Abstract
Combining the specificity of tumor-targeting bacteria with the sensitivity of biomarker detection would create a screening method able to detect small tumors and metastases. To create this system, we genetically modified an attenuated strain of Salmonella enterica to release a recombinant fluorescent biomarker (or fluoromarker). Salmonella expressing ZsGreen were intravenously administered to tumor-bearing mice and fluoromarker production was induced after 48 hr. The quantities and locations of bacteria and ZsGreen were measured in tumors, livers and spleens by immunofluorescence, and the plasma concentration of ZsGreen was measured using single-layer ELISA. In the plasma, the ZsGreen concentration was in the range of 0.5–1.5 ng/ml and was dependent on tumor mass (with a proportion of 0.81 ± 0.32 ng·ml−1g·−1). No adverse reaction to ZsGreen or bacteria was observed in any mice. ZsGreen was released at an average rate of 4.3 fg·CFU−1·hr−1 and cleared from the plasma with a rate constant of 0.259 hr−1. ZsGreen production was highest in viable tissue (7.6 fg·CFU−1·hr−1) and lowest in necrotic tissue (0.47 fg·CFU−1·hr−1). The mass transfer rate constant from tumor to blood was 0.0125 hr−1. Based on these measurements, this system has the capability to detect tumors as small as 0.12 g. These results demonstrate four essential mechanisms of this method: (i) preferential tumor colonization by bacteria, (ii) fluoromarker release in vivo, (iii) fluoromarker transport through tumor tissue and (iv) slow enough systemic clearance to enable measurement. This bacteria-based blood test would be minimally invasive and has the potential to identify previously undetectable microscopic tumors.
Keywords: tumor detection, intratumoral diffusion, bacterial fluoromarker release
Introduction
Detecting cancer when tumor burden is low would improve treatment outcomes and increase a patient’s chance of remission.1 A detection method that combines the benefits of bacterial tumor-targeting and biomarker detection (Fig. 1) would complement current screening techniques because it would be both specific and sensitive. Facultative anaerobes, such as Salmonella enterica, specifically replicate in tumors and metastases of many different types of cancers, including bone, brain, lung, colon, mammary and ovarian2–11 at densities thousands of times greater than healthy organs.7,12–14 Biomarker detection is sensitive because proteins in the blood can be detected at low concentrations (picomolar and below).15 Sensitivity would be limited only by the detection limit of the molecular maker. A screening method that integrates these capabilities has the potential to detect small malignant lesions, identify recurrence sooner and improve patient survival.
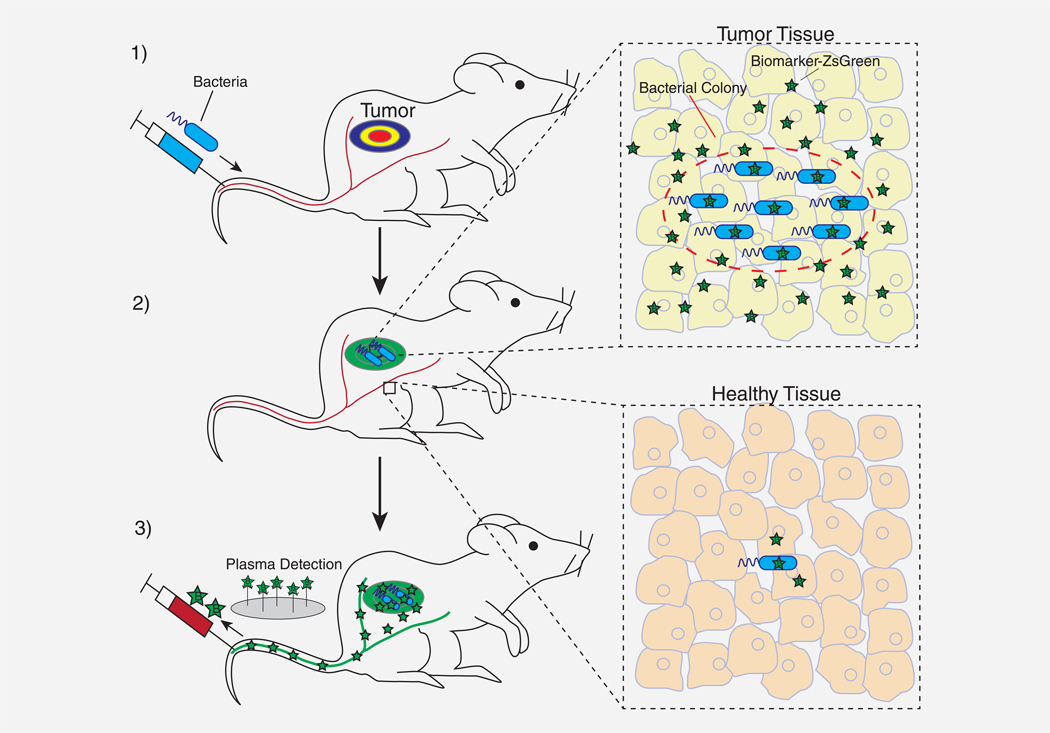
Figure 1.
Tumor detection with fluoromarker-releasing bacteria. Tumor identification would occur via three steps. (1) After intravenous injection, detecting bacteria would disperse throughout the circulatory system. (2) Bacteria would preferentially accumulate in tumors over healthy tissue; replicate and increase density; and express a fluoromarker (stars) that diffuses through the tumor interstitium. (3) The fluoromarker would accumulate in the plasma for immunological detection.
In the first step of this strategy, specifically engineered bacteria would be injected into the blood (Fig. 1; Step 1). The bacteria would proliferate specifically in tumor tissue, where they would express and release a recombinant exogenous fluorescent marker, or fluoromarker (Step 2). The released fluoromarker would diffuse through the tumor interstitium to the bloodstream, where it would be measured (Step 3). The concentration of the fluoromarker in the plasma would indicate the presence of cancer.
The specificity of facultative anaerobes to cancer has been attributed to several mechanisms. These include entrapment in tumor vasculature;14 increased influx of bacteria into the tumor site after systemic injection due to inflammation and tumor necrosis factor (TNF)-α response;16,17 protection from clearance by the immune system;18 chemotaxis into tumor sites;19,20 and preferential growth in the nutrient-rich tumor microenvironment.13,19,21 In our study, we utilized a nonpathogenic strain of Salmonella (VNP20009) that is 10,000-fold less toxic than its wild-type counterpart because of a partial deletion of the msbB gene.6,22 This mutation reduces the TNF immune response to bacterial lipopolysaccharides and prevents septic shock.2,23 This mutation strategy is similar to other groups that have shown that blocking autophagy and modifying the structure of lipopolysaccharide increases the safety, tumor tropism, and efficacy of bacteria for cancer therapy.4,8,13,24,25 In preclinical and clinical trials, this nonpathogenic strain of Salmonella had few side effects in mice, dogs and humans.26–28
A bacteria-based detection technique would complement established detection methods. After initial treatment of a localized tumor (in the breast, for example), routine screening is performed to detect local recurrence or metastatic spread.29 When clinical symptoms appear, imaging techniques and blood tests are used to diagnose recurrence.30 High concentrations of biomarkers, e.g., cancer antigen (CA)-15–3, CA 27–29 or carcinoembryonic antigen (CEA) in the blood are indicators of the existence of malignant tissue.31 Tomographic imaging techniques (e.g., magnetic resonance imaging [MRI] and positron emission tomography [PET]) are used to locate and stage recurrent or metastatic tumors. A bacterial/fluoromarker system would complement these techniques because it (i) would not require specialized equipment; (ii) be based on an exogenous marker that may be less likely to produce false-positive results than endogenous biomarkers15,32,33; and (iii) could potentially detect tumors beneath the detection limit of tomographic techniques.34–36
To date, several strategies for detecting cancer with engineered bacteria have been developed. Escherichia coli have been used to enhance imaging techniques (MRI and PET) using genetic engineering strategies to increased signal magnitude.37,38 Bioluminescent imaging strategies have been developed to identify tumor colonization using Escherichia coli, S. enterica, Vibrio cholera and Listeria monocytogenes.39–43 A recent technique utilized oral delivery of probiotic E. coli to produce an enzyme in liver metastases that cleaves a precursor signal.44 Detection of the cleaved signal in the urine indicates the presence of liver metastases. A bacterial/fluoromarker system (Fig. 1) would complement these strategies because it would enable detection of tumors and metastases that are accessible to intravenously injected Salmonella; it would not be dependent on the resolution of the tomographic techniques; and it would enable detection of lesions farther from the skin than the penetration depth of light.
To create tumor-detecting bacterial system, Salmonella were engineered to express and release ZsGreen, a fluorescent marker protein. We hypothesized that, when colonized in subcutaneous tumors, Salmonella release ZsGreen at rates sufficient to be detected in the plasma. To test this hypothesis, fluoromarker-releasing Salmonella were administered to tumor-bearing mice. The effectiveness of this strategy was tested by quantifying four mechanisms that are essential for it to be effective: (i) the extent of bacterial colonization, (ii) the rate of protein release, (iii) the transport rate through tumor interstitium and (iv) the systemic clearance rate. The plasma concentration of ZsGreen was quantified using single-layer enzyme-linked immunosorbent assay (ELISA). The locations of bacterial colonization and produced ZsGreen were measured by immunofluorescence. The clearance rate of ZsGreen was measured by injecting purified recombinant ZsGreen into mice. A mathematical model of protein production and diffusion was used to interpret immunofluorescence images of ZsGreen in tumors and quantify (i) the rate of protein release in vivo, (ii) the transfer rate from tumors into the blood and (iii) the locations of greatest production. Fluoromarker-producing bacteria have the potential to improve the detection of cancer and enable more effective treatment of cancer.
Methods
Bacterial strains, plasmids and purified ZsGreen
Attenuated S. enterica serovar Typhimurium (strain VNP20009, ΔmsbB, ΔpurI, Δxyl)23 was transformed with pDF02, which contains the ZsGreen gene under control of the pBAD promoter.45 These bacteria release ZsGreen after induction with L-arabinose.1 To produce purified ZsGreen, a 6x-histidine tag was fused to the c-terminus of ZsGreen to create pDF02–6xHis. E. coli (DH5α) were transformed with pDF02–6xHis, grown in 1 l of LB, centrifuged and lysed. His-tagged ZsGreen was purified with nickel (Ni-NTA) chromatography. Buffer exchange was performed to solubilize purified ZsGreen crystals into an excipient formulation suitable for systemic injection, which was 200 mM glycine, 8 mM L-histidine, 40 mM sodium chloride and 0.8% sucrose in sterile water for injection.
Clearance of ZsGreen
The rate of clearance was measured by intravenously injecting purified ZsGreen into mice and measuring the plasma concentration over time. One hundred nanograms of purified ZsGreen-6x-his was injected into the tail vein of 20, 8- to 10-week-old female BALB/c (an inbred mouse strain) mice. Blood samples were collected at 1, 3, 9 and 24 hr by cardiac puncture with a 21 gauge needle after euthanasia with CO2. Collecting 3 ml syringes were coated with 0.5 M ethylenediaminetetraacetic acid as an anticoagulant and stored at 4°C upon retrieval. Samples were centrifuged to separate plasma from heavy components and the ZsGreen concentration was determined with single-layer ELISA. The clearance rate constant was determined by fitting measured plasma concentrations to a model of single-compartment clearance: C = Ci·e−kt, where k is the clearance rate constant, t is time, C is concentration and Ci is the initial concentration.
Single-layer ELISA
The plasma concentration of ZsGreen was measured with single-layer antibody ELISA dots, as described previously.1 Plasma was incubated overnight in 96-well plates containing circular nitrocellulose membranes coated with anti-ZsGreen antibodies (Clontech, Madison, WI). The fluorescence intensity of each dot was quantified by averaging the intensity over the entire area at 1 sec exposure with an epi-fluorescence microscope (Olympus, Center Valley, PA) and a cooled charge-coupled device camera (Hamamatsu Photonics K. K., Hamamatsu City, Japan). The background fluorescence of antibody dots treated with bovine plasma was subtracted from all measured intensities. Antibody-dot fluorescence measurements were calibrated with a concentation range of purified ZsGreen-6x-his in bovine plasma.
Fluoromarker production by tumor-colonized bacteria
Tumors were formed by subcutaneously injecting 50,000 4T1 mammary carcinoma cells into the flank of 8- to 10-week-old, female BALB/c mice. Implantation occurred 27 and 16 days prior to bacterial injection to create two groups of mice with a range of tumor sizes. After tumor growth, Salmonella with pDF02 was injected intravenously at a density of 10,000 colony forming units (CFU)·g−1 into the two groups of tumor-bearing mice and a group of tumor-free mice. As a precaution, ampicillin (10 mg/kg) was subcutaneously injected on the opposite flank every 8 hr to promote plasmid retention. Previously, we have shown that this bacterial system retains plasmid ZsGreen expression under in vitro culture for over 58 hr without antibiotics.1 At 48 hr after the bacterial injection, L-arabinose was injected every 8 hr (for three doses), to induce ZsGreen expression. At 72 hr after bacterial injection, blood was collected from the mice by cardiac puncture and the tumor, liver, and spleen were collected. The plasma ZsGreen concentration was determined with single-layer ELISA. Tumors were fixed in 10% neutral buffered formalin, embedded in paraffin and cut into 5 μm sections.
Bacterial density was determined by colony plate counting of harvested organs. Harvested organs (tumor, liver and spleen) were weighed, minced, suspended in equal volumes of phosphate buffered saline. Four dilutions of tissue homogenates were applied to LB agar plates with 100 mg/ml ampicillin. Liver and splenic tissue were plated at dilutions of 1, 1:10, 1:100 and 1:1000 and tumors at 1:103, 1:104, 1:105 and 1:106. After overnight incubation at 37°C, plates were imaged and colonies were counted using a Fiji/ImageJ particle analyzer.
Immunofluorescent staining of Salmonella and ZsGreen in tumors
The locations of bacteria and ZsGreen in tumors was determined with immunofluorescence. Paraffin-embedded tumor sections were rehydrated and treated with sodium citrate buffer (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0) for 20 min at 65°C for antigen retrieval. Sections were blocked with protein block (Dako, Santa Clara, CA); stained with two primary antibodies, 1:100 rabbit anti-RCFP polyclonal antibody (Clontech) and 1:10 FITC-conjugated, anti-Salmonella antibody (Abcam, Cambridge, MA); and washed with TBS-T. A secondary antibody, Alexa Fluor 546-donkey-antirabbit IgG (Life Technologies, Carlsbad, CA), was applied at 1:100. 4′,6-Diamidino-2-phenylindole (DAPI) was applied to stain cell nuclei and identify necrosis. Immunofluorescent images were acquired with an inverted epifluorescent microscope (Olympus, Tokyo, Japan) with a 10x Plan-APO fluorescence objective and an automated script written in IPLab (BD Biosciences, Rockville, MD). The script assembled a tiled montage of individual images acquired using three fluorescent filters (Chroma, Bellows Falls, VT): D350/50x (DAPI), D546/10x (ZsGreen) and D455/70x (Salmonella).
Image analysis of ZsGreen localization in tumors
Tissue sections were analyzed for amounts of bacteria and ZsGreen as functions of tumor location based on the immunofluorescent staining. Monochrome images were background subtracted and thresholded to identify bacteria and ZsGreen. Individual binary images were aligned by maximizing the area of overlap through sequential rigid transformation. Necrotic tissue was identified by the size and density of nuclei in DAPI-stained images. The relative amount of tumor necrosis was determined by the ratio of the area of necrotic tissue to the ratio of healthy tissue in cross-sectional images of each tumor. The mass of necrotic tissue was calculated by multiplying this ratio by the tumor mass. The amount of ZsGreen per tumor was calculated based on immunofluorescent staining assuming a minimum detectable concentration of 1.0 μmol/l and a pixel volume of 0.6452 × 0.6452 × 10 μm3.
The transition region was defined as an area within 500 μm of the boundary between viable and necrotic tissue. Location relative to the transition boundary was determined by multiplying binary bacteria and ZsGreen images by Euclidean distance maps and quantifying the number of pixels as a function of distance. Extracellular ZsGreen was identified by subtracting the binary bacteria image from the binary ZsGreen image and applying the distance map. ZsGreen per bacterium was determined by dividing the number of ZsGreen pixels by the number of bacteria pixels, after binning by distance.
Colony-specific ZsGreen production
The production of ZsGreen by bacterial colonies was quantified for whole tumors and as a function of distance from the tumor edge. Individual bacterial colonies were identified by particle analysis of binary bacteria images (Fiji/ImageJ). To determine tumor-average values, six images were randomly selected from 27-day tumors. Images containing tissue edges, folds or over 75% of blank space were excluded. For each image, the colony size distribution and the average ZsGreen diffusion distance were quantified. Binary images of bacterial colonies were sequentially dilated by one pixel at a time. The union of the dilated image and the ZsGreen binary image gave the coverage at that distance from the colony edge. Dilation continued until the union contained no ZsGreen pixels. The fractional coverage was the ratio of ZsGreen pixel area to the theoretical area of the circular annulus.
To determine values as a function of position in tumors, adjacent images were identified from tumor edges to the centers. From colony particle analysis and ZsGreen binary images, the percentage of colonies expressing ZsGreen and the average colony size was measured. Colony diameter was estimated from colony area, assuming colonies were perfect circles. Fractional coverage of ZsGreen around individual colonies was measured by serial dilation.
Calculation of ZsGreen release rates
ZsGreen release rates were calculated using profiles of fractional coverage around tumor-colonized bacteria and a mathematical model of ZsGreen diffusion. ZsGreen production from a colony was described by a spherical one-dimensional diffusion balance with generation from a single point source.
| (1) |
In this balance, the extracellular ZsGreen concentration (C), is a function of time (t) and radial distance (r). At the edge of the colony, , which has volume , ZsGreen is released at rate (moles/time). Initially and at infinite distance, the ZsGreen concentration was zero. Equation 1 has a complementary error function solution, which simplifies at steady state.
| (2) |
In this equation, dimensionless concentration is a function of dimensionless production (M) and dimensionless radius . The minimum detectable ZsGreen concentration () was estimated from average values for paraffin embedded proteins.46 The effective diffusion coefficient in tumor interstitium (D) was estimated47 using the hydrodynamic radius of a globular protein48 with a similar molecular weight as tetrameric ZsGreen (104.2 g/mol). The coverage fraction was assumed to be proportional to dimensionless concentration. Dimensionless colony production (M) was calculated for every colony by fitting the ZsGreen fractional coverage profiles to Eq. 2. Values were averaged over entire tumors and were calculated as a function of tumor position. For each of these, the ZsGreen release rate () was calculated from M using the minimum detectable concentration () and the effective diffusion coefficient (D).
Calculation of mass transfer rates
ZsGreen production by tumor-colonized bacteria was described by a coupled pharmacokinetic, mass-transfer model.
| (3) |
| (4) |
ZsGreen accumulation in tumors, , was the result of production by tumor-colonized bacteria, , and mass transfer to the plasma, . ZsGreen accumulation in the plasma, , was the result of mass transfer from tumors and clearance from the circulation, . In this system, and are the concentrations of ZsGreen in the tumor and the plasma (ng·ml−1); is the tumor volume (ml); is the rate of ZsGreen production per bacterium (fg·CFU−1·hr−1); is the tumor bacterial density (CFUml−1); is the plasma volume (ml); and is the plasma clearance rate constant (hr−1). The mass transfer rate constant , is composed of the mass transfer coefficient for flux across the vascular surface, k (cm·hr−1), and the average surface area per volume of tumor, v (cm2·cm−3). This rate constant was determined from the analytical solution (Eqns. S5 and S7 in Supporting Information) to the model of ZsGreen pharmacokinetics and mass-transfer (Eqns. 3 and 4) using least-squares regression and the measured values of , , and the plasma ZsGreen concentration () as a function of tumor volume (). The plasma volume () was assumed to be 2.0 ml. Tumor weights were converted to volume using a tissue density of 1.0 g·ml−1. The number of bacteria in healthy tissue was determined from the plasma concentration of ZsGreen in tumor-free mice and the production rate of ZsGreen per tissue, which was linear with time (Supporting Information). Integrating factors were used to determine the relationship between bacteria number and plasma concentration (Eq. S12). The minimum detectable tumor volume was the volume that produced a plasma concentration of 0.5 ng/ml at long times. Minimum volumes were determined at 485.3 CFU·mg−1, which is the bacterial density measured in these experiments.
Statistical analysis
Statistical analysis was performed using Student’s t-test, with p < 0.05 considered as significant. The correlation between tumor mass and ZsGreen content was determined by comparing the slope of the linear fit to a slope of zero.
Results
Bacteria in tumors produced detectable ZsGreen
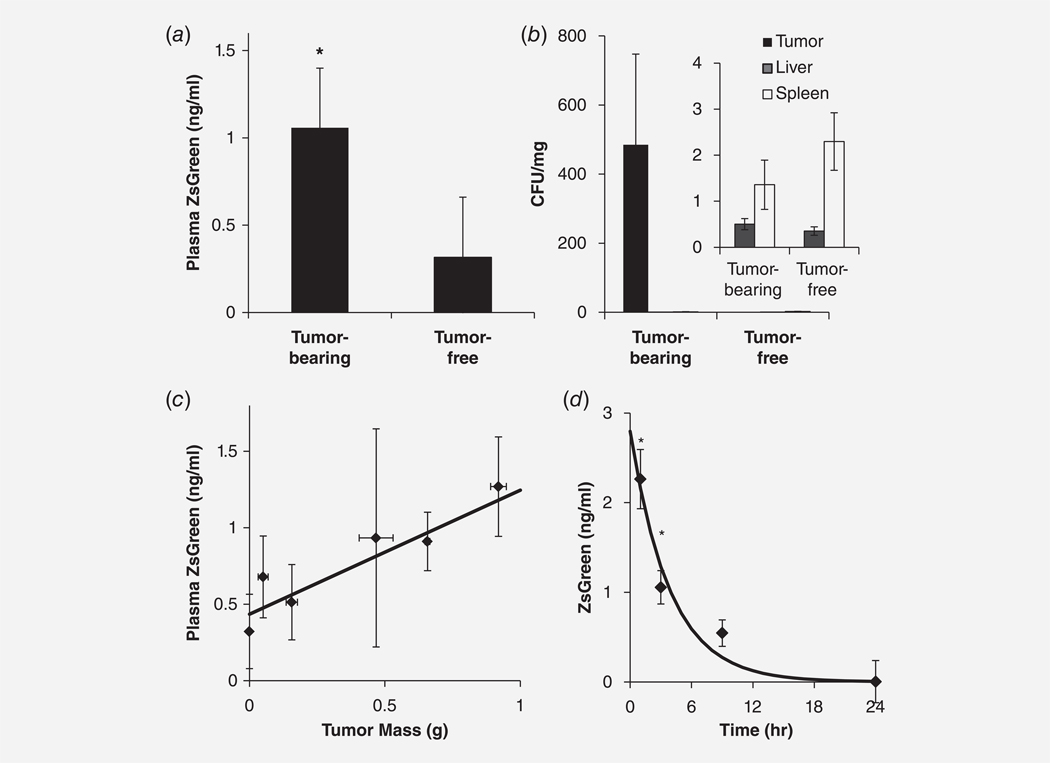
After systemic injection, tumor-colonized bacteria produced detectable levels of ZsGreen that increased with tumor size (Fig. 2). Three days after injection, the plasma concentration of ZsGreen from mice with tumors (n = 8) was significantly (p < 0.05) higher than controls without tumors (n = 9; Fig. 2a). The average bacterial density in tumors was 357 times greater than in healthy tissues (Fig. 2b). The livers and spleens of all mice contained less than 3 CFU/mg bacteria and had an average density of 1.57 ± 0.35 CFU/mg (Fig. 2b). The plasma concentration of ZsGreen increased with tumor mass at a proportion of 0.81 ± 0.32 ng·ml−1·g−1 (Fig. 2c). This increase in mass was significantly greater than zero (p < 0.05). The average tumor was 0.56 ± 0.10 g. The ZsGreen produced in healthy tissue (at mass = 0 g) was 0.43 ± 0.16 ng/ml (Fig. 2c). The clearance rate constant for purified intravenously injected ZsGreen was 0.259 hr−1 in mice, which is equivalent to a half-life of 2.68 ± 0.44 hr (n = 5 per time point, Fig. 2d). Systemic administration of ZsGreen-expressing Salmonella to mice was safe. All mice were healthy, had high activity levels, and had appropriate body weights (14.0–21.0 g) for the duration of the experiment (Fig. S1). After injection, some mice had ruffled fur, but a healthy coat was recovered within 24 hr. Similarly, intravenous injection of ZsGreen did not have any observable adverse effects on the health of mice and no changes in activity level, coat, posture or weight was observed.
Figure 2.
ZsGreen was produced by bacteria in tumors. (a) Mice with tumors were intravenously injected with ZsGreen-releasing bacteria. After 72 hr, the plasma ZsGreen concentration was higher in mice with tumors (n = 8) than tumor-free control mice (n = 9; *, p < 0.05). (b) In tumor-bearing mice, the average bacterial density was 357 times the density in the spleen and 966 times the density in the liver. Inset: The liver and spleen densities were similar in control and tumor-bearing mice. (c) The plasma ZsGreen concentration increased linearly with tumor weight (p < 0.05), with a slope of 0.81 ± 0.32 ng·ml−1g·−1. (d) After intravenous injection of purified ZsGreen, the plasma concentration decreased exponentially with a half-life of 2.68 ± 0.44 hr. The concentrations at 1 and 3 hr were significantly greater than after 9 hr in circulation (*p < 0.05).
Bacteria and ZsGreen were primarily in located the transition region
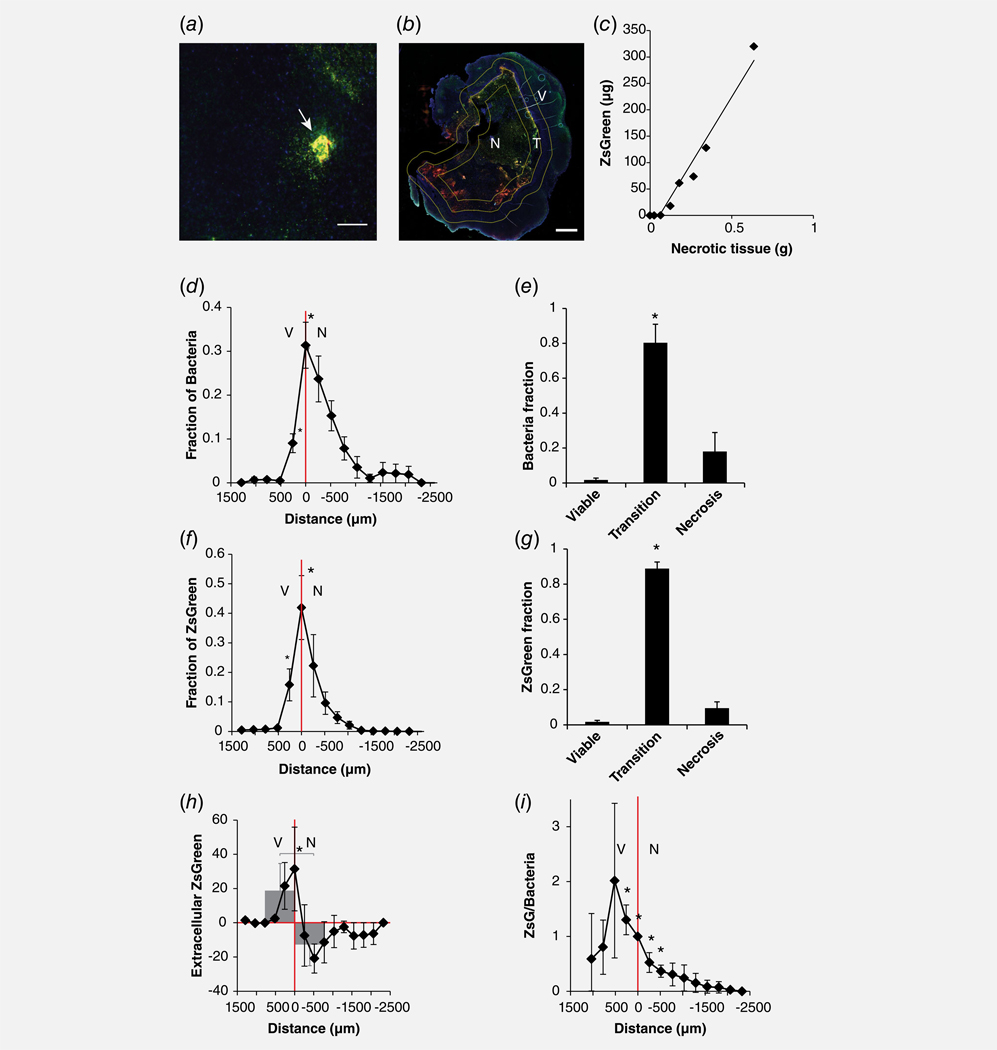
In tumors, bacterial colonies (red, merged to yellow) were surrounded by released ZsGreen (green; Fig. 3a). The amount of ZsGreen was dependent on location. To quantify the distribution, tumors were divided into three regions (Fig. 3b) based on nuclear morphology: viable (V), necrotic (N) and transition (T). The transition region was defined as an area 500 μm thick on either side of the boundary between viable and necrotic tissue (Fig. 3b). Tumors with more necrosis had more intratumoral ZsGreen (p < 0.001; Fig. 3c). The amount of ZsGreen increased 320 ± 25 (μg) per gram of necrotic tissue. This correlation was predominantly caused by preferential colonization of bacteria in the transition region. The greatest number of bacteria and the most ZsGreen were found at the viable/necrotic boundary (p < 0.05) and decreased with distance into viable and the necrotic tissue (Figs. 3d–3g). Little ZsGreen or bacteria were detected more than 1,500 μm from this boundary. Across all tumors, over 80% of bacteria and ZsGreen molecules were found within transition regions (p < 0.01; n = 5; Figs. 3e and 3g).
Figure 3.
Localization of ZsGreen in tumors. (a) In stained tumor sections, bacterial colonies (red merged to yellow; white arrow) were surrounded by ZsGreen (green). Sections were counterstained with DAPI (blue). Scale bar is 100 μm. (b) Tumors were divided into three regions based on nuclear morphology: viable (V), necrotic (N) and transition (T). The transition region was defined as 1,000 μm thick band around the boundary (yellow line) between viable and necrotic tissue. Scale bar is 1 mm. (c) Tumors with a greater mass of necrosis contained more ZsGreen (p < 0.05). (d–g) Bacteria (d and e) and ZsGreen (f and g) were primarily located at the boundary between viable and necrotic tissue. The fractions at the boundary and 250 μm into viable tissue were significantly greater (n = 5; *p < 0.05) than deep in necrosis (>1,500 μm from the boundary). The majority of bacteria (e) and ZsGreen (g) were in the transition region (*p < 0.05). (h) The amount of extracellular ZsGreen (minus bacteria) was greatest at the boundary between viable and necrotic tissue. Negative values indicate that not all bacteria produce ZsGreen in necrotic tissue. Overall, more extracellular ZsGreen was present in viable compared to necrotic tissue (gray bar graph; *p < 0.05). (i) The ratio of ZsGreen per bacteria decreased with distance from the transition boundary into necrosis. At distances 250 to −500, the ZsGreen produced per bacterium was significantly greater (*p < 0.05) than deep in necrosis (>1500 μm from the boundary).
The amount of ZsGreen produced per bacterium changed with location in the tumor. Viable regions contained more extracellular ZsGreen than necrotic regions (Fig. 3h), indicated by a positive difference between the areas of tissue stained for ZsGreen and bacteria (p < 0.05). The amount of extracellular ZsGreen was greatest immediately adjoining the transition boundary. The negative difference in stained areas indicates that not all bacteria in necrosis produced ZsGreen. The amount of ZsGreen expressed per bacterium was greatest in viable regions (p < 0.05), and had a maximum 500 μm into viable tissue from the transition boundary (Fig. 3i). The amount of ZsGreen per bacterium decreased with distance into necrosis (p < 0.05; Fig. 3i).
ZsGreen diffused into the tumor interstitium from bacterial colonies
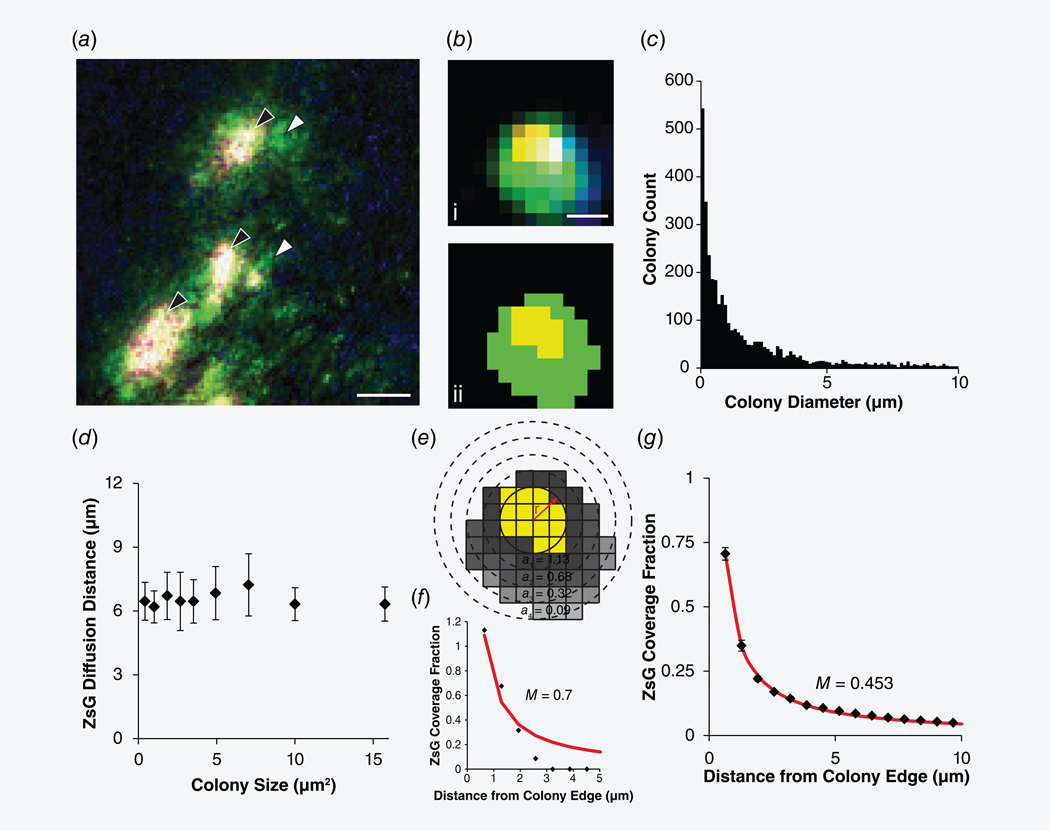
Bacteria expressed and released ZsGreen, which diffused into surrounding tumor tissue (Fig. 4). In tumors, bacteria accumulated as colonies that were surrounded by ZsGreen (Figs. 4a and 4b). Most colonies in tumors were small; 75% were less than 5 μm in diameter (n = 19,408 colonies across five tumors; Fig. 4c). The average colony diameter across all tumors was 4.3 ± 0.3 μm. Approximately half of the bacteria in the tumors (53 ± 8%; n = 5) produced ZsGreen. The average distance that ZsGreen diffused from the edge of colonies was 6.5 ± 1.0 μm (Fig. 4d), which did not change with colony size.
Figure 4.
Salmonella release ZsGreen in tumors. (a) In tumors, Salmonella form colonies (red, merged to yellow; black arrows) and release ZsGreen (green; white arrows). Sections were counterstained with DAPI (blue). Scale bar is 50 μm. (b) Images of colonies (i) were converted to binary (ii) to determine colony size and distance of ZsGreen diffusion. Scale bar is 2 μm. (c) Colony size distribution. Most colonies were small, <5 μm in diameter. (d) Colony size did not affect diffusion distance. (e) The coverage fraction is the number of ZsGreen pixels (gray) in a circular annulus (a1, a2, a3 and a4) around a colony (yellow) normalized by the area of the annulus. (f) For the example colony in (b) and (e), the coverage fraction as a function of distance yields a dimensionless production rate, M, of 0.7, where The average extracellular ZsGreen coverage fraction of the entire population of colonies across five tumors decreased inversely with distance from the colony (black diamonds). From the continuous point source model (Eq. 2, red line), the average dimensionless production rate, M, was 0.453.
The rates of ZsGreen release were calculated using profiles of fractional coverage around tumor-colonized bacteria and a mathematical model of ZsGreen diffusion (Figs. 4e–4g). The distance of ZsGreen from colony edges was dependent on the rate of ZsGreen production and diffusion through tissue. For each colony, the fractional coverage of ZsGreen decreased with distance from the colony border (Fig. 4e). Fractional coverage is the amount of ZsGreen (gray) in concentric rings around colony edges (yellow; Fig. 4e) and is equivalent to normalized concentration, . The decrease in fractional coverage with increasing radius (Fig. 4f) is equivalent to dimensionless ZsGreen production, M, which is given by (Eq. 2, left). For this example colony, M is 0.70 (Fig. 4f). The dimensionless parameter M is the ratio of bacterial protein production to diffusion (Eq. 2, right).
For the entire population of ZsGreen-producing colonies, the fractional coverage decreased with radius at a consistent rate (Fig. 4g). The average dimensionless production rate, M, for these colonies was 0.453. Based on a diffusion coefficient, D, for ZsGreen of 1.94 × 10−6 cm2/s47 and a minimum detectable concentration, C0, of 1.0 μmol/l,46 this dimensionless production is equivalent to a bacterial ZsGreen production rate, , of 4.3 fg·CFU−1·hr−1.
ZsGreen production was greatest in viable tumor regions
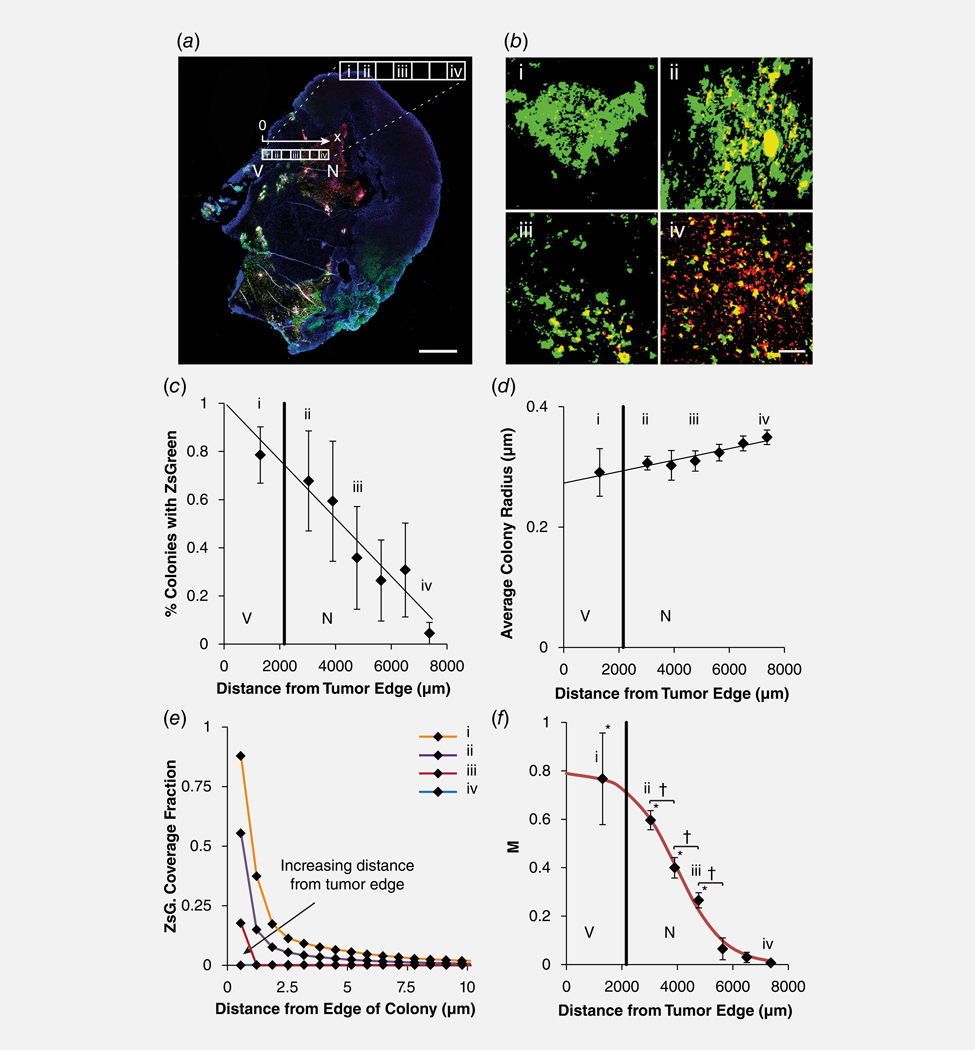
ZsGreen production per bacterium was higher at the viable edge of tumors than in the necrotic center (Fig. 5). In viable regions close to the tumor edge (region i, Fig. 5a), bacterial colonies were surrounded by large areas of ZsGreen (Fig. 5b). At the interface between viable and necrotic tissue (region ii), colonies were surrounded by less ZsGreen. Beyond the viable/necrotic boundary (region iii), bacteria were surrounded by almost no extracellular ZsGreen. Deep in the necrotic core of tumors (region iv), most bacteria did not express ZsGreen. The number of colonies surrounded by extracellular ZsGreen decreased with distance from the tumor edge to the center (p < 0.05; Fig. 5c). The number of colonies with extracellular ZsGreen decreased 1.1% every 100 μm into the tumor. Near the periphery (region i), 80% had extracellular ZsGreen, which dropped to less than 10% in the necrotic core (region iv). Position within tumors did not affect average colony size (Fig. 5d).
Figure 5.
ZsGreen production rate decreases with distance into tumors. (a) Inadjacent regions across tumors, the production of ZsGreen decreased from the viable edge (V) to the necrotic center (N). Distances (x) are relative to the tumor edge. In this section, the distance from the edge to the center was spanned by seven adjacent regions. The first, second, fourth and seventh regions were labeled i, ii, iii and iv, respectively (see expansion in the inset). Scale bar is 1 mm. (b) The amount of extracellular ZsGreen (green) surrounding colonies (red, merged to yellow) decreased for colonies in viable tissue (I; in panel a), at the viable/necrotic boundary (ii), innecrosis (iii) and at the necroticcenter (iv). Scalebar is 100 μm. (c) The percentage of colonies with ZsGreen decreased with distance into the tumors (p < 0.05; n =3 tumors). (d) The average colony size did not change significantly as a function of distance from the tumor edge.(e) Fractional coverage of ZsGreen around colonies in the tumor section in (a). Fractional coverage decreased with distance from the colony edge inversely proportional to radial distance. Over all magnitude of the concentration decreased with depth into the tumor. (f) Dimensionless production rate, M, determined from fractional coverage profiles (e.g.,panel e), decreased as a function of distance from the edge of tumors (n =3). At distances ≤5,000 μm, the production rate was greater than in necrosis (*p < 0.05). Between 3,000 and 5,000 um, M decreased for every 1,000 μm of increased depth (†p < 0.05).
In each tumor region (i–iv), the fractional coverage decreased from the edge of colonies (Fig. 5e), similar to the average decrease for all colonies (Fig. 4g). With distance from the tumor edge, the overall magnitude of the coverage also decreased (Fig. 5e). The decrease of coverage with radius is indicative of dimensionless ZsGreen production, M, where (Eq. 2, left). The dimensionless production rate, M, decreased significantly with increasing depth into tumors (Fig. 5f). Viable regions (region i) had the highest production rate (), compared to necrotic regions, (; region iv; p < 0.05). In necrosis, M decreased by 0.1 for every 591 μm of depth (p < 0.05). At depths greater than 6,000 μm into tumors, M was <0.05 (Fig. 5f).
Transfer of released ZsGreen into the plasma
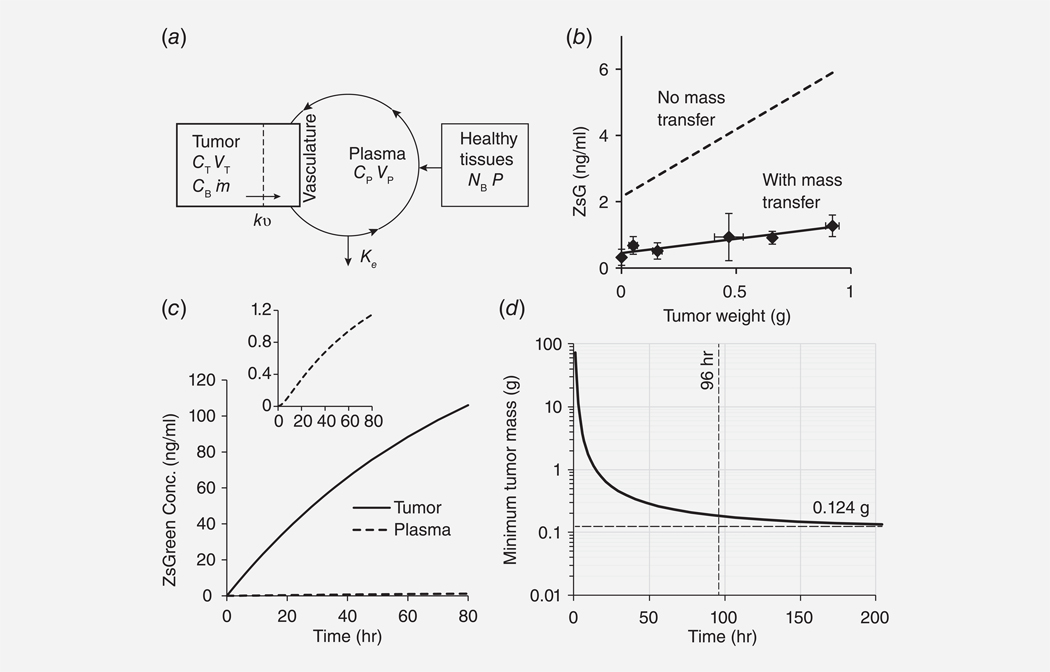
The performance of tumor-detecting bacteria was determined by incorporating the measured rates of ZsGreen production, transfer to the plasma and systemic clearance into a coupled pharmacokinetic, mass-transfer model (Fig. 6a). This model describes the average concentrations of ZsGreen in the tumor and consists of two coupled differential equations (Eqns. 3 and 4). Based on the concentration of plasma ZsGreen as a function of tumor volume (Fig. 6b), the rate of ZsGreen production per bacterium (Fig. 4g) and the clearance rate constant (, Fig. 2d), the mass transfer rate constant, kv, was calculated to be 0.0125 hr−1. Note that in the absence of mass transfer, the predicted plasma concentration of ZsGreen did not match measured values (Fig. 6b). Bacterial growth between induction and measurement would reduce the average number of bacteria in tumors and would increase the predicted value of the mass transfer rate constant. Production in healthy tissue resulted in a nonzero plasma concentration in control, tumor-free mice (tumor weight = 0 g; Fig. 6b). Based on this measured ZsGreen concentration and the production rate per gram of tissue (Supporting Information), the number of bacteria in healthy tissue () was 7.30 × 104 CFU.
Figure 6.
Transfer of ZsGreen from tumors into blood plasma. (a) Coupled mass-transfer, pharmacokinetic model of ZsGreen transfer in mice. The pharmacokinetic model contains three compartments: tumor, plasma and healthy tissue. Parameters in tumors are ZsGreen concentration (), volume (), bacterial density () and ZsGreen production rate (). In the plasma and healthy tissue, parameters are ZsGreen concentration (), volume (), number of bacteria () and ZsGreen production rate (P). The clearance rate constant is and kv is the mass transfer rate constant. (b) The mass transfer rate constant was determined by the increase of plasma ZsGreen concentration with tumor mass (solid line) and the colony production rate. In the absence of mass transfer, the predicted concentration (dashed line) was greater than was measured. (c) The predicted concentration profiles of ZsGreen was greater in tumors than in the plasma. Profiles are for a 0.468g tumor. Inset: Expansion of plasma concentration profile. With time, the rate of increase slowed and eventually saturated in both compartments. (d) The minimum detectable tumor size decreased with time as the concentration of plasma ZsGreen accumulated. At a bacterial density of 485.3CFU/mg, the minimum detectable size would be 0.124g after approximately 150hr. By 96hr after induction, the minimum detectable size is within 0.3% of its theoretical minimum.
ZsGreen was transferred from tumors considerably slower than it was cleared from the blood (Fig. 6c). The calculated value for kv was 20.7 times lower than the clearance rate, (the ratio of these parameters is the dimensionless parameter in Supporting Information). Slow transfer caused a buildup in tumors, where the predicted concentration was greater than in the plasma (Fig. 6c and inset). In these predicted profiles, the concentrations increased almost linearly at early times and begin to saturate toward the end of the experiment. In the first few minutes, the plasma appearance rate was zero but quickly increased (Fig. 6c inset). At 72 hr, the predicted concentration in tumors was 92.8 times greater than in the plasma, which plateaued at 1.88 ng/ml. The buildup of ZsGreen in tumors was observed in histological sections as ZsGreen surrounding colonized bacteria (Figs. 4a and 5b). The minimum detectable tumor volume, at the measured bacterial density of 485.3 CFU/mg, was 0.124 g (Fig. 6d).
Discussion
A detection method was created using genetically modified bacteria to colonize tumors and release a unique fluoromarker to detect tumor burden. Experiments presented here answered critical questions about the mechanisms of bacterial tumor detection in vivo. To be effective, four properties were required. Detecting bacteria needed to (i) specifically colonize tumors and (ii) release ZsGreen within tumors. The released fluoromarker needed to (iii) transport from the tumor interstitium into the blood and (iv) be cleared slowly enough to be detected. All of these phenomena were observed and rates were measured for each. Tumor-detecting bacteria colonized tumors at a density of 485.3 CFU·mg−1 (Fig. 2a) and released ZsGreen at a rate of 4.3 fg·CFU−1·hr−1 (Fig. 4). The rate constant for ZsGreen mass transfer from tumors to blood was 0.0125 hr−1 (Fig. 6) and the clearance rate constant was 0.259 hr−1 (Fig. 1d). Based on these measured rates, this system could detect tumors that are 0.124 g with a bacterial density 485.3 CFU·mg−1 (Fig. 6d). This size is comparable to the current limit of tomographic imaging, which is approximately 0.1 g. With a bacterial density of 100,000 CFU·mg−1, which has been observed in many animal experiments,2,3,6,14 the minimal detectable volume would be 200 times less (6.01 × 10−4 g). This tumor size may be difficult to detect because of poor bacterial colonization and limited tumor connection to vasculature. It was also determined that the plasma concentration of ZsGreen correlated with tumor mass (Fig. 2c). Critically, no adverse effects were observed in mice after administration of detecting bacteria or purified ZsGreen. These results demonstrate that bacterial detection has the potential to detect small tumor masses via a minimally invasive blood test.
The location of colonization within tumors affected ZsGreen production. Most bacteria colonized the transition region between viable and necrotic tissue (Fig. 3). Most ZsGreen was also present in this region. However, the fastest rate of production was in viable tissue (Fig. 5). At the edge of tumors, the production rate, 7.59 fg·CFU−1·hr−1, was 51% of the value measured in vitro in tumor cell masses, 14.8 fg·CFU−1·hr−1 (Ref 1). The similarity of these values suggests that colonization in tumors in mice did not affect protein production rates. In necrosis, however, production was considerably slower, 0.475 fg·CFU−1·hr−1, which was 3.2% of the in vitro rate. This reduction in productivity was most likely caused by the local environment in necrosis. Because of this difference in productivity, most ZsGreen detected in the blood was produced by bacteria colonized in viable tissue. This suggests that this technique would be more effective in detecting smaller tumors, which typically have less necrotic tissue. The ability to predominantly identify viable tumor tissue would be advantageous for detecting metastases, which contain a higher proportion of viable tissue than subcutaneous tumors in mice.
Measurement of individual rate parameters and incorporating them into a computational model gave a fairly complete description of the mechanisms of fluoromarker release from tumors. Appearance in the plasma (Fig. 2a) shows that ZsGreen diffused through tumor interstitium into the blood. Although the in vivo release rate was comparable to rates in vitro (Figs. 4 and 5), transport through tumor tissue was slow. This difference caused a buildup until the concentration was high enough to overcome transport resistance (Fig. 6c) and more ZsGreen was pushed into the blood. With time, clearance would catch up with the rate of ZsGreen appearance from tumors and the plasma concentration would stabilize. This balance sets the minimum detectable tumor volume, which asymptotically approaches an overall minimum at about 4 days (96 hr; Fig. 6d).
An important advantage of this detection method is that bacteria target malignant tissue with high specificity. Identifying the location of malignant tissue is not necessary for this technique to be effective. Because of its specificity to human tumors, detecting bacteria could be used as a primary screen for metastatic recurrence. In the clinic, high-risk patients would receive doses at regular time intervals after surgical resection to detect recurrence or metastatic spread. If tumor tissue was detected, then other modalities (e.g., MRI or PET) would be used to locate it. Currently, metastatic disease is predominantly treated with systemic therapy. Early detection would increase the efficacy of these therapies, which are more effective on smaller tumor masses. The observed correlation between tumor size and plasma ZsGreen concentration (Fig. 2c) suggests that this technique could be used to measure the amount of malignant tissue in the body. It could also enable monitoring of changes in tumor size in response to treatment.
Combining the tumor-targeting capabilities of Salmonella with the release of a detectable fluoromarker could safely detect tumors and measure cancer burden. These results show that detecting Salmonella produce measurable levels of ZsGreen when injected into tumor-bearing mice. No adverse effects were observed. This method would be fast and minimally invasive. Detection would simply require bacterial injection and a subsequent blood draw several days later. Using fluoromarker-releasing bacteria for early detection has the potential to improve life expectancy by enabling earlier and more effective treatment.
Supplementary Material
What’s new?
Facultative anaerobic bacteria readily colonize solid tumors. The diffusion into the bloodstream of a fluorescent marker released by these organisms as they proliferate inside a tumor could facilitate cancer detection. Here, an engineered bacterial system employing a fluoromarker-producing attenuated strain of Salmonella enterica demonstrated capability in detecting tumor volumes of 0.12 grams in mice. This tumor size is comparable to the smallest tumors detected by tomographic imaging. The experiments provide insight into mechanisms underlying bacterial tumor colonization, fluoromarker release in vivo, and fluoromarker transport through tumor tissue. Fluoromarker tissue transport and systemic clearance was gradual, enabling measurement in blood plasma.
Acknowledgements
We gratefully acknowledge financial support from the National Institutes of Health (Grant No. R01CA120825), the National Science Foundation (Grant No. 1159689) and the Friends for Earlier Breast Cancer Test (Grant No. 1140866).
Grant sponsor:
Friends for Earlier Breast Cancer Test; Grant number: 1140866; Grant sponsor: National Institutes of Health; Grant number: R01CA120825; Grant sponsor: National Science Foundation; Grant number: 1159689
Abbreviations:
- CFU
colony forming units
- DAPI
4′,6-diamidino-2-phenylindole
- ELISA
enzyme-linked immunosorbent assay
- MRI
magnetic resonance imaging
- PET
positron emission tomography
- TNF
tumor necrosis factor
- C B
tumor bacterial density (CFU·ml−1)
- C P
concentrations of ZsGreen in the plasma (ng·ml−1)
- C T
concentrations of ZsGreen in the tumor (ng·ml−1)
normalized concentration
minimum detectable ZsGreen concentration
- D
effective diffusion coefficient in tumor interstitium
- k
mass transfer coefficient for flux across the vascular surface (cm·h−1)
plasma clearance rate constant (h−1)
- kv
mass transfer rate constant (h−1)
rate of ZsGreen production per bacterium (fg·CFU−1·h−1)
- M
dimensionless ZsGreen production
- v
average surface area per volume of tumor (cm2·cm−3)
number of bacteria in healthy tissue
dimensionless radius
plasma volume (ml)
tumor volume (ml)
Footnotes
Additional Supporting Information may be found in the online version of this article
References
- 1.Panteli JT, Forkus BA, Van Dessel N, et al. Genetically modified bacteria as a tool to detect microscopic solid tumor masses with triggered release of a recombinant biomarker. Integr Biol (Camb) 2015;7:423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Low KB, Ittensohn M, Le T, et al. Lipid a mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol 1999;17:37–41. [DOI] [PubMed] [Google Scholar]
- 3.Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res 1997;57:4537–44. [PubMed] [Google Scholar]
- 4.Matsumoto Y, Miwa S, Zhang Y, et al. Efficacy of tumor-targeting Salmonella typhimurium A1-R on nude mouse models of metastatic and disseminated human ovarian cancer. J Cell Biochem 2014; 115:1996–2003. [DOI] [PubMed] [Google Scholar]
- 5.Uchugonova A, Zhang Y, Salz R, et al. Imaging the different mechanisms of prostate cancer cell-killing by tumor-targeting Salmonella typhimurium A1-R. Anticancer Res 2015;35:5225–9. [PubMed] [Google Scholar]
- 6.Clairmont C, Lee KC, Pike J, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis 2000;181:1996–2002. [DOI] [PubMed] [Google Scholar]
- 7.Coutermarsh-Ott SL, Broadway KM, Scharf BE, et al. Effect of Salmonella enterica serovar Typhimurium VNP20009 and VNP20009 with restored chemotaxis on 4T1 mouse mammary carcinoma progression. Oncotarget 2017;8:33601–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi K, Zhao M, Yamauchi K, et al. Systemic targeting of primary bone tumor and lung metastasis of high-grade osteosarcoma in nude mice with a tumor-selective strain of Salmonella typhimurium. Cell Cycle 2009;8:870–5. [DOI] [PubMed] [Google Scholar]
- 9.Kawaguchi K, Murakami T, Suetsugu A, et al. High-efficacy targeting of colon-cancer liver metastasis with Salmonella typhimurium A1-R via intra-portal-vein injection in orthotopic nudemouse models. Oncotarget 2017;8:19065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Miwa S, Zhang N, et al. Tumor-targeting Salmonella typhimurium A1-R arrests growth of breast-cancer brain metastasis. Oncotarget 2015;6:2615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao M, Suetsugu A, Ma HY, et al. Efficacy against lung metastasis with a tumor-targeting mutant of Salmonella typhimurium in immunocompetent mice. Cell Cycle 2012;11:187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganai S, Arenas RB, Sauer JP, et al. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther 2011;18:457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao M, Yang M, Li XM, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A 2005;102:755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes NS, Munn LL, Fukumura D, et al. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res 2003;63: 5188–93. [PubMed] [Google Scholar]
- 15.Brooks JD. Translational genomics: the challenge of developing cancer biomarkers. Genome Res 2012;22:183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frahm M, Felgner S, Kocijancic D, et al. Efficiency of conditionally attenuated Salmonella enterica serovar Typhimurium in bacterium-mediated tumor therapy. MBio 2015;6:e00254–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Swofford CA, Forbes NS. Lipid A controls the robustness of intratumoral accumulation of attenuated Salmonella in mice. Int J Cancer 2014;135:647–57. [DOI] [PubMed] [Google Scholar]
- 18.Sznol M, Lin SL, Bermudes D, et al. Use of preferentially replicating bacteria for the treatment of cancer. J Clin Invest 2000;105:1027–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasinskas RW, Forbes NS. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol Bioeng 2006;94:710–21. [DOI] [PubMed] [Google Scholar]
- 20.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res 2007;67:3201–9. [DOI] [PubMed] [Google Scholar]
- 21.Silva-Valenzuela CA, Desai PT, Molina-Quiroz RC, et al. Solid tumors provide niche-specific conditions that lead to preferential growth of Salmonella. Oncotarget 2016;7: 35169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KC, Zheng LM, Luo X, et al. Comparative evaluation of the acute toxic effects in monkeys, pigs and mice of a genetically engineered Salmonella strain (VNP20009) being developed as an antitumor agent. Int J Toxicol 2000;19: 19–25. [DOI] [PubMed] [Google Scholar]
- 23.Low KB, Ittensohn M, Luo X, et al. Construction of VNP20009: a novel, genetically stable antibiotic-sensitive strain of tumor-targeting Salmonella for parenteral administration in humans. Methods Mol Med 2004;90:47–60. [PubMed] [Google Scholar]
- 24.Felgner S, Kocijancic D, Frahm M, et al. Optimizing Salmonella enterica serovar Typhimurium for bacteria-mediated tumor therapy. Gut Microbes 2016;7:171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Jiang Y, Dong T, et al. Blockage of autophagy pathway enhances Salmonella tumor-targeting. Oncotarget 2016;7:22873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo X, Li ZJ, Lin S, et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol Res 2001;12:501–8. [DOI] [PubMed] [Google Scholar]
- 27.Thamm DH, Kurzman ID, King I, et al. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin Cancer Res 2005;11:4827–34. [DOI] [PubMed] [Google Scholar]
- 28.Toso JF, Gill VJ, Hwu P, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol 2002;20:142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senkus E, Kyriakides S, Penault-Llorca F, et al. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24(Suppl 6):vi7–23. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet 2007;8:341–52. [DOI] [PubMed] [Google Scholar]
- 31.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007;25:5287–312. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee SK, Zetter BR. Cancer biomarkers: knowing the present and predicting the future. Future Oncol 2005;1:37–50. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 2005;5:845–56. [DOI] [PubMed] [Google Scholar]
- 34.Behjatnia B, Sim J, Bassett LW, et al. Does size matter? Comparison study between MRI, gross, and microscopic tumor sizes in breast cancer in lumpectomy specimens. Int J Clin Exp Pathol 2010;3:303–9. [PMC free article] [PubMed] [Google Scholar]
- 35.Schoder H, Gonen M. Screening for cancer with PET and PET/CT: potential and limitations. J Nucl Med 2007;48(Suppl 1):4S–18S. [PubMed] [Google Scholar]
- 36.Erdi YE. Limits of tumor detectability in nuclear medicine and PET. Mol Imaging Radionucl Ther 2012;21:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brader P, Stritzker J, Riedl CC, et al. Escherichia coli Nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clin Cancer Res 2008;14:2295–302. [DOI] [PubMed] [Google Scholar]
- 38.Hill PJ, Stritzker J, Scadeng M, et al. Magnetic resonance imaging of tumors colonized with bacterial ferritin-expressing Escherichia coli. PLoS One 2011;6:e25409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cronin M, Akin AR, Collins SA, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One 2012;7:e30940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min JJ, Kim HJ, Park JH, et al. Noninvasive real-time imaging of tumors and metastases using tumor-targeting light-emitting Escherichia coli. Mol Imaging Biol 2008;10:54–61. [DOI] [PubMed] [Google Scholar]
- 41.Min JJ, Nguyen VH, Kim HJ, et al. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat Protoc 2008;3:629–36. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen VH, Kim HS, Ha JM, et al. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res 2010;70:18–23. [DOI] [PubMed] [Google Scholar]
- 43.Yu YA, Shabahang S, Timiryasova TM, et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol 2004;22:313–20. [DOI] [PubMed] [Google Scholar]
- 44.Danino T, Prindle A, Kwong GA, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med 2015;7:289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai YM, Toley BJ, Swofford CA, et al. Construction of an inducible cell-communication system that amplifies Salmonella gene expression in tumor tissue. Biotechnol Bioeng 2013;110:1769–81. [DOI] [PubMed] [Google Scholar]
- 46.Davis MD, Plager DA, George TJ, et al. Interactions of eosinophil granule proteins with skin: limits of detection, persistence, and vasopermeabilization. J Allergy Clin Immunol 2003;112:988–94. [DOI] [PubMed] [Google Scholar]
- 47.Pluen A, Boucher Y, Ramanujan S, et al. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc Natl Acad Sci U S A 2001;98:4628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narang P, Bhushan K, Bose S, et al. A computational pathway for bracketing native-like structures fo small alpha helical globular proteins. Phys Chem Chem Phys 2005;7:2364–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.