Abstract
EZH1, a polycomb repressive complex-2 component, is involved in a myriad of cellular processes. EZH1 represses transcription of downstream target genes through histone 3 lysine27 (H3K27) trimethylation (H3K27me3). Genetic variants in histone modifiers have been associated with developmental disorders, while EZH1 has not yet been linked to any human disease. However, the paralog EZH2 is associated with Weaver syndrome. Here we report a previously undiagnosed individual with a novel neurodevelopmental phenotype identified to have a de novo missense variant in EZH1 through exome sequencing. The individual presented in infancy with neurodevelopmental delay and hypotonia and was later noted to have proximal muscle weakness. The variant, p.A678G, is in the SET domain, known for its methyltransferase activity, and an analogous somatic or germline mutation in EZH2 has been reported in patients with B-cell lymphoma or Weaver syndrome, respectively. Human EZH1/2 are homologous to fly Enhancer of zeste (E(z)), an essential gene in Drosophila, and the affected residue (p.A678 in humans, p.A691 in flies) is conserved. To further study this variant, we obtained null alleles and generated transgenic flies expressing wildtype [E(z)WT] and the variant [E(z)A691G]. When expressed ubiquitously the variant rescues null-lethality similar to the wildtype. Overexpression of E(z)WT induces homeotic patterning defects but notably the E(z)A691G variant leads to dramatically stronger morphological phenotypes. We also note a dramatic loss of H3K27me2 and a corresponding increase in H3K27me3 in flies expressing E(z)A691G, suggesting this acts as a gain-of-function allele. In conclusion, here we present a novel EZH1 de novo variant associated with a neurodevelopmental disorder. Furthermore, we found that this variant has a functional impact in Drosophila.
Keywords: Drosophila melanogaster, EZH1, E(z), H3K27-trimethylation, rare undiagnosed disease, polycomb repressive complex 2, patterning defect, missense, developmental delay
Human EZH1 has not been previously linked to human Mendelian disease. Here, Jangam, Briere, Jay et al. describe an individual with a de novo missense change in EZH1. An identical missense substitution in the paralog EZH2 has also been found in human Weaver syndrome. While Drosophila null E(z) mutants are lethal, the authors use rescue and overexpression experiments to demonstrate that this EZH1 substitution appears to have biochemical gain-of-function characteristics.
Introduction
EZH1 (Enhancer of Zeste, homolog 1), encodes an enzyme with Histone-lysine N-methyltransferase activity, which is a component of the polycomb repressive complex 2 (PRC2) (Jacobs and van Lohuizen 1999; Shen et al. 2008). The PRC2 plays a crucial role in compacting and maintaining heterochromatin resulting in gene repression through methylation of Histone 3 (H3) at lysine 27 (K27) (Jacobs and van Lohuizen 1999; Kuzmichev et al. 2002; Margueron and Reinberg 2011). H3K27 trimethylation (H3K27me3) leads to the reorganization of chromatin and the resultant repression of downstream genes (Rea et al. 2000; Bannister et al. 2002). The H3K27me3 mark is recognized by polycomb repressive complex 1, which further modifies the histone tail to repress gene expression (Luo et al. 2021). The Polycomb (Pc) group (PcG) genes were initially identified in the fruit fly Drosophila melanogaster and found to repress expression of HOX genes in the antennapedia and bithorax complex to maintain anterior to posterior segmentation and thus direct proper patterning of the animal (Lewis 1978). Importantly, PcG repressor genes function antagonistically to the gene-activating trithorax (trx) group of genes to regulate HOX gene expression (Schuettengruber and Cavalli 2009). HOX genes are evolutionarily conserved across multicellular organisms and, in humans, maintain cell identity to mediate regional patterning along the body axis, including the nervous system (Mark et al. 1997; Gonçalves et al. 2020). The PRC2 is comprised of multiple proteins, including the histone methyltransferase EZH1 or its paralog EZH2 (Enhancer of Zeste, homolog 2), EED (Embryonic Ectoderm Development) that functions as a scaffolding protein that also binds to H3K27me3, SUZ12 (Suppressor of Zeste 12) that supports complex formation, and several accessory histone binding proteins including RBBP7 and RBBP4 (Retinoblastoma binding protein 7 and 4, a.k.a RbAp46 and RbAp48) (Margueron and Reinberg 2011; Cao et al. 2014; Lee et al. 2022). In humans, EZH1 and its paralog EZH2 share 63% amino acid identity (Yu et al. 2019) and can function redundantly in the PRC2 complex (Kook et al. 2017).
EZH1 and EZH2 are homologous to the fly Enhancer of zeste (E(z)) gene. EZH1 and E(z) share 55% amino acid identity and 70% similarity (Abel et al. 1996). EZH1 is involved in the di- and trimethylation of H3K27 through its SET domain (Shen et al. 2008; Hidalgo and Gonzalez 2013). The SET domain consists of 130–140 amino acids and was first characterized in the fly proteins Su(var)3-9, E(z), and Trithorax, thus named the SET domain (Rastelli et al. 1993; Jones et al. 1998). EZH1 and E(z) have high homology in the SET domain with 79% identity and 89% similarity, suggesting a conserved function between species (Abel et al. 1996). Indeed, SET domain-containing genes have been among the prominent genes identified by classic Drosophila studies on position-effect variegation, which is a phenotypic readout for changes in the euchromatin and heterochromatin border (Dillon et al. 2005; Elgin and Reuter 2013; Herz et al. 2013). Complete loss of E(z) in flies results in early pupal lethality (Jones and Gelbart 1990). Clonal analysis has been used to identify developmental processes that are regulated by E(z). For example, mutant clones of E(z) in the third instar larval brain result in reduced mitotic activity impacting neuroblast size (Bello et al. 2007). In the adult brain, loss of E(z) results in misguided axons and/or ectopic glia-like cells (Wang et al. 2006).
Human Mendelian disorders have been reported for multiple members of the PRC2 complex (Deevy and Bracken 2019). Cohen–Gibson syndrome (MIM #617561) is an overgrowth syndrome with neurodevelopmental abnormalities and dysmorphic features caused by de novo missense variants in the EED gene (Cohen and Gibson 2016). These variants disrupt EED-EZH1/2 protein interactions (Imagawa et al. 2017). Imagawa–Matsumoto syndrome (MIM #618786) is an overgrowth syndrome caused by variants in SUZ12 (Imagawa et al. 2017), and EZH2 is responsible for Weaver syndrome (MIM #277590), an overgrowth and developmental condition (Weaver et al. 1974).
While EZH2 and other members of the complex have been linked to diseases with overgrowth and developmental delay, EZH1 is not yet associated with human disease. EZH1/2 has the potential to function redundantly biochemically, however, their ability to modulate downstream gene expression to execute developmental pathways may be compromised upon loss of one paralog. Indeed, their functions in vivo are divergent (Shen et al. 2008). Knock-out of Ezh2 is lethal in mice (O’Carroll et al. 2001), while heterozygotes have increased growth and body weight, an interesting correlation to the human Weaver overgrowth syndrome (Béguelin et al. 2013). Ezh1 in mice is not essential and does not associate with obvious overgrowth, but it has been shown to be essential for hematopoietic stem cell maintenance (Shen et al. 2008; Hidalgo et al. 2012). This suggests that EZH1 and EZH2 have unique functions in vivo, thus resulting in distinct disorders upon dysregulation of protein function.
Gain-of-function mutations in EZH2 are also implicated in tumorigenesis and poor prognosis in cancer. EZH2 p.Y641N, p.Y641F, and p.A677G are among the most frequently found somatic mutations in diffuse large B-cell lymphoma and result in increased H3K27me3 (McCabe et al. 2012a). McCabe et al. (2012b) furthermore identified that inhibition of EZH2 results in a significant reduction in H3K27me3 in the context of B-cell lymphoma. Importantly, a de novo variant in EZH2 p.A677G (in a shorter isoform) or p.A682G (in a longer isoform) was reported in a patient with Weaver syndrome (Tatton-Brown et al. 2011; Cohen et al. 2016; Diets et al. 2018), suggesting that this variant as a somatic or germline mutation can cause cancer or a Mendelian disease, respectively. Interestingly, here we identify the analogous variant to EZH2 p.A677G in the paralog EZH1 (p.A678G) as a de novo germline variant in an individual with a neurodevelopmental phenotype. Here, we report the clinical findings of this case and further present the results of variant-specific functional studies using the D. melanogaster model organism to investigate the mechanism of the observed variant.
Materials and methods
Genome sequencing and analysis
Proband, parent, and sibling clinical quad genome sequencing were performed by HudsonAlpha Clinical Services Lab as part of the family's participation in the Undiagnosed Diseases Network (UDN) study. Genomic DNA was extracted from blood and, after quality control and fragmenting, sequenced using the Illumina HiSeqX platform to generate 150 bp paired-end reads. The reads were then aligned to GRCh37/hg19 (BWA-mem v0.7.12). After quality control steps, including removal of fragments mapping to multiple locations, duplicate fragments, and fragments with low-quality scores (SMAbamba v0.5.4, markdup, GATKv3.3), variant calling was performed (GATKv3.3). Variant annotation and prioritization were performed using CarpeNovo v6.0.1 and/or Codicem, a custom software analysis application. The pipeline used is expected to yield 40× coverage of 90–95% of the human reference genome.
Research-based analysis of the data by HudsonAlpha and by Brigham Genomic Medicine (Haghighi et al. 2018) independently identified the de novo EZH1 variant as a candidate. This variant was confirmed via dideoxy (Sanger) sequence analysis by HudsonAlpha.
Fly stocks
E(z)731 from Bloomington Stock Center #24470
w*; E(z)731 p{1xFRT.G}2A/TM6C,Sb1 Tb1.
E(z)63 kindly gifted by Dr. Richard Jones, University of Dallas, Texas: sc z1 wis; E(z)63 e11/TM6B, Tb Hu.
p{UAS-lacZ} gift from Hugo Bellen Lab.
p{Actin-GAL4} balanced over CyO-Tb was generated in our lab.
p{GawB}elav[C155] gift from Hugo Bellen Lab.
p{daughterless-GAL4} gift from Hugo Bellen Lab.
p{nubbin-GAL4} gift from Hugo Bellen Lab.
y w; ptub::E(z)WT/SM6a (generated in the lab).
y w; ptub::E(z)A691G/SM6a (generated in the lab).
Fly husbandry
Stocks were reared on standard fly food (water, yeast, soy flour, cornmeal, agar, molasses, and propionic acid) at room temperature (∼22°C) and routinely maintained. Unless otherwise noted, all flies used in experiments were grown in a temperature and humidity-controlled incubator at 25°C and 50% humidity on a 12-hr light/dark cycle.
Generation of E(z) transgenic flies
Generation of the UAS-E(z)WT and UAS-E(z)A691 transgenic flies (Supplementary Fig. 1): The original E(z)-cDNA construct—pGEX-2T{E(z)cDNA e32} was kindly gifted by Dr. Richard Jones, University of Dallas (Jones and Gelbart 1990). The attB-E(z) primers were designed to excise from pGEX-2T and clone into the pDONR-221 vector using the Gateway BP Clonase II Enzyme mix (Thermo Fisher-11789100). New England Biolabs NEBase changer was used to generate primers to create the p.A691G variant from the E(z) wildtype construct. Variant primers were prepared (Sigma-Aldrich), and the variant was generated with a Q5 mutagenesis (NEB-E0554S) as previously described (Tsang et al. 2016). After confirming variant conversion via Sanger sequencing, pENTR-221 E(z)cDNAWT and pENTR-221-E(z)A691G were cloned into the pGW.attB destination vector with Gateway LR Clonase II Enzyme mix (Thermo Fisher-11791020). Sanger sequence-verified wildtype and variant constructs were then microinjected in ∼200 embryos at the VK00037 attP docking site to generate pBac{UASg-E(z)WT}VK00037 and pBac{UASg-E(z)A691G}VK00037 transgenic lines.
attB-E(z) primers: (5′GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGAATAGCACTAAAGTGCCGC-3′ and 5′GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATCAAACAATTTCCATTTCACGCTCT-3′)
Q5-mutagenesis variant primers:
(5′-GTTGTGGATGgCACTCGGAAG-3′ and 5′-AAAATCGTTGTTCAGATTGAAAAGG-3′)
b) Generation of the E(z)A691 transgenic flies under the constitutively active tubulin promoter (Supplementary Fig. 2): pwmc{ptub:EGFP::E(z)} construct and flies were gifted by Dr. Leonie Ringrose, Professor, Humboldt-Universität zu Berlin, IRI Life Sciences, Berlin, Germany. Mutagenesis primers were designed with NEBase changer and Q5 mutagenesis was used to create the p.A691G variant from the wildtype construct (NEB-E0554S). Both the wildtype and variant constructs underwent restriction digestion with NotI (NEB-R3189S) and AvrII (NEB-R0174S) to remove the constructs from the pwmc vector. The destination vector was digested with NotI (NEB-R3189S) and XbaI (NEB-R0145S). Reference and variant constructs were then ligated into the pattB-Basler vector with the T4 DNA ligase (NEB M0202S) for injection. Sanger sequenced verified pattB{ptub:EGFP::E(z)WT}, and pattB{ptub:EGFP::E(z)A691G} were then microinjected in ɸc31 mediated VK00037 docking site embryos to generate pBac{ptub:EGFP::E(z)WT}VK00037 and pBac{ptub:EGFP::E(z)A691G}VK00037 transgenic flies.
Longevity assay
The ptub::E(z)WT and the ptub::E(z)A691G transgenic flies were crossed to y w to avoid any phenotypic interference because of the balancer—SM6a-CyO. The F1 flies were then raised at 25°C and transferred every 3–4 days. Rescued flies were also treated using the same technique. Any lethality observed was plotted in Prism software (Survival plot).
Bang sensitivity assay
Flies to be used were isolated 1–3 days post-eclosure and were single-housed in isolation vials until assessment. On the day of the trial, flies were transferred into an empty polystyrene vial with no food. The flies were vortexed at full speed (Fisher STD Vortex Mixer, Cat. No. 02215365) for 10 s and recovery times were recorded using a digital stopwatch. Bang assay was performed on a minimum of 20 flies.
Over-expression assay (assessment of lethality and morphological phenotypes)
Lethality and morphological phenotyping assays were performed by crossing GAL4 drivers as indicated in the text, using 5–10 virgin females crossed to a similar number of males. Parents were transferred into a new vial after every 3–5 days to collect multiple F1-progenies. Flies were collected after most pupae were eclosed, and the total number of flies was scored based on the presence or absence of balancers. For the lethality assessment, a minimum of 70 flies were scored. Viability was calculated by evaluating the number of observed progenies compared to the number of expected progeny based on the Mendelian ratio. Animals were classified as lethal if the O/E ratio was <0.15, and semilethality is classified as an O/E ratio <0.8. Morphological phenotypes were assessed only for animals lacking balancers, and phenotypes were noted if they appeared in >70% of the progeny.
Western blot assay
Western blots were performed using whole larvae, whole adult flies, or adult heads [5 whole larvae in 100 µL, 6 adult heads (n ≥ 3 males and 3 females) in 30 µL, or single whole adult flies in 30 µL of sample buffer] were used and transferred directly to ice-cold sample buffer (4× XT Sample Buffer Bio-Rad #1610791 + 10% Beta-mercaptoethanol + ddH2O). The larvae/whole flies/adult heads were homogenized for 1 min, transferred to ice, and boiled at 94°C for 5 min. The boiled homogenate was centrifuged for 5 min at 14,000 RPM to remove the debris. Protein (10 µL) was loaded on tris-glycine precast gels (4–20% precast TGX gels Bio-Rad #4561094). PVDF membrane was activated by applying 100% methanol for 2–3 s. The gels were transformed into the 0.2 µm activated PDVF membrane using the trans-blot turbo transfer unit. This membrane was then blocked using a 5% blocking solution (1× TBST with 0.1% TWEEN-20 and 2.5 g nonfat dry milk) for 1 h at room temperature with rotation. The membrane was then incubated overnight at 4°C with trimethyl-histone H3 (Lys27) (CST #9733, 1:2,000) or Histone H3 (CST #9715, 1:5,000) and E(z) (Agrisera: AS16 3935, 1:5,000) primary antibodies in the blocking solution. The membrane was washed at least 3 times with 0.1%TBST before the secondary antibody incubation—Goat anti-Rabbit IgG (H + L) secondary Antibody, HRP (Thermo Fisher #31460, 1:5,000 in the blocking solution). SuperSignal West Pico PLUS (34580, Thermo) was used to develop the membrane. Images of the developed blot were taken in the Chemiblot imager and analyzed using the ImageJ software.
Drug feeding assay
ptub::E(z)WT and ptub::E(z)A691G transgenic flies were supplemented with 2 EZH2 inhibitors, GSK126 (MedChemExpress: HY-13470) and EPZ-6438 (Tazemetostat) (Selleckchem: 1403254-99-8) and feeding assays were performed both on adults or and on developing animals.
Female fertility testing assay
We crossed 3–4 days old virgins of the ptub::E(z)WT; E(z)63/E(z)731 and ptub::E(z)A691G; E(z)63/E(z)731 with 3–4 days old males of [y w] and quantified the resulting progenies produced.
Websites accessed
DenovoDB: https://denovo-db.gs.washington.edu/denovo-db/index.jsp
UniProt consortium: https://www.uniprot.org/
GETx: https://gtexportal.org/home/
NEBase changer: https://nebasechanger.neb.com/
BrainSpan Developmental Transcriptome: https://brainspan.org/static/home
Results
Case presentation
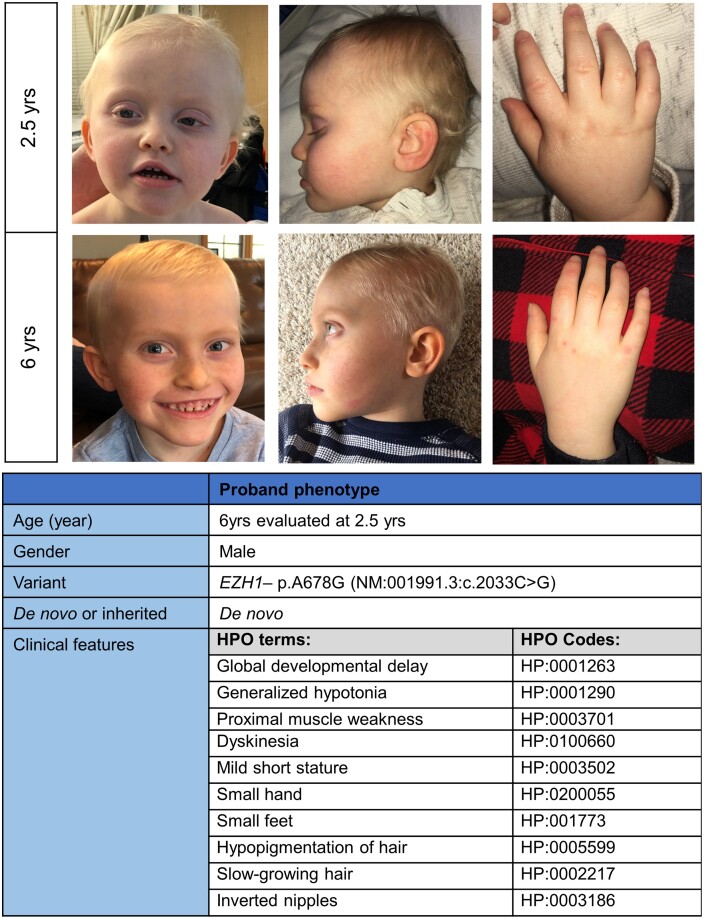
The patient is a 6-year-old boy initially evaluated in the UDN (Splinter et al. 2018) at age 2.5 years (31 months) (Fig. 1). He presented with severe global developmental delay, proximal muscle weakness, intermittent exotropia, and mild dysmorphic features. He has brachycephaly and a flattened occiput. His eyes are mildly deep-set. He has mild prognathia, widely spaced teeth, and torus palatinus. His ears are mildly low set, with thickened helices and underdeveloped tragus and antitragus bilaterally. When evaluated at 31 months of age, he had inverted nipples and a prominent suprapubic fat pad. In addition, his skin and particularly his hair were felt to be unusually fair for his family. The developmental delay included gross motor milestone delay such as achieving moderate head control at 18 months of age, independent sitting at 36 months, and at 6 years he is not able to pull to stand. In fine motor areas, he is unable to use utensils at age 6. Speech delay is also present and at age 6 he is not using 2-word sentences. He also has a number of neurological abnormalities such as considerable difficulty in initiating saccades, frequent turning of the head to track objects, low axial muscle tone, and he exhibits frequent complex dyskinetic movements of the upper extremities as well as occasional jerking movements (see Supplementary Text for entire case history). He has not had any seizures to date. His head circumference and weight have consistently been in the average range. His height was low-normal at birth and in early childhood, but he now has short stature (−2.3 SD).
Fig. 1.
Clinical information for EZH1 proband. Photos (top row- 2.5 years; bottom row- 6 years), and clinical features (bottom table) of the patient at 31 months of age and approximately 5 years of age (top row right). Craniofacial features include flattened occiput, brachycephaly, mild prognathia, mildly deep-set eyes, widely spaced teeth, mildly low-set ears with mildly thickened helices and underdevelopment of the tragus and antitragus. His fingers are tapered, and his coloring, particularly his hair, is unusually fair for his family. HPO, Human Phenotype Ontology.
He had extensive imaging laboratory evaluations, including brain magnetic resonance imaging studies at 11 months and 3.5 years. These showed mild prominence of the ventricular system and extra-axial spaces but were otherwise unremarkable. Electroencephalograms at 9 months and 4 years of age showed mild to moderate background slowing, indicative of mild diffuse cerebral dysfunction. Electromyography and nerve conduction velocity studies at 10 months of age were unremarkable, showing no evidence of neuropathy or myopathy. Muscle biopsy (Vastus lateralis) revealed moderate excess variation in fiber size with smaller type I fibers than type II fibers. Electron microscopy of the muscle did not reveal any diagnostic abnormalities but noted clusters of mitochondria and occasional large mitochondria. Extensive mitochondrial, metabolic, and genetic testing was negative. Clinical quad exome sequencing (a clinical test done on proband, parents, and sibling) did not reveal pathogenic or likely pathogenic variants that would explain the phenotype; however, subsequent research-based analysis (a research activity aimed at identifying new disease genes) of the exome data highlighted a novel de novo missense variant (not reported in the parents or sibling but in our proband) on the clinical report because it was found in a gene not previously associated with disease risk. This variant in EZH1 [NM_001991.3:c.2033G > C (p.A678G)], was Sanger confirmed.
EZH1 candidate disease-causing variant
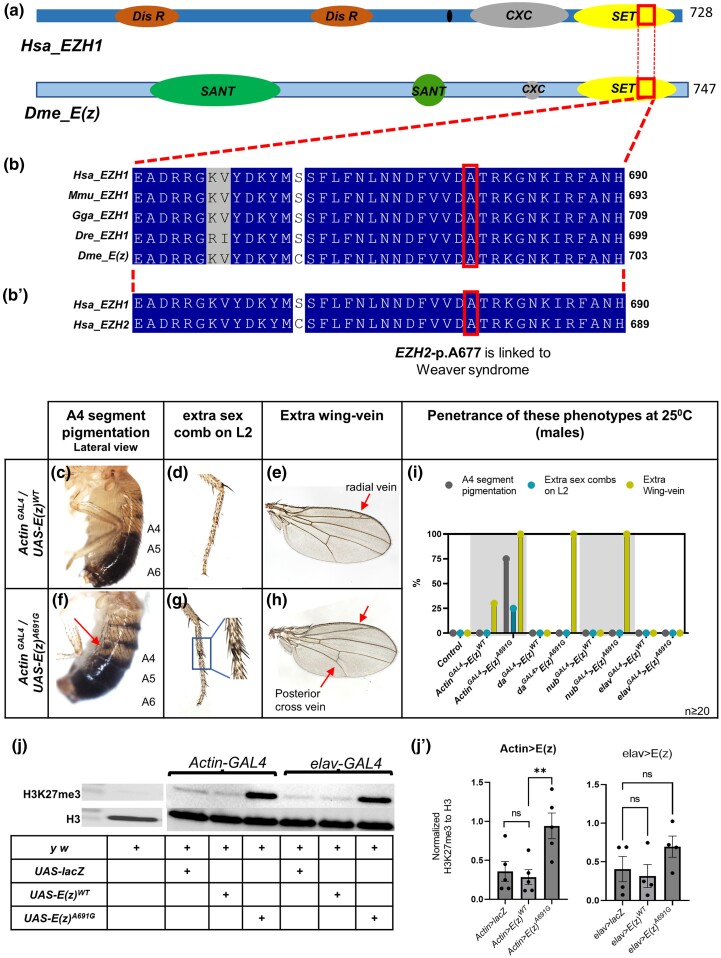
The de novo missense variant in EZH1 is considered a strong novel disease candidate for this case for multiple reasons. First, EZH1 is highly expressed in neuronal cells (Uhlén et al. 2015). Second, the variant is not found in control populations in the Genome Aggregation Database (gnomAD), a database generally used as a control for rare disease studies (Karczewski et al. 2020). EZH1 does not have a known relationship to human disease, and within the gnomAD database has a probability of loss-of-function intolerance (pLI) of 0.04, indicating no constraint in control populations for loss-of-function variants. However, the missense Z-score of 4.2 indicates a missense constraint in gnomAD control populations (Karczewski et al. 2020). One preliminary interpretation could be that missense changes are therefore under higher constraint than loss-of-function in this gene. Third, computational predictions suggest the missense variant is deleterious (SIFT = 0.0) (Ng and Henikoff 2001) and probably damaging (Polyphen = 1) (Adzhubei et al. 2010), with a CADD score of 32 also supporting a deleterious prediction (Rentzsch et al. 2021). Forth, the p.A678G variant is located within the SET domain of EZH1, and the region and residue are highly conserved across species (Fig. 2, a and b). Fifth, the equivalent variant (p.A677G in EZH2, aligned with p.A678G in EZH1, Fig. 2b′) is a known disease-causing variant in Weaver syndrome (p.A677G {isoform EZH2-Q15910-1} or p.A682G {isoform EZH2-Q15910-2}) (Diets et al. 2018; UniProt Consortium 2022). Moreover, another variant impacting the same amino acid has also been identified as disease-causing in Weaver syndrome (EZH2 p.A677T or EZH2 p.A682T) (Tatton-Brown et al. 2011, 2013). Taken together the EZH1 variant is a strong candidate variant for this case and was therefore submitted to the UDN Model Organisms Screening Center (MOSC) and evaluated in Drosophila (Baldridge et al. 2021).
Fig. 2.
Conservation, phenotypes, and methylation pattern for UAS-E(z) variant. a) Domain structure schematic of human EZH1(Hsa_EZH1) and Drosophila melanogaster E(z)(Dmel_E(z)) b-b′) Conservation of EZH1-pA678 across species and also in human paralogs EZH1 and EZH2 (The structures are derived from UniProt) [Hsa_EZH1: Homo Sapiens EZH1; Dis R: Disordered Region; SANT: SANT-Myb- Protein binding Domain, CXC: cysteine-rich DNA binding domain; Dme_E(z): D. melanogaster Enhancer of zeste [E(z)]. c–i) Overexpression assay of UAS-E(z) constructs with different GAL4 lines. The progenies of GAL4 driven UAS-E(z) flies are evaluated for patterning defects, such as A4 segment pigmentation (lateral view), presence of extra sex combs on the second pair of legs (L2) and extra wing veins. Actin-GAL4/UAS-E(z)WT flies are represented in c–e. The phenotypes observed in Actin-GAL4/UAS-E(z)A691G flies are represented in f–h. The penetrance of patterning variation seen in male progeny of different ubiquitous or tissue-specific GAL4-driven UAS-E(z) flies is represented in i (n ≥ 20). The extra wing-vein patterning defect is also present in females. j–j′): Western blot assay for H3K27me3 of UAS-E(z)WT and UAS-E(z)A691G overexpression with Actin-GAL4 and elav-GAL4. Trimethylation of H3K27 was evaluated in the Actin-GAL4 and elav-GAL4 driven UAS-E(z)WT and UAS-E(z)A691G adult progenies (j). j′) is the graphical representation of the H3K27me3 values normalized to H3 values. ActinGAL4 P = 0.0087, elavGAL4 P = 0.1091. UAS-lacZ was used as a positive control. Unpaired t-tests determined P-values.
EZH1/EZH2 are human homologs of Drosophila enhancer of Zeste E(z)
The EZH1 variant was accepted for modeling in the MOSC Fly Core, and previous model organism phenotypes were reviewed using MARRVEL (Wang et al. 2017, 2019a, 2019b). Human EZH1 and EZH2 have a single ortholog, E(z), in D. melanogaster. E(z) has greater sequence similarity to EZH2 than to EZH1 (DIOPT scores 14/15, and 11/16, respectively) (Hu et al. 2011). Based on the information collected from MARRVEL and FlyBase (Jenkins et al. 2022) we obtained genetic resources from prior Drosophila E(z) studies. We obtained E(z)731 (Müller et al. 2002) and E(z)63 (Carrington and Jones 1996), both of which are null alleles. Strong loss-of-function alleles of E(z) in flies have been shown to be lethal (Steffen et al. 2013). The de novo p.A678G variant in EZH1 is found within a highly conserved region within a stretch of the SET domain, corresponding to E(z)-p.A691G (E(z)A691G) in Drosophila (Fig. 2b).
E(z)A691G variant induces patterning defects and hyper H3K27 trimethylation in Drosophila
We generated the Drosophila E(z) cDNA wildtype and variant transgenic flies under an upstream activation sequence (UAS) containing promoter (Brand and Perrimon 1993). The UAS-E(z)WT and UAS-E(z)A691G fly constructs allow the expression of these transgenes in the presence of a GAL4 transcriptional activator protein which could be expressed in different tissues (Supplementary Fig. 1). Ubiquitous overexpression with Actin-GAL4 of UAS-E(z)A691G resulted in an array of homeotic abdominal, leg, and wing patterning defects in adult flies compared to UAS-E(z)WT. Expression of UAS-E(z)WT results in appropriate patterning of the abdominal segments (Fig. 2c), normal bristle formation on the second pair of legs (L2) (Fig. 2d), and mild ectopic wing vein formation on the radial vein (Fig. 2e). In contrast, when UAS-E(z)A691G is expressed, abnormal pigmentation on the A4 lateral abdominal segment is present (Fig. 2f), ectopic sex combs are present on the L2 leg pair (Fig. 2g), and the ectopic wing vein patterning defect is more pronounced with ectopic veins present on the radial vein and posterior cross-vein (Fig. 2h). These phenotypes are not fully penetrant with 30% of UAS-E(z)WT having ectopic wing veins whereas 100% of UAS-E(z)A691G adult flies are affected with ectopic wing vein formation; A4 pigmentation is not present in UAS-E(z)WT but seen in 75% of UAS-E(z)A691G; and extra sex combs on L2 are not present in UAS-E(z)WT but are seen in 25% of UAS-E(z)A691G (Fig. 2i). Overexpression of UAS-E(z)WT with a weaker ubiquitous driver (daughterless-GAL4), wing pouch driver (nubbin-GAL4), and neuronal driver (elav-GAL4) did not result in any obvious phenotypes, but expression of UAS-E(z)A691G resulted in fully penetrant ectopic wing veins with daughterless- and nubbin-GAL4 expression (Fig. 2i).
Next, we evaluated H3K27me3 in adult flies expressing E(z) under the control of ubiquitous (Actin-) and neuronal-specific (elav-) GAL4 drivers. H3K27 methylation is an obvious target in these flies because E(z) is the only methyltransferase capable of H3K27 methylation (Cao et al. 2002; Stepanik and Harte 2012). We evaluated H3K27me3 with western blot assays and noted significantly increased H3K27me3 in Actin-GAL4/UAS-E(z)A691G flies (Fig. 2j-j′). We did not note a significant change in H3K27me3 using elav-GAL4. Interestingly the overexpression of reference and variant E(z) using elav-GAL4 driver also did not show the dramatic phenotypes as seen in ubiquitous expression. In conclusion, expression of the p.A691G variant E(z) leads to increased H3K27me3 and a number of patterning phenotypes in adult flies that are not seen when wildtype E(z) is overexpressed.
E(z)A691G rescues E(z) heteroallelic lethality with homeotic patterning defects
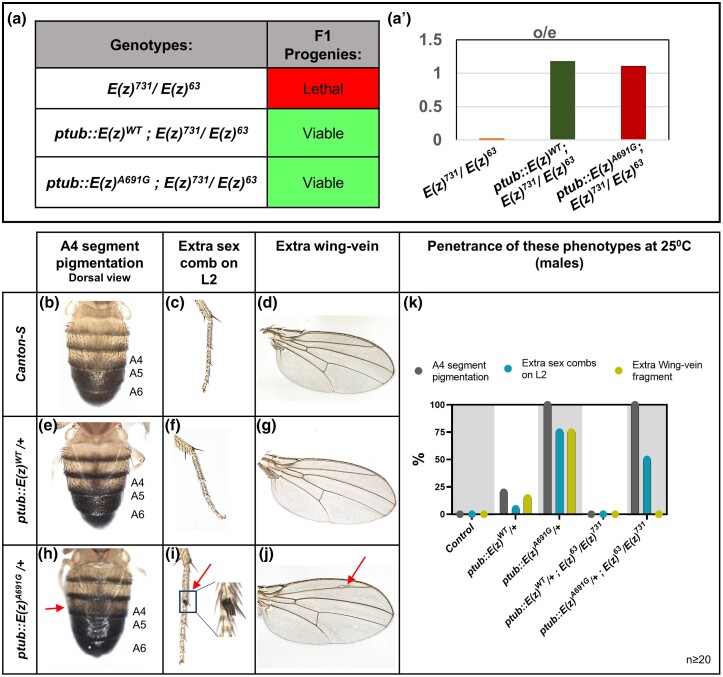
Previous work generated an EGFP tagged E(z)-cDNA construct that is under the control of the tubulin promoter ptub::EGFP::E(z) (Steffen et al. 2013). While the transgene is ubiquitously expressed under the tubulin promoter, the protein is likely expressed at a different level compared to ubiquitous overexpression with the GAL4/UAS system. Steffen et al. showed that the heteroallelic lethality of E(z) mutants can be rescued with this ptub::EGFP::E(z) construct. We obtained this construct and used site-directed mutagenesis to create the p.A691G variant (analogous to EZH1 p.A678G) and generated transgenic animals expressing wildtype and variant constructs (Supplementary Fig. 2). We used these constructs to test whether the introduction of the p.A691G variant in this construct would affect its ability to rescue the null alleles previously obtained [E(z)731 (Müller et al. 2002) and E(z)63 (Carrington and Jones 1996)]. These alleles cause homozygous lethality and fail to complement each other, thus resulting in heteroallelic lethality (Fig. 3a). Interestingly, we found that both the transgenic flies ptub::E(z)WT or ptub::E(z)A691G can fully rescue lethality in a heteroallelic background (E(z)731/E(z)63) (Fig. 3a, a′). Moreover, the Mendelian ratios of inheritance are similar when either ptub::E(z)WT or ptub::E(z)A691G constructs are expressed in the E(z) null background (Fig. 3a′). We also noted no defects in fertility in the rescued animals using ptub::E(z)WT or ptub::E(z)A691G. Therefore, these results indicate that E(z)A691G is unlikely to be a loss-of-function allele.
Fig. 3.
Phenotypes and methylation pattern for E(z)cDNA constructs under tubulin promotor (ptub). a–a′) Genetic crosses for the rescue of heteroallelic lethality: Under constitutively active tubulin promotor (ptub) with EGFP-tag at the N terminal, the ptub::E(z)WT and the ptub::E(z)A691G constructs are both able to rescue the E(z)731/E(z)63 heteroallelic lethality in a. a′) Graphical representation of the observed/expected ratio of the F1 progenies in a. For E(z)731/E(z)63 total F1 = 94, o/e = 0/47; for ptub::E(z)WT; E(z)731/E(z)63 total F1 = 411, o/e = 97/82 = 1.18; for ptub::E(z)A691G; E(z)731/E(z)63 total F1 = 167, o/e = 37/33 = 1.10. b–k) Phenotypical assessment of males of ptub::E(z) transgenic lines: males were observed for A4 segment pigmentation (Dorsal view), presence of extra sex combs on the second pair of legs (L2), and extra wing veins. Control images are from the Canton-S strain in b–d. ptub::E(z)WT/+ flies are represented in e–g. The phenotypes observed in ptub::E(z)A691G/+ flies are represented in h–j. Penetrance of the phenotypes seen in 20 different transgenic males in the fly wildtype background and the fly null background were scored and graphically represented in k.
Similar to what is seen upon GAL4-induced overexpression, the constitutively active ptub::E(z)A691G allele also induces A4 abdominal segment defects (clearly visible on the dorsal side), ectopic sex comb expression on L2 legs, and ectopic wing veins. Wildtype phenotypes were evaluated using the laboratory control strain Canton-S (Fig. 3, b–d). The vast majority of ptub::E(z)WT expressing animals appear wildtype (Fig. 3, e–g), with a few animals exhibiting abnormal A4 segment pigmentation, extra L2 sex combs, and/or extra wing vein. In flies expressing ptub::E(z)A691G, we observed highly penetrant ectopic pigmentation of the dorsal A4 abdominal segment, ectopic sex combs on the L2 leg pair, and ectopic wing veins (Fig. 3, h–j). A much more penetrant phenotype is observed upon expression of ptub::E(z)A691G with 100% of flies having A4 segment pigmentation, 75% with extra sex combs on L2, and 75% with an extra wing vein fragment (Fig. 3k). Moreover, the penetrance of the A4 segment pigmentation and the L2 extra sex combs were not significantly affected when either ptub::E(z)WT or ptub::E(z)A691G was expressed in the E(z) null background (Fig. 3k). This suggests that although the constructs rescue lethality, the removal of endogenous E(z) does not affect the A4 segment pigmentation or extra sex comb phenotypes produced by the E(z)A691G transgene.
E(z)A691G causes hyper trimethylation of H3K27
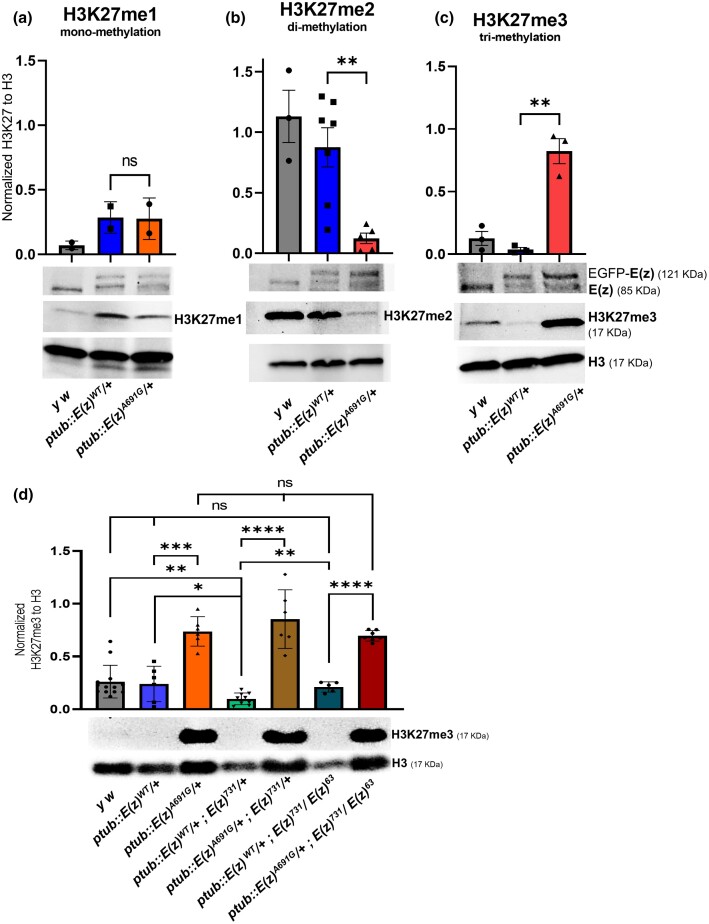
Given the role of E(z) in H3K27me3 and the phenotypes observed in the p.A691G variant lines, we performed histone H3K27 methylation profiling of heterozygous ptub::E(z)WT and ptub::E(z)A691G third instar larvae. We examined mono-, di-, and tri-H3K27 methylation using western blot analysis. Expression of heterozygous ptub::E(z)WT does not induce significant changes in mono- (me1), di- (me2), or me3, however the expression of ptub::E(z)A691G results in a significant decrease in H3K27me2 (P = 0.0035) and increase in H3K27me3 (P = 0.0015) (Fig. 4, a–c). We next compared the levels of E(z) to test the hypothesis that the p.A691G might affect total protein levels. Using an anti-E(z) antibody (Agrisera AS16 3935) we determined that the heterozygous ptub::E(z)WT and heterozygous ptub::E(z)A691G have roughly equivalent levels of the tagged (121 kDa) E(z) (Fig. 4, a–c). In summary, the E(z)A691G leads to a dramatic reduction in di-methylation and a corresponding increase in trimethylation of H3K27, consistent with it being a gain-of-function allele.
Fig. 4.
Methylation profiling of the ptub::E(z) transgenic larvae and adults. a–c) Methylation Profiling of H3K27 in larvae: larvae of the heterozygous ptub::E(z)WT and ptub::E(z)A691G in wildtype fly background are used for this western blot assay. Bands for endogenous E(z) (85 kDa) and N-terminal-EGFP tagged E(z) (121 kDa) (anti-E(z) antibody—Agrisera: AS16 3935) from these constructs can be seen in the transgenic lines. In the ptub::E(z)A691G variant, mono-methylation-H3K27me1 levels did not show any significant change when compared to the ptub::E(z)WT/+ wildtype (a, P = 0.9525). When assayed for di-methylation-H3K27me2, the levels are significantly reduced in the variant compared to the wildtype (b, P = 0.0035). Whereas, when assayed for trimethylation-H3K27me3, the levels are significantly increased in the variant compared to the wildtype (c, P = 0.0015). d) H3K27-trimethylation assay in adults: In adults, the H3K27me3 levels in the variant ptub::E(z)A691G is significantly increased compared to ptub::E(z)WT (P = 0.0002). In the sensitized background, when one copy of E(z) is removed using an allele (E(z)731), the variant continues to show hyper trimethylation as compared to WT (P < 0.0001). In the heteroallelic null background, the variant maintains hyper-trimethylation when compared with WT (P < 0.0001). Unpaired t-tests determined P-values.
In adult flies, expression of ptub::E(z)WT [ptub::E(z)WT/+] again does not impact H3K27me3, but expression of the variant ptub::E(z)A691 [ptub::E(z)A691G/+] leads to an approximate 2-fold increase in H3K27me3 (Fig. 4d, P = 0.0002). When we remove one functional copy of E(z), using E(z)731, this difference is more pronounced. We refer to this as a “sensitized background.” When the wildtype transgenic construct in a sensitized background was evaluated [ptub::E(z)WT/+; E(z)731/+] with control fly strain [y w], it appeared to have slightly reduced trimethylation [P = 0.073]. But the variant ptub::E(z)A691G in the same background [ptub::E(z)A691G/+; E(z)731/+] continues to cause increased H3K27me3 compared to wildtype (Fig. 4d, P < 0.0001). Additionally, as both the transgenes rescue the E(z)731/E(z)63 null lethality, we quantified the difference in H3K27me3 levels in these flies. The wildtype [ptub::E(z)WT/+; E(z)731/E(z)63] shows no significant increase in trimethylation levels compared to that of a laboratory control strain [y w], but the variant [ptub::E(z)A691G/+; E(z)731/E(z)63] continues to show significantly increased trimethylation compared to wildtype (Fig. 4d, P < 0.0001). Expression of the variant transgene ptub::E(z)A691G maintains hyper-trimethylation regardless of the E(z) genetic background. The ptub::E(z)A691G construct continues to induce more trimethylation as wildtype E(z) copies are removed, indicating this phenotype is not suppressible and suggests a gain-of-function mechanism for the E(z)A691G variant in this biochemical assay.
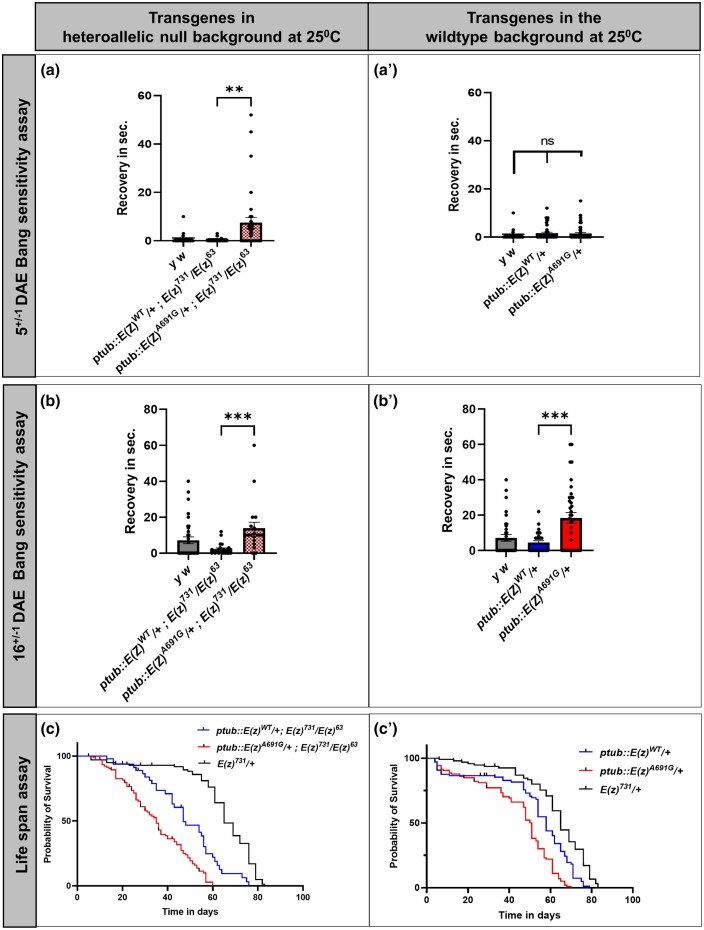
E(z)A691G-rescued flies display bang sensitivity and shortened lifespan
During normal fly husbandry, we observed a phenotype reminiscent of bang sensitivity (abnormally slow recovery during transfers of flies) in the ptub::E(z)A691G animals. We decided to quantify and study this phenotype in more detail. We performed a classical bang sensitivity assay in young adult flies at 5 days after eclosion (DAE) and in aged adults at 16 DAE (±1 days). Control [y w] flies only begin to exhibit bang-sensitive phenotypes after 16 DAE in the condition we tested. When we tested the flies in which the E(z) null animals were rescued by E(z) transgenes expressed using a tubulin promotor at 5 DAE, the variant ptub::E(z)A691G [ptub::E(z)A691G/+; E(z)731/E(z)63] shows significant bang-sensitivity when compared to ptub::E(z)WT [ptub::E(z)WT/+; E(z)731/E(z)63] (Fig. 5a, P = 0.0041) or control [y w] animals. To test if these transgenes have any effect on bang sensitivity by themselves at this early age, the same assay was performed in which the E(z) was overexpressed in a wildtype background. At 5 DAE, neither the ptub::E(z)A691G [ptub::E(z)A691G/+] nor the ptub::E(z)WT [ptub::E(z)WT/+] flies exhibit bang sensitivity compared to y w (Fig. 5a′, p = ns). When the bang sensitivity assay was performed at 16 DAE in the variant in the fly null background ptub::E(z)A691G [ptub::E(z)A691G/+; E(z)731/E(z)63] again shows significant bang sensitivity compared to ptub::E(z)WT [ptub::E(z)WT/+; E(z)731/E(z)63] (Fig. 5b, P = 0.0004). Interestingly, at 16 DAE, transgenic ptub::E(z)A691G flies in a wildtype background [ptub::E(z)A691G/+] are also bang sensitive when compared to ptub::E(z)WT [ptub::E(z)WT/+] (Fig. 5b′, P = 0.0002).
Fig. 5.
Bang sensitivity and life span of the transgenic flies. a–a′) Bang sensitivity assays were performed at 5 DAE: ptub::E(z)A691G transgenic flies in the heteroallelic null background show bang sensitivity whereas ptub::E(z)WT flies are not bang sensitive (a, P = 0.0041). When bang sensitivity assays are carried out in the wildtype background, ptub::E(z)A691G does not produce a significantly different bang sensitive phenotype than the wildtype ptub::E(z)WT (a′, P = ns). Unpaired t-tests determined P-values. b–b′) Bang sensitivity performed at 16 DAE: ptub::E(z)A691G transgenic flies in the heteroallelic null background continue to show bang sensitivity at 16 DAE when compared to ptub::E(z)WT (b, P = 0.0004). In the wildtype background, the ptub::E(z)A691G shows bang sensitivity when compared to ptub::E(z)WT (b′, P = 0.0002). Unpaired t-tests determined P-values. c–c′) Life span assay: ptub::E(z)A691G transgenic flies in the heteroallelic null background live shorter than the ptub::E(z)WT (c, Median Survival: E(z)WT = 47, E(z)A691G = 35, E(z)731 = 65, P < 0.0001). In the fly wildtype background, ptub::E(z)A691G transgenic flies again show a similar life span defect when compared to ptub::E(z)WT (c′, Median Survival: E(z)WT = 58, E(z)A691G = 51, E(z)731 = 65, P < 0.0001). Curve comparison was used to determine P-values.
Reduced lifespan is often observed in flies that are bang-sensitive (Reynolds 2018). When we quantified the overall lifespan upon expression of the ptub::E(z)WT transgene in the E(z) null background, [ptub::E(z)WT/+; E(z)731/E(z)63] flies show reduced longevity when compared to heterozygous null E(z)731[E(z)731/+] flies. The lifespan of flies expressing the ptub::E(z A691G transgene in a E(z) null background [ptub::E(z)A691G/+; E(z)731/E(z)63] is even further reduced (Fig. 5c, P < 0.0001). Additionally, when the same transgenes were tested in the fly wildtype background, the ptub::E(z)A691G [ptub::E(z)A691G/+] flies show a significant reduction in life span compared to ptub::E(z)WT [ptub::E(z)WT/+] and E(z) heterozygous [E(z)731/+] animals (Fig. 5c′, P < 0.0001).
Previous research determined that pharmacological inhibition of EZH2 (the paralog of EZH1) results in a repression of H3K27me3 (McCabe et al. 2012b; Harding et al. 2018). Since the fly E(z) is orthologous to EZH2, we attempted to suppress the bang sensitivity and increased H3K27me3 phenotypes induced by the expression of p.A691G pharmacologically. Flies were supplemented with two EZH2 inhibitors, GSK126 and EPZ-6438 (Tazemetostat), and feeding assays were performed both on adults or on developing animals. Adults expressing ptub::E(z)A691G/+ at 13 DAE exhibit bang-sensitive phenotypes. When adult ptub::E(z)A691G/+ flies are fed with 100 or 500 mM of GSK126 or EPZ-6438, we observe a decreasing trend in bang sensitivity which is not statistically significant (Supplementary Fig. 3a). In ptub::E(z)WT/+ flies, we observe no bang sensitivity at 13 DAE. When these adult flies are fed with 100 or 500 mM GSK126, we do not observe any differences. We found that 500 mM of EPZ-6438 induces slightly increased bang sensitivity, which is not statistically significant (Supplementary Fig. 3a). To evaluate the effect on trimethylation, we quantified H3K27me3 compared to overall H3 in adults at 8 DAE. Neither GSK126 nor EPZ-6438 is capable of suppressing the increased H3K27me3 induced by expression of ptub::E(z)A691G/+ in flies (Supplementary Fig. 3b). When fed with GSK126 or EPZ-6438 throughout development, we again observed no decrease in H3K27me3 levels upon expression of ptub::E(z)A691G/+ at 2 DAE (Supplementary Fig. 3c). Increasing the concentration of GSK126 and EPZ-6438 throughout development to 1,000 mM causes pupal lethality in both ptub::E(z)WT/+ and ptub::E(z)A691G/+. EZH2 inhibitors have been previously shown to suppress E(z) mediated H3K27me3 in flies (Xia et al. 2016). However, in this overexpression study, inhibitors of EZH2 do not appear to affect the trimethylation differences we observe in the E(z)A691G.
Discussion
Here we report a de novo EZH1 missense variant identified in an individual with severe global developmental delay, proximal muscle weakness, acquired short stature, and dysmorphic features. While this represents an n = 1 case, the evidence for pathogenicity includes: 1. A de novo variant with damaging predictions that is absent from large population databases and lacks of another compelling gene candidate, 2. The fact that the identical germline variant in the human paralog EZH2 is a known disease-causing variant in patients with Weaver syndrome (Diets et al. 2018), and 3. Drosophila studies show the variant leads to increased H3K27-trimethylation and morphological phenotypes.
The Drosophila model presented is within the E(z) homolog and could be considered an animal model for both this new rare disease and for EZH2-related Weaver syndrome. Indeed, de novo Weaver syndrome missense variants in EZH2 have been reported to result in increased H3K27 trimethylation based on biochemical experiments (McCabe et al. 2012a; Cohen et al. 2015). While this is strong evidence for pathogenicity, additional patients and studies are required to determine which aspects of the affected individual's phenotype are due to EZH1.
We provide evidence that, with respect to H3K27me3, the EZH1 variant is a gain-of-function allele. Consistent with our conclusion, within the EZH1 paralog EZH2, the analogous variant has been proposed to act through a gain-of-function mechanism (McCabe et al. 2012a). Furthermore, in our study, H3K27me3 is increased when the E(z)A691G variant is expressed in the E(z) null, wildtype, and heterozygous backgrounds. Also, the variant produces morphological phenotypes including A4 segment pigmentation, L2 extra sex combs, and wing vein abnormalities. We observe these phenotypes when the E(z)A691G variant is expressed in null and wildtype backgrounds, also providing further evidence for a gain-of-function mechanism for p.A691G, which is analogous to the EZH1A678G allele identified in the affected individual reported in this study. Importantly, the E(z)A691G can fully rescue the lethality of the E(z) null flies, providing evidence against a loss-of-function mechanism.
Moreover, the significant bang-sensitivity and shortened lifespan observed upon expression of E(z)A691G but not the E(z)WT in an E(z) null background further demonstrates the functional difference between the variant and wildtype alleles. Indeed, E(z) heterozygous loss-of-function animals show increased lifespan, so an altered lifespan is expected with gain-of-function variants (Moskalev et al. 2019). Furthermore, we have shown the loss of H3K27me2 and drastic increase of H3K27me3 in flies expressing E(z)A691G, suggesting that target loci that should be di-methylated are instead trimethylated, which likely leads to dysregulation of downstream target genes. It is possible that there are multiple functions of EZH1 and that the bang sensitivity and survival phenotypes may not be coupled entirely to the hyper-trimethylation phenotypes. The Drosophila model could be an important in vivo tool for assessing the function of genetic variants in EZH1 genes beyond their direct effect on histone methylation.
The Drosophila phenotypes reported in this study may provide mechanistic insights into disease pathogenesis. We observed extra sex combs and A4 segment pigmentation patterning defects in flies expressing E(z)A691G, suggesting a possible impact of E(z) gain-of-function alleles on HOX gene expression. Fly E(z) mutant clones were previously associated with improper axon guidance (Wang et al. 2006), which may be related to the bang sensitivity defect seen upon expression of E(z)A691G. Since E(z)A691G increases H3K27me3, expression of neurodevelopmental genes could be dysregulated resulting in neurological deficits in affected individuals with variants in EZH1. Considering that bang sensitivity in flies is observed in a wide range of mutants with deficits in excitatory or inhibitory signaling, it would be interesting to assess how the neuronal specification or wiring of the nervous system is affected in animals that express E(z)A691G.
Weaver syndrome is primarily known as an overgrowth syndrome, with increased height and weight, macrocephaly, and large hands. In contrast, our patient has a low normal height with normal weight and head circumference and small hands and feet. Retrognathia is a common feature in Weaver syndrome, while our patient had mild prognathia. Our patient is also not known to have any of the skeletal features described in Weaver syndrome. Our patient's developmental delay and hypotonia are more severe than what is typically seen in Weaver syndrome. In addition, muscle weakness and hypopigmentation have not been described in Weaver syndrome to our knowledge. The shared features between Weaver syndrome and our patient are relatively nonspecific. These include developmental delay, hypotonia, strabismus, inverted nipples, and inguinal hernia.
We hypothesize that the different phenotypes between Weaver syndrome and our patient may result from differences in expression patterns between EZH2 and EZH1. EZH2 is most abundant in proliferating tissues, particularly in lymphocytes, while EZH1 is most highly expressed in the cerebellum and tibial nerves (GTEx, and Margueron et al. 2008). Compared to EZH2, EZH1 is also more highly expressed in the cerebral cortex, the frontal cortex, and skeletal muscle (GTEx), consistent with our patient's more severe neurodevelopmental phenotype and muscle weakness. The timing of expression in the developing brain also differs between the two genes. EZH2 expression in the brain is moderately high early in the first trimester of pregnancy and then trends downward, with very low expression from late gestation through 40 years of age (BrainSpan Developmental Transcriptome). By contrast, EZH1 expression in the brain is relatively low in the early first trimester but increases steadily throughout development (BrainSpan Developmental transcriptome). This suggests that EZH1 may be maintaining H3K27me3 after initial embryonic development. Thus, the EZH1 variant identified in this patient may be suppressing critical genes during later stages of embryonic development.
Undiagnosed diseases present opportunities to identify novel genetic determinants of disease. Members of the PRC2 complex have well-reported disease associations, including the p.A677 amino acid of EZH2. Here the equivalent amino acid substitution of the alternative subunit and paralog, EZH1 is identified in an undiagnosed individual with a neurodevelopmental phenotype. Drosophila models of the variant show increased H3K27me3 and resultant behavioral phenotypes, providing some molecular metrics to elucidate the underlying mechanism of this potential new disease. Due to the homology between E(z), EZH1, and EZH2, this model is incidentally relevant to Weaver syndrome. Further studies will be needed to delineate the full human phenotypic spectrum for EZH1-related disorders.
“Note added in revision”
During the review for this manuscript, we noted a preprint of a separate manuscript that includes the EZH1 subject and variant reported and provides evidence for gain-of-function activity (https://www.medrxiv.org/content/10.1101/2022.08.09.22278430v1).
Patient 4 in that publication is the same individual we describe here.
Supplementary Material
Acknowledgments
We thank Dr. Leonie Ringrose, Berlin, Germany, and Dr. Richard Jones, Dallas, Texas, USA for kindly providing the study's cDNA and fly reagents. We thank Dr. Hugo J. Bellen for providing reagents and Hongling Pan for generating our patient variant transgenics, BCM, Houston, Texas, USA. Also, we are thankful for the resources provided by the Bloomington Drosophila Stock Center.
Contributor Information
Sharayu V Jangam, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston, TX 77030, USA.
Lauren C Briere, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA.
Kristy L Jay, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston, TX 77030, USA.
Jonathan C Andrews, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston, TX 77030, USA.
Melissa A Walker, Department of Neurology, Division of Neurogenetics, Child Neurology, Massachusetts General Hospital, Boston, MA 02114, USA.
Lance H Rodan, Department of Neurology, Boston Children's Hospital, Boston, MA 02115, USA.
Frances A High, Division of Medical Genetics and Metabolism, Massachusetts General Hospital for Children, Boston, MA 02114, USA.
Shinya Yamamoto, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston, TX 77030, USA; Department of Neuroscience, Baylor College of Medicine, Houston, TX 77030, USA.
David A Sweetser, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Division of Medical Genetics and Metabolism, Massachusetts General Hospital for Children, Boston, MA 02114, USA.
Michael F Wangler, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston, TX 77030, USA.
Data availability
The authors confirm that the data supporting the findings of this research are available within the manuscript. The deidentified genome and transcriptome sequencing data and phenotype data for the patient/family described are available in the National Center for Biotechnology (NCBI) database of Genotypes and Phenotypes (dbGap; http://www.ncbi.nlm.nih/gov/gap) under the accession numbers phs001232.v2.p1 and phs001232.v3.p1, respectively. The variant c.2033C > G (p.Ala678Gly) has been submitted to ClinVar and can be found under the accession number VCV000977755.2.
Supplemental material available at GENETICS online.
Ascertainment and ethics
The patient was ascertained through participation in the Undiagnosed Diseases Network (UDN). The family described herein was studied through the UDN. This study was approved by the National Institutes of Health Intramural Intuitional Review Board, protocol number 15HG0130. Informed consent was obtained for the proband and his first-degree relatives for research and publication. The proband's parents consented to the publication of his identifiable photos.
Funding
This study was supported by NIH-NINDS grants U54 NS 093793 (The UDN MOSC) to M.F.W. and S.Y.
Literature cited
- Abel KJ, Brody LC, Valdes JM, Erdos MR, McKinley DR, Castilla LH, Merajver SD, Couch FJ, Friedman LS, Ostermeyer EA, et al. . Characterization of EZH1, a human homolog of Drosophila enhancer of zestenearBRCA1. Genomics. 1996;37(2):161–171. doi: 10.1006/GENO.1996.0537. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge D; Undiagnosed Diseases Network, Wangler MF, Bowman AN, Yamamoto S, Schedl T, Pak SC, Postlethwait JH, Shin J, Solnica-Krezel L, et al. . Model organisms contribute to diagnosis and discovery in the undiagnosed diseases network: current state and a future vision. Orphanet J Rare Dis. 2021;16(1):206. doi: 10.1186/s13023-021-01839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109(7):801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, et al. . EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B, Holbro N, Reichert H. Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila. Development. 2007;134(6):1091–1099. doi: 10.1242/dev.02793. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y, Poliakov A, Yocum AK, Li Y, Chen W, et al. . The central role of EED in the orchestration of polycomb group complexes. Nat Commun. 2014;5(1):3127. doi: 10.1038/ncomms4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carrington EA, Jones RS. The Drosophila enhancer of zeste gene encodes a chromosomal protein: examination of wildtype and mutant protein distribution. Development. 1996;122(12):4073–4083. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- Cohen AS, Gibson WT. EED-associated overgrowth in a second male patient. J Hum Genet. 2016;61(9):831–834. doi: 10.1038/jhg.2016.51. [DOI] [PubMed] [Google Scholar]
- Cohen ASA, Tuysuz B, Shen Y, Bhalla SK, Jones SJM, Gibson WT. A novel mutation in EED associated with overgrowth. J Hum Genet. 2015;60(6):339–342. doi: 10.1038/jhg.2015.26. [DOI] [PubMed] [Google Scholar]
- Cohen ASA, Yap DB, Lewis MES, Chijiwa C, Ramos-Arroyo MA, Tkachenko N, Milano V, Fradin M, McKinnon ML, Townsend KN, et al. . Weaver syndrome-associated EZH2 protein variants show impaired histone methyltransferase function in vitro. Hum Mutat. 2016;37(3):301–307. doi: 10.1002/humu.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deevy O, Bracken AP. PRC2 functions in development and congenital disorders. Development. 2019;146(19):dev181354. doi: 10.1242/dev.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diets IJ, Waanders E, Ligtenberg MJ, van Bladel DAG, Kamping EJ, Hoogerbrugge PM, Hopman S, Olderode-Berends MJ, Gerkes EH, Koolen DA, et al. . High yield of pathogenic germline mutations causative or likely causative of the cancer phenotype in selected children with cancer. Clin Cancer Res. 2018;24(7):1594–1603. doi: 10.1158/1078-0432.ccr-17-1725. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5(8):a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves CS, Le Boiteux E, Arnaud P, Costa BM. HOX gene cluster (de)regulation in brain: from neurodevelopment to malignant glial tumours. Cell Mol Life Sci. 2020;77(19):3797–3821. doi: 10.1007/s00018-020-03508-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi A, Krier JB, Toth-Petroczy A, Cassa CA, Frank NY, Carmichael N, Fieg E, Bjonnes A, Mohanty A, Briere LC, et al. . An integrated clinical program and crowdsourcing strategy for genomic sequencing and Mendelian disease gene discovery. NPJ Genom Med. 2018;3(1):21. doi: 10.1038/s41525-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T, Swanson J, Van Ness B. EZH2 inhibitors sensitize myeloma cell lines to panobinostat resulting in unique combinatorial transcriptomic changes. Oncotarget. 2018;9(31):21930–21942. doi: 10.18632/oncotarget.25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H-M, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38(12):621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo I, Gonzalez S. New epigenetic pathway for stemness maintenance mediated by the histone methyltransferase Ezh1. Cell Cycle. 2013;12(3):383–384. doi: 10.4161/cc.23550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nuñez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11(5):649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12(1):357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa E, Higashimoto K, Sakai Y, Numakura C, Okamoto N, Matsunaga S, Ryo A, Sato Y, Sanefuji M, Ihara K, et al. . Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum Mutat. 2017;38(6):637–648. doi: 10.1002/humu.23200. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Cellular memory of transcriptional states by Polycomb-group proteins. Semin Cell Dev Biol. 1999;10(2):227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- Jenkins VK, Larkin A, Thurmond J; FlyBase Consortium . Using FlyBase: a database of Drosophila genes and genetics. Methods Mol Biol. 2022;2540(3):1–34. doi: 10.1007/978-1-0716-2541-5_1. [DOI] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in Polycomb group-mediated repression. Mol Cell Biol. 1998;18(5):2825. doi: 10.1128/MCB.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126(1):185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook H, Seo S-B, Jain R. EZ Switch from EZH2 to EZH1: histone methylation opens a window of cardiac regeneration. Circ Res. 2017;121(2):91–94. doi: 10.1161/CIRCRESAHA.117.311351. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Li Y, Kim H, Eum S, Park K, Lee C-H. The role of EZH1 and EZH2 in development and cancer. BMB Rep. 2022;55(12):595–601. doi: 10.5483/bmbrep.2022.55.12.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Luo X, Schoch K, Jangam SV, Bhavana VH, Graves HK, Kansagra S, Jasien JM, Stong N, Keren B, Mignot C, et al. . Rare deleterious de novo missense variants in Rnf2/Ring2 are associated with a neurodevelopmental disorder with unique clinical features. Hum Mol Genet. 2021;30(14):1283–1292. doi: 10.1093/hmg/ddab110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42(4):421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. . Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A. 2012a;109(8):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della PietraA, III, Diaz E, et al. . EZH2 Inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012b;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Shaposhnikov MV, Zemskaya NV, Koval LА, Schegoleva EV, Guvatova ZG, Krasnov GS, Solovev IA, Sheptyakov MA, Zhavoronkov A, et al. . Transcriptome analysis of long-lived Drosophila melanogaster E(z) mutants sheds light on the molecular mechanisms of longevity. Sci Rep. 2019;9(1):9151. doi: 10.1038/s41598-019-45714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The Polycomb -group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–4336. doi: 10.1128/mcb.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and polycomb group proteins in Drosophila and their dependence on enhancer of zeste function. EMBO J. 1993;12(4):1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. . Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13(1):31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ER. Shortened lifespan and other age-related defects in bang sensitive mutants of Drosophila melanogaster. G3 (Bethesda). 2018;8(12):3953–3960. doi: 10.1534/g3.118.200610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu Y-J, Fujiwara Y, Kim J, Mao X, Yuan G-C, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter K, Adams DR, Bacino CA, Bellen HJ, Bernstein JA, Cheatle-Jarvela AM, Eng CM, Esteves C, Gahl WA, Hamid R, et al. . Effect of genetic diagnosis on patients with previously undiagnosed disease. N Engl J Med. 2018;379(22):2131–2139. doi: 10.1056/NEJMoa1714458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Fonseca JP, Gänger C, Dworschak E, Kockmann T, Beisel C, Ringrose L. Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res. 2013;41(10):5235. doi: 10.1093/nar/gkt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanik VA, Harte PJ. A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes. Dev Biol. 2012;364(2):249–258. doi: 10.1016/j.ydbio.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte SDV, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S, et al. . Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2(12):1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Murray A, Hanks S, Douglas J, Armstrong R, Banka S, Bird LM, Clericuzio CL, Cormier-Daire V, Cushing T, et al. . Weaver syndrome and EZH2 mutations: clarifying the clinical phenotype. Am J Med Genet A. 2013;161A(12):2972–2980. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- Tsang YH, Dogruluk T, Tedeschi PM, Wardwell-Ozgo J, Lu H, Espitia M, Nair N, Minelli R, Chong Z, Chen F, et al. . Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat Commun. 2016;7(1):10500. doi: 10.1038/ncomms10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. . Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium . UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2022;46(5):2699. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Al-Ouran R, Hu Y, Kim S-Y, Wan Y-W, Wangler MF, Yamamoto S, Chao H-T, Comjean A, Mohr SE, et al. . MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am J Hum Genet. 2017;100(6):843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee C-HJ, Lin S, Lee T. Steroid hormone-dependent transformation of polyhomeotic mutant neurons in the Drosophila brain. Development. 2006;133(7):1231–1240. doi: 10.1242/dev.02299. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu Z, Bellen HJ, Yamamoto S. Navigating MARRVEL, a web-based tool that integrates human genomics and model organism genetics information. J Vis Exp. 2019a;(150). doi: 10.3791/59542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mao D, Fazal F, Kim S-Y, Yamamoto S, Bellen H, Liu Z. Using MARRVEL v1.2 for bioinformatics analysis of human genes and variant pathogenicity. Curr Protoc Bioinformatics. 2019b;67(1):e85. doi: 10.1002/cpbi.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DD, Graham CB, Thomas IT, Smith DW. A new overgrowth syndrome with accelerated skeletal maturation, unusual facies, and camptodactyly. J Pediatr. 1974;84(4):547–552. doi: 10.1016/s0022-3476(74)80675-x. [DOI] [PubMed] [Google Scholar]
- Xia B, Gerstin E, Schones DE, Huang W, Steven de Belle J. Transgenerational programming of longevity through E(z)-mediated histone H3K27 trimethylation in Drosophila. Aging. 2016;8(11):2988–3008. doi: 10.18632/aging.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-R, Lee C-H, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33(15–16):903–935. doi: 10.1101/gad.325050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this research are available within the manuscript. The deidentified genome and transcriptome sequencing data and phenotype data for the patient/family described are available in the National Center for Biotechnology (NCBI) database of Genotypes and Phenotypes (dbGap; http://www.ncbi.nlm.nih/gov/gap) under the accession numbers phs001232.v2.p1 and phs001232.v3.p1, respectively. The variant c.2033C > G (p.Ala678Gly) has been submitted to ClinVar and can be found under the accession number VCV000977755.2.
Supplemental material available at GENETICS online.