Abstract
Eosinophil/basophil (Eo/B) progenitor phenotype and function in cord blood (CB) are associated with atopic risk at birth and infant clinical outcomes. Molecular analyses of eosinophil–basophil differentiation events could identify clinically predictive biomarkers. To determine CB kinetic patterns of Eo/B lineage-associated gene expression (GATA-1, MBP1 and IL-5Rα) after IL-5 stimulation, CB non-adherent mononuclear cells were isolated from random fresh and frozen samples and incubated in the presence of recombinant human interleukin-5. Some underwent CD34+ positive selection using magnetic cell separation. At various time-points, mRNA expression of GATA-1, MBP1 and IL-5Rα (total transcripts) was determined utilizing multiplex quantitative polymerase chain reaction (Q-PCR). Relative expression levels of the IL-5Rα soluble vs. transmembrane isoforms were also analyzed. Stimulation of the non-adherent mononuclear cells with IL-5 resulted in early up-regulation of GATA-1, peaking at 48 h, followed by decreasing expression and down-regulation by 96 h. The CD34+ enriched population demonstrated an equivalent expression pattern (r = 0.963, p = 0.0349). MBP1 mRNA expression [non-adherent mononuclear cells (NAMNCs) and CD34+ alike; r = 0.988, p = 0.012] was slowly up-regulated in response to IL-5, maximal at 96 h. Total IL-5Rα expression appeared stable over the time-course, mediated by differential expression of the soluble and transmembrane isoforms (i.e., initial increase in the transmembrane contribution followed by a predominance of the soluble isoform by 48–72 h). Multiplex Q-PCR analysis of mRNA from CB demonstrates expression of critical eosinophil-basophil lineage-specific events that are consistent with current understanding of eosinophil differentiation and maturation. The non-adherent mononuclear cell population provides a surrogate signal for the CD34+ progenitor population.
Keywords: cord blood, eosinophils, hematopoiesis, IL-5R, transcription factors, major basic protein-1
The prevalence of allergic disease has increased dramatically in the developed world in the last quarter century. Allergic airway conditions are associated with high morbidity and significant burden to the affected individuals, (1) and importantly, because of these recent epidemiologic changes, they have emerged as the most common chronic illness in childhood (3). The reasons for this are not yet fully elucidated, but it has become apparent that there is an inflammatory basis for these conditions, and thus, efforts to understand the development of these inflammatory responses, their basis in immunity and factors underlying their increased prevalence have intensified over the last decade and are critical to the goal of slowing the ‘Allergy Epidemic’.
Identification of a biomarker in cord blood (CB) that is strongly predictive of later allergy could allow for early intervention with appropriate treatment and/or avoidance strategies that could potentially slow or halt the ‘atopic march’ (the recognized tendency toward atopic progression over time in an affected patient from atopic dermatitis → food allergy → asthma/rhinitis) (5).
Several birth cohort studies in which we have recently undertaken CB analyses have shown that the number and functional phenotype of CD34+CB stem cells (hematopoietic progenitors) are altered in infants at high atopic risk compared to low-risk infants (6–9), particularly in the eosinophil/basophil (Eo/B) lineage-committed progenitor cell population (10). It was additionally noted that differences in these CD34+ populations were associated with increased risk of fever and wheeze with respiratory illness in the first year of life (11). The collective results of these studies demonstrate that in infants at risk of atopy (by parental history and/or maternal allergy skin tests), these CB progenitors are reduced in number and show reduced expression of cell surface cytokine receptors such as IL-5Rα or GM-CSFRα than those at low risk of atopy. These findings have been interpreted as a possible consequence of in utero exposure of progenitors to comparatively higher ambient levels of Th2 cytokines and suggest that the resultant CB progenitor phenotype may be predictive of future atopic outcomes (7, 10).
Rationale for current study
All studies to date have evaluated these progenitors via a labor-intensive, 14-day methylcellulose colony assay for functional outcomes, and by flow cytometry for phenotypic outcomes, but not by analyses of the molecular biomarkers involved in eosinophil lineage commitment and differentiation. Potential molecular targets of interest include GATA-1, a pivotal transcription factor required for eosinophil development (12), the IL-5Rα subunit of the high affinity IL-5 receptor that binds IL-5 (13) and the eosinophil major basic protein-1 (MBP1), a major constituent of the large specific (secondary) granule in the terminally differentiated eosinophil (14). In a very preliminary evaluation of kinetic changes in the expression of GATA-1 over time, our laboratory was able to show that GATA-1 was up-regulated early in CB non-adherent mononuclear cells (NAMNCs) stimulated with recombinant human interleukin-5 (rhIL-5) in a SYBR Green-based RT-PCR, peaking at 48 h, with subsequent diminishments in expression (9), but the other aforementioned target molecular biomarkers had not been investigated. Of relevance to the IL-5Rα subunit, the full-length IL-5Rα gene can produce two mRNA species that correspond to alternatively spliced soluble and transmembrane isoforms (sol-IL-5Rα, Tm-IL-5Rα) (15–18). The Tm-IL-5Rα is the functional isoform that allows for signal transduction and initiation of responses mediated via the IL-5Rα and βc chain shared with IL-3 and GM-CSF receptors. The sol-IL-5Rα subunit binds IL-5 and neutralizes the biologic activity of the cytokine in vitro (15–17).
Developing and applying an understanding of early molecular events in Eo/B lineage commitment with specific focus on mRNA expression in CB progenitors of GATA-1, IL-5Rα and MBP1 would not only enhance our current understanding of hematopoietic (especially Eo/B progenitor) development in the neonate but could also potentially provide a very early marker of atopic risk in the newborn.
Materials and methods
This study was reviewed and approved by the Hamilton Health Sciences & McMaster Health Sciences Research Ethics Board. All CB donors gave written informed consent to participate.
Materials
Materials were purchased as follows: Accuprep® was obtained from Accurate Chemicals Inc. (Westbury, NY, USA); McCoys 5A medium, penicillin, streptomycin and FBS were obtained from Gibco BRL (Gaithersburg, MD, USA); heparin, dextran (molecular weight 260,000), from Sigma-Aldrich (Oakville, ON, Canada); and 2-mercaptoethanol from BDH Inc. (Toronto, ON, Canada).
Preparation of cord blood cells
Immediately after delivery, umbilical CB was collected in a heparin-containing syringe. For this preliminary evaluation to verify the validity of our proposed multiplex quantitative polymerase chain reaction (Q-PCR), we only recruited anonymous, random CB donors with unknown atopic status. The CB was depleted of red cells by unit gravity sedimentation in 1% (vol/vol) dextran at 37°C for 30 min, followed by isolation of mononuclear cells on an Accuprep™ density gradient (2000 rpm for 20 min at 20°C). After washing, cells were re-suspended in McCoy’s 5A supplemented with 15% FBS, penicillin/streptomycin and 2-mercaptoethanol and depleted of adherent cells by incubation in plastic flasks for 2 h at 37°C. Cell viability was assessed by the trypan blue staining method. The NAMNCs were then incubated in the presence of rhIL-5 (1 ng/ml). Half of the samples utilized in these experiments were from frozen CB samples prepared as mentioned earlier but stored in liquid nitrogen after appropriate cryopreservation with freezing media. Frozen NAMNCs were flash-thawed at 37°C for 30 s then washed with ice-cold McCoy’s 5A and similarly incubated with IL-5 after viability assessment and enumeration. Average recovery was 38% with 96% viability from the frozen samples. At 0, 24, 48, 72 and 96 h post-stimulation, cells were lysed in Buffer RPE (Qiagen, Valencia, CA, USA) with added 2-mercaptoethanol and stored at −80°C for future batch processing to ensure consistency of processing.
In addition to the NAMNC analyses, some CB samples underwent positive selection for CD34+ hematopoietic progenitors. This was achieved by taking the mononuclear layer and processing it through a CD34+ magnet-activated positive selection cocktail (EasySep; Stem Cell Technologies, Vancouver, BC, Canada). A cell count was performed at the end of the last purification step to determine the number of time-points that could be analyzed, aiming for a total of 4. The cells were re-suspended in an appropriate volume of media (complete RPMI), the zero-hour samples were immediately treated with lysis buffer (Buffer RLT with beta-mercaptoethanol), and the others were incubated with IL-5. At 24, 48 and 72 h, and for some experiments, 1-wk post-stimulation, cells were lysed and frozen as described earlier.
RNA isolation and reverse transcription
RNA was extracted from each cell lysate sample using RNeasy® Mini-kit columns (Qiagen) according to manufacturer’s instructions. DNA contamination was removed using a DNA Free kit® containing DNAse-1 buffer and DNAse-1 mix (Ambion, Austin, TX, USA) according to manufacturer’s instructions. Total RNA in each sample was quantified using a UV spectrophotometer, and a fixed amount of RNA was reverse-transcribed for each sample (determined via lowest concentration of RNA per sample, as calculated via total RNA quantification). Reverse transcription (RT) was completed using a Stratascript cDNA synthesis kit (Stratagene, Cedar Creek, TX, USA), with 2.97 ml random hexamer primers and 0.03 ml oligo (dT) primer (both 100 ng/ml).
Multiplex Q-PCR analysis
mRNA expression of GATA-1, IL-5Rα (total) and MBP1 was determined utilizing comparative Q-PCR in a multiplex reaction (Stratagene MX4000; Stratagene). The housekeeping gene was GAPDH, which was selected using the TaqMan Human Endogenous Control Plate and had been established previously in our laboratory as a successful reference gene for GATA-1 in the NAMNC population (9). Primer/probe sets (including housekeeping gene) and master mix were obtained from Qiagen. Standard curves were run in duplicate using a commercially available total RNA (Stratagene) that was reverse-transcribed in tandem with the experimental samples, along with no RT and no template controls. Each gene was analyzed in separate individual reactions first, and then efficiencies and standard curves compared when run in concert in a multiplex reaction, with no evidence of inhibition or competition as a result of multiplexing. The sequences utilized for GATA-1, MBP1, IL-5Rα (total) and GAPDH are detailed in Table 1. The IL-5Rα soluble (Sol) and transmembrane (Tm) isoforms were then analyzed using a SYBR green-based assay and a BioRad iCycler (iQ5; BioRad, Hercules, CA, USA) with beta-2-microglobulin as the housekeeping gene as previously standardized and described for CD34+ cells differentiated to eosinophils (19). Standard curves were run in duplicate using RNA purified from the IL-5Rα+AML14.3D10 eosinophil myelocyte cell line (19). The primer sequences for the Tm-, Sol-IL-5Rα and beta-2-microglobulin are similarly detailed in Table 1. If the efficiency of the reaction (either for the housekeeping gene or any of the given genes of interest) was less than 85%, the data were rejected. Similarly, only reactions with r2 values ≥0.97 were considered acceptable for analysis. Primer pairs used for both multiplex and individual real-time Q-PCR for the genes of interest (Table 1) were first analyzed by standard RT-PCRs for 40 cycles. We then used total RNA purified from the AML14.3D10 eosinophilic myelocyte cell line to confirm their amplification of single cDNA (mRNA) products of predicted size (Table 1) with standard 1.5% agarose DNA gel electrophoresis (Fig. S1). As well, melting curves were performed using real-time Q-PCR to confirm that these primer sets produced single peaks at high efficiencies with total cDNA generated from commercially available RNA (BioRad).
Table 1.
Primer/probe sequences for housekeeping genes and genes of interest
| Gene | Sequence | Amplicon (bp) |
|---|---|---|
| Multiplex reactions | ||
| GAPDH | Probe: AAGGCTGAGAACGGG Forward primer: GACCTCAACTACATGGTTT Reverse primer: CTGGAAGATGGTGATGGGATTT |
238 |
| GATA-1 | Probe: ACTCTCCCCCTGCCTC Forward primer: AGCCTATTCCTCTCCCAA Reverse primer: TCCGCAGTTCACACACTC |
78 |
| MBP1 | Probe: GAGGAAGAGGAGGAGTG Forward primer: AGCAGGAGATGGAGGAGA Reverse primer: CTGGCACTGAGATAGACTCAAC |
113 |
| IL-5Rα | Probe: TTGGTTGTTGG*GAGTT Forward primer: TAATGGCTATCTGGACGAG Reverse primer: AAGCAAAT*AT*GGA*GGAGGA |
110 |
| SYBR green-based reactions (no probe required) | ||
| IL-5Rα soluble | Forward primer: GCAGCAGTGAGCTCCATGTG Reverse primer: TTCAGATACGGTGTGGGGCAG |
124 |
| IL-5Rα transmembrane | Forward primer: GCAGCAGTGAGCTCCATGTG Reverse primer: AGGGCTTGTGTTCATCATTTCCC |
87 |
| β-2-microglobulin | Forward primer: CCTGAATTGCTATGTGTCTGGG Reverse primer: TGATGCTGCTTACAGTCTCGA |
245 |
Denotes superbases.
Statistical considerations
All data were normalized to pre-treatment controls with an equal quantity of viable cells; no medium-only controls were analyzed. Normalized relative expression ratios (RERs) between stimulated and un-stimulated cells were calculated using the delta-delta Ct method (2−ΔΔCt), with the zero-hour time-point as reference. Statistical analyses were completed using SigmaStat v2.0 for Windows (Systat Software, Inc. Chicago, IL, USA). The mean RERs were compared using the Mann–Whitney U test and/or ANOVA on ranks for non-parametric data, as appropriate. Correlation analyses between mRNA expression and Eo/B colony forming units (CFUs) were completed with Pearson’s and/or Spearman’s correlation coefficients, as the normality of the data dictated.
Results
Cord blood NAMNC population results
We were able to successfully determine the mRNA expression of each of the genes of interest from CB NAMNCs utilizing a multiplex reaction (Fig. 1). Stimulation of CB NAMNC with rhIL-5 resulted in the up-regulation of GATA-1 mRNA expression that peaked at 48 h (mean fold-increase of 10.3), followed by decreasing expression by 72 h, and clear down-regulation by 96 h (mean fold-decrease of −8.3). These kinetic changes were statistically significantly different over time; p = 0.04. MBP1 gene expression was up-regulated in a slowly progressive pattern, with maximal up-regulation at 96 h (mean fold-increase of 2.3). The MBP1 mRNA kinetic changes were also statistically different; p < 0.001. When the pattern of GATA-1 up-regulation and subsequent down-regulation was examined alongside changes in MBP1 mRNA expression, GATA-1’s role as a co-promoter of MBP1 expression was apparent (Fig. 2), as GATA-1 expression preceded MBP1 expression.
Fig. 1.
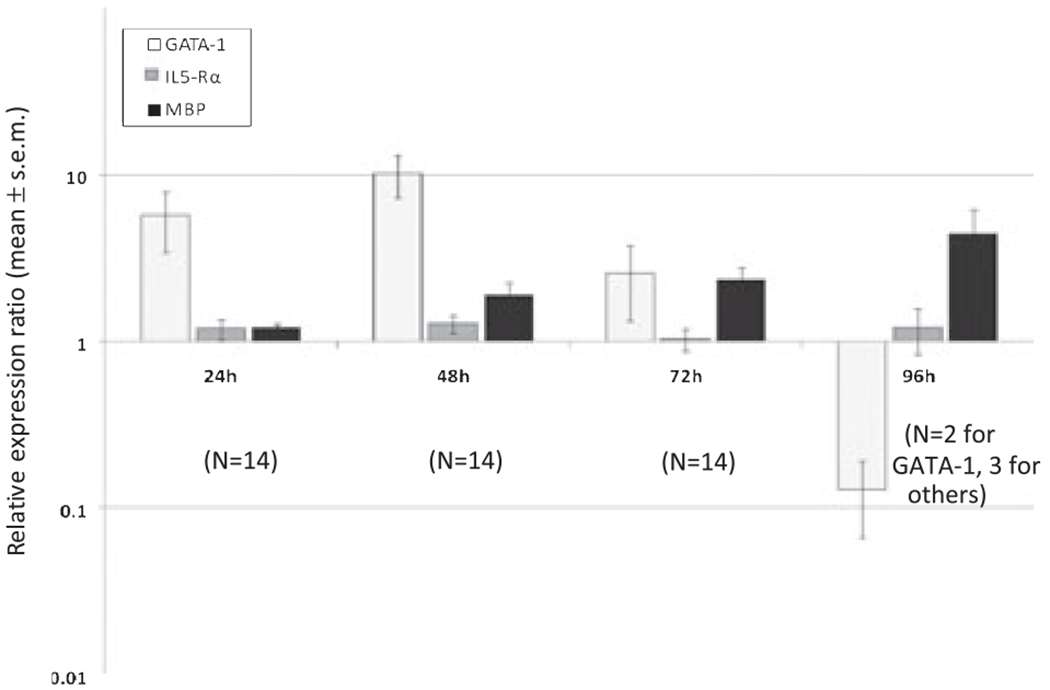
Kinetic mRNA expression of GATA-1, MBP1, and IL-5Rα (total) from cord blood (CB) non-adherent mononuclear cells (NAMNCs) stimulated with IL-5. NAMNCs from random fresh and frozen CB samples were incubated in the presence of rhIL-5, and removed at various time points for cell lysis, RNA extraction, reverse-transcription and quantitative polymerase chain reaction (Q-PCR) analysis using a multiplex reaction. Displayed are relative expression ratios (normalized to housekeeping gene, GAPDH), calculated using the standard 2(−ΔΔCt) formula (GATA-1: p < 0.001; MBP1: p < 0.001; IL-5Rα means not different: p = 0.488; anova on ranks).
Fig. 2.
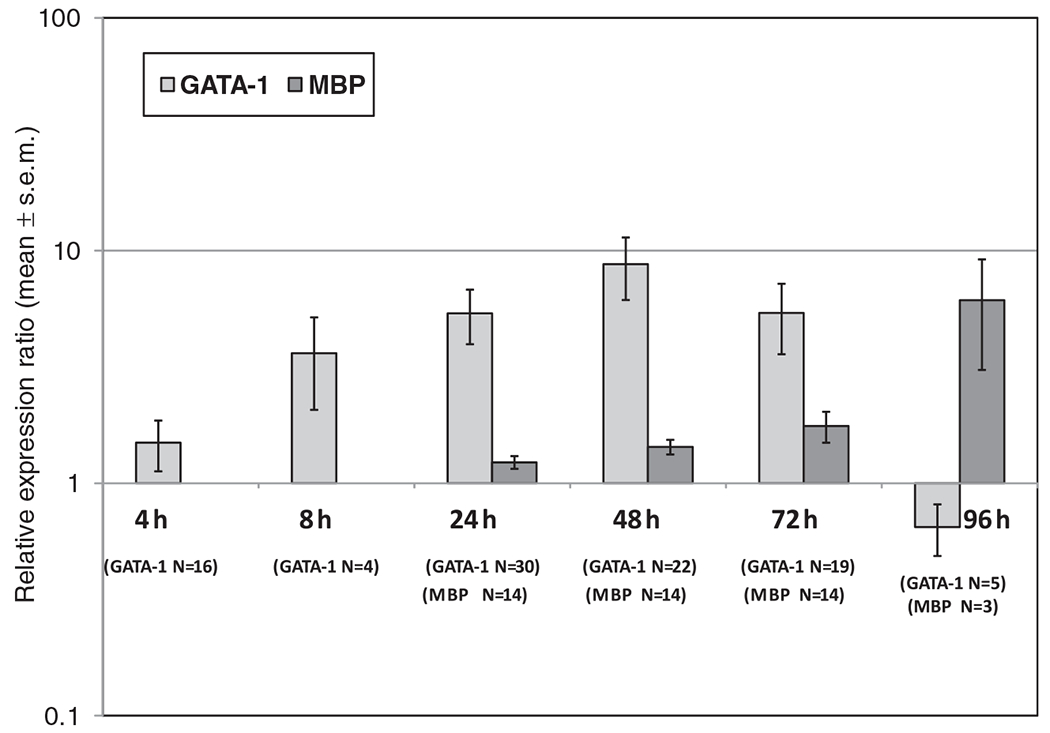
Kinetic mRNA expression of GATA-1 vs. MBP1 from cord blood (CB) non-adherent mononuclear cells (NAMNCs) stimulated with IL-5: presence of GATA-1 precedes MBP gene transcription (p < 0.001, ANOVA on ranks). NAMNCs from random fresh and frozen CB samples were incubated in the presence of rhIL-5, and removed at various time points for cell lysis, RNA extraction, reverse-transcription and quantitative polymerase chain reaction (Q-PCR) analysis using a multiplex reaction. Displayed are relative expression ratios (normalized to housekeeping gene, GAPDH), calculated using the standard 2(−ΔΔCt) formula.
There was an inverse correlation between early MBP1 expression and total number of Eo/B CFUs in the CB samples. This relationship existed at both the 24-h and 48-h time-points (r = −0.602, p = 0.046; and r = −0.660, p = 0.0275, respectively).
Although the expression of total IL-5Rα transcripts appeared comparatively stable over all time-points studied (Fig. 1), with no significant differences in the means (p = 0.488), there was differential (reciprocal) expression of the TM- and sol-IL-5Rα isoforms over time (Fig. 3). Specifically, we observed an initial up-regulation followed by progressive decrease in the proportion of the transmembrane (functional) isoform compared to the soluble (antagonistic) isoform.
Fig. 3.
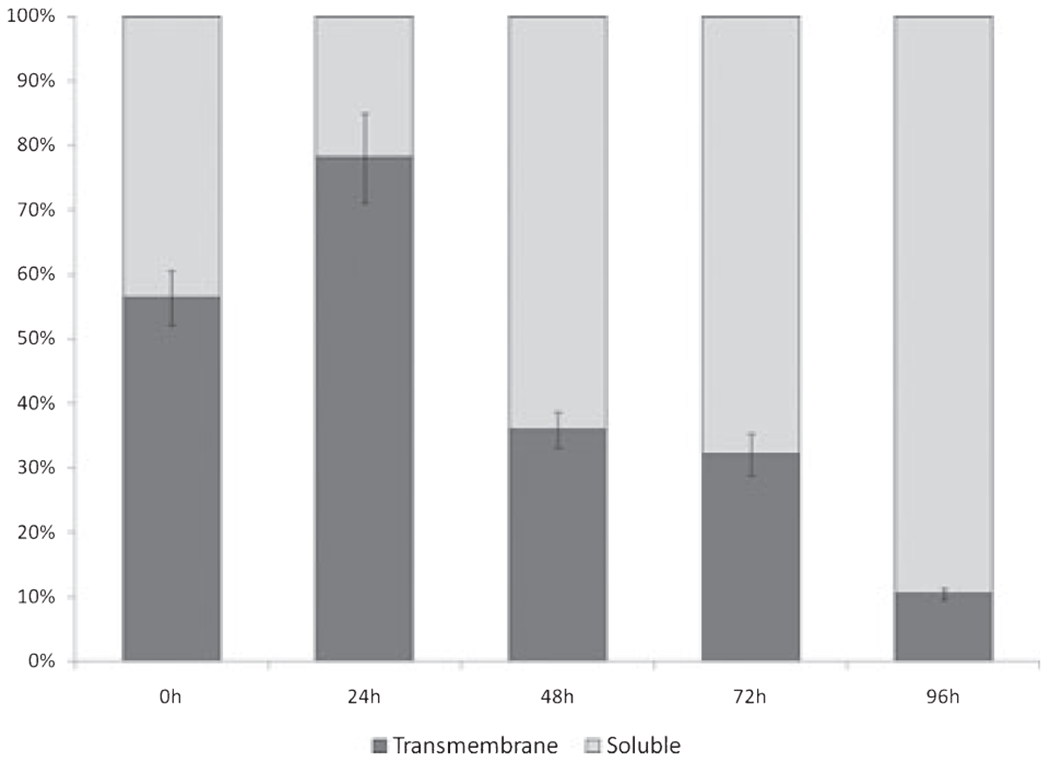
Kinetic changes in the mRNA expression of the Tm- & Sol-IL-5Rα isoforms in cord blood (CB) non-adherent mononuclear cells (NAMNCs) incubated with IL-5 (shown as percent of total IL-5Rα). NAMNCs from random fresh and frozen CB samples were incubated in the presence of rhIL-5, and removed at various time points for cell lysis, RNA extraction, reverse-transcription and determination of Tm-IL-5Rα and Sol-IL-5Rα mRNA expression patterns using SYBR green based quantitative polymerase chain reaction (Q-PCR) analysis.
Cord blood CD34+ progenitor cell studies
Subjecting the CB samples to CD34+ cell purification resulted in a significant decrease in the number of cells available for culture (average number per time-point = 9.3 × 104). As a result, the total RNA available for analysis was reduced almost 100-fold for these studies. Despite this, we indeed found that stimulation of a purified fraction of CD34+ cells resulted in a pattern of GATA-1, IL-5Rα and MBP1 expression that mirrored that seen in the non-adherent fraction (Fig. 4). These expression patterns in CD34+ cells were highly correlated to the patterns seen with the NAMNC population (r = 0.963, p = 0.035 for GATA-1; r = 0.988, p = 0.012 for MBP1). The expression of the IL-5Rα subunit was again comparatively unchanged over time when total IL-5Rα transcripts (Sol + Tm) were assessed.
Fig. 4.
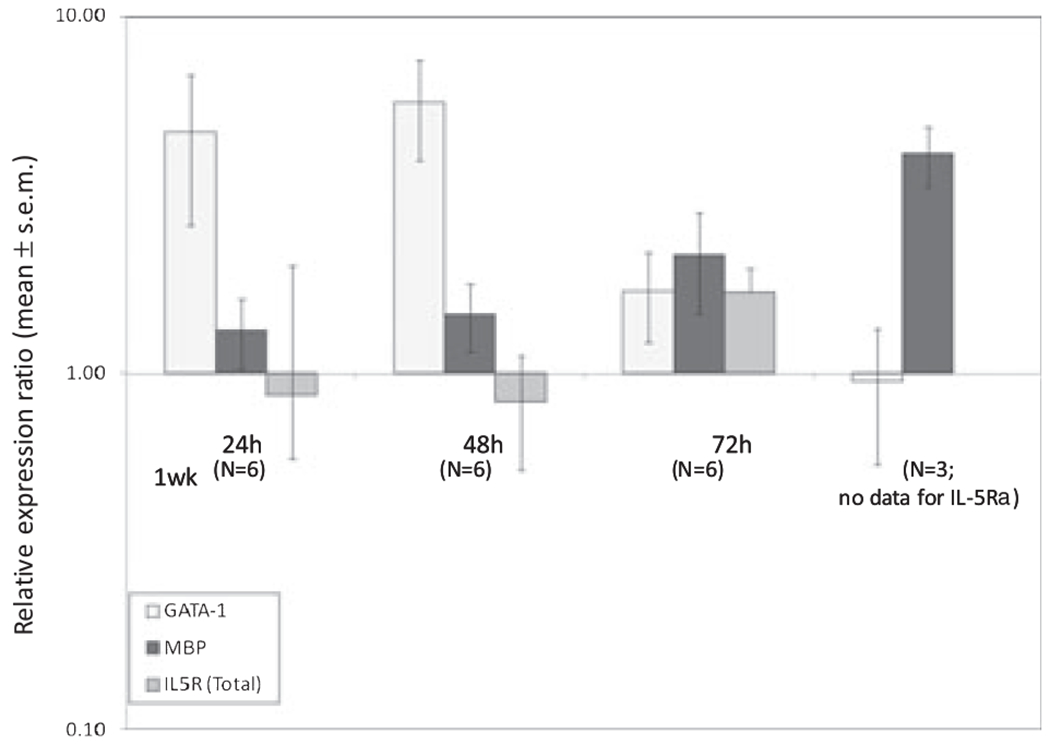
Kinetic mRNA expression patterns of GATA-1, MBP1 and IL-5Rα (Total) from cord blood (CB) CD34+ purified cell populations in response to stimulation with IL-5. Mononuclear cells from random fresh and frozen CB samples underwent magnet-activated cell separation using a CD34+ selection kit. CB CD34+ cells were incubated in the presence of rhIL-5, and removed at various time points for cell lysis, RNA extraction, reverse-transcription and determination of GATA-1, MBP1 and IL-5Rα (Total) mRNA expression using a multiplex quantitative polymerase chain reaction (Q-PCR) reaction. Displayed are relative expression ratios (normalized to housekeeping gene, GAPDH), calculated using the standard 2(−ΔΔCt) formula (GATA-1: p = 0.013; MBP1: p = 0.033; IL5Ra (total) means not different: p = 0.68; anova on ranks).
As seen in the CB NAMNC samples, the mRNA for the two isoforms of the IL-5Rα was differentially expressed in the cultured CD34 + cells over time, such that we observed an initial increase in the percentage of the expression of the transmembrane isoform, which was quickly replaced by an up-regulation in the percentage of the soluble IL-5Rα splice variant (Fig. 5).
Fig. 5.
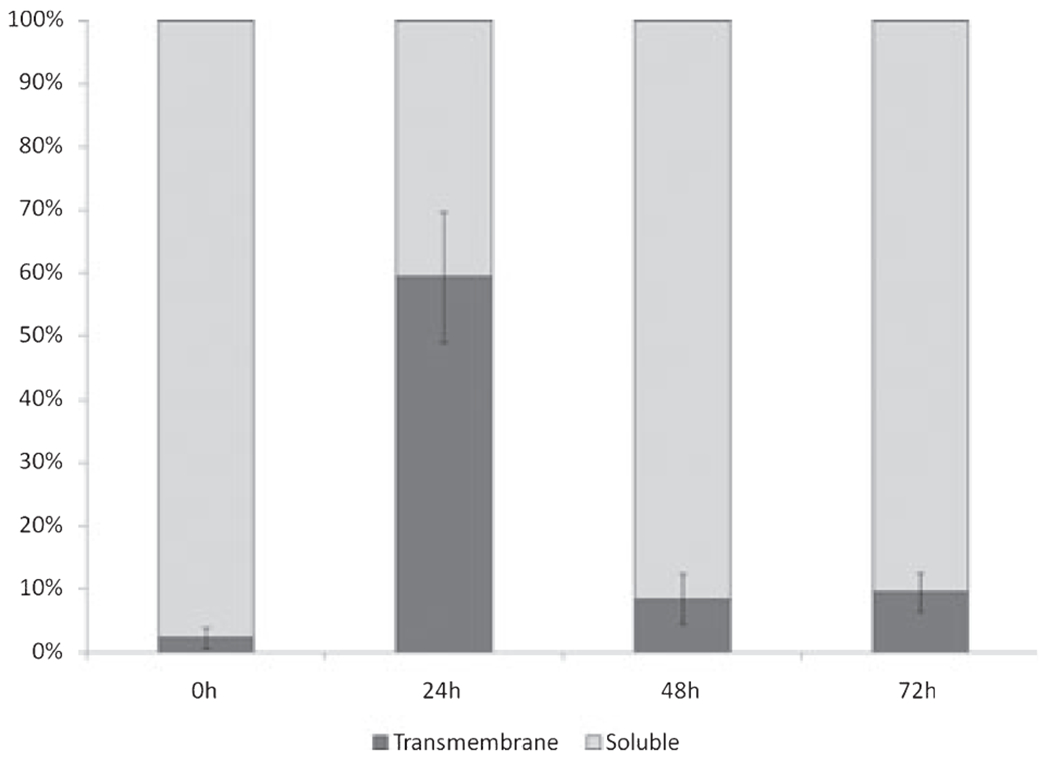
Kinetic changes in the mRNA expression of the Tm- & Sol-IL-5Rα isoforms in cord blood (CB) CD34+ cells incubated with IL-5 (shown as percent of total IL-5Rα). Non-adherent mononuclear cells (NAMNCs) from random fresh and frozen CB samples underwent magnet-activated cell separation using a CD34+ selection kit. CB CD34+ cells were incubated in the presence of rhIL-5, and removed at various time points for cell lysis, RNA extraction, reverse-transcription and determination of Tm-IL-5Rα and Sol-IL-5Rα mRNA expression patterns using SYBR green based quantitative polymerase chain reaction (Q-PCR) analysis.
Discussion
In this study, we were able to successfully measure kinetic patterns of expression of three key mRNA species: GATA-1, IL-5Rα and MBP1, as biomarkers of eosinophil lineage commitment and maturation, in a multiplex Q-PCR in both CB non-adherent cells and a CD34+ purified progenitor fraction.
GATA-1 is not only necessary and sufficient for eosinophilopoiesis (20), it has been clearly established as a pivotal transcription factor for eosinophil lineage specification, as revealed by the loss of the eosinophil lineage in mice harboring a targeted transgenic deletion of the high-affinity GATA-binding site in the HS-2 region of the murine GATA-1 promoter (21) and in the GATA-1 knockout mouse (12). GATA-1 is critical in the early stage of eosinophil differentiation (12), and furthermore, others have shown that GATA-1 can transactivate the expression of the eosinophil-specific MBP1 P2 promoter (22, 23).
In this study, we saw an appropriate up-regulation of GATA-1 mRNA expression upon stimulation of the non-adherent mononuclear cell population with IL-5, suggesting activation of cellular events leading to eosinophil development. Our results suggest that IL-5 is responsible for driving the maturation of the eosinophil lineage, with early peaking of GATA-1 followed by later up-regulation of MBP1, a secondary granule protein product produced almost exclusively by eosinophils. Importantly, while undergoing maturation, eosinophil progenitors will express detectable mRNA levels of MBP1 (24), but mature eosinophils do not express any mRNA for MBP1 or other granule cationic proteins (25). Additionally, mature blood eosinophils no longer express GATA-1 protein, so its expression is also down-regulated as eosinophils complete their terminal differentiation into mature cells. Our results support the observations of others that GATA-1 is a potent transcriptional activator of MBP1 gene expression (22, 23), as the up-regulation of MBP1 followed the initial expression of GATA-1, and was closely associated with peak GATA-1 expression (Fig. 2).
Full in-vitro differentiation of eosinophils from CB CD34+ cells takes around 15–17 days, so the fact that the increase in MBP1 mRNA expression by 72 h was only two-fold is not particularly surprising. The pilot data extending to 1 wk of incubation showed a mean up-regulation of ~four-fold, suggesting that a plateau may not yet be reached. Presumably then, the silencing of MBP1 mRNA does not occur until the end stages of terminal differentiation, as it is known that once fully matured, eosinophils no longer express mRNA for MBP1 (25). The parameters of this evaluation do not allow us to comment on when precisely this plateau in kinetic expression occurs.
IL-5 is unquestionably an important factor in the functional maturation of eosinophils in terms of priming and activation, and their tissue survival (27, 28). Cellular responses to IL-5 are mediated through the high affinity IL-5R, a heterodimer consisting of a ligand-binding 60 kDa IL-5-specific κ subunit and a 130 kDa signal-transducing β subunit (also referred to as the common β chain, or βc), which is identical to the signal-transducing subunit of the IL-3R and GM-CSFR (29).
The IL-5Rα subunit is mainly restricted to mature eosinophils and basophils (30), but this IL-5-binding subunit of the receptor has also been shown to be present on late-lineage-committed CD34–CD33+ cells and has also recently been demonstrated to be on the surface of some CD34+ cells, suggesting its presence to be a marker of an early hematopoietic event that can indicate commitment to the eosinophil-basophil lineages (31). Human eosinophil progenitors (EoP) were recently re-defined as IL-5Rα+ CD34+CD38+IL-3Ra+CD45RA− constituents of the common myeloid progenitor (CMP) pool (32). The full-length IL-5Rα gene can produce two mRNA species that correspond to alternatively spliced soluble and transmembrane isoforms (sol-IL-5Rα, Tm-IL-5Rα) (15–18). The Tm-IL-5Rα is the functional isoform that allows for signal transduction and initiation of responses mediated via the IL-5Rα and βc chain. The sol-IL-5Rα subunit binds IL-5 and neutralizes the biologic activity of the cytokine in vitro (15–17), suggesting that the sol-IL-5Rα subunit may play a role in the immunoregulation of eosinophilia in vivo. It has been proposed that eosinophils are able to control their responsiveness to IL-5 by regulated expression of the IL-5Rα isoforms (17). In the current evaluation, the mRNA expression of total IL-5Rα transcripts in CB NAMNCs and CD34+ populations remained apparently stable after stimulation with rhIL-5, but subsequent analysis demonstrated reciprocal contributory roles of the soluble and transmembrane variants of the IL-5Rα subunit, with progressive down-regulation of the Tm-IL-5Rα isoform concomitant with progressive up-regulation of the soluble isoform. This is in keeping with the work from others who have shown a similar switch from the predominantly soluble isoform to Tm-IL-5Rα in early IL-5-driven eosinophil development from human umbilical cord CD34+ cells (34), but predominant expression of the sol-IL-5Rα isoform in mature eosinophils (35). We would expect these findings to be specific to IL-5 stimulation (as opposed to IL-3 or GM-CSF stimulation), as signaling through the common beta-chain shared by the IL-3R, IL-5R and GM-CSFR is still dependent upon specific activation of the relevant alpha subunits (29).
The Eo/B CFU correlations in this study identified an inverse relationship between early MBP1 up-regulation and the number of Eo/B CFUs in the CB samples. This has biologic plausibility, as earlier peaking of MBP1 expression would indicate a population of a more further-differentiated eosinophil progenitor population in the sample, as opposed to one with a higher number of undifferentiated CD34+ cells (indicated by a larger number of Eo/B CFUs in the methylcellulose colony assay), which would need to follow the stimulation → GATA-1 → MBP cycle before showing an increase in MBP mRNA expression.
The results obtained in the experiments utilizing a purified CD34+ fraction mirrored those found in the less purified NAMNC population, indicating that these studies can be pursued using the NAMNC cell population as a surrogate for the responses of purified CD34+ progenitors. This results in a less complicated, less laborintensive assay that could more easily be adopted by other laboratories.
We are currently applying these techniques to CB samples from infants with increased risk of the development of atopy to determine whether differences in either the kinetic patterns or the maximal expression of these biomarkers can be applied as predictors of atopy at birth.
Relevance of this study
Based on our newly enhanced knowledge of, and assays for, these hematopoietic biomarkers, it may be possible to elucidate molecular mechanisms and patterns that are associated with atopic risk and potentially influence the subsequent development of atopy and/or asthma.
Supplementary Material
Figure S1. Standard 1.5% agarose DNA gel electrophoresis of all amplicons studied (IL-5R total, IL-5RTm, IL-5Rsol, MBP1, GATA-1).
Acknowledgments
This study was funded by grants from the Canadian Institute for Health Research and AllerGen NCE Inc to Dr Denburg. Dr Ellis was funded by an AllerGen/Bayer/CA-AIF Clinician-Scientist Research Fellowship Award and a Hamilton Health Sciences Career Award. Dr Ackerman was funded by a grant from the National Institutes of Health (R01AI33043). Richa Bedi was supported in part by an Institutional NIH pre-doctoral training grant from the NIH/NIDDK ((Training Program in Signal Transduction and Cellular Endocrinology, T32 DK007739 to SJA.
Abbreviations:
- C/W
compared with
- CB
cord blood
- CD34
cluster of differentiation marker 34
- CFU
colony forming unit
- Eo/B
eosinophil/basophil
- GM-CSFRα
granulocyte-macrophage colony stimulating factor receptor α-subunit
- IL-5Rα
interleukin-5 receptor α-subunit
- MBP1
major basic protein-1
- MNCs
mononuclear cells
- NAMNCs
non-adherent mononuclear cells
- PCR
polymerase chain reaction
- Q-PCR
quantitative polymerase chain reaction
- RER
relative expression ratio
- rhIL-5
recombinant human interleukin-5
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Nathan RA. The burden of allergic rhinitis. Allergy Asthma Proc 2007: 28: 3–9. [DOI] [PubMed] [Google Scholar]
- 2.Bousquet J Global Surveillance, Prevention and Control of Chronic Respiratory Disease: A Comprehensive Approach. Bousquet J and Khaltaev N Geneva: World Health Organization, 2007. [Google Scholar]
- 3.Hahn EL, Bacharier LB. The atopic march: the pattern of allergic disease development in childhood. Immunol Allergy Clin North Am 2005: 25: 231–46, v. [DOI] [PubMed] [Google Scholar]
- 4.Cyr MM, Hatfield H, Dunstan JA, Prescott SL, Holt PG, Denburg JA. Relationship of maternal skin test responses to infant cord-blood progenitor cytokine receptor expression. J Allergy Clin Immunol 2004: 113: S162. [Google Scholar]
- 5.Denburg JA, Hatfield HM, Cyr MM, et al. Fish oil supplementation in pregnancy modifies neonatal progenitors at birth in infants at risk of atopy. Pediatr Res 2005: 57: 276–81. [DOI] [PubMed] [Google Scholar]
- 6.Upham JW, Hayes LM, Lundahl J, Sehmi R, Denburg JA. Reduced expression of hemopoietic cytokine receptors on cord blood progenitor cells in neonates at risk for atopy. J Allergy Clin Immunol 1999: 104: 370–5. [DOI] [PubMed] [Google Scholar]
- 7.Cyr MM, Baatjes AJ, Dorman SC, et al. In vitro effects of budesonide on eosinophil-basophil lineage commitment. Open Respir Med J 2008: 2: 60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denburg JA, Fernandes R, Ellis AK, et al. Eosinophil progenitors at birth: intimations of future atopy and inflammation. Allergy Clin Immunol Int 2007: 2: 29–31. [Google Scholar]
- 9.Fernandes R, Kusel M, Cyr M, et al. Cord blood hemopoietic progenitor profiles predict acute respiratory symptoms in infancy. Pediatr Allergy Immunol 2008: 19: 239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirasawa R, Shimizu R, Takahashi S, et al. Essential and instructive roles of GATA factors in eosinophil development. J Exp Med 2002: 195: 1379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, Yergeau DA, Wong IC, et al. Interleukin-5 receptor alpha subunit gene regulation in human eosinophil development: identification of a unique cis-element that acts like an enhancer in regulating activity of the IL-5R alpha promoter. Curr Top Microbiol Immunol 1996: 211: 173–87. [DOI] [PubMed] [Google Scholar]
- 12.Tavernier J, Devos R, Cornelis S, et al. A human high affinity interleukin-5 receptor (IL5R) is composed of an IL5-specific α chain and a β chain shared with the receptor for GM-CSF. Cell 1991: 66: 1175–84. [DOI] [PubMed] [Google Scholar]
- 13.Murata Y, Takaki S, Migita M, Kikuchi Y, Tominaga A, Takatsu K. Molecular cloning and expression of the human interleukin 5 receptor. J Exp Med 1992: 175: 341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown PM, Tagari P, Rowan KR, et al. Epitope-labeled soluble human interleukin-5 (IL-5) receptors. J Biol Chem 1995: 270: 29236–43. [DOI] [PubMed] [Google Scholar]
- 15.Tavernier J, Tuypens T, Plaetinck G, Verhee A, Fiers W, Devos R. Molecular basis of the membrane-anchored and two soluble isoforms of the human interleukin 5 receptor α subunit. Proc Natl Acad Sci USA 1992: 89: 7041–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedi R, Du J, Sharma AK, Gomes I, Ackerman SJ. Human C/EBP-epsilon activator and repressor isoforms differentially reprogram myeloid lineage commitment and differentiation. Blood 2009: 113: 317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNagny K, Graf T. Making eosinophils through subtle shifts in transcription factor expression. J Exp Med 2002: 195: F43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C, Cantor AB, Yang H, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med 2002: 195: 1387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi Y, Ackerman SJ, Minegishi N, Takiguchi M, Yamamoto M, Suda T. Mechanisms of transcription in eosinophils: GATA-1, but not GATA-2, transactivates the promoter of the eosinophil granule major basic protein gene. Blood 1998: 91: 3447–58. [PubMed] [Google Scholar]
- 20.Du J, Stankiewicz MJ, Liu Y, et al. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem 2002: 277: 43481–94. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki H, Mizuno S, Mayfield R, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med 2005: 201: 1891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruart V, Truong MJ, Plumas J, et al. Decreased expression of eosinophil peroxidase and major basic protein messenger RNAs during eosinophil maturation. Blood 1992: 79: 2592–7. [PubMed] [Google Scholar]
- 23.Gleich GJ. Mechanisms of eosinophil-associated inflammation. J Allergy Clin Immunol 2000: 105: 651. [DOI] [PubMed] [Google Scholar]
- 24.Liu LY, Sedgwick JB, Bates ME, et al. Decreased expression of membrane IL-5 receptor alpha on human eosinophils: II. IL-5 down-modulates its receptor via a proteinase-mediated process. J Immunol 2002: 169: 6459–66. [DOI] [PubMed] [Google Scholar]
- 25.Geijsen N, Koenderman L, Coffer PJ. Specificity in cytokine signal transduction: lessons learned from the IL-3/IL-5/GM-CSF receptor family. Cytokine Growth Factor Rev 2001: 12: 19–25. [DOI] [PubMed] [Google Scholar]
- 26.Geijsen N, Uings IJ, Pals C, et al. Cytokine-specific transcriptional regulation through an IL-5Ralpha interacting protein. Science 2001: 293: 1136–8. [DOI] [PubMed] [Google Scholar]
- 27.Upham JW, Sehmi R, Hayes LM, Howie K, Lundahl J, Denburg JA. Retinoic acid modulates IL-5 receptor expression and selectively inhibits eosinophil-basophil differentiation of hemopoietic progenitor cells. J Allergy Clin Immunol 2002: 109: 307–13. [DOI] [PubMed] [Google Scholar]
- 28.Mori Y, Iwasaki H, Kohno K, et al. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med 2009: 206: 183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavernier J, Van der Heyden J, Verhee A, et al. Interleukin 5 regulates the isoform expression of its own receptor alpha-subunit. Blood 2000: 95: 1600–7. [PubMed] [Google Scholar]
- 30.Gevaert P, Bachert C, Holtappels G, et al. Enhanced soluble interleukin-5 receptor alpha expression in nasal polyposis. Allergy 2003: 58: 371–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Standard 1.5% agarose DNA gel electrophoresis of all amplicons studied (IL-5R total, IL-5RTm, IL-5Rsol, MBP1, GATA-1).


