Abstract
For many members of the Picornaviridae family, infection of cells results in a shutoff of host protein synthesis. For rhinoviruses and enteroviruses, the shutoff has been explained in part by the cleavage of eukaryotic initiation factor 4GI (eIF4GI), a component of the cap-binding protein complex eIF4F. The cleavage of eIF4GI is mediated by the virus-specific proteinase 2Apro and results in inhibition of cap-dependent, but not cap-independent, translation. The inhibition of host protein synthesis after infection with human rhinovirus 14 (HRV-14) lags behind the cleavage of eIF4GI. Recently, we discovered a functional homolog of eIF4GI, termed eIF4GII, and showed that cleavage of eIF4GII coincides with the shutoff of host cell protein synthesis after poliovirus infection (Gradi et al., Proc. Natl. Acad. Sci. USA 95:11089–11094, 1998). We wished to determine whether eIF4GII cleavage kinetics could also explain the lack of correlation between the kinetics of eIF4GI cleavage and the shutoff of host protein synthesis after rhinovirus infection. In this study, we examined the correlation between human rhinovirus-induced shutoff of host protein synthesis and cleavage of eIF4GI and eIF4GII. In HRV-14-infected HeLa cells, almost no intact eIF4GI could be detected by 4 h postinfection, while only 4% of eIF4GII was cleaved at this time. By 6 h, however, 67% of eIF4GII was cleaved, and this cleavage coincided with a significant (60%) decline of host translation. These results suggest that cleavage of both eIF4GI and eIF4GII is required for HRV-mediated inhibition of host cell protein synthesis and that the cleavage of eIF4GII is the rate-limiting step in the shutoff of host cell protein synthesis after rhinovirus infection.
The genome of human rhinoviruses (HRV), similar to that of other picornaviruses, contains a single long open reading frame which encodes a large polyprotein precursor. The polyprotein is processed, partially in the nascent state, by two virus-specific proteases. Protease 2Apro performs the initial cleavage of the nascent polyprotein that liberates the capsid protein precursor (P1), while 3Cpro is responsible for the remaining cleavage events (33).
To establish an efficient infection, picornaviruses have evolved mechanisms to inhibit host cell protein synthesis. Translation of the capped cellular mRNAs is blocked at the initiation step, while translation of the uncapped virus mRNA proceeds efficiently via a cap-independent mechanism (2, 11). The mechanism of this selective inhibition of host protein synthesis varies depending on the particular virus-host system. In rhinovirus- and enterovirus-infected cells, a cap-binding protein complex, the eukaryotic initiation factor 4F (eIF4F), is altered, both structurally and functionally (11). eIF4F is composed of the following three subunits: (i) eIF4E, which specifically recognizes the cap structure (44, 45); (ii) eIF4A, which possesses an RNA helicase activity (39, 42); and (iii) eIF4G (formerly p220), which serves as a scaffold to bring together eIF4E, eIF4A, and eIF3, a factor which links eIF4G to the ribosome (23, 28, 43).
A large body of evidence supports the idea that rhinoviruses and enteroviruses inactivate eIF4F by cleavage of the eIF4G subunit in a process mediated by 2Apro (3, 22, 27). Poliovirus 2Apro mutants defective in eIF4G cleavage are also defective in inducing the shutoff of host translation (3, 32). 2Apro of human rhinovirus 2 (HRV-2) and coxsackievirus B4 cleave eIF4G directly (24, 25). Poliovirus 2Apro is also capable of directly cleaving eIF4G (7, 17); however, it was concluded that the major eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is not associated with a 2Apro (8). The C-terminal cleavage product of eIF4G interacts with eIF4A and eIF3 and supports cap-independent translation (6, 31, 37). However, it lacks the eIF4E recognition domain and is unable to support cap-dependent translation. In addition, cap-independent viral protein synthesis is stimulated by the C-terminal cleavage product of eIF4G of several (6, 31, 47, 48), but not all (5, 41), picornaviruses.
Although eIF4G proteolysis provides an attractive explanation for the shutoff phenomenon, several earlier findings obtained with poliovirus are inconsistent with such a model. For example, there is no good temporal correlation between the proteolysis of eIF4G and the shutoff of host translation in poliovirus-infected HeLa cells (13). While the cleavage of eIF4G occurs rapidly, i.e., within an average of 2 h postinfection, the shutoff is observed at 2.5 h or even later. Moreover, a complete cleavage of eIF4G occurs in the presence of guanidine-HCl and other inhibitors of poliovirus replication, which strongly mitigate the inhibition of host cell protein synthesis (4, 21, 36). It was suggested, therefore, that complete inhibition of host cell protein synthesis following poliovirus infection requires a second event in addition to the cleavage of eIF4G (4). An alternative interpretation of these data is that eIF4G cleavage is irrelevant to the shutoff of host translation (21). These apparent discrepancies may now be explained by recent findings. We have recently identified a functional homolog of eIF4G, termed eIF4GII (16). eIF4GII is 46% identical to the original eIF4G (46), which was renamed eIF4GI. eIF4GII is cleaved both in vivo and in vitro by poliovirus 2Apro (17). However, after poliovirus infection, the cleavage of eIF4GII is slower than that of eIF4GI and, in contrast to cleavage of eIF4GI, it coincides with the shutoff of host protein synthesis (17).
In human rhinovirus 14 (HRV-14)-infected HeLa cells, as in poliovirus-infected cells, there is a discrepancy between the time course of eIF4GI cleavage and the shutoff of host protein synthesis (12). It is possible that this discrepancy can be explained for rhinovirus, as was shown for poliovirus, by the slower kinetics of cleavage of eIF4GII. Here, we addressed this possibility.
HeLa-I cells (Ohio strain) (1) were grown in monolayers in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS). HRV-14 was kindly provided by R. Rueckert. Virus stock was generated by passages in HeLa cells at 34°C in DMEM containing 2% FBS. Virus titers were determined in four replicates in a 50% tissue culture infective dose (TCID50) assay on HeLa cells, using 10-fold virus dilutions in 96-well microtiter plates. Virus dilutions were mixed with 104 cells in DMEM containing 2% FBS in a total volume of 0.3 ml. The plates were incubated at 34°C for 4 days. Following incubation, the cytopathic effect was determined, and the titers were estimated according to the method of Reed and Muench (40). For a one-step infection, HeLa cells were grown to ∼80% confluency in 10-cm petri dishes. The cells were infected in serum-free medium at a multiplicity of infection of 50 TCID50 per cell. Following virus adsorption at room temperature for 30 min, the cells were washed with phosphate-buffered saline (PBS) and further incubated in 5 ml of methionine-free DMEM without serum at 34°C. At the times indicated in the figure legends, the cells were incubated for 30 min with [35S]methionine (10 μCi/ml; 1 Ci = 37 GBq), washed with PBS, and scraped on ice into 0.2 ml of cold buffer containing 20 mM HEPES-KOH (pH 7.6), 150 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, and a protease inhibitor cocktail (Boehringer Mannheim). Cells were lysed by three cycles of freezing and thawing. Cell debris was pelleted by centrifugation, and protein concentration in the supernatant was measured by the Bio-Rad assay. The protein concentration was adjusted to 4 mg/ml by diluting with PBS. Proteins were denatured with an equal volume of 2× Laemmli sample buffer and boiled for 2 min. Aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide). The gels were processed for fluorography with En3Hance (Dupont). Antibodies and the protocol for Western blotting have been described previously (16, 17). Briefly, 40 μg of HeLa cell protein extract was subjected to SDS-PAGE (6% acrylamide). The proteins were transferred onto 0.45-μm-pore-size nitrocellulose membranes (Schleicher & Schuell). The membranes were blocked overnight with 5% skim milk in PBS at 4°C and probed with the indicated antibodies for 3 to 4 h at 4°C in 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Tween 20, and 5% skim milk. After washing, the blots were incubated in the same buffer, but without skim milk, with donkey anti-rabbit immunoglobulin-horseradish peroxidase (Amersham) at a 1:5,000 dilution for 45 min at room temperature. Following extensive washing, the blots were developed with the Renaissance ECL system (Amersham). The intensities of the bands were quantified by using a BioImage densitometer (Milligen/Biosearch).
HRV-14 infection results in rapid cleavage of eIF4GI between 1 and 2 h after infection (12, 29). However, the shutoff of host protein synthesis lags considerably behind the cleavage of eIF4GI, as significant inhibition of host protein synthesis occurs only after 3 h of infection (12). To determine whether eIF4GII is more resistant to proteolysis than eIF4GI, we examined the integrity of eIF4GII following HRV-14 infection. The state of eIF4GI and the pattern of protein synthesis in HRV-14-infected HeLa cells were analyzed in parallel. Samples of mock- or virus-infected HeLa cells (50 TCID50 per cell) were grown in monolayers in 10-cm petri dishes and pulse-labeled for 30 min with [35S]methionine at different times after infection. Aliquots of cytoplasmic extracts were processed for SDS-PAGE followed by autoradiography or Western blotting using polyclonal anti-eIF4GI or anti-eIF4GII antibody as specified in the figure legends.
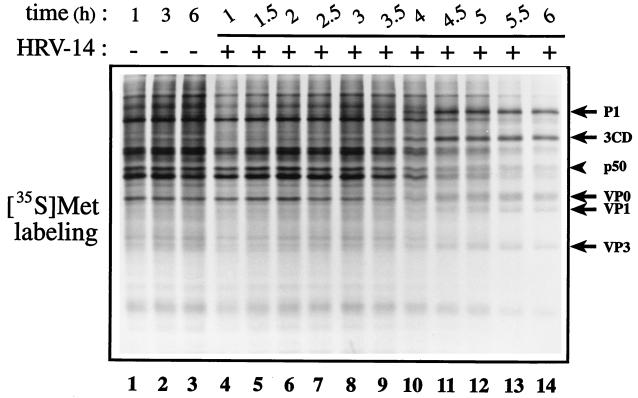
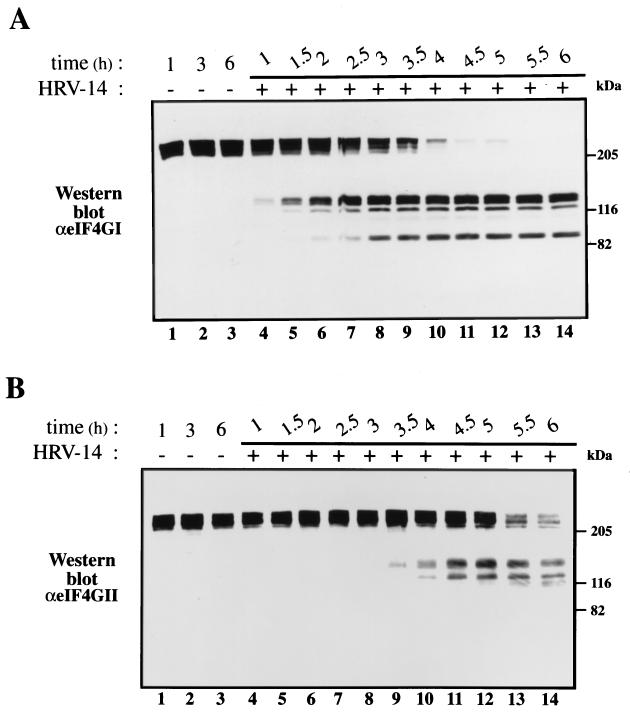
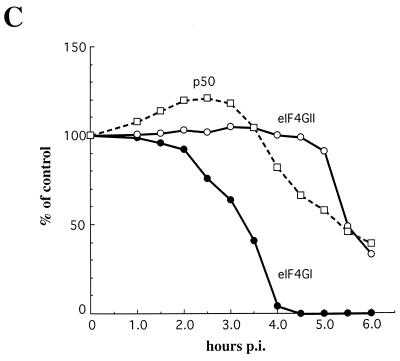
HRV-14 infection resulted in a feeble inhibition of host cell protein synthesis (Fig. 1; for quantitation see Fig. 2C). The partial inhibition of host translation (∼60% at 5.5 to 6 h postinfection) occurred concomitantly with the synthesis of the bulk of virus proteins. Inhibition of [35S]methionine incorporation into cellular proteins has been assessed by quantification of the amount of radioactivity in an arbitrarily chosen representative cellular polypeptide (p50). Western blot analysis of infected cell extract revealed that while both eIF4GI and eIF4GII were cleaved during infection, the cleavage kinetics of the two isoforms varied dramatically. Cleavage of eIF4GI was detectable at 1.0 to 1.5 h after infection and was almost complete by 4 h (Fig. 2A). The disappearance of the full-size eIF4GI was accompanied by accumulation of the characteristic cleavage products that have been described for both rhinovirus and poliovirus (12, 13). Cleavage of eIF4GII occurred much later than that of eIF4GI (Fig. 2B). Strikingly, only a minute fraction of eIF4GII was cleaved at 4 h postinfection as judged by the first appearance of the characteristic cleavage products. Quantification of intact eIF4GII showed that about one-third of eIF4GII remained intact even at 6 h after infection (Fig. 2C). The low rate of eIF4GII cleavage is consistent with the inefficient inhibition of host mRNA translation after HRV-14 infection (Fig. 1 and 2C).
FIG. 1.
Protein synthesis in HRV-14-infected cells. HeLa cells were mock infected or infected with HRV-14 (50 TCID50 per cell) and pulse-labeled for 30 min with [35S]methionine at the indicated times after infection. Equal amounts of cytoplasmic protein extract (5 μg) were analyzed by SDS–15% PAGE, and the gel was fluorographed. Arrows indicate virus-specific proteins. The arrowhead indicates an arbitrary cellular protein (p50).
FIG. 2.
(A and B) eIF4GI and eIF4GII cleavage. Proteins (40 μg) of the samples indicated in the legend for Fig. 1 were separated by SDS–6% PAGE, blotted onto nitrocellulose paper, and treated with polyclonal antibodies against eIF4GI (A) or eIF4GII (B) (16, 17). Positions of molecular mass standards are shown. (C) Quantitative analysis of the results shown in Fig. 1 and 2. The intensity of the p50 cellular protein band (Fig. 1) was determined by using a Bas 2000 phosphorimager and is presented as a percentage of the signal in p50 of mock-infected extracts (there was no significant change in the labeling of p50 in mock-infected cells at different times). The amount of intact eIF4GI and eIF4GII present in each lane is presented as a percentage of that of uncleaved eIF4Gs in mock-infected cells (no change was observed in the amount of eIF4Gs in mock-infected cells upon incubation). The intensities of the bands were determined with a BioImage densitometer (Milligan Bioresearch). p.i., postinfection.
Our results show a coincidence between the cleavage of eIF4GII and the inhibition of host protein synthesis in HRV-14-infected cells. eIF4GI is cleaved before eIF4GII, and its cleavage is most probably required for the inhibition of host protein synthesis. Thus, the decrease in translation during HRV-14 infection correlates with eIF4GII cleavage, because this is the last event to occur. Several factors could account for the differential cleavage of eIF4GI and eIF4GII by HRV-14, as is the case for poliovirus infection (17). First, eIF4GII might be a poorer substrate for HRV-14 2Apro than eIF4GI. Second, the fraction of eIF4GI and eIF4GII in the cell that is associated with eIF4E might be different. The susceptibility of eIF4GI and eIF4GII to HRV-2 2Apro proteolysis is dramatically increased when these factors are bound to eIF4E (16, 18, 30). It would, therefore, be important to determine what fraction of eIF4GI and eIF4GII is present as a complex with eIF4E. It is also possible that eIF4GI-eIF4E and eIF4GII-eIF4E complexes differ in their sensitivities to cleavage by 2Apro.
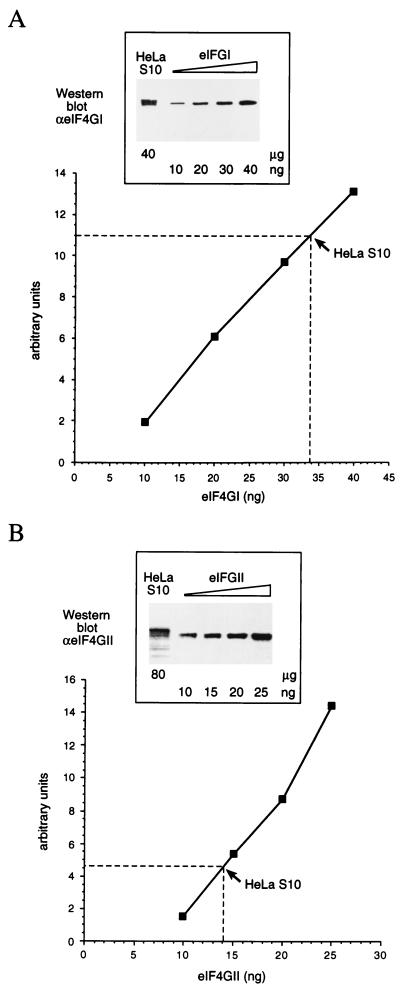
An important issue in addressing the correlation between eIF4GII cleavage and the inhibition of host translation concerns the relative abundance of eIF4GI and eIF4GII in cells. We determined the concentration of eIF4GI and eIF4GII in HeLa cell extracts by quantitative Western immunoblotting (35) using a range of eIF4GI and eIF4GII concentrations. Purified recombinant eIF4GI and eIF4GII proteins expressed by the baculovirus system (16) were used as standards. The eIF4Gs possess an Xpress epitope (Invitrogen) within their N-terminal portions. The concentrations of the eIF4G isoforms were estimated after SDS-PAGE and Coomassie blue staining. The intensities of the recombinant eIF4GI and eIF4GII bands were compared to those of bovine serum albumin over a range of concentrations. Using these estimations as well as the results of Western blot analysis with an Anti-Xpress antibody (Invitrogen) the concentrations of eIF4GI and eIF4GII protein standards were determined and normalized. For eIF4GI quantification, 40 μg of HeLa cell S10 fraction was analyzed together with known amounts of eIF4GI (Fig. 3A). Note that the cytoplasmic eIF4GI migrates as several bands, possibly due to posttranslational modification or proteolytic cleavage. Also, the recombinant eIF4GI migrates somewhat faster than the native forms, presumably because it possesses a 156-amino-acid (aa) truncation at the N-terminal portion while containing an extra sequence of 46 aa from the pBlueBacHis2 C baculovirus vector (Invitrogen) (16, 20). The ECL signal of the HeLa S10 corresponded to that elicited by 34 ng of eIF4GI. We calculated that eIF4GI constitutes about 0.085% of HeLa S10 and eIF4GII constitutes 0.018% (Fig. 3B). Similar to eIF4GI, endogenous eIF4GII migrates as several forms and does so more slowly than recombinant eIF4GII, which lacks 158 aa but contains 46 aa derived from the vector (16). Quantification of eIF4GI and eIF4GII in two other batches of HeLa S10 resulted in similar values. The mean content of eIF4GI in the HeLa S10 fraction is 0.08 ± 0.005% and the content of eIF4GII is 0.019 ± 0.002%. Assuming similar molecular weights for eIF4GI and eIF4GII, the molar ratio of eIF4GI to eIF4GII is 4.2. Thus, eIF4GII constitutes about 20% of the total amount of eIF4G in HeLa-I cells. Although eIF4GII is less abundant than eIF4GI, its amount in the cell is sufficient to maintain host cell protein synthesis at late times of HRV-14 infection, when eIF4GI is cleaved. Thus, eIF4GI is not rate limiting for translation. Indeed, it is believed that the availability of eIF4E, rather than eIF4GI, limits translation in eukaryotic cells. First, with the exception of one report (38), eIF4E was found to be present in most cells at a lower molar concentration than eIF4GI (10, 15, 19). Second, eIF4E may undergo functional inactivation through sequestering into complexes with repressor proteins, the 4E-BPs (26, 34).
FIG. 3.
Quantification of eIF4GI and eIF4GII in HeLa S10 cell extracts. (A) HeLa cell protein (40 μg) was analyzed side by side with known amounts of recombinant eIF4GI by SDS–8% PAGE and Western immunoblotting using anti-eIF4GI antibodies (insert). The signals of the eIF4GI standards were used to generate a calibration curve. The dashed line indicates the signal of the HeLa cell S10 fraction. (B) HeLa cell protein (80 μg) was analyzed together with known amounts of recombinant eIF4GII as described above, with anti-eIF4GII antibodies (insert). The signals of the eIF4GII standards were used to generate a calibration curve. The dashed line indicates the signal of the HeLa cell S10 fraction.
Taken together, our results show that after poliovirus (17) and HRV-14 infections (this report), cleavage of eIF4GII lags behind that of eIF4GI. Interestingly, this pattern might not be general for all picornavirus infections, as our preliminary data do not reveal such a difference after aphthovirus infection (45b). It is possible that aphthovirus L protease does not discriminate between eIF4GI and eIF4GII cleavage sites in a manner similar to poliovirus and HRV-14 2Apro proteases. Finally, it should be noted that although the cleavage of eIF4G might be the sole cause of the inhibition of host translation after rhinovirus infection, the contributions of other mechanisms, such as 4E-BP1 dephosphorylation (14) or changes in ionic conditions to favor virus versus host mRNA translation (9), cannot be excluded. We, however, were unable to detect any 4E-BP1 dephosphorylation in the course of HRV-14 or HRV-16 infections (45a).
Acknowledgments
We thank R. Rueckert for HRV-14 and F. G. Hayden, E. Arruda, and C. E. Crump for HeLa-I (Ohio) cells.
This work was supported by grants from the Medical Research Council of Canada to N.S. N.S. is a Medical Research Council Distinguished Scientist and a Howard Hughes Medical School International Scholar.
REFERENCES
- 1.Arruda E, Crump C E, Rollins B S, Ohlin A, Hayden F G. Comparative susceptibilities of human embryonic fibroblasts and HeLa cells for isolation of human rhinoviruses. J Clin Microbiol. 1996;34:1277–1279. doi: 10.1128/jcm.34.5.1277-1279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein H D, Sonenberg N, Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985;5:2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonneau A-M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A M, Kean K M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 6.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 7.Bovee M L, Lamphear B J, Rhoads R E, Lloyd R E. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology. 1998;245:241–249. doi: 10.1006/viro.1998.9172. [DOI] [PubMed] [Google Scholar]
- 8.Bovee M L, Marissen W E, Zamora M, Lloyd R E. The predominant eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco L, Smith A E. Sodium ions and the shut-off of host cell protein synthesis by picornavirus. Nature. 1976;264:807–809. doi: 10.1038/264807a0. [DOI] [PubMed] [Google Scholar]
- 10.Duncan R, Milburn S C, Hershey J W B. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 11.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 12.Etchison D, Fout S. Human rhinovirus 14 infection of HeLa cells results in the proteolytic cleavage of the p220 cap-binding complex subunit and inactivates globin mRNA translation in vitro. J Virol. 1985;54:634–638. doi: 10.1128/jvi.54.2.634-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 14.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goss D J, Carberry S E, Dever T E, Merrick W C, Rhoads R E. Fluorescence study of the binding of m7GpppG and rabbit globin mRNA to protein synthesis initiation factors 4A, 4E, and 4F. Biochemistry. 1990;29:5008–5012. doi: 10.1021/bi00473a002. [DOI] [PubMed] [Google Scholar]
- 16.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiremath L S, Webb N R, Rhoads R E. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- 20.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krausslich H G, Nicklin M J H, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 24.Lamphear B J, Yan R, Yang F, Waters D, Liebig H-D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 25.Liebig H-D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 26.Lin T A, Kong X M, Haystead T, Pause A, Belsham G, Sonenberg N, Lawrence J C. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd R E, Grubman M J, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J Virol. 1988;62:4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosenkis J, Daniels-McQueen S, Janovec S, Duncan R, Hershey J W B, Grifo J A, Merrick W C, Thach R E. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eucaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J Virol. 1985;54:643–645. doi: 10.1128/jvi.54.2.643-645.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlmann T, Pain V M, Wood W, Rau M, Morley S J. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E-binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 1997;16:844–855. doi: 10.1093/emboj/16.4.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neill R E, Racaniello V R. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the alpha subunit of eucaryotic initiation factor 2. J Virol. 1989;63:5069–5075. doi: 10.1128/jvi.63.12.5069-5075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 34.Pause A, Belsham G J, Gingras A C, Donzé O, Lin T A, Lawrence J C, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 35.Pause A, Méthot N, Svitkin Y V, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 37.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rau M, Ohlmann T, Morley S J, Pain V M. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 39.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 40.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 41.Roberts L O, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells: influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 44.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 45.Sonenberg N, Morgan M A, Merrick W C, Shatkin A J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Svitkin, Y. V., and A. Gradi. Unpublished observations.
- 45b.Svitkin, Y. V., A. Gradi, and G. Belsham. Unpublished observations.
- 46.Yan R, Rychlik W, Etchison D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 47.Ziegler E, Borman A M, Deliat F G, Liebig H D, Jugovic D, Kean K M, Skern T, Kuechler E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/s0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 48.Ziegler E, Borman A M, Kirchweger R, Skern T, Kean K M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]