Abstract
Purpose:
Paclitaxel is a widely used anti-cancer therapeutic. Peripheral neuropathy is the dose-limiting toxicity and negatively impacts quality of life. Rare germline gene markers were evaluated for predicting severe taxane induced peripheral neuropathy (TIPN) in the patients of European ancestry. In addition, the impact of Cytochrome P450 (CYP) 2C8, CYP3A4, and CYP3A5 metabolizer status on likelihood of severe TIPN was also assessed.
Experimental design:
Whole exome sequencing (WES) analyses were performed in 340 patients of European ancestry who received a standard dose and schedule of paclitaxel in the adjuvant, randomized phase III breast cancer trial, E5103. Patients who experienced grade 3-4 (n=168) TIPN were compared to controls (n=172) who did not experience TIPN. For the analyses, rare variants with a minor allele frequency ≤3% and predicted to be deleterious by protein prediction programs were retained. A gene-based, case-control analysis using SKAT was performed to identify genes that harbored an imbalance of deleterious variants associated with increased risk of severe TIPN. CYP star alleles for CYP2C8, CYP3A4, and CYP3A5 were called. An additive logistic regression model was performed to test the association of CYP2C8, CYP3A4, and CYP3A5 metabolizer status with severe TIPN.
Results:
Cytochrome P450 oxidoreductase, POR, was significantly associated with severe TIPN (p-value=1.8 ×10−6). Six variants were predicted to be deleterious in POR. There were no associations between CYP2C8, CYP3A4 or CYP3A5 metabolizer status with severe TIPN.
Conclusion:
Rare variants in POR, predict an increased risk of severe TIPN in patients of European ancestry who receive paclitaxel.
Keywords: Whole exome sequencing, paclitaxel, Peripheral neuropathy, CYP2C8, CYP 3A4, CYP3A5
INTRODUCTION
Paclitaxel is a standard therapeutic used to improve outcomes for patients with early and metastatic breast cancer.1,2 However, taxane-induced peripheral neuropathy (TIPN) is a common, and occasionally dose-limiting neurotoxic side-effect that impacts long-term quality of life.3 TIPN can be severe and irreversible and has been designated as one of the most important cancer survivorship issues by the American Society of Clinical Oncology.4,5 The mechanisms of TIPN are not fully understood and there are no highly effective agents to prevent or treat this toxicity.6,7
We and others have previously investigated the association of germline genetic variants with TIPN to uncover potential predictive genetic markers with variable findings.8-12 These prior studies largely focused on the impact of common genetic variants, most commonly with minor allele frequency (MAF) greater than 3%. Rare variants, however, are more likely to associate with phenotypes of larger effect size and/or severity; serving as the rationale for this approach.13
In addition, germline variability in genes encoding the drug metabolizing enzymes are known to impact disposition of substrate drugs. Unfortunately, variability in the CYP450 system is complex and not readily interpretable from comprehensive approaches, such as GWAS. Paclitaxel is predominantly metabolized by CYP3A4, CYP3A5, and CYP2C814,15 and genetic variability of CYP2C8 impacts the pharmacokinetics.16. Prior studies have revealed an association of CYP2C8, CYP3A4, CYP3A5 polymorphisms with the risk of TIPN,17-19serving as the rationale for this analysis.
METHODS
Severe TIPN cases and controls from ECOG-ACRIN E5103
E5103 was a phase III adjuvant breast cancer trial that randomized 4,994 patients with node-positive or high-risk node-negative breast cancer to intravenous doxorubicin and cyclophosphamide every 2 or 3 weeks (at the discretion of the treating physician) for four cycles followed by 12 weeks of weekly paclitaxel (80 mg/m2) alone (Arm A), or to the same chemotherapy with either concurrent bevacizumab (Arm B) or concurrent plus sequential bevacizumab (Arm C).20
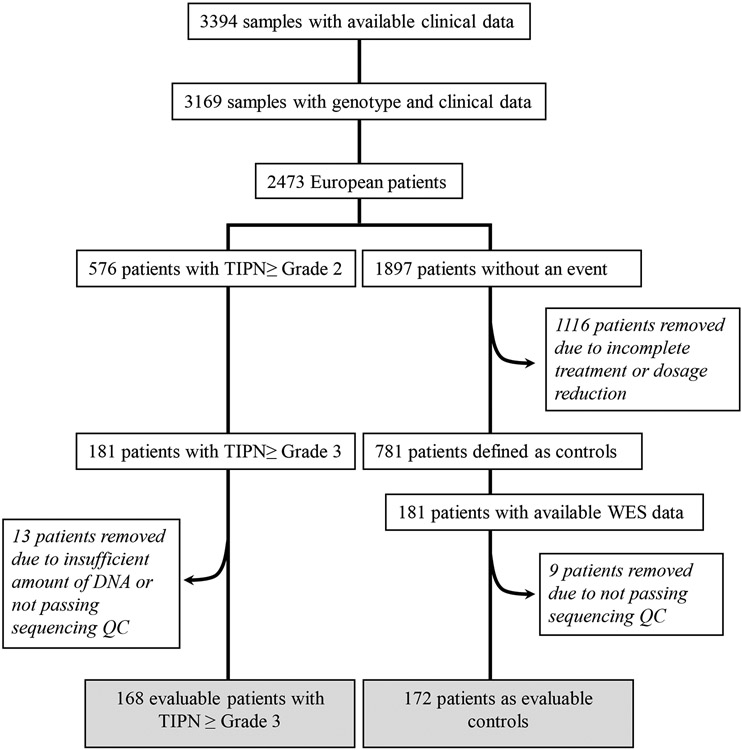
Severe TIPN cases were defined as grade 3-4 TIPN (n=181) by the Common Toxicity Criteria Adverse Events (CTCAE) version 3.0. Cases included patients that received at least one dose of paclitaxel and experienced a neuropathy event during treatment or within 3 months of the last dose of therapy. Controls included patients who met all the following: 1). received all planned doses of paclitaxel; 2) had follow-up for at least 3 months after the last dose of drug; 3) did not meet any of the case definitions as outlined above; and 4) had either paclitaxel or bevacizumab held or modified for any reason (i.e. disease progression or other toxicity) excluded, as previously described (Figure 1).
Figure 1.
Consort diagram
Genome-wide genotyping, whole-exome sequencing, and TaqMan™ assay
Germline DNA from whole blood and companion clinical data were available from 3,394 patients, including 2,473 patients who were genetically determined to be of European ancestry (Figure 1). Genome-wide genotyping and WES were performed as previously described.8,21 Briefly, genome-wide SNP arrays (either Illumina HumanOmni1-Quad or Human OmniExpress) were performed. A principal component analysis (PCA) was performed using Eigenstrat and reference data from 11 HapMap phase III populations. Clusters were identified using the first two eigenvectors computed using common SNPs from these two arrays. Samples that clustered with those of European ancestry were used in subsequent analyses. For whole-exome sequencing, DNA concentrations were determined by fluorometry (Qubit, Life Technologies, Eugene, USA). Exomes were enriched from 50-100 ng of genomic DNA using the AmpliSeq™ Exome RDY kit. Library preparation included adaptor and barcode attachment, as well as magnetic bead-based cleanups. Libraries were quantified by qPCR before sequencing. Typically, sequencing runs generated 50-90 million reads, yielding on average over 100X coverage of the exome with >90% uniformity. Exomes were aligned to the human genome version GRCh37.3. Variants (SNPs, indels, MNPs) were identified by the Torrent VariantCaller 4.2 software. Variants with missing rates > 20% were filtered out of the dataset. Samples with missing rates > 30% were excluded. ANNOVAR (http://annovar.openbioinformatics.org/) was used to annotate chromosome location, functional consequences and allele frequencies on public datasets of each variant. To focus on rare variants, those with MAF ≤ 3% in EA population in ESP 6500 (http://evs.gs.washington.edu/EVS/), 1000G (http://www.1000genomes.org/) or ExAc (http://exac.broadinstitute.org/) were considered. Only SNPs defined by RefGene as one of the following were retained: frameshift substitution, nonsynonymous, stop gains, stop loss, or unknown. Retained SNPs were predicted to be deleterious by at least one of the following: 1). SIFT, 2). POLYPHEN2, or 3). CADD (with a score ≥20), as previously described TaqMan™ assays were performed as previously described.22
CYP star allele calling
The ALDY v3.3 tool 23 was used to extract CYP star alleles for CYP2C8 and CYP3A4*2 from the WES BAM files. Samples with no star-defining variants were considered *1’s for each of the CYP genes, CYP3A4 and CYP3A5*1. The two intronic star alleles, CYP3A5*3 and CYP3A4*22, were obtained from genome-wide genotyping and TaqMan™ assay, respectively.
CYP2C8, CYP3A4, and CYP3A5 metabolizer status
The metabolizer status of CYP2C8, CYP3A4, and CYP3A5 for each individual was predicted based on the functional classification of the alleles they carried (Table 1).24,25 Patients who inherited two functional copies of the CYP2C8, CYP3A4, or CYP3A5 genes were classified as “normal metabolizers” for the respective enzyme. Patients who carried either one or two copies of a reduced-function variant were classified as “intermediate metabolizers” or “poor metabolizers,” respectively.
Table 1.
Diplotype and metabolizer status for CYP2C8, CYP3A4, and CYP3A5
| Diplotype | N | Frequency (%) | Predicted metabolizer status | |
|---|---|---|---|---|
| CYP2C8 | *1/*1 | 227 | 68.4 | Normal |
| *1/*2 | 1 | 0.3 | Intermediate | |
| *1/*3 | 63 | 19.0 | Intermediate | |
| *1/*4 | 28 | 8.4 | Intermediate | |
| *3/*3 | 6 | 1.8 | Poor | |
| *3/*4 | 4 | 1.2 | Poor | |
| *4/*4 | 3 | 0.9 | Poor | |
| Total | 332 | 100 | ||
| CYP3A4 | *1/*1 | 297 | 90.8 | Normal |
| *1/*2 | 1 | 0.3 | Intermediate | |
| *1/*22 | 28 | 8.7 | Intermediate | |
| *22/*22 | 1 | 0.3 | Poor | |
| Total | 327 | 100 | ||
| CYP3A5 | *1/*1 | 2 | 0.6 | Normal |
| *1/*3 | 48 | 14.1 | Intermediate | |
| *3/*3 | 290 | 85.3 | Poor | |
| Total | 340 | 100 |
Statistical analysis
For the rare variant approach, a gene-based case-control analysis (SKAT; http://www.hsph.harvard.edu/skat/) was performed to identify genes associated with a risk of severe TIPN. We followed the manual of SKAT package (https://cran.r-project.org/web/packages/SKAT/) by including a covariates matrix in the SKAT_Null_Model, with covariates selected from the stepwise logistic regression. Study arm, body surface area (BSA), and age were considered as covariates in this analysis. Samples were sequenced separately; therefore, a batch indicator was also included to adjust potential effect of different experiments. The stepwise logistic regression was used, and p-value threshold was < 0.05 for inclusion of covariates in the regression model. Only genes having at least two retained variants were included in the gene-based analysis. The significance threshold (6.9 × 10−6) for the SKAT analysis was determined by correcting for the number of genes tested using Bonferroni correction. Given the rarity of the individual variants as well as the focus on a gene-based analysis, we do not report individual odds ratios for each variant on TIPN risk. For assessment of metabolizer status (CYP2C8, CYP3A4, and CYP3A5) with TIPN, an additive logistic regression model was performed with age, BSA, treatment arm, and batch as covariates, as previously described
Study approval
This study involves secondary research of coded biological samples and clinical data for which patient consent was not required. It was determined exempt from Institutional Review Board (IRB) review as described in 45CFR 46.104 from the Indiana University IRB (application# 1311859668).
Data availability
Raw sequence data and unidentified TIPN clinical data can be accessed at dbGaP (accession number phs003201.v1. p1 https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003201.v1.p1).
RESULTS
Rates of severe TIPN and significant covariates
In the parent trial E5103, 8-9% patients (depending on treatment arm) experienced severe TIPN.20 For the subset of 3,169 patients who had analyzable genetic data, the risk of severe TIPN was 9.6 %.8 Within the previously genotyped patients of European ancestry (N=2,473), 7.3% experienced severe TIPN (Figure 1). In the current study, we evaluated covariates for this sub-population of patients of European ancestry that underwent WES. Older age (OR=2.2 for decade years; p=9.0 × 10−8), increased BSA (OR= 12.7; p=2.2 × 10−4), and study arm (for arm B vs arm A, OR=0.11; p=7.2×10−6; for arm C vs arm A, OR= 0.23; p= 2.7×10−3) were found to be significant risk factors for severe TIPN and appropriate corrections were made (Supplemental table 1).
Top associations for whole-exome sequencing in E5103
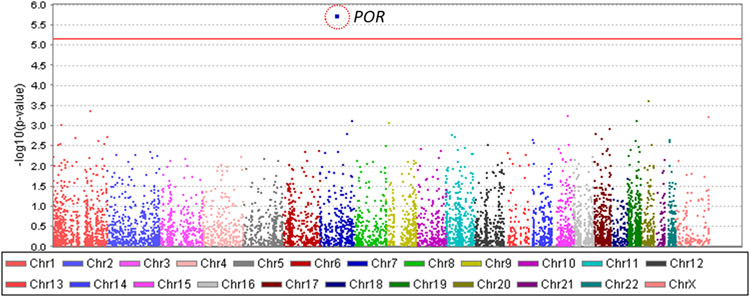
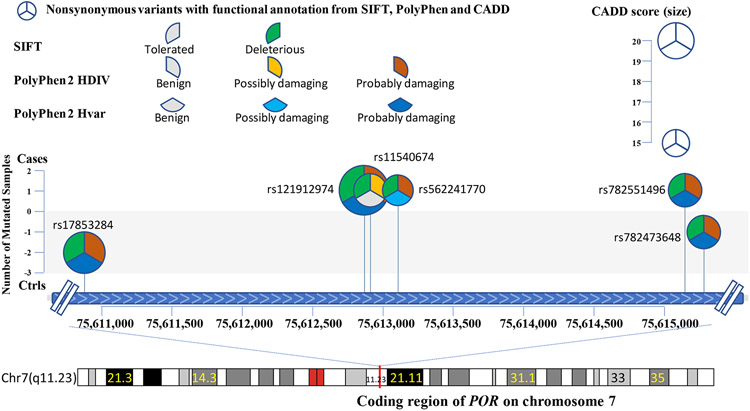
A total of 340 samples (168 with severe TIPN and 172 controls) passed sequencing quality control and were evaluable for further analysis (Figure 1). In total, 414,951 variants were identified. After removing variants with a missing rate > 20% and those which were monomorphic, 258,957 variants were used for annotation by ANNOVAR. 31,699 rare variants predicted to be deleterious were considered for gene-based analyses. Gene-based analysis also required at least two variants to be retained in a given gene for that gene to be included. 7,278 genes were represented by two or more variants, thus setting the threshold for significance at p-value < 6.9 x10−6. The results of the gene-based association analyses are shown in Table 2 and Figure 2. Nine genes had a p-value < 10−3 and the top association was with cytochrome P450 oxidoreductase (POR); (p-value=1.8 × 10−6). Six mutations were predicted to be deleterious in POR (Figure 3 and Table 3).
Table 2.
Top genes associated with the risk of grade 3/4 TIPN
| Gene | P-value |
|---|---|
| POR | 1.82E-06 |
| TTI1 | 0.000237 |
| FMO4 | 0.000431 |
| TBC1D21 | 0.000577 |
| HCFC1 | 0.000599 |
| NOS3 | 0.000742 |
| CEACAM5 | 0.000751 |
| FREM1 | 0.000851 |
| EPHA10 | 0.000947 |
Figure 2.
Manhattan plot for grade 3/4 TIPN from patients of European ancestry in E5103. X-axis indicates the chromosomal position of each gene analyzed; Y-axis denotes magnitude of the evidence for association, shown as −log10(p-value); Each dot represents an evaluable gene. The solid red line indicates the exome-wide significance threshold.
Figure 3.
Comprehensive representation of the six rare variants detected in the cases from patients of European Ancestry with grade 3/4 TIPN. The X-axis denotes the chromosomal location of each mutation and the Y-axis denotes the number of patients with TIPN that had a given mutation. Each pie-wedge shape & direction represents the program prediction method and the color represents the type of mutation. The size of the pie represents the CADD score.
Table 3.
Counts of POR rare deleterious variants and variant carriers in treatment arm
| Variant | Grade 3/4TIPN | Treatment arm | |
|---|---|---|---|
| Cases | Controls | ||
| rs17853284 | 0 | 2 | C |
| rs121912974 | 1 | 0 | B |
| rs11540674 | 1 | 0 | C |
| rs562241770 | 1 | 0 | B |
| rs782551496 | 1 | 0 | B |
| rs782473648 | 0 | 1 | A |
Association with CYP2C8, CYP3A4, and CYP3A5 metabolizer status
Star alleles were successfully identified from 327, 332, and 340 samples for CYP2C8, CYP3A4, and CYP3A5, respectively (Table 1). Seven diplotypes for CYP2C8, four for CYP3A4, and three for CYP3A5 were identified separately. Of note, the CYP2C8*3 allele is mapped to both rs11572080 and rs10509681, which are in complete linkage disequilibrium. The genotype frequency and the major/minor allele frequencies of CYP2C8, CYP3A4, and CYP3A5 in this study sample set of European ancestry are shown in Supplemental Table 2. There was no association between CYP2C8, CYP3A4, or CYP3A5 metabolizer status and severe TIPN.
DISCUSSION
TIPN is a commonly experienced toxicity for cancer patients receiving paclitaxel.26,27 TIPN can impact dose intensity which can subsequently compromise treatment efficacy and profoundly affect quality life. Currently, effective therapies for TIPN are not available. Previously, we and others have used GWAS to investigate common genetic variants for predicting TIPN.8 Herein, we applied WES to evaluate the effect of rare, deleterious coding variants across the genome and the impact of CYP2C8 and CYP3A4/5 metabolizer status on risk of severe TIPN.
Prior work has largely focused on the impact of common germline variants and variants in genes that impact metabolism of paclitaxel. Paclitaxel is metabolized by CYP2C8 and CYP3A4 enzymes.14 Prior studies have been conflicting, with some reporting an association between CYP2C8, CYP3A4, and CYP3A5 metabolizer status and TIPN, 19,28,29 while others finding no association.30,31
Using a gene-based approach for our rare variant analysis, we identified a significant association between those with deleterious mutations in POR and severe TIPN. POR, located on chromosome 7q11.2, encodes for cytochrome P450 oxidoreductase. This enzyme is required to transfer electrons from NADPH to other cytochrome p450 family members, which is essential for the catalytic activation.32 While we didn’t find genetically altered CYP2C8, CYP3A5 or CYP3A4 to be associated with TIPN, their activity may have been impacted by a suboptimal functioning POR instead.33
Previously, we reported a significant association of SET binding factor 2 (SBF2) with severe TIPN in patients of African descent using a similar approach. We further demonstrated that decreasing SBF2 expression exacerbated the effect of paclitaxel on the sensory neurons in an ex vivo model, providing mechanistic support for this finding.34 In the current study focused on patients of European descent, however, SBF2 was excluded from the analysis as only one variant was detected.21 These results demonstrate that genetic associations can differ based on the population evaluated. In addition, it highlights the need for work to unravel the mechanistic underpinning for POR.
A major strength of this study is that it was conducted in a large, randomized, phase III breast cancer clinical trial with rigorous data collection and the delivery of a uniform dose and schedule of paclitaxel. Whole-exome analyses performed in this study allowed for a comprehensive and unbiased evaluation of rare coding variants to discover the most impactful genes associated with severe TIPN. Some limitations include the inability to detect potentially important variants in the non-coding region of the genome and the drop-out of genes represented by less than two variants. Despite the analysis from a large clinical trial, the correlative association study was unplanned and relatively underpowered. We also acknowledge that validation and/or mechanistic studies are necessary before these identified variants and genes can be developed for TIPN predictive biomarkers and target therapy.
The discordance and variable findings across dataset in discovering TIPN genetic predictors require careful consideration. Reasons for the lack of cross-validation are likely multi-factorial. One clear reason is the relatively small sample size for each of these studies. Others include the lack of unified drug exposure and schedule, differences in phenotype definitions, and variable search space for candidate variants, among others. While all prior and current findings may represent false positive associations, given the strength of statistical significance, and in some cases strong biological plausibility, this seems unlikely. Rather, a more likely explanation is that TIPN is a biologically complex pathology impacted by number host-specific factors (e.g. obesity, diabetes, age) and regulated by a number of genes making reproducibility in underpowered datasets difficult. This is even more complicated when considering metabolizer status as it can also be influenced by concurrent medications 35,36 which are often not accounted for. As outlined above, we previously demonstrated African ancestry to be associated with increased risk of TIPN and the predictors were unique to patients of African descent despite the same dataset. The discordance here may be a function of different frequencies of key variants, differences in host-gene interactions, or nuanced differences in the mechanism of causation. The impact of germline variation on TIPN in Black breast cancer patients is currently being studied in the NCI-Cooperative group trial, EAZ171 which hopes to provide prospective validation for these germline predictors.
The discovery of predictive genetic markers for severe TIPN has major potential clinical consequences, including the immediate application of optimal patient counseling and monitoring. Perhaps equally important, genetic associations may lend insight to the underlying biological mechanism for this toxicity and lead to targets for development of therapies designed to either prevent or treat TIPN.
Supplementary Material
STATEMENT OF CLINICAL RELEVANCE.
The taxanes are commonly employed chemotherapeutic agents for a variety of malignancies. The most common and clinically relevant toxicity is peripheral neuropathy (TIPN). However, there are no clinically available biomarkers for predicting TIPN. We have previously reported common germline variants associated with TIPN in the adjuvant breast cancer trial, ECOG-ACRIN E5103. Herein, we used whole exome sequencing (WES) to comprehensively evaluate the impact of rare, deleterious coding variants across the genome. Cytochrome P450 oxidoreductase (POR) was significantly associated with an increased risk of TIPN. This work provides the immediate translational potential of improving personalized counseling and monitoring for onset of TIPN and ultimately may lend insight to underlying biological mechanisms.
FINANCIAL SUPPORT
This study was conducted by the ECOG-ACRIN Cancer Research Group (Peter J. O'Dwyer, MD and Mitchell D. Schnall, MD, PhD, Group Co-Chairs) and supported by the National Cancer Institute of the National Institutes of Health under award number U10CA180820. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Susan G. Komen for the Cure (SAC110056) (BPS) and Vera Bradley Foundation Center (BPS). Bevacizumab and matching placebo were provided free of charge by Roche/Genentech.
Footnotes
CONFLICT OF INTEREST STATEMENT:
CD has received research funds from Roche/Genentech, Puma and Daiichi, and reported personal fees from Roche/Genentech, PUMA, Daiichi, Lilly, Evicore, Pfizer and Seagen. BPS has received research support from Roche/Genentech and Pfizer for drug supply, Epic Sciences for CTC assessment, and Foundation medicine for sequencing provision, and reported less than $5K personal fees from Lilly. The remaining authors declare no competing financial interests.
REFERENCES
- 1.Sparano JA: Taxanes for breast cancer: an evidence-based review of randomized phase II and phase III trials. Clin Breast Cancer 1:32–40; discussion 41-2, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Capri G, Tarenzi E, Fulfaro F, et al. : The role of taxanes in the treatment of breast cancer. Semin Oncol 23:68–75, 1996 [PubMed] [Google Scholar]
- 3.Schneider BP, Hershman DL, Loprinzi C: Symptoms: Chemotherapy-Induced Peripheral Neuropathy. Adv Exp Med Biol 862:77–87, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Derman BA, Davis AM: Recommendations for Prevention and Management of Chemotherapy-Induced Peripheral Neuropathy. JAMA 326:1058–1059, 2021 [DOI] [PubMed] [Google Scholar]
- 5.Loprinzi CL, Lacchetti C, Bleeker J, et al. : Prevention and Management of Chemotherapy-Induced Peripheral Neuropathy in Survivors of Adult Cancers: ASCO Guideline Update. J Clin Oncol 38:3325–3348, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Staff NP, Fehrenbacher JC, Caillaud M, et al. : Pathogenesis of paclitaxel-induced peripheral neuropathy: A current review of in vitro and in vivo findings using rodent and human model systems. Exp Neurol 324:113121, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colvin LA: Chemotherapy-induced peripheral neuropathy: where are we now? Pain 160 Suppl 1:S1–S10, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider BP, Li L, Radovich M, et al. : Genome-Wide Association Studies for Taxane-Induced Peripheral Neuropathy in ECOG-5103 and ECOG-1199. Clin Cancer Res 21:5082–5091, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leandro-Garcia LJ, Inglada-Perez L, Pita G, et al. : Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J Med Genet 50:599–605, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Baldwin RM, Owzar K, Zembutsu H, et al. : A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res 18:5099–109, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adjei AA, Lopez CL, Schaid DJ, et al. : Genetic Predictors of Chemotherapy-Induced Peripheral Neuropathy from Paclitaxel, Carboplatin and Oxaliplatin: NCCTG/Alliance N08C1, N08CA and N08CB Study. Cancers (Basel) 13, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chhibber A, Mefford J, Stahl EA, et al. : Polygenic inheritance of paclitaxel-induced sensory peripheral neuropathy driven by axon outgrowth gene sets in CALGB 40101 (Alliance). Pharmacogenomics J 14:336–42, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bomba L, Walter K, Soranzo N: The impact of rare and low-frequency genetic variants in common disease. Genome Biol 18:77, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cresteil T, Monsarrat B, Dubois J, et al. : Regioselective metabolism of taxoids by human CYP3A4 and 2C8: structure-activity relationship. Drug Metab Dispos 30:438–45, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Stage TB, Bergmann TK, Kroetz DL: Clinical Pharmacokinetics of Paclitaxel Monotherapy: An Updated Literature Review. Clin Pharmacokinet 57:7–19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann TK, Brasch-Andersen C, Green H, et al. : Impact of CYP2C8*3 on paclitaxel clearance: a population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J 11:113–20, 2011 [DOI] [PubMed] [Google Scholar]
- 17.Hertz DL, Motsinger-Reif AA, Drobish A, et al. : CYP2C8*3 predicts benefit/risk profile in breast cancer patients receiving neoadjuvant paclitaxel. Breast Cancer Res Treat 134:401–10, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kus T, Aktas G, Kalender ME, et al. : Polymorphism of CYP3A4 and ABCB1 genes increase the risk of neuropathy in breast cancer patients treated with paclitaxel and docetaxel. Onco Targets Ther 9:5073–80, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leskela S, Jara C, Leandro-Garcia LJ, et al. : Polymorphisms in cytochromes P450 2C8 and 3A5 are associated with paclitaxel neurotoxicity. Pharmacogenomics J 11:121–9, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Miller KD, O'Neill A, Gradishar W, et al. : Double-Blind Phase III Trial of Adjuvant Chemotherapy With and Without Bevacizumab in Patients With Lymph Node-Positive and High-Risk Lymph Node-Negative Breast Cancer (E5103). J Clin Oncol 36:2621–2629, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider BP, Lai D, Shen F, et al. : Charcot-Marie-Tooth gene, SBF2, associated with taxane-induced peripheral neuropathy in African Americans. Oncotarget 7:82244–82253, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider BP, Wang M, Radovich M, et al. : Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol 26:4672–8, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ly RC, Shugg T, Ratcliff R, et al. : Analytical Validation of a Computational Method for Pharmacogenetic Genotyping from Clinical Whole Exome Sequencing. J Mol Diagn, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Ingelman-Sundberg M, Lauschke VM: Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin Pharmacol Ther 102:688–700, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.PharmGKB.: Gene-specific information tables for CYP3A5. https://www.pharmgkb.org/page/cyp3a5RefMaterials [Google Scholar]
- 26.Sparano JA, Wang M, Martino S, et al. : Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med 358:1663–71, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershman DL, Weimer LH, Wang A, et al. : Association between patient reported outcomes and quantitative sensory tests for measuring long-term neurotoxicity in breast cancer survivors treated with adjuvant paclitaxel chemotherapy. Breast Cancer Res Treat 125:767–74, 2011 [DOI] [PubMed] [Google Scholar]
- 28.de Graan AJ, Elens L, Sprowl JA, et al. : CYP3A4*22 genotype and systemic exposure affect paclitaxel-induced neurotoxicity. Clin Cancer Res 19:3316–24, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apellaniz-Ruiz M, Lee MY, Sanchez-Barroso L, et al. : Whole-exome sequencing reveals defective CYP3A4 variants predictive of paclitaxel dose-limiting neuropathy. Clin Cancer Res 21:322–8, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Lee MY, Apellaniz-Ruiz M, Johansson I, et al. : Role of cytochrome P450 2C8*3 (CYP2C8*3) in paclitaxel metabolism and paclitaxel-induced neurotoxicity. Pharmacogenomics 16:929–37, 2015 [DOI] [PubMed] [Google Scholar]
- 31.Bergmann TK, Green H, Brasch-Andersen C, et al. : Retrospective study of the impact of pharmacogenetic variants on paclitaxel toxicity and survival in patients with ovarian cancer. Eur J Clin Pharmacol 67:693–700, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Pandey AV, Fluck CE: NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol Ther 138:229–54, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Agrawal V, Choi JH, Giacomini KM, et al. : Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase. Pharmacogenet Genomics 20:611–8, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham GM, Shen F, Wu X, et al. : The impact of SBF2 on taxane-induced peripheral neuropathy. PLoS Genet 18:e1009968, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stage TB, Mortensen C, Khalaf S, et al. : P-Glycoprotein Inhibition Exacerbates Paclitaxel Neurotoxicity in Neurons and Patients With Cancer. Clin Pharmacol Ther 108:671–680, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Barroso L, Apellaniz-Ruiz M, Gutierrez-Gutierrez G, et al. : Concomitant Medications and Risk of Chemotherapy-Induced Peripheral Neuropathy. Oncologist 24:e784–e792, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data and unidentified TIPN clinical data can be accessed at dbGaP (accession number phs003201.v1. p1 https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003201.v1.p1).