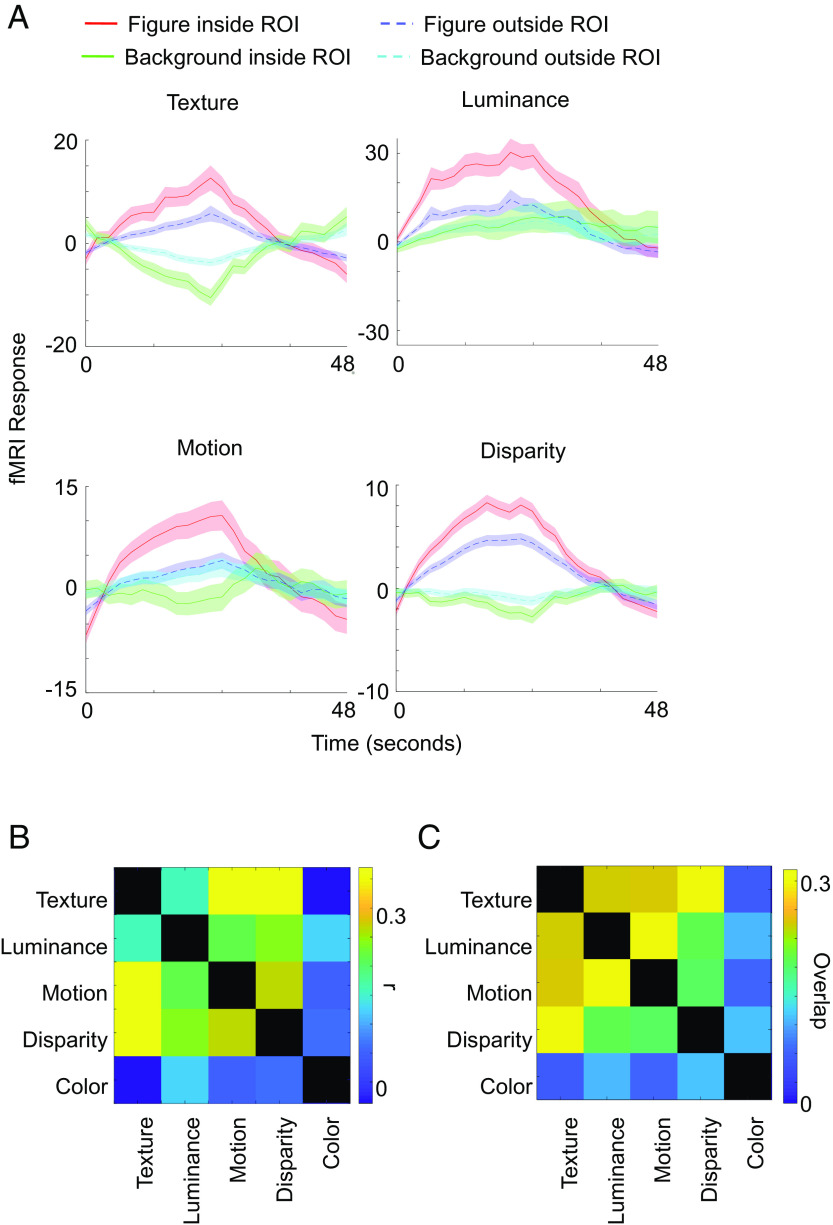
Fig. 2.
fMRI voxels in segmentation patches prefer figure over background across modalities. (A) Visual cortex voxels that significantly preferred texture stimuli containing figures compared to background stimuli (purple outlines in Fig. 1) were defined as a region of interest (ROI). Red and green traces show fMRI responses averaged across voxels inside the ROI to stimuli containing figures and stimuli containing background, respectively, defined by different modalities. Blue and yellow traces show fMRI responses averaged across voxels from the rest of visual cortex to stimuli containing figures and stimuli containing background, respectively. Each response was baseline subtracted, i.e., we subtracted the average response from the previous gray block. Time courses are shown across 48 s from onset of the respective block, i.e., they also show the response during the subsequent gray block, which is still selective due to the delay of the fMRI signal. We injected MION as contrast agent and hence inverted the signal. Shaded areas are based on SEM across runs, repeated blocks, and monkeys. (B) Correlation maps between different modalities or color. Modulation indices were computed for each voxel inside visual areas V1-V4A for figure versus background (texture, luminance, motion, and disparity) and color versus grayscale, respectively (see Materials and Methods for details). For each pair of conditions, we computed correlation coefficients across voxels from three monkeys. Correlation coefficients are indicated by color of the squares. (C) Spatial overlap of significantly activated ROIs for different modalities (shown in Fig. 1). For each pair of modalities, we quantified the spatial overlap of activated regions as the area of their intersection divided by the area of their union.
Since we can clearly segment objects across the visual field, a natural question is how retinotopy in visual areas is related to locations of segmentation patches. Therefore, we performed an experiment where we compared fMRI activations to figure versus background where figures were presented at specific retinotopic positions (SI Appendix, Fig. S1). The activations of these location-specific contrasts turned out to be mostly contained within the segmentation patches revealed by our original stimulus, in which figures were shown across the entire visual field. This shows that segmentation patches, while not occupying the entirety of visual cortex, can be observed across the entire visual field.
We computed average fMRI time courses inside ROIs that were significantly activated by texture-defined figures versus background to quantify the consistency of figure-ground activation across different modalities. When computing activations to texture stimuli inside the texture-defined ROI, to avoid circularity, we defined the ROI based on one half of runs and plotted activations based on the other half of runs. For other modalities, we incorporated all runs. Activations in the texture-defined ROIs turned out to be higher for stimuli containing figures compared to background stimuli for all modalities (Fig. 2A). This difference was significant ( for texture, for luminance, for motion, for disparity) and more pronounced than in visual cortex voxels outside of the ROI. For visual cortex voxels outside of the ROI, only texture and disparity showed a significant, albeit weaker than inside the ROI, preference for stimuli containing figures, whereas luminance and motion did not reach significance ( for texture, for luminance, for motion, for disparity). Although each individual voxel outside of the ROI by definition did not reach the significance threshold for texture, after averaging across these voxels, population activity did reach significance for texture and disparity. This suggests that the sensitivity of our method was not high enough to capture all figure-ground selective voxels, and some processing of segmentation may occur outside the fMRI-defined segmentation patches.
Color-selective patches containing high concentrations of hue-selective cells, dubbed “globs,” have been reported in area V4 (20). Activations to color were mostly separate from segmentation patches, with the exception of a patch in V4 in a subset of monkeys. To quantify the overlap between different modalities of segmentation or color, we computed modulation indices of figure versus background and color versus grayscale, respectively, and computed correlation coefficients between modulation indices of different modalities across all voxels inside visual areas (Fig. 2B). Figure-ground modulation indices of different modalities were positively correlated, consistent with the observed overlap in Fig. 1. Notably, correlations between segmentation and color were lower than all pairwise correlations between segmentation modalities. Texture-defined modulation indices and color modulation indices even showed a negative correlation. Similarly, we computed the spatial overlap of regions that showed activation in Fig. 1 between pairs of different contrasts by computing the area of their intersection divided by the area of their union (Fig. 2C). This yielded the qualitatively same result that overlap between segmentation-activated regions, defined by different modalities, was strong while there was very little overlap between segmentation-activated regions and color-activated regions.
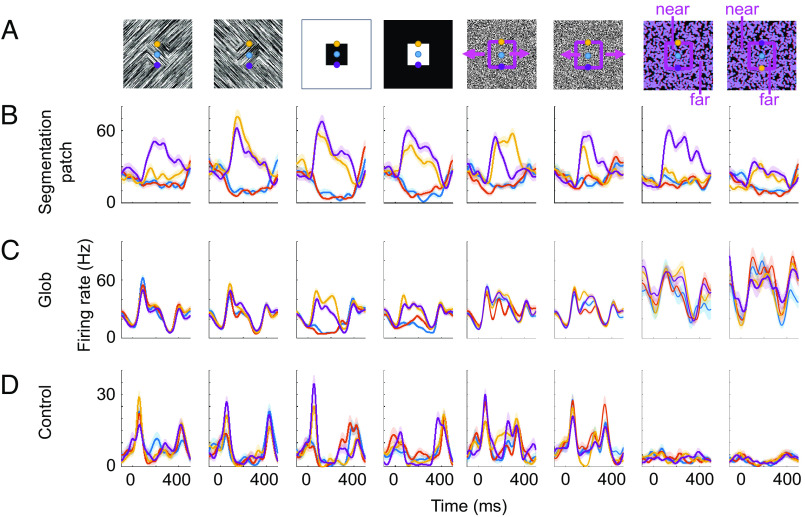
We targeted a subset of segmentation patches for electrophysiological recordings to compare segmentation encoding by single cells inside versus outside these regions. We recorded cells from 1) segmentation patches, 2) color-selective patches, and 3) control regions outside of either 1) or 2). During these electrophysiology experiments, we showed stimuli defined by texture, luminance, motion, or disparity, similar to the fMRI stimuli; however, in contrast to the latter, the stimuli during electrophysiology contained only a single square (Fig. 3). For each modality, stimuli were presented at four locations, such that either the center (blue), top edge (yellow), or bottom edge (purple) of a single square was centered on the receptive field of a cell; as a control, we also showed just the background without a square (red) (Fig. 3A). For each stimulus, there was also a stimulus with inverted polarity, e.g., white square on a black background, and black square on a white background for luminance. Note that for disparity, the far square may be perceived as cutout in an occluder with nonsquare shape (21). For each recorded cell, stimuli were rotated to the preferred orientation of the cell. We recorded a total of 338 (328 visually responsive, see Materials and Methods) cells from segmentation patches, 58 (47 visually responsive) cells from globs, and 274 (274 visually responsive) cells from control regions in two monkeys (monkey F and monkey M). Three example cells from segmentation patches, globs, and control regions, respectively, illustrate the diversity in responses to the different stimuli (Fig. 3 B–D). The segmentation patch cell (Fig. 3B) consistently preferred edges over figure or background, regardless of modality, and for most modalities, the response to bottom edge was higher than to top edge. In control regions and globs, consistent responses like these were exceedingly rare. The example cell from a glob preferred luminance-defined top or bottom edges over luminance-defined figure or background but did not show this selectivity for stimuli defined by other modalities (Fig. 3C). The cell recorded from a control region was visually responsive but did not show any clear selectivity (Fig. 3D).
Fig. 3.
Example cells from segmentation patches, globs, and control regions. (A) Stimuli consisted of the center (blue), top edge (yellow), or bottom edge (purple) of a single square presented on the receptive field or just background without a square (red, stimuli not shown). Columns correspond to different modalities (texture, luminance, motion, and disparity) and contrast polarities where the quality of figure and background were swapped. (B) Trial-averaged responses from an example segmentation patch cell. X axis and Y axis correspond to time aligned to stimulus onset (t=0) and firing rate, respectively. Different colored lines correspond to response time courses when the receptive field was on the square center (blue), top edge (yellow), bottom edge (purple), or background (red). Shaded areas indicate SEM across trials. (C) Same as (B) for an example glob cell. (D) Same as (B) for an example control cell.
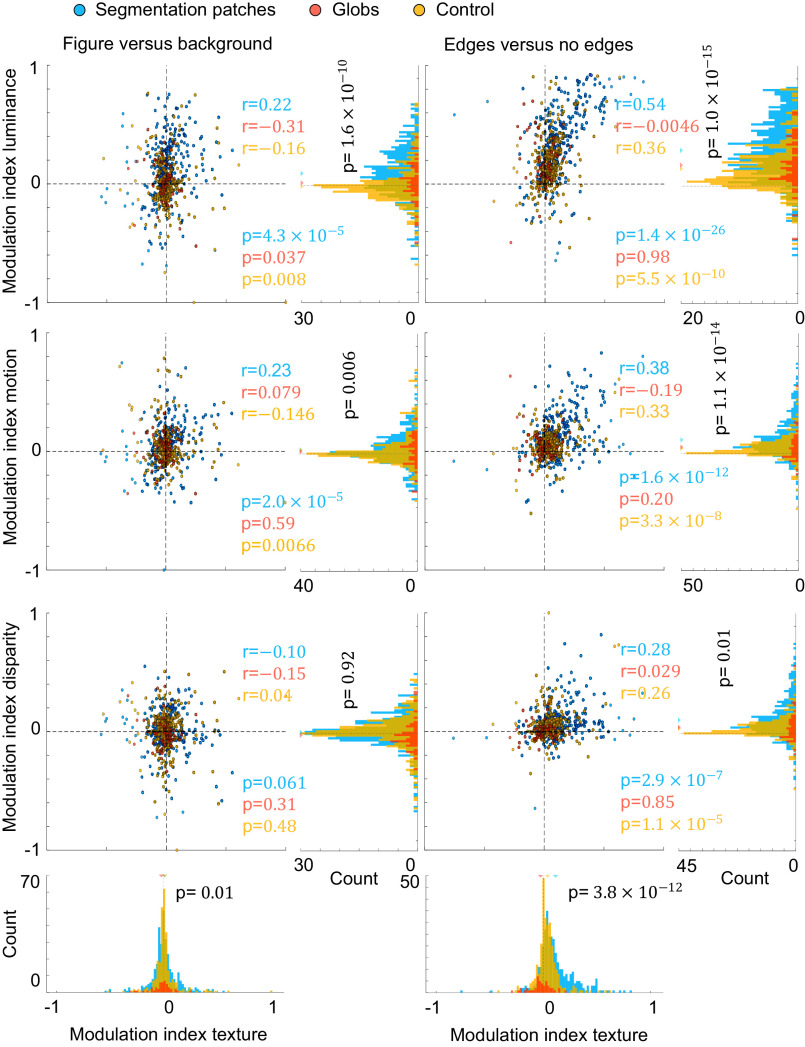
We next measured consistency of segmentation coding across modalities on a population level, specifically for coding of 1) figure-ground, 2) borders, and 3) border ownership. First, we compared how preference for figure over background for texture-defined figures transferred to other modalities (Fig. 4, left column). Only visually responsive cells were included in this analysis ( ). Inside segmentation patches, cells that tended to prefer texture-defined figures over background also tended to prefer figure over background for luminance-defined stimuli (see Fig. 4 for correlation coefficients and P values). Surprisingly, for control regions and globs, this trend was reversed, i.e., cells that preferred figure over background for texture stimuli tended to prefer background over figure for luminance stimuli. Similarly, for motion-defined stimuli, segmentation patch cells tended to show consistent figure-ground tuning with texture-defined stimuli, whereas glob cells did not show a significant correlation, and control cells were inconsistent. None of the regions (segmentation patches, globs, or control) reached significance of correlations for figure-ground in disparity. To conclude, only segmentation patches showed consistent figure-ground tuning across texture, luminance, and motion.
Fig. 4.
Single cells in segmentation patches have consistent segmentation signals. For the left panel, we computed the modulation indices for stimuli where a figure was centered on the receptive field versus background stimuli. For the right panel, we computed the preference for stimuli where a border was centered on the receptive field versus stimuli where no border was in the receptive field. We computed these modulation indices based on average responses for a given modality, namely, texture on the x-axes, and luminance, motion, or disparity on the y-axes. For scatter plots, each circle represents one cell from segmentation patches (blue), globs (red), or control regions (yellow), and its position represents the modulation indices for a pair of modalities of that cell. Correlation coefficients r and corresponding P values are provided in blue, red, and yellow, for segmentation patches, globs, and control region, respectively. Histograms represent marginal distributions of modulation indices for a single modality, using cells from segmentation patches (blue bars), globs (red bars), or control regions (yellow bars). For each histogram, P values for unpaired t tests comparing absolute values of modulation indices for cells from segmentation patches and cells from globs/control regions are shown in black.
We next turn our attention to edge tuning to see whether cells could distinguish borders from figural or background regions, which is a critical segmentation-related feature. We compared responses to edges versus nonedges, i.e., when either the top or bottom edge was on the receptive field versus when the square center or background was on the receptive field (Fig. 4, right column). Segmentation patch cells that preferred edges over nonedges for texture stimuli also tended to consistently prefer edges for luminance, motion, and disparity stimuli. Cells from control regions also showed consistency in edge tuning; however, correlations were weaker than in segmentation patches for all modalities. Cells from globs did not show significant consistency in edge tuning for any modality. To summarize, segmentation patch cells showed the most consistent tuning to edges, followed by control region cells, and there was no significant consistency in globs. Besides consistency between modalities, it is also important to note that the magnitude of edge versus nonedge modulation indices was higher for segmentation patch cells versus glob/control region cells, for all four modalities (unpaired t test, see P values next to marginal distributions in Fig. 4). This was also true for figure versus background modulation indices, except for disparity figure versus background.
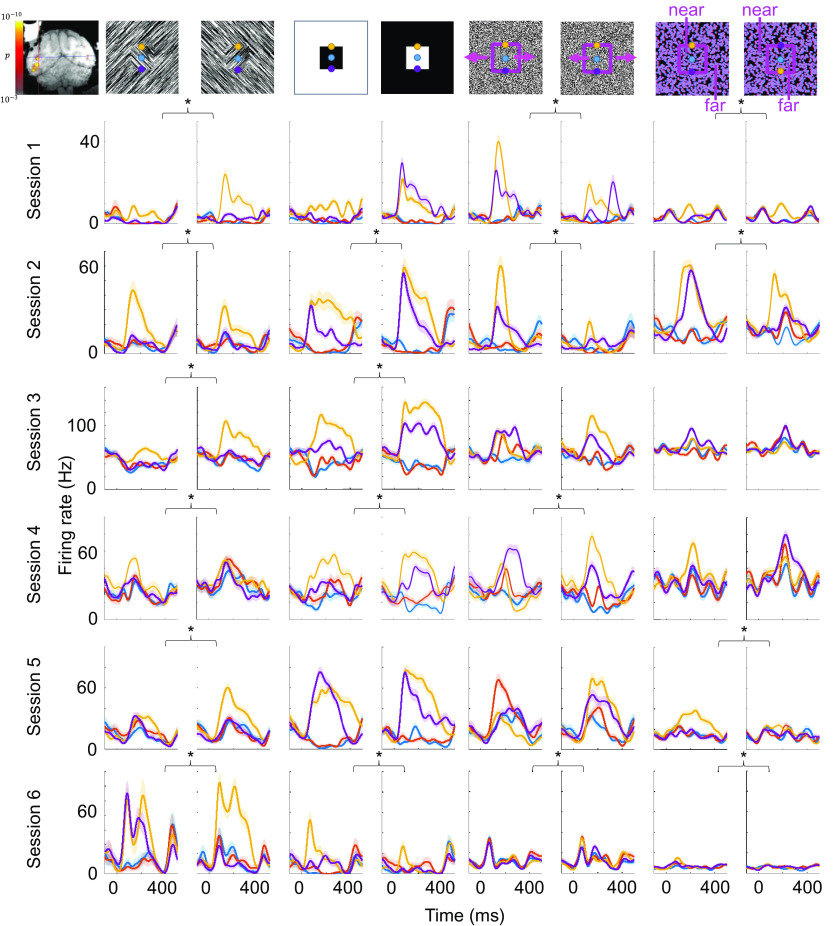
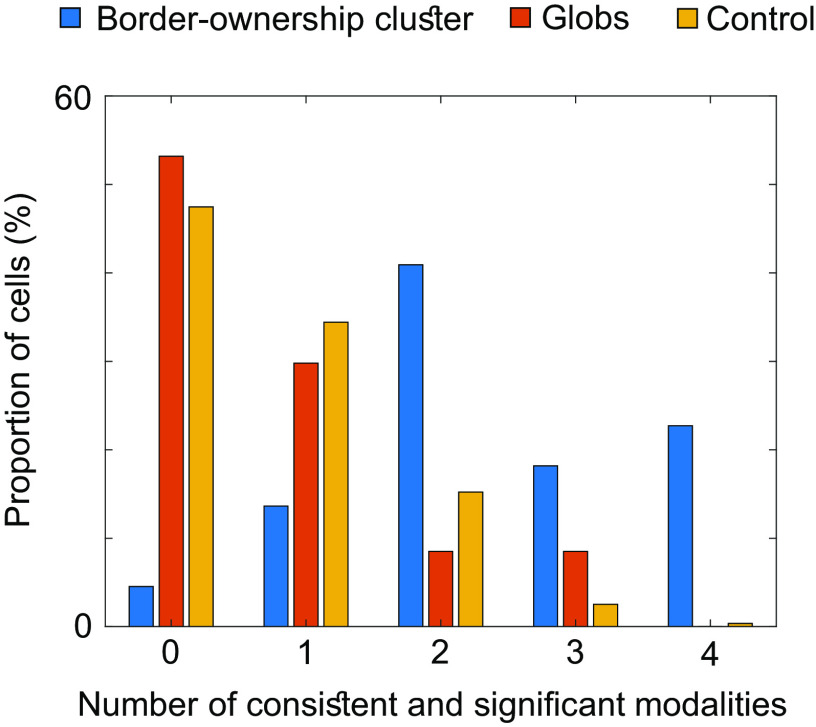
We next investigated the occurrence of consistent border-ownership cells, e.g., cells that consistently preferred the top edge over the bottom edge of a square for all four modalities. We found that even in segmentation patches, cells that showed consistent and significant border-ownership tuning across all four modalities were rare (1.8% of all segmentation patch cells), and even rarer in globs and control regions (0% of glob cells and 0.3% of control region cells). However, we did find a site inside a segmentation patch where consistent border-ownership cells were clustered. We recorded a total of 22 cells from this site. Reproducibly across days, we were able to find cells within 1 mm of this site that were consistent across all four modalities (Fig. 5). We note that the location of this cluster coincided with the region inside the lunate fundus in V3d where Adams and Zeki had discovered disparity columns (22). The distribution of cells that were significantly and consistently tuned to border ownership (Fig. 6) differed greatly between cells recorded from inside this site and globs or control regions ( , one-way ANOVA). In monkey T, we were not able to perform a segmentation localizer scan and hence targeted a similar location in the lunate fundus based on anatomy. In this monkey we also found a site where we reproducibly recorded border-ownership cells with consistent tuning across four sessions (SI Appendix, Fig. S2).
Fig. 5.
Cluster of consistent border-ownership cells across days. Same conventions as in Fig. 3 but every row corresponds to a cell recorded from the same site in different sessions in monkey F. Inset on the top left shows electrode targeting the patch with fMRI contrast overlaid. Asterisks indicate which of the four modalities showed a significant and consistent border-ownership preference for each cell.
Fig. 6.
Much higher proportions inside the border-ownership cluster of monkey F (cf. Fig. 5) show consistent coding of border ownership. Bars indicate proportions of cells within the border-ownership cluster (blue), globs (red), and control regions (yellow), respectively, that were significantly and consistently tuned to border-ownership across a given number of modalities (shown on the X axis). For a given modality, responses across all trials of top edge and bottom edge, respectively, were averaged to determine if the cell preferred top edge or bottom edge for that modality. To determine if this preference was significant, two-sample t tests across trials were performed (threshold P<0.05). For each cell the number of modalities where the cell was significantly tuned to side-of-figure and where the cell preferred the same side-of-figure as for texture, was computed, to produce the histogram. For texture, only significance was required to reach the significant and consistent criterion.
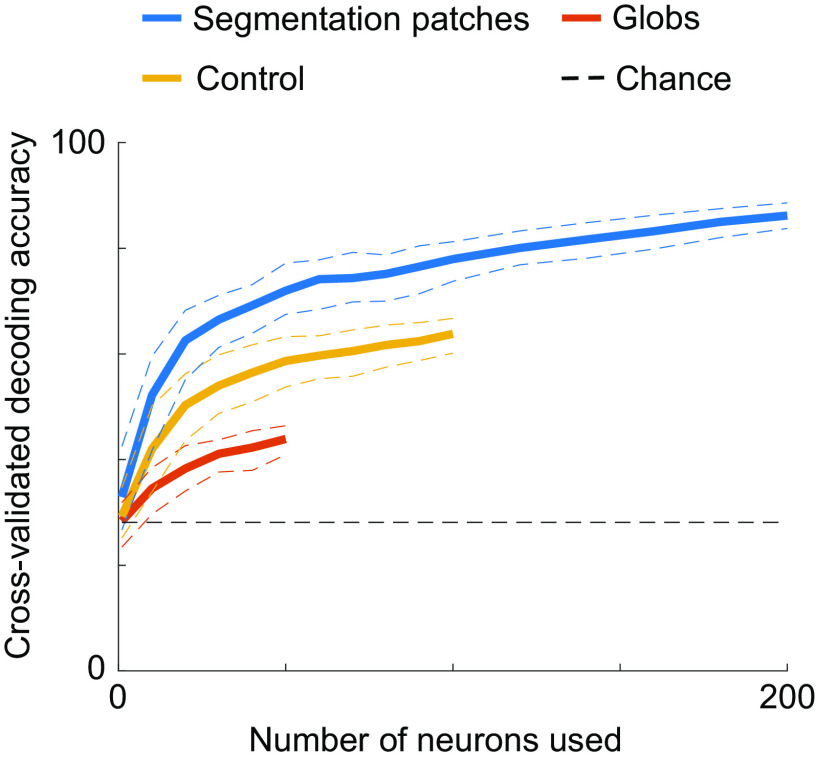
Even without single cells that consistently encode figure-ground assignment, it is possible that the segmentation of a scene can be read out from a population code. We therefore trained a linear classifier using all cells from segmentation patches, globs, or control regions, to discriminate between figure, background, top edge, or bottom edge (Fig. 7; expected chance decoding performance = 25%). The linear classifier’s objective was to correctly assign one of the four labels to a given trial, on which texture, luminance, motion, or disparity stimuli were presented, using the firing rates of a given number of neurons on that trial. We cross-validated decoding accuracies by training on one half of all trials and testing on the other half. Decoding accuracies were significantly higher when using cell populations from segmentation patches (reaching up to 86.1%) compared to control regions and globs, with globs yielding the lowest decoding accuracies. It is notable that the decoding accuracy using 200 segmentation patch cells was ~2.6 times higher compared to the average decoding accuracy using single segmentation patch cells (Fig. 7; leftmost data point), underscoring the power of the population decoding approach.
Fig. 7.
Segmentation can be decoded with higher accuracy from segmentation patches. We used a linear classifier to discriminate between four classes (figure, background, top edge, bottom edge; chance performance: 25%). For each data point, we performed 100 iterations of randomly selecting a given number of neurons (X axis) from all cells recorded from segmentation patches (blue), globs (red), or control regions (yellow) and randomly selecting one half of the trials for training the classifier and the other half for testing to determine cross-validated decoding accuracies (Y axis). Firing rates averaged across 0 ms to 250 ms after stimulus onset for each neuron were used as features for the classifier. Solid lines indicate cross-validated decoding accuracies averaged across iterations. Dashed lines indicate the 95% CI across the 100 iterations.
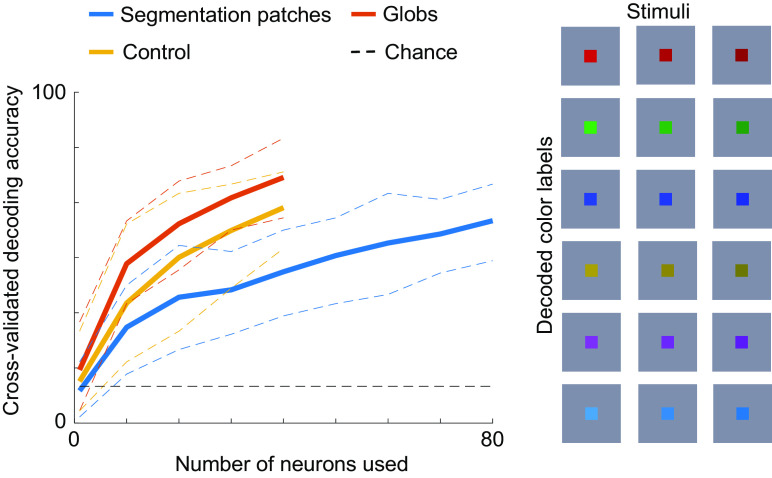
As a control, we also tried to decode color (Fig. 8). For a subset of sessions, we presented colored squares centered on the receptive field of the cell. The square in each stimulus had one of three luminances (dark, regular, and bright), and one of six equiluminant colors, i.e., 18 stimuli total. Using the same procedures as for figure-ground segmentation, we trained a decoder to discriminate between the six colors (chance: 16.7%). This time, globs had the highest decoding accuracy, followed by control regions, and then segmentation patches. This shows that segmentation patches are not better at encoding visual properties in general but are rather specialized for encoding segmentation features.
Fig. 8.
Color is preferentially encoded outside of segmentation patches. Here, the same conventions are used as in Fig. 7, but instead of decoding segmentation, we decoded color from responses in a separate experiment where we presented colored squares centered on the receptive field. The decoder had to discriminate between six color labels (red, green, blue, brown, purple, and cyan) invariant to luminance, as shown on the right. This stimulus set was shown in a subset of sessions and hence the maximum number of neurons that could be used for decoding is less than in Fig. 7.
Discussion
We found that fMRI activations evoked by stimuli containing figures compared to ground significantly overlap for different types of stimuli but are mostly distinct from color-activated regions of visual cortex. This suggests that visual cortex possesses modules that are specialized for segmentation and can use a variety of different cues, including texture, luminance, motion, and disparity, to segment the visual scene. While earlier studies examined figure-ground and border-ownership signals in V1 and V2, respectively, here we also found segmentation patches in V3, V3A, V4, and V4A. Signals obtained during fMRI are only an indirect measure of neural activity, however, and we therefore targeted a subset of these figure-activated regions with electrophysiological recordings to determine whether single units support this hypothesis. We found that both figure-ground and figure borders are encoded more consistently across modalities inside segmentation patches compared to outside the patches. Across cortex, only a small percentage of cells showed consistent border-ownership tuning across modalities. However, remarkably, we found that border-ownership cells with consistent tuning were clustered in a segmentation patch and could be recorded from reliably across days from the same site. Going beyond coding of single cells, we found that segmentation could be decoded better from a population inside the segmentation patches than in either globs or control regions. Finally, the improved decoding of segmentation features could not be explained by better signal-to-noise ratio or more selective tuning in general within segmentation patches, as decoding of color turned out to be significantly better in globs compared to segmentation patches.
Earlier studies reported cells scattered across retinotopic areas tuned to segmentation features such as figure-ground and border ownership for a specific stimulus such as luminance-defined squares (2–5, 23, 24). However, critics may dismiss these cells as a chance byproduct of, e.g., a simple feedforward network trained on object recognition that does not actually perform scene segmentation (25), analogous to how a completely untrained neural network with random weights may exhibit units with selectivity to certain face features (26). In fact, a previous study that used a larger battery of stimulus types found that ostensible border-ownership cells actually encoded side-of-figure inconsistently across stimuli and that none of the recorded cells encoded border ownership consistently across all stimulus types (12). However, the clustering of segmentation coding into discrete modules and the existence of cells therein that encode border-ownership invariant to cue, as described in the present study, gives credence to the hypothesis that border-ownership cells indeed play a functional role for segmenting the visual scene. The fact that figure-ground and border-ownership modulation were defined by modulation to stimulus pairs where the local contrast was identical (e.g., a white/black square on a black/white background compared to a full field white/black square) suggests that cells were modulated by segmentation features and not just low-level attributes within the receptive field.
In a recent complementary study, Yamane et al. also employed a decoding approach to figure-ground using natural images while recording from a population of V4 neurons (27) and reported a similar decoding accuracy of ~85%. Compared to our study, however, there were several important differences: First, Yamane et al. only trained the decoder to discriminate between two labels, figure and background, which is an easier task than discriminating between the four labels (figure, background, top edge, bottom edge) in the present study (chance performance 50% instead of 25%); when we tried decoding only figure versus background using 200 segmentation patch cells, average decoding accuracy was 90.6%. In addition, the natural images used by Yamane et al. had abundant local information within the receptive field that could be used by the decoder. In contrast, our stimuli were designed so that every stimulus was matched by a complementary stimulus with the same content within the receptive field but opposite figure-ground label, and hence, our decoder could not have relied solely on local cues within the receptive field to reach the observed decoding accuracy. Instead, cells must have used the global configuration of the stimulus to compute figure-ground segmentation. Future studies may investigate whether recording cells from segmentation patches improves decoding of figure-ground information for natural images.
In a parallel recent study, Franken and Reynolds reported clusters of border-ownership cells showing the same border-ownership preference, confirming our finding of modularity (11). Importantly, while Franken and Reynolds targeted random locations in V4 and used only one stimulus type (namely, luminance-defined squares), here we report clustering of border-ownership coding inside fMRI-defined locations that was consistent across stimulus types. Furthermore, our study shows that segmentation patches are found not only in V4 but also in V2, V3, V3A, and V4A. Finally, we find that these segmentation patches encode not only border ownership but also figure-ground.
Previous accounts of modularity in retinotopic cortex focused on segregation of coding for relatively low-level features of an image such as color, motion, and disparity (17). The present study extends this concept to modules coding features with a specific computational function, segmentation of a visual scene. The existence of specialized modules for segmentation is predicted by computational models of object representation which separate shape (i.e., the outline or segmentation mask of an object) from appearance (i.e., the texture map of an object) (13–16). In computer vision, there has been a large amount of work exploring the contributions of texture versus shape to visual recognition. For example, in the recursive cortical network model used by George et al. to solve “captchas,” objects are factorized into a surface and a contour representation, enabling objects with highly distinct surface textures but the same contour structure to be readily recognized (15). Similarly, it has been found that conventional deep networks strongly rely on texture, but if trained on a diet of texture-poor images, they can learn to exploit shape, which can yield improved object detection performance (16). These studies suggest that there is a computational advantage to separate the computation of object texture and object shape, invariant to texture. Thus, there are strong theoretical reasons to expect the existence of segmentation modules dedicated to computing cue-invariant shape. The current results suggests that primate retinotopic cortex exploits this strategy. Segmentation-related features computed by segmentation patches may be used by later areas beyond retinotopic cortex to compute, e.g., cue-invariant shape in inferotemporal cortex (28).
We note that the targeted segmentation patches represent merely the strongest regions of fMRI activation to figure versus background. We by no means claim that the regions we sampled are exhaustive: There may very well be regions outside of our targeted regions that encode segmentation. As a further limitation, we only showed square-shaped figures so units involved in segmentation but selective for nonsquare shapes may have been missed. Moreover, the spatial resolution of fMRI is coarse, so it is possible that a finer spatial organization within segmentation patches, such as border-ownership columns, exists. The finding of clusters of border-ownership cells that could be found across days within a <1 mm site is consistent with this possibility. In the present study, we found only one cluster each in two monkeys, so future experiments are needed to characterize the prevalence and spatial organization of these clusters, e.g., using optical imaging (29, 30). Overall, we believe that using an imaging-guided approach as described here could help make future studies of segmentation more tractable, as it allows recording from regions that preferentially encode segmentation signals rather than sampling randomly from the visual cortex. Finally, showing that cells inside functionally defined patches preferentially encode segmentation signals is suggestive but does not conclusively show that the brain actually uses these signals to segment the visual scene. The discovery of segmentation patches opens up the possibility for targeted perturbation experiments in the future to investigate whether, e.g., inhibiting segmentation patches causes behavioral deficits related to scene segmentation.
Materials and Methods
All animal procedures used in this study complied with local and NIH guidelines including the US NIH Guide for Care and Use of Laboratory Animals. All experiments were performed with the approval of the Caltech Institutional Animal Care and Use Committee (IACUC).
fMRI Scanning.
We implanted four male rhesus macaques with fMRI-compatible head posts and trained them to maintain fixation on a dot in the center of the screen for a juice reward. Subsequently, monkeys were scanned in a 3T TIM magnet (Siemens). We followed the same scanning and analysis procedures as described in Tsao et al. (31), Freiwald and Tsao (32), and Ohayon and Tsao (33). Monkeys were injected with monocrystalline iron oxide nanoparticle (MION) contrast agent to improve signal-to-noise ratio (34) and passively viewed stimuli on a screen of 45-degree diameter while in the scanner. Different types of stimuli were presented in blocks of 12 repetition times (TRs) (=24 s) length each. Stimulus blocks were interleaved with gray blocks of 12 TRs where no stimuli were presented.
Electrophysiology.
Cylindrical recording chambers (Crist) were implanted using dental acrylic on the left hemisphere of monkey F, the right hemisphere of monkey A, and the right hemisphere of monkey T. Custom grids were printed and inserted into chambers to record from targets defined by fMRI. Chamber positioning and grid design were planned using the software Planner (33). Guide tubes were cut and inserted through the grid to extend 2 mm beyond the dura. Single tungsten electrodes (FHC) with 1 MΩ impedance were inserted through guide tubes and used for recording. An oil hydraulic microdrive (Narishige) was used to advance electrodes through the brain. Neural signals were recorded using an Omniplex system (Plexon). Local field potentials were low-pass filtered at 200 Hz and recorded at 1,000 Hz, while spike data were high-pass filtered at 300 Hz and recorded at 40 kHz. Only well-isolated units were considered for further analysis. During electrophysiology, stimuli were presented on an LCD screen (Acer) of 47-degree diameter.
Task.
Monkeys were head fixed and passively viewed the stimuli presented on a screen in the dark. In the center of the screen, a fixation spot of 0.25-degree diameter was presented and monkeys received juice reward for properly maintaining fixation for 3 s. Eye position was monitored using an infrared eye tracking system (ISCAN). Images were presented in random order using custom software. For the main segmentation fMRI localizer, stimuli consisted of either 8 large (10-degree diameter) or 72 small (3-degree diameter) rounded squares that formed a grid covering the entire screen (see Fig. 1 B–E for examples of the large square version). Squares were laid out interleaved on a checkerboard, i.e., filling up every second position of a 4×4 grid (for large squares) or 12×12 grid (for small squares), see Fig. 1. The stimulus set also contained a version of each stimulus where the locations of squares and empty spaces were swapped, by shifting the squares by one square width, so that every position of the visual field was occupied by figure equally as often as it was occupied by background. The background control contained no shapes. For both the main (full-field) segmentation localizer (Fig. 1 A–E) and segmentation retinotopy localizer (SI Appendix, Fig. S1), figure shapes were defined by luminance, texture, motion, or disparity. For the segmentation retinotopy localizer, stimulus shapes were the same as for the standard retinotopy localizer, i.e., wedges for polar angle and rings for eccentricity. For luminance, figures and background were black or white; for texture, figures and background were created from lines of two different orientations at random positions; for motion, figures and background consisted of dots at random positions moving left or right; for disparity, figures and background consisted of random dots viewed through red-cyan goggles that had near or far disparity. For all stimuli, we also showed stimuli with switched figure and background assignment, e.g., for luminance, we also showed the same stimuli where figures were white and background was black. During fMRI experiments, stimuli were presented in a block design. Stimuli of each modality (luminance, texture, motion, and disparity) were presented in different blocks. Moreover, for each modality there was a block of stimuli that contained figures and a separate control block of stimuli that contained only background. Each block was presented for 24 s, and stimuli of each block were presented in pseudorandom order with 500 ms ON time, 0 ms OFF time.
Stimuli presented during electrophysiology experiments were similar to the main segmentation fMRI localizer, in that figures were defined by luminance, texture, motion, or disparity; however, they only contained a single (nonrounded) square that had a size and orientation determined by the receptive field and orientation tuning (see Online analysis). The square position was shifted giving rise to the four conditions described in the Results (square centered on receptive field, top edge of square centered on receptive field, bottom edge of square centered on receptive field, only background without a square). During electrophysiology, stimuli were presented for 250 ms ON time and 50 ms OFF time. Emission spectra were measured using a PR-650 SpectraScan colorimeter (Photo Research) and for the color experiment colors were adjusted to be equiluminant. For disparity stimuli, random dots were slightly shifted to the left and right, respectively, depending on whether they were on the square or in the background, by a distance of 3% of the square diameter, leading to horizontal disparities on the order of 0.2 degrees.
Online Analysis.
Spikes were isolated and sorted online using the software PlexControl (Plexon). We first computed receptive fields and orientation tuning of each cell. After determining an approximate location of the receptive field by manually sweeping a small blinking square (0.2 degrees) across the screen, we mapped receptive fields by computing the spike-triggered average (STA) in response to a random stimulus of size 8 degrees that was centered on the hand-mapped location as described in Hesse and Tsao (12). A 2-dimensional Gaussian was fitted to the STA to determine the center and size of the receptive field. Subsequently, stimulus position and size were adjusted so that the receptive field was centered on the stimulus and the square contained in the stimulus was larger than the receptive field. Moreover, the preferred orientation of the cell was determined from the sine grating orientation that elicited the highest response, and subsequent stimuli were rotated so that edges centered on the receptive field were presented in the preferred orientation.
Offline Analysis.
fMRI data were analyzed using FS-FAST and Freesurfer (35) as well as custom code written in Matlab. For computing fMRI time courses, we defined ROI based on voxels that either were a) significantly activated by the texture-defined figures minus background contrast ( , t test, computed using FS-FAST) and inside retinotopically defined visual areas V1, V2, V3, V3A, V4, or V4A, as defined by retinotopy, or b) not significantly activated by the texture-defined contrast and in one of the visual areas. We plotted time courses averaged across these voxels for each run for the first 48 s after stimulus onset (and hence, also including the following gray block). Baselines, computed from the average response in the preceding gray block, were subtracted from responses (Fig. 2A). For statistical testing of fMRI time courses, we computed the average response across a time window from 8 s after block onset to 8 s after block offset and performed two-sample t tests. For computing modulation indices (Fig. 2B), we computed average responses across the same time window, without baseline subtraction to ensure nonnegativity of responses.
For electrophysiology data, spikes were resorted offline using OfflineSorter (Plexon). Trials in which monkeys broke fixation were discarded (using a 1-degree eccentricity fixation window). Peristimulus time histograms were computed and smoothed with a Gaussian kernel (σ=100 ms) for plotting. To determine whether a cell is visually responsive, we computed each trial’s visual response (averaged spike count from 50 ms to 250 ms after trial onset) and baseline (averaged spike count from 50 ms before trial onset to 50 ms after trial onset) and performed a paired t test across trials (threshold: ). Since mean spike counts are not expected to satisfy a Gaussian distribution, we also applied an Anscombe transform to spike counts and repeated the t test which yielded the same results on visual responsiveness for 99.9% of all cells. To compute modulation indices and determine consistency of cells’ selectivity, we used average spike counts 50 ms to 250 ms after trial onset. For figure versus background modulation indices, we compared responses for stimuli where a figure was centered on the receptive field (i.e., square center) versus background stimuli, i.e., . Note that for disparity, we defined the center of the far-disparity square on a near-disparity background (i.e., a hole) as background, so that only the center of the near-disparity square on a far-disparity background was labeled as figure. For edge versus nonedge modulation indices, we computed the preference for stimuli where a border was centered on the receptive field (i.e., top edge or bottom edge) versus stimuli where no border was in the receptive field (i.e., square center or background), i.e., . For decoding, we trained a support vector machine (libsvm package in Matlab (36) with a linear kernel function and otherwise default parameters) to discriminate between the four classes “Figure,” “Background,” “Top edge,” and “Bottom edge” on a single-trial-basis using spike counts from 50 ms to 250 ms after trial onset of each recorded neuron as features. Since neurons were recorded across different sessions, we created a pseudopopulation [similar to Yamane et al. (27), see Discussion for a detailed comparison] with the requirement that for every included neuron at least 40 trials were collected for each stimulus. We varied the number of neurons to be included for training (X axis in Figs. 7 and 8) and constructed a feature matrix with number of neurons as columns and total number of trials (40 per stimulus) as rows. We performed twofold cross-validation by randomly splitting all trials into two halves, with one half being used for training and the other half being used for testing. For each number of neurons to include (value on the X axis in Figs. 7 and 8), we performed 100 iterations of randomly selecting neurons from the pseudopopulation and randomly selecting training and testing trials to get a distribution of decoding accuracies. All analysis was performed in Matlab (Mathworks).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The work was supported by the Howard Hughes Medical Institute and the Simons Foundation.
Author contributions
J.K.H. and D.Y.T. designed research; J.K.H. performed research; J.K.H. analyzed data; and J.K.H. and D.Y.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: G.A.O., Universita degli Studi di Parma; and D.T., State University of New York Upstate Medical University.
Contributor Information
Janis K. Hesse, Email: janishesse@googlemail.com.
Doris Y. Tsao, Email: tsao.doris@gmail.com.
Data, Materials, and Software Availability
The data that were analyzed in this manuscript are available on Dryad (https://doi.org/10.6078/D1313P) (37).
Supporting Information
References
- 1.Qiu F. T., Von Der Heydt R., Figure and ground in the visual cortex: V2 combines stereoscopic cues with Gestalt rules. Neuron 47, 155–166 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou H., Friedman H. S., Von Der Heydt R., Coding of border ownership in monkey visual cortex. J. Neurosci. 20, 6594–6611 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zipser K., Lamme V. A., Schiller P. H., Contextual modulation in primary visual cortex. J. Neurosci. 16, 7376–7389 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamme V. A., The neurophysiology of figure-ground segregation in primary visual cortex. J. Neurosci. 15, 1605–1615 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roelfsema P. R., Cortical algorithms for perceptual grouping. Annu. Rev. Neurosci. 29, 203–227 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Carlson E. T., Rasquinha R. J., Zhang K., Connor C. E., A sparse object coding scheme in area V4. Curr. Biol. 21, 288–293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasupathy A., Connor C. E., Responses to contour features in macaque area V4. J. Neurophysiol. 82, 2490–2502 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Mysore S. G., Vogels R., Raiguel S. E., Orban G. A., Processing of kinetic boundaries in macaque V4. J. Neurophysiol. 95, 1864–1880 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Marcar V., Raiguel S., Xiao D., Orban G., Processing of kinetically defined boundaries in areas V1 and V2 of the macaque monkey. J. Neurophysiol. 84, 2786–2798 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Marcar V., Xiao D., Raiguel S., Maes H., Orban G., Processing of kinetically defined boundaries in the cortical motion area MT of the macaque monkey. J. Neurophysiol. 74, 1258–1270 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Franken T. P., Reynolds J. H., Columnar processing of border ownership in primate visual cortex. Elife 10, e72573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesse J. K., Tsao D. Y., Consistency of border-ownership cells across artificial stimuli, natural stimuli, and stimuli with ambiguous contours. J. Neurosci. 36, 11338–11349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang L., Tsao D. Y., The code for facial identity in the primate brain. Cell 169, 1013–1028.e1014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cootes T. F., Edwards G. J., Taylor C. J., “Active appearance models” in European Conference on Computer Vision, Burkhardt H., Neumann B., Eds., Computer Vision - ECCV'98 (Springer, Berlin, Heidelberg, 1998), pp. 484–498. [Google Scholar]
- 15.George D., et al. , A generative vision model that trains with high data efficiency and breaks text-based CAPTCHAs. Science 358, eaag2612 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Geirhos R., et al. , ImageNet-trained CNNs are biased towards texture; increasing shape bias improves accuracy and robustness. arXiv [Preprint] (2018). 10.48550/arXiv.1811.12231 (Accessed 12 July 2023). [DOI]
- 17.Livingstone M., Hubel D., Segregation of form, color, movement, and depth: Anatomy, physiology, and perception. Science 240, 740–749 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Kolster H., Janssens T., Orban G. A., Vanduffel W., The retinotopic organization of macaque occipitotemporal cortex anterior to V4 and caudoventral to the middle temporal (MT) cluster. J. Neurosci. 34, 10168–10191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julesz B., Binocular depth perception of computer-generated patterns. Bell System Tech. J. 39, 1125–1162 (1960). [Google Scholar]
- 20.Conway B. R., Moeller S., Tsao D. Y., Specialized color modules in macaque extrastriate cortex. Neuron 56, 560–573 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovács G., Vogels R., Orban G. A., Selectivity of macaque inferior temporal neurons for partially occluded shapes. J. Neurosci. 15, 1984–1997 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams D. L., Zeki S., Functional organization of macaque V3 for stereoscopic depth. J. Neurophysiol. 86, 2195–2203 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Chen M., et al. , Incremental integration of global contours through interplay between visual cortical areas. Neuron 82, 682–694 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Zarella M. D., Ts’o D. Y., Cue combination encoding via contextual modulation of V1 and V2 neurons. Eye Brain 8, 177–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luongo F. J., et al. , Mice and primates use distinct strategies for visual segmentation. Elife 12, e74394 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek S., Song M., Jang J., Kim G., Paik S.-B., Face detection in untrained deep neural networks. Nat. Commun. 12, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamane Y., et al. , Population coding of figure and ground in natural image patches by V4 neurons. Plos One 15, e0235128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sáry G., Vogels R., Orban G. A., Cue-invariant shape selectivity of macaque inferior temporal neurons. Science 260, 995–997 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Ghose G. M., Ts’o D. Y., Form processing modules in primate area V4. J. Neurophysiol. 77, 2191–2196 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Zarella M. D., Ts’o D. Y., Contextual modulation revealed by optical imaging exhibits figural asymmetry in macaque V1 and V2. Eye Brain 9, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao D. Y., Freiwald W. A., Tootell R. B., Livingstone M. S., A cortical region consisting entirely of face-selective cells. Science 311, 670–674 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freiwald W. A., Tsao D. Y., Functional compartmentalization and viewpoint generalization within the macaque face-processing system. Science 330, 845–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohayon S., Tsao D. Y., MR-guided stereotactic navigation. J. Neurosci. Methods 204, 389–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leite F. P., et al. , Repeated fMRI using iron oxide contrast agent in awake, behaving macaques at 3 Tesla. Neuroimage 16, 283–294 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Fischl B., FreeSurfer. Neuroimage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C.-C., Lin C.-J., LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 1–27 (2011). [Google Scholar]
- 37.Hesse J. K., Tsao D. Y., Functional modules for visual scene segmentation in macaque visual cortex data set. Dryad. 10.6078/D1313P. Deposited 15 July 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The data that were analyzed in this manuscript are available on Dryad (https://doi.org/10.6078/D1313P) (37).