Abstract
Experiments were conducted to analyze the equine herpesvirus 1 (EHV-1) gene 68 product which is encoded by the EHV-1 US2 homolog. An antiserum directed against the amino-terminal 206 amino acids of the EHV-1 US2 protein specifically detected a protein with an Mr of 34,000 in cells infected with EHV-1 strain RacL11. EHV-1 strain Ab4 encodes a 44,000-Mr Us2 protein, whereas vaccine strain RacH, a high-passage derivative of RacL11, encodes a 31,000-Mr Us2 polypeptide. Irrespective of its size, the US2 protein was incorporated into virions. The EHV-1 US2 protein localized to membrane and nuclear fractions of RacL11-infected cells and to the envelope fraction of purified virions. To monitor intracellular trafficking of the protein, the green fluorescent protein (GFP) was fused to the carboxy terminus of the EHV-1 US2 protein or to a truncated US2 protein lacking a stretch of 16 hydrophobic amino acids at the extreme amino terminus. Both fusion proteins were detected at the plasma membrane and accumulated in the vicinity of nuclei of transfected cells. However, trafficking of either GFP fusion protein through the secretory pathway could not be demonstrated, and the EHV-1 US2 protein lacked detectable N- and O-linked carbohydrates. Consistent with the presence of the US2 protein in the viral envelope and plasma membrane of infected cells, a US2-negative RacL11 mutant (L11ΔUS2) exhibited delayed penetration kinetics and produced smaller plaques compared with either wild-type RacL11 or a US2-repaired virus. After infection of BALB/c mice with L11ΔUS2, reduced pathogenicity compared with the parental RacL11 virus and the repaired virus was observed. It is concluded that the EHV-1 US2 protein modulates virus entry and cell-to-cell spread and appears to support sustained EHV-1 replication in vivo.
Equine herpesvirus type 1 (EHV-1), a member of the family Alphaherpesvirinae, causes late-time abortions, respiratory disease, and neurological disorders in equines (1). EHV-1 possesses a type D herpesvirus genome which is composed of a unique long (UL) region and a unique short (US) region, the latter being bracketed by two inverted repeat regions (IRS and IRT) (16, 47). Comparison of the nucleotide sequences of the US-IRS region of strains Ab4p, KyA, and RacL11 and attenuated modified live vaccine (MLV) strain RacH revealed some genomic variation, and a 0.85-kbp deletion affecting genes 67 and 68 of RacH (the IR6 gene and the US2 homolog) was identified (5, 19, 32, 43). The unique IR6 protein has been shown to form rod-like structures in infected cells, and ability to form these structures is correlated with the virulence of EHV-1 Rac strains (32–34).
EHV-1 gene 68 is the homolog of the US2 gene of herpes simplex virus type 1 (HSV-1) (5, 11, 26). According to nucleotide sequences and the deduced amino acid sequences, EHV-1 strain Ab4p would encode a 47,000-Mr Us2 protein, whereas wild-type strain RacL11 would encode a 34,000-Mr US2 protein. This difference in Mr is caused by a frameshift mutation in strain Ab4p at position 125,540 (43) compared to RacL11 (19).
US2 homologs have been described in various members of the subfamily Alphaherpesvirinae, e.g., in HSV-2 (13), bovine herpesvirus 1 (24), canine herpesvirus (15), EHV-4 (42), and pseudorabies virus (PrV) (38, 44), as well as in the avian members of the subfamily, i.e., Marek’s disease virus (MDV) (7, 8, 40), herpesvirus of turkeys (49), and infectious laryngotracheitis virus (48). In contrast, varicella-zoster virus does not encode a US2 homolog (12). Analyses of a US2-US3-negative HSV-1 mutant showed that deletion of the respective genes resulted in an attenuated phenotype in a mouse infection model (27). In addition, a US2-negative HSV-1 mutant exhibited a small-plaque phenotype and grew to slightly reduced titers after infection of cells at a low multiplicity of infection (MOI) compared with the wild-type virus (46). The Bartha and Norden strains of PrV contain genomic deletions that encompass the US2-homologous gene (28K gene), yet they are able to replicate efficiently in tissue culture and in the natural host (28, 38). A 28K-negative mutant was not attenuated in pigs (21), but the mutant virus exhibited delayed kinetics of penetration into cells of the nasal mucosa (45). US2-negative MDV retained the abilities to establish latent infection and cause oncogenic transformation (36, 37).
The aims of this study were to characterize the US2 homolog of EHV-1 and to analyze the function of the US2 protein for virus replication in vitro and in vivo. This was achieved by generating a US2-specific antiserum which was used to examine the localization of the protein in infected cells and virions. The intracellular localization of the EHV-1 US2 protein was further assessed by analysis of constructs of the green fluorescent protein (GFP) fused to full-length or the amino-terminally truncated US2 protein. Lastly, by engineering and testing of US2-negative EHV-1, the role of the US2 gene product in virus replication was examined.
Identification of the EHV-1 US2 protein.
Previous studies had shown that US2-encoding sequences vary between individual EHV-1 strains. To confirm previously published sequences of strains Ab4p (43), RacL11 (19), RacH (19), and KyA (5), the US2 gene region of strains Ab4 (not plaque purified; kindly provided by N. Edington), wild-type RacL11, MLV strain RacH, and KyA (kindly provided by D. J. O’Callaghan) was amplified by a standard PCR (39) using Pfu polymerase (Stratagene) and the primers listed in Table 1. The amplified sequences were cloned in pTZ18R (Amersham-Pharmacia) and sequenced by cycle sequencing using fluorescent M13 primers (MWG-Biotech) and Taq polymerase (Amersham-Pharmacia). The gene 68 (US2) sequence of strain Ab4 did not differ from that reported for the corresponding sequence of plaque-purified isolate Ab4p (43). Also, the reported RacL11 and KyA US2 sequences (5, 19) were confirmed. MLV strain RacH exhibited an 853-bp deletion in the IRS-US junction. This deletion leads to a frameshift in the US2 open reading frame (ORF) compared with the wild-type RacL11 US2 sequence, such that the carboxy-terminal 27 amino acids of the RacL11 US2 gene product are replaced with 11 missense amino acids in strain RacH (data not shown; 19).
TABLE 1.
Primers used for PCR amplification of the EHV-1 US2 gene to construct different plasmids
| Plasmid | Positiona | 5′ primer | Position | 3′ primer |
|---|---|---|---|---|
| pQUS2 | 126,274 | 5′-acgtggatccATGGGTGTGGTAATTAC-3′ | 125,654 | 5′-acgtgaattcGAGCTATGTTTGG-3′ |
| pUS2-GFP | 126,274 | 5′-acgtggatccATGGGTGTGGTAATTAC-3′ | 125,365 | 5′-tatactgcagACGTGTGGATGTCCGGCC-3′ |
| phyUS2-GFP | 126,212 | 5′-tataggatccATGAGTTCCATCGACGTCG-3′ | 125,365 | 5′-tatactgcagACGTGTGGATGTCCGGCC-3′ |
| US2 genes in pTZ18R | 126,274 | 5′-acgtggatccATGGGTGTGGTAATTAC-3′ | 125,002 | 5′-CGGGTTGAACAGGTGCTTAC-3′ |
| 124,942 (RacH) | 5′-AGCTCTGGAGATCCAGCCGC-3′ |
Nucleotide positions are given in accordance with Telford et al. (43). Primers consisted of overhangs (lowercase letters), restriction enzyme cleavage sites (boldface lowercase letters), and US2 sequences (capital letters).
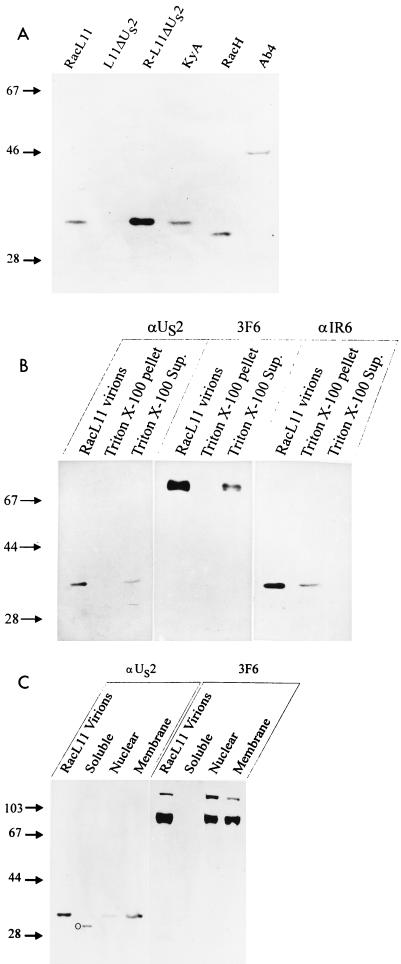
To specifically detect the EHV-1 US2 protein in infected cells and purified virions, sequences encoding the amino-terminal 206 amino acids of the RacL11 US2 ORF were amplified by PCR (Table 1) and cloned into plasmid pQE30 (Qiagen) (Fig. 1). The resulting recombinant plasmid, pQUS2, was transformed into Escherichia coli M15 cells. The His-tagged US2 protein was overexpressed and purified by Ni2+ affinity chromatography in accordance with the manufacturer’s instructions. After elution of the fusion protein using imidazole, a 24,000-Mr protein could be identified by Coomassie blue staining or Western blot analysis using the MRGS-His antibody (Qiagen) (data not shown). The pQUS2 protein was dialyzed against phosphate-buffered saline (PBS), emulsified in complete (first immunization) or incomplete (booster immunizations) Freund’s adjuvant, and injected intramuscularly a total of five times into a New Zealand White rabbit (Charles River). The EHV-1 US2-specific antiserum obtained was used to investigate US2 expression in infected-cell lysates and purified virions by Western blotting (22, 34). Rabbit kidney Rk13 cells were infected with EHV-1 at an MOI of 5, and cell lysates were prepared at 2, 4, 6, 8, 10, 14, 16, or 24 h postinfection (p.i.), adjusted to equal protein concentrations by using the BCA kit (Pierce), and separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (PAGE) (23, 34, 41). From 8 to 24 h p.i., a 34,000-Mr protein was detected with the anti-US2 serum in cells infected with EHV-1 strains RacL11, RacM24, RacM36 (19, 32, 33), and KyA. Consistent with the altered nucleotide sequences, a 44,000-Mr US2-specific protein was detected in Ab4-infected cell lysates, and the US2 protein of MLV EHV-1 strain RacH exhibited an apparent Mr of 31,000. In contrast, no US2-specific reactivity was detected in lanes containing L11ΔUS2-infected cell proteins (Fig. 1) or in mock-infected Rk13 cells (data not shown). Western blot analyses using virions purified by sucrose gradient centrifugation (32) were performed to analyze whether the EHV-1 US2 protein is a structural component of virions. US2-specific reactivity was detected in all of the virus strains investigated, irrespective of the size of the US2 protein (Fig. 2A). However, the electrophoretic mobilities of the US2 proteins were slightly decreased compared with those of infected-cell lysates, and a 35,000-Mr protein was detected in RacL11 and KyA virions, a 32,000-Mr protein was detected in RacH virions, and a 45,000-Mr polypeptide was detected in Ab4. No specific reaction with the anti-US2 antibody was observed in lanes containing L11ΔUS2 virion proteins (Fig. 2A).
FIG. 1.
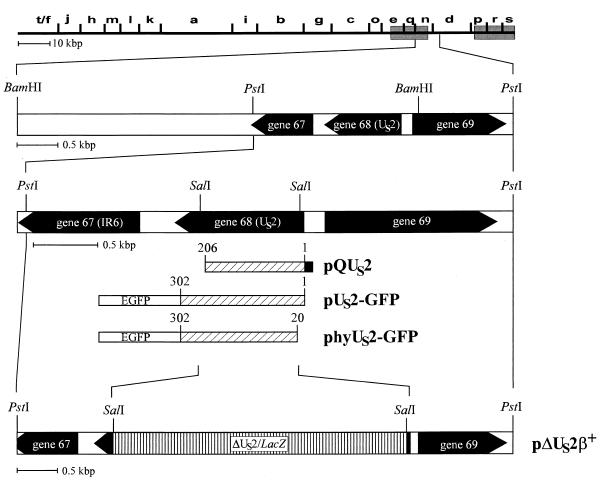
Diagram of the recombinant plasmids and viruses used in this study. Shown is a BamHI map of EHV-1 strain RacL11 and a magnification of the US-IRS junction. ORFs present at the US-IRS junction are indicated. The IR6 gene (gene 67) is a unique gene, gene 68 represents the US2 homolog, and gene 69 encodes the US protein kinase (5, 31, 43). The construction of recombinant plasmids pQUS2, pUS2-GFP, phyUS2-GFP, and pΔUS2β+ is detailed in the text. The His tag of pQUS2 is shown as a black box, and the 24,000-Mr pQUS2 protein was purified by Ni2+ affinity chromatography. The enhanced GFP (EGFP) sequences of pUS2-GFP and phyUS2-GFP are indicated. Restriction enzyme sites are given.
FIG. 2.
Western blot analysis of the US2 protein. Purified virions, as well as subcellular or subviral fractions, were separated by sodium dodecyl sulfate–12% PAGE (23), transferred to nitrocellulose (22), and probed with the indicated antibodies. (A) Detection of the US2 protein with the generated US2-specific antiserum in virions purified by sucrose gradient centrifugation. The US2-negative RacL11 virus (L11ΔUS2) served as a negative control. (B) Analysis of Triton X-100-treated virions. Purified RacL11 virions were incubated with 2% Triton X-100 and subjected to ultracentrifugation. Pellet and supernatant (Sup.) fractions were loaded onto individual lanes of a polyacrylamide gel and analyzed with anti-US2 serum, MAb 3F6 (2), or an anti-IR6 antibody (31). (C) Cells infected with RacL11 were harvested at 14 h p.i., and subcellular fractionation (4) was performed. Isolated fractions were separated and probed with anti-US2 serum. Anti-EHV-1 gB MAb 3F6 served as a control antibody. An unspecific 30,000-Mr band reacting in the cytoplasmic fraction with the anti-US2 antibody is marked with a circle. Prestained molecular weight marker (Gibco-BRL) sizes are indicated in thousands.
In a series of experiments, the putative N- or O-glycosylation of the EHV-1 US2 protein was analyzed because the amino acid sequence of the EHV-1 US2 protein contains two N-glycosylation consensus sites (5, 19, 43). Enzymatic deglycosylation by using peptide-N-glycosidase F and endo-β-N-acetylglucosaminidase H (Roche Molecular Biochemicals) or incubation of infected cells in the presence of tunicamycin (Roche Molecular Biochemicals) did not result in increased electrophoretic mobility of the US2 protein, whereas increased electrophoretic mobility of EHV-1 glycoprotein B (gB) was readily detected in both experiments. Similarly, incubation of purified RacL11 virions in the presence of O-glycosidase and neuraminidase (Roche Molecular Biochemicals) did not result in alteration of the apparent Mr of the US2 protein, whereas the apparent Mr of EHV-1 gD was reduced by approximately 2 kDa after de-O-glycosylation (data not shown). From these experiments, we concluded that the US2 protein does not contain detectable N- or O-linked carbohydrates.
The EHV-1 US2 protein is membrane associated.
Two approaches were taken to address the localization of the EHV-1 US2 protein. Firstly, viral envelopes were separated from nucleocapsids by incubation of purified RacL11 virions with 2% (final concentration) Triton X-100 for 20 min at 45°C (31). The resulting suspension was centrifuged for 30 min at 100,000 × g. The pellet containing viral nucleocapsids and the supernatant (viral envelopes) were examined by Western blot analysis using the US2-specific antiserum. Anti-EHV-1 gB monoclonal antibody (MAb) 3F6 (2; kindly provided by G. P. Allen) and an IR6-specific antiserum which detects the nucleocapsid-associated IR6 protein (31) were used to control the prepared fractions. gB- and US2-specific reactivities were detected in lanes containing the Triton X-100-soluble proteins (Fig. 2B), whereas the IR6 protein localized to the nucleocapsid fraction (Fig. 2B), as was demonstrated previously (32). The reduced signal intensities of the detected viral proteins in subviral fractions relative to whole virion lysates were probably caused by the incubation at 45°C, because identical amounts of lysates were loaded on the gel. Secondly, to investigate the subcellular localization of the EHV-1 US2 protein, infected cells were fractionated (4). Briefly, 107 Rk13 cells were infected with RacL11 at an MOI of 5 and harvested at 14 h p.i. Cells were washed with PBS, scraped into fractionation buffer (5 mM Na phosphate [pH 7.5], 2 mM MgCl2, 0.5 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride), and broken with a Dounce homogenizer. After addition of sucrose (final concentration, 0.3 M), still-intact cells, nuclei, and large membrane fragments were pelleted (P-1) by low-speed centrifugation (800 × g, 10 min). Supernatants were collected and centrifuged (10 min, 10,000 × g) to eliminate residual nuclei and cellular debris. The resulting supernatant was centrifuged (100,000 × g, 1 h, 4°C). The supernatant from this centrifugation step represented the soluble components of the cytoplasm, whereas the pellet contained plasma membrane fragments and vesicles from the endoplasmic reticulum and Golgi network. To enrich infected-cell nuclei, P-1 was redissolved in fractionation buffer containing 0.3 M sucrose and centrifuged twice through a 1.62 M sucrose cushion (2,100 × g, 15 min). The resulting pellet was dissolved in fractionation buffer containing 0.5% Triton X-100 and centrifuged for 15 min at 1,000 × g and contained mainly infected-cell nuclei.
All fractions were adjusted to the same protein concentration, and the fractions—soluble cytoplasmic proteins and nuclear and membrane fractions—were subsequently analyzed by Western blotting. MAb 3F6 and the IR6-specific antiserum served as control antibodies to confirm the identities of the fractions. The anti-US2 antibody specifically reacted with the 34,000-Mr US2 protein in membrane and nuclear fractions only (Fig. 2C). The same subcellular distribution, i.e., reactivity with gB-specific bands in nuclear and membrane fractions but not in the soluble cytoplasmic fraction, was observed with anti-gB MAb 3F6 (Fig. 2C). IR6-specific reactivity was not detected in the membrane fraction, whereas the 33,000-Mr IR6 protein was detected in the soluble cytoplasmic fraction and the nuclear fraction, as reported previously (32; data not shown). Taken together, these results indicated that the EHV-1 US2 protein is associated with viral and cellular membranes. Because the EHV-1 US2 protein could be solubilized from pelleted membranes with 1% Triton X-100 and with 0.1 M Na2CO3 (pH 11.0) (9), we concluded that the EHV-1 US2 protein most likely represents a peripheral membrane protein (data not shown). To examine whether the US2 protein is secreted from infected cells, Rk13 cells were grown in medium without addition of fetal calf serum and infected at an MOI of 5 with wild-type strain RacL11. At 16 h p.i., the cell culture supernatant was harvested and cleared from cellular debris and virions by ultracentrifugation, and the protein concentration was enriched by acetone precipitation. Western blot analyses with the US2-specific antiserum did not detect US2 protein in the supernatant of infected cells (data not shown).
Analysis of US2-GFP and hyUS2-GFP fusion proteins.
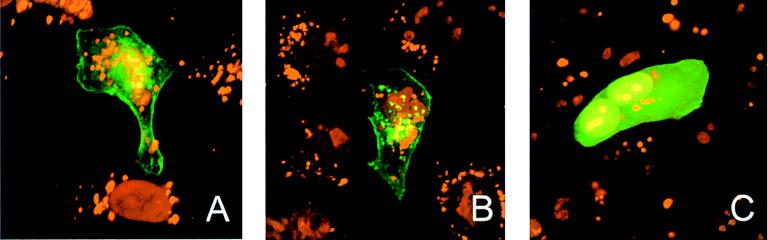
Plasmids encoding fusion proteins consisting of GFP and the full-length US2 protein or a US2 mutant protein lacking the amino-terminal 20 amino acids containing a possible transmembrane sequence (amino acids 1 to 16) (5, 26) were constructed to monitor the intracellular trafficking of the US2 protein. Full-length or truncated US2 genes were produced by PCR (39) (Table 1) and cloned into vector pEGFP-N1 (Clontech). The resulting plasmids, pUS2-GFP and phyUS2-GFP (Fig. 1), were transfected into Rk13 cells seeded on coverslips (33, 34). At different times posttransfection (p.tr.), cells were fixed with 3% paraformaldehyde in PBS–0.3% Triton X-100. Nuclear DNA was stained with 10−6 M propidium iodide (PI), and indirect immunofluorescent staining using an anti-γ-adaptin MAb (Sigma) or anti-EHV-1 gB MAb 3F6 was done as previously described (34). Coverslips were analyzed by using a confocal laser scanning microscope (Zeiss LSM 510). Green (GFP) and red (PI, Cy3) fluorescences were recorded separately by using appropriate filters (34). Fluorescence signals were readily detected in cells transfected with either plasmid starting at 8 h p.tr. At 24 h p.tr., both the US2-GFP and hyUS2-GFP fusion proteins accumulated in the vicinity of nuclei and at the plasma membrane of transfected cells. Intranuclear staining, however, was not observed (Fig. 3). The distribution of the fusion proteins within transfected cells did not significantly change with time, and at 48 h p.tr., the same pattern of fluorescence was observed in cells transfected with either pUS2-GFP or phyUS2-GFP. In pEGFP-N1-transfected cells, bright and homogeneously distributed nuclear and cytoplasmic fluorescence was observed at 24 h (Fig. 3) and also at 48 h p.tr. To examine the possible presence of the GFP fusions in the secretory pathway of transfected cells, colocalization studies using a γ-adaptin antibody, a marker of the trans-Golgi network, were performed. Colocalization of US2-GFP or hyUS2-GFP signals with γ-adaptin was not observed at any time p.tr. Similarly, colocalization of the GFP fusion proteins with gB was not observed after infection of previously transfected cells with RacL11 (data not shown).
FIG. 3.
Confocal laser scanning image of Rk13 cells transfected with pUS2-GFP (A), phyUS2-GFP (B), or pEGFP-N1 alone (C). Cells were transfected with 5 μg of the indicated plasmid and fixed at 24 h after transfection. Cellular DNA was counterstained with 10−6 M PI. Green and red fluorescences were recorded separately by using appropriate filters, merged, and printed. Magnification, ×1,000.
Growth characteristics of US2-negative EHV-1 in vitro.
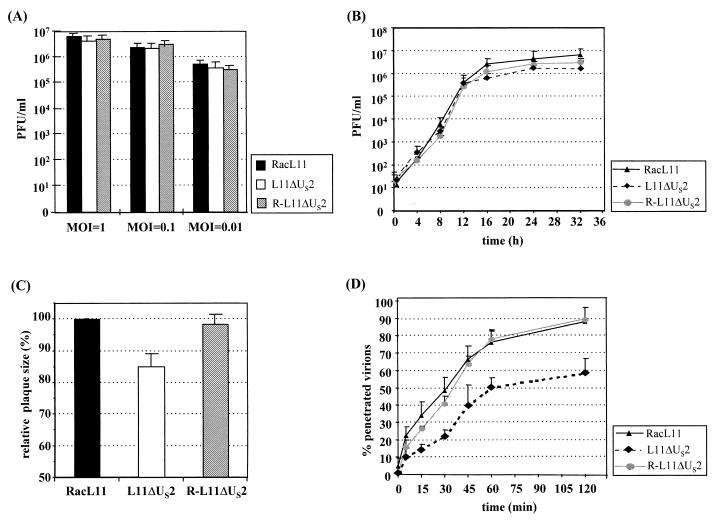
The US2-negative EHV-1 mutant L11ΔUS2 was constructed (30, 33) by cotransfection of 1 μg of RacL11 DNA and 10 μg of recombinant plasmid pΔUS2β+ (Fig. 1). Plasmid pΔUS2β+ contains a 3.3-kbp PstI fragment comprising the US-IRS junction, the entire US2 gene, and flanking sequences of EHV-1 strain RacL11, in which the US2-coding sequences for amino acids 12 to 221 were replaced with a β-galactosidase expression cassette (29) (Fig. 1). After addition of Bluo-Gal (Gibco-BRL), blue-staining virus plaques were picked and purified to homogeneity. The repaired virus R-L11ΔUS2 was isolated after cotransfection of L11ΔUS2 DNA and the 3.3-kbp PstI fragment (Fig. 1) by purification of white virus plaques in the presence of Bluo-Gal. Recombinant viruses were tested by Southern blot analyses using a US2 probe and a lacZ probe (Fig. 1), and the expected genotypes of both recombinant viruses were confirmed. Isolation of L11ΔUS2 on noncomplementing Rk13 cells demonstrated that the US2 gene is nonessential for virus replication in vitro. To characterize the growth properties of L11ΔUS2 in detail, a series of experiments was performed. Growth kinetics of the mutant virus after infection with different MOIs were determined. Rk13 cells (106) were infected at an MOI of 1, 0.1, or 0.01 with L11ΔUS2, RacL11, or R-L11ΔUS2. At 24 h p.i., infectious virus was harvested by freeze-thawing and titrated on Rk13 cells. In all cases, L11ΔUS2 grew to titers which were comparable to those produced by RacL11 and R-L11ΔUS2 (Fig. 4A). To determine single-step growth kinetics, 106 Rk13 cells were infected with the indicated viruses at an MOI of 5. After 1.5 h of incubation at 37°C, extracellular viruses were inactivated with a citrate buffer (pH 3.0) for 2 min (17). At different times p.i., supernatants were harvested and virus titers were determined on Rk13 cells. The results of the single-step growth kinetics corroborated the findings reported above, and no significant differences in the amount of infectious viruses produced were observed among RacL11, L11ΔUS2, or the repaired virus (Fig. 4B). Plaque sizes of L11ΔUS2, however, were slightly but significantly smaller than those observed in the case of either RacL11 or R-L11ΔUS2 (Fig. 4C). To test whether the US2 gene plays a role in viral entry, virus penetration assays were performed (17, 30). It could be shown that whereas 50% of RacL11 or R-L11ΔUS2 infectivity was protected from acid treatment at 35 min after a temperature shift to 37°C, only 30% of L11ΔUS2 infectivity was protected at this time point. After 2 h of incubation at 37°C, approximately 90% of the input RacL11 and R-L11ΔUS2 viruses, but only 60% of L11ΔUS2 virions, had entered the cells (Fig. 4D). It should be noted that virus adsorption assays using purified virions labeled with [methyl-3H]thymidine did not reveal significant differences in adsorption kinetics between L11ΔUS2 and parental virus RacL11 or R-L11ΔUS2 (data not shown). From these experiments, it was concluded that the US2 gene product modulates viral penetration and cell-to-cell spread but does not influence virus titers, even at a low MOI.
FIG. 4.
Analysis of US2-negative EHV-1 in vitro. (A) Replication of RacL11, L11ΔUS2, and R-L11ΔUS2 at different MOIs. Rk13 cells (106) were infected at an MOI of 1, 0.1, or 0.01 and treated for 2 min with citrate buffer (pH 3.0) at 2 h p.i. (17). Viral titers were determined at 24 h p.i. The values are means of three independent experiments, and standard deviations (error bars) are given. (B) Single-step growth kinetics of the different viruses. Rk13 cells (106) were infected at an MOI of 5, and at the indicated times p.i., infected-cell supernatants were harvested and titrated. The values are means and standard deviations of three independent experiments. (C) Rk13 cells seeded in six-well plates were infected with the indicated viruses (200 PFU/well), and plaque sizes were determined. The values are means of 150 individual plaques each. RacL11 plaque diameters were set to 100%. Error bars indicate standard deviations. (D) Penetration kinetics of RacL11, L11ΔUS2, and R-L11ΔUS2 into Rk13 cells. The values are means of four independent experiments. The error bars indicate standard deviations.
Importance of the EHV-1 US2 protein in vivo.
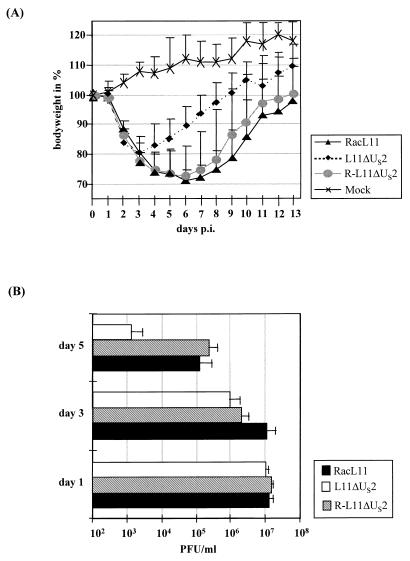
To analyze the influence of the US2 protein on EHV-1 virulence, experiments in a murine model of infection were performed (3, 19, 35). Three- to 4-week-old BALB/c mice (Charles River) were divided into groups of 14 and infected intranasally with 105 PFU (in 30 μl) of RacL11, L11ΔUS2, or R-L11ΔUS2 virus per mouse. Six control mice were inoculated with 30 μl of supernatant of uninfected Rk13 cells. The results are summarized as follows (Fig. 5). (i) RacL11- and R-L11ΔUS2-infected mice lost approximately 30% of their mean body weights until day 7 p.i., and three animals died as a consequence of the infection. (ii) In contrast, none of the L11ΔUS2-infected mice died, although they also showed typical signs of EHV-1 infection, such as ruffled fur and dyspnea from day 2 p.i. A decrease in mean body weight of up to 20% was observed in this group. However, L11ΔUS2-infected animals recovered significantly faster than RacL11- and R-L11ΔUS2-infected mice and gained body weight from day 4 p.i. When pairwise testing of groups was done by the method of Bonferroni (18), a significant difference between the groups infected with RacL11 or R-L11ΔUS2 and the animals infected with L11ΔUS2 could be demonstrated from days 4 to 8 p.i. Mock-infected animals did not show any signs of illness and gradually gained weight during the duration of the experiment (Fig. 5A). Determination of viral titers of individual mice necropsied on days 1 to 5 p.i. demonstrated that infected mice exhibited viral titers of up to 107 per lung on day 1 p.i., irrespective of the inoculum. Therefore, viral replication must have occurred, since only 105 PFU were administered to each animal (Fig. 5B). Viral titers in lungs of mice infected with L11ΔUS2 were reduced on day 3 after infection compared to those of mice infected with RacL11 or R-L11ΔUS2. On day 5 p.i., a 100-fold decrease in virus titers was determined in lungs of L11ΔUS2-infected mice compared to mice infected with the other viruses (Fig. 5B). In addition, blood was obtained by cardiac puncture on day 3 p.i. and viremia could be detected in all infected animals, although the viral load in the peripheral blood of L11ΔUS2-infected animals was approximately 50-fold lower than that of RacL11- or R-L11ΔUS2-infected mice. Virus could never be isolated from blood of mock-infected animals (data not shown). From these data, we concluded that the EHV-1 US2 gene is nonessential for EHV-1 replication in vivo but is required for a sustained virus load in murine lungs at later times after infection.
FIG. 5.
Analysis of US2-negative EHV-1 in vivo. (A) Fourteen BALB/c mice per group were infected with RacL11, L11ΔUS2, or R-L11ΔUS2, and six mice were mock infected. Body weights of individual mice were recorded daily from the day of infection (day 0) until day 13 after infection, and mean body weights on day 0 were set to 100%. Mean body weights (in relation to day 0) and standard deviations (error bars) are shown. Decreasing numbers of mice were weighed on days 0 and 1 (14), days 2 and 3 (12), days 4 and 5 (9), and days 6 to 13 (6). Two mice in the RacL11 group and one in the R-L11ΔUS2 group died. (B) Virus titers of the lungs of two (days 1 and 5) or three (day 3) mice necropsied on day 1, 3, or 5 after infection. Mean virus titers and standard deviations (error bars) are given.
Conclusions.
In this communication, an initial characterization of the EHV-1 US2 protein is presented. The salient findings are that the EHV-1 US2 protein (i) varies in size between selected EHV-1 strains, (ii) predominantly localizes to the membrane fraction of infected cells, (iii) is present in the envelope fraction of purified virions, (iv) is nonessential for virus growth in cultured cells but contributes to virus penetration and efficient cell-to-cell spread, and (v) plays a role in sustained virus replication in vivo. Sequence analyses of selected EHV-1 strains had suggested that the size of the EHV-1 US2 ORF is variable (5, 19, 43). It was shown that strain Ab4 encodes a 44,000-Mr US2 protein in infected cells, whereas another wild-type EHV-1 strain, RacL11, encodes a 34,000-Mr US2 protein. The fact that the Ab4 sequence determined here and that reported previously for plaque-purified Ab4p (43) are identical indicates that the frameshift mutation relative to wild-type strain RacL11 was not caused by serial passage, as is the case for MLV strain RacH (19, 25). Mutations in both Ab4 and RacH are located in the carboxy-terminal half of the molecule, which exhibits much less homology within the subfamily Alphaherpesvirinae than does the amino-terminal portion (7, 26, 42, 43). Because no differences in the behavior of the different EHV-1 US2 proteins with regard to subcellular localization or virion incorporation were observed, the functional domains appear to map to the amino-terminal 224 amino acids of the US2 protein. The EHV-1 US2 protein localized to the membrane fraction of infected cells and the envelope fraction of purified virions. All US2 homologs share a highly conserved stretch of 16 hydrophobic amino acids at the extreme amino terminus, and it was suggested that the US2 homologs may encode a secreted or an N-terminally anchored transmembrane (glyco)protein (5, 26). However, several lines of evidence do not support the hypothesis that the US2 protein is translated at membrane-bound ribosomes. Firstly, the US2 protein lacks carbohydrate modification. Secondly, localization of the EHV-1 US2 protein to the secretory pathway could not be demonstrated by using a US2-GFP fusion protein and a marker of the trans-Golgi network (γ-adaptin). Thirdly, a fusion protein consisting of GFP and US2 lacking the amino-terminal 20 amino acids of the protein did not behave differently from US2-GFP after transient expression: both fusion proteins were detectable at the plasma membrane and appeared clustered around the nuclei of transfected cells but did not exhibit the prominent granular appearance that is typical of endoplasmic reticulum or Golgi vesicles. It should be noted that the US2-GFP and hyUS2-GFP fusion proteins appeared in the same subcellular fractions as the native US2 protein and were also present in the Triton X-100-soluble fraction of purified virions after infection of transfected cells with L11ΔUS2. This was demonstrated by Western blotting using the US2-specific antiserum and anti-GFP antibodies (data not shown). So far, however, the exact posttranslational processing and trafficking of the EHV-1 US2 protein is not entirely clear, and localization to the secretory pathway in infected cells cannot be formally excluded. We were not able to visualize the native US2 protein in infected cells because the antiserum did not react in indirect immunofluorescence assays or immunoprecipitations. Therefore, GFP fusions were used to monitor the intracellular trafficking of the US2 protein. Previous reports on herpesviral proteins fused to GFP have demonstrated that the properties of the viral proteins, especially their subcellular localization, did not differ from those of the native protein (6, 14).
Nevertheless, it is possible that the US2 protein needs another viral protein for translocation to the secretory pathway, although a disulfide-linked complex of the US2 protein with a viral or cellular partner is very unlikely, as concluded from analyses using two-dimensional PAGE under nonreducing and reducing conditions (10, 20; data not shown). The HSV-1 US2-encoded protein and the PrV and MDV homologs are nonessential for virus growth in vitro (36, 37, 45). Similarly, the EHV-1 US2 protein is nonessential for virus growth, but the US2-negative RacL11 mutant is slightly impaired in virus penetration and cell-to-cell spread. These findings correlate well with the presence of the US2 protein in the membrane fraction of infected cells and in the Triton X-100-soluble fraction of purified virions. In addition, restricted growth of L11ΔUS2 in infected animals at late times p.i. was observed. Despite these properties of US2-negative RacL11, it is very unlikely that the alteration of the last 27 amino acids of the US2 protein of MLV strain RacH accounts for its apathogenicity because (i) RacH entirely lacks the direct neighbor of US2, the IR6 gene, which has been shown to be involved in virulence (33); (ii) EHV-1 strains RacM24 and RacM36 expressing a wild-type US2 protein but carrying mutations in the IR6 gene are completely apathogenic (19, 25); and (iii) preliminary results indicate that a RacH virus in which the wild-type US2 gene has been inserted is still completely apathogenic for mice. In contrast to the effects of the deletion of the US2 gene reported here, HSV-1, PrV, and MDV mutants with deletions in the US2 homologous genes retain their pathogenicity (27, 36, 37, 45, 46). Whether these differences are caused by distinct functions of the US2 proteins in the context of both different viruses and different hosts remains to be analyzed.
Acknowledgments
We thank George P. Allen, University of Kentucky, Lexington, for generously providing MAbs 3F6 and 20C4; Neil Edington, Royal Veterinary College, London, United Kingdom, for EHV-1 strain Ab4; and Dennis J. O’Callaghan, LSUMC, Shreveport, La., for EHV-1 strain KyA and the anti-IR6 serum.
A.M. was supported by a grant from the Studienstiftung des Deutschen Volkes. Part of this study was financed by a grant from the Mehl-Mülhens-Stiftung to N.O.
REFERENCES
- 1.Allen G P, Bryans J T. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog Vet Microbiol Immunol. 1986;2:78–144. [PubMed] [Google Scholar]
- 2.Allen G P, Yeargan M R. Use of λgt11 and monoclonal antibodies to map the genes for the six major glycoproteins of equine herpesvirus 1. J Virol. 1987;61:2454–2461. doi: 10.1128/jvi.61.8.2454-2461.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awan A R, Chong Y C, Field H J. The pathogenesis of equine herpesvirus type 1 in the mouse: a new model for studying host responses to the infection. J Gen Virol. 1990;71:1131–1140. doi: 10.1099/0022-1317-71-5-1131. [DOI] [PubMed] [Google Scholar]
- 4.Bogner E, Reschke M, Reis B, Reis E, Britt W, Radsak K. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch Virol. 1992;126:67–80. doi: 10.1007/BF01309685. [DOI] [PubMed] [Google Scholar]
- 5.Breeden C A, Yalamanchili R R, Colle C F, O’Callaghan D J. Identification and transcriptional mapping of genes encoded at the IR/Us junction of equine herpesvirus type 1. Virology. 1992;191:649–660. doi: 10.1016/0042-6822(92)90240-p. [DOI] [PubMed] [Google Scholar]
- 6.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunovskis P, Velicer L F. The Marek’s disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 8.Cantello J L, Anderson A S, Francesconi A, Morgan R W. Isolation of a Marek’s disease virus (MDV) recombinant containing the lacZ gene of Escherichia coli stably inserted within the MDV US2 gene. J Virol. 1991;65:1584–1588. doi: 10.1128/jvi.65.3.1584-1588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cockrell A S, Muggeridge M I. Herpes simplex virus 2 UL45 is a type II membrane protein. J Virol. 1998;72:4430–4433. doi: 10.1128/jvi.72.5.4430-4433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colle C F, O’Callaghan D J. Transcriptional analyses of the unique short segment of EHV-1 strain Kentucky A. Virus Genes. 1995;9:257–268. doi: 10.1007/BF01702881. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J. DNA sequence of the US component of the varicella-zoster virus genome. EMBO J. 1983;2:2203–2209. doi: 10.1002/j.1460-2075.1983.tb01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 15.Haanes E J, Tomlinson C C. Genomic organization of the canine herpesvirus US region. Virus Res. 1998;53:151–162. doi: 10.1016/s0168-1702(97)00152-4. [DOI] [PubMed] [Google Scholar]
- 16.Henry B E, Robinson R A, Dauenhauer S A, Atherton S S, Hayward G S, O’Callaghan D J. Structure of the genome of equine herpesvirus type 1. Virology. 1981;115:97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- 17.Highlander S L, Cai W, Person S, Levine M, Glorioso J C. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol. 1988;62:1881–1888. doi: 10.1128/jvi.62.6.1881-1888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 19.Hübert P H, Birkenmaier S, Rziha H J, Osterrieder N. Alterations in the equine herpesvirus type-1 (EHV-1) strain RacH during attenuation. J Vet Med B. 1996;43:1–14. doi: 10.1111/j.1439-0450.1996.tb00282.x. [DOI] [PubMed] [Google Scholar]
- 20.Jöns A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimman T G, DeWind N, Oei-Lie N, Pol J M A, Berns A J M, Gielkens A L J. Contribution of single genes within the unique short of Aujeszky’s disease virus (Suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 22.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without tank for rapid transfer of proteins from polyacrylamide gels to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–210. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Leung-Tack P, Audonnet J C, Riviere M. The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpesvirus type 1 (ST strain) Virology. 1994;199:409–421. doi: 10.1006/viro.1994.1139. [DOI] [PubMed] [Google Scholar]
- 25.Mayr A, Pette J, Petzoldt K, Wagener K. Untersuchungen zur Entwicklung eines Lebendimpfstoffes gegen die Rhinopneumonitis (Stutenabort) der Pferde. J Vet Med B. 1968;15:406–418. [PubMed] [Google Scholar]
- 26.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 27.Meignier B, Longnecker R, Mavromara Nazos P, Sears A E, Roizman B. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 28.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mettenleiter T C, Rauh I. A glycoprotein gX–β-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 30.Neubauer A, Braun B, Brandmüller C, Kaaden O R, Osterrieder N. Analysis of the contributions of the equine herpesvirus 1 glycoprotein gB homolog to virus entry and direct cell to cell spread. Virology. 1997;227:281–294. doi: 10.1006/viro.1996.8336. [DOI] [PubMed] [Google Scholar]
- 31.O’Callaghan D J, Colle C F, Flowers C C, Smith R H, Benoit J N, Bigger C A. Identification and initial characterization of the IR6 protein of equine herpesvirus 1. J Virol. 1994;68:5351–5364. doi: 10.1128/jvi.68.9.5351-5364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osterrieder N, Holden V R, Brandmüller C, Neubauer A, Kaaden O R, O’Callaghan D J. The equine herpesvirus 1 IR6 protein is nonessential for virus growth in vitro and modified by serial virus passage in cell culture. Virology. 1996;217:442–451. doi: 10.1006/viro.1996.0138. [DOI] [PubMed] [Google Scholar]
- 33.Osterrieder N, Neubauer A, Brandmüller C, Kaaden O R, O’Callaghan D J. The equine herpesvirus 1 IR6 protein influences virus growth at elevated temperature and is a major determinant of virulence. Virology. 1996;226:243–251. doi: 10.1006/viro.1996.0652. [DOI] [PubMed] [Google Scholar]
- 34.Osterrieder N, Neubauer A, Brandmüller C, Kaaden O-R, O’Callaghan D J. Equine herpesvirus 1 IR6 protein that colocalizes with nuclear lamins is involved in nucleocapsid egress and migrates from cell to cell independently of virus infection. J Virol. 1998;72:9806–9817. doi: 10.1128/jvi.72.12.9806-9817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osterrieder N, Wagner R, Brandmüller C, Schmidt P, Wolf H, Kaaden O R. Protection against EHV-1 challenge infection in the murine model after vaccination with various formulations of recombinant glycoprotein gp14 (gB) Virology. 1995;208:500–510. doi: 10.1006/viro.1995.1181. [DOI] [PubMed] [Google Scholar]
- 36.Parcells M S, Anderson A S, Cantello J L, Morgan R W. Characterization of Marek’s disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames. J Virol. 1994;68:8239–8253. doi: 10.1128/jvi.68.12.8239-8253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parcells M S, Anderson A S, Morgan T W. Retention of oncogenicity by a Marek’s disease virus mutant lacking six unique short region genes. J Virol. 1995;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrovskis E A, Post L E. A small open reading frame in pseudorabies virus and implications for evolutionary relationships between herpesviruses. Virology. 1987;159:193–195. doi: 10.1016/0042-6822(87)90368-0. [DOI] [PubMed] [Google Scholar]
- 39.Saiki A K, Gelfand D H, Stoffel S, Scharf S, Higuchi R, Horn G T, Mullis K B, Ehrlich H A. Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 40.Sakaguchi M, Urakawa T, Hirayama Y, Miki N, Yamamoto M, Hirai K. Sequence determination and genetic content of an 8.9-kb restriction fragment in the short unique region and the internal inverted repeat of Marek’s disease virus type 1 DNA. Virus Genes. 1992;6:365–378. doi: 10.1007/BF01703085. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch D F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Telford E A, Watson M S, Perry J, Cullinane A A, Davison A J. The DNA sequence of equine herpesvirus-4. J Gen Virol. 1998;79:1197–1203. doi: 10.1099/0022-1317-79-5-1197. [DOI] [PubMed] [Google Scholar]
- 43.Telford E A, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 44.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. Identification of two genes in the unique short region of pseudorabies virus: comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]
- 45.Wagenaar F, Pol J M A, Peeters B, Gielkens A L J, DeWind N, Kimman T G. The US3 encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 46.Weber P C, Levine M, Glorioso J C. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science. 1987;236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]
- 47.Whalley J M, Robertson G R, Davison A J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981;57:307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]
- 48.Wild M A, Cook S, Cochran M. A genomic map of infectious laryngotracheitis virus and the sequence and organization of genes present in the unique short and flanking regions. Virus Genes. 1996;12:107–116. doi: 10.1007/BF00572949. [DOI] [PubMed] [Google Scholar]
- 49.Zelnik V, Darteil R, Audonnet J C, Smith G D, Riviere M, Pastorek J, Ross L J. The complete sequence and gene organization of the short unique region of herpesvirus of turkeys. J Gen Virol. 1993;74:2151–2162. doi: 10.1099/0022-1317-74-10-2151. [DOI] [PubMed] [Google Scholar]