Abstract
In the last decade, researchers using Drosophila melanogaster have made extraordinary progress in uncovering the mysteries underlying learning and memory. This progress has been propelled by the amazing toolkit available that affords combined behavioral, molecular, electrophysiological, and systems neuroscience approaches. The arduous reconstruction of electron microscopic images resulted in a first-generation connectome of the adult and larval brain, revealing complex structural interconnections between memory-related neurons. This serves as substrate for future investigations on these connections and for building complete circuits from sensory cue detection to changes in motor behavior. Mushroom body output neurons (MBOn) were discovered, which individually forward information from discrete and non-overlapping compartments of the axons of mushroom body neurons (MBn). These neurons mirror the previously discovered tiling of mushroom body axons by inputs from dopamine neurons and have led to a model that ascribes the valence of the learning event, either appetitive or aversive, to the activity of different populations of dopamine neurons and the balance of MBOn activity in promoting avoidance or approach behavior. Studies of the calyx, which houses the MBn dendrites, have revealed a beautiful microglomeruluar organization and structural changes of synapses that occur with long-term memory (LTM) formation. Larval learning has advanced, positioning it to possibly lead in producing new conceptual insights due to its markedly simpler structure over the adult brain. Advances were made in how cAMP response element-binding protein interacts with protein kinases and other transcription factors to promote the formation of LTM. New insights were made on Orb2, a prion-like protein that forms oligomers to enhance synaptic protein synthesis required for LTM formation. Finally, Drosophila research has pioneered our understanding of the mechanisms that mediate permanent and transient active forgetting, an important function of the brain along with acquisition, consolidation, and retrieval. This was catalyzed partly by the identification of memory suppressor genes—genes whose normal function is to limit memory formation.
Keywords: learning, memory, forgetting, mushroom body, memory suppressor genes, CREB, Orb2, FlyBook
Introduction
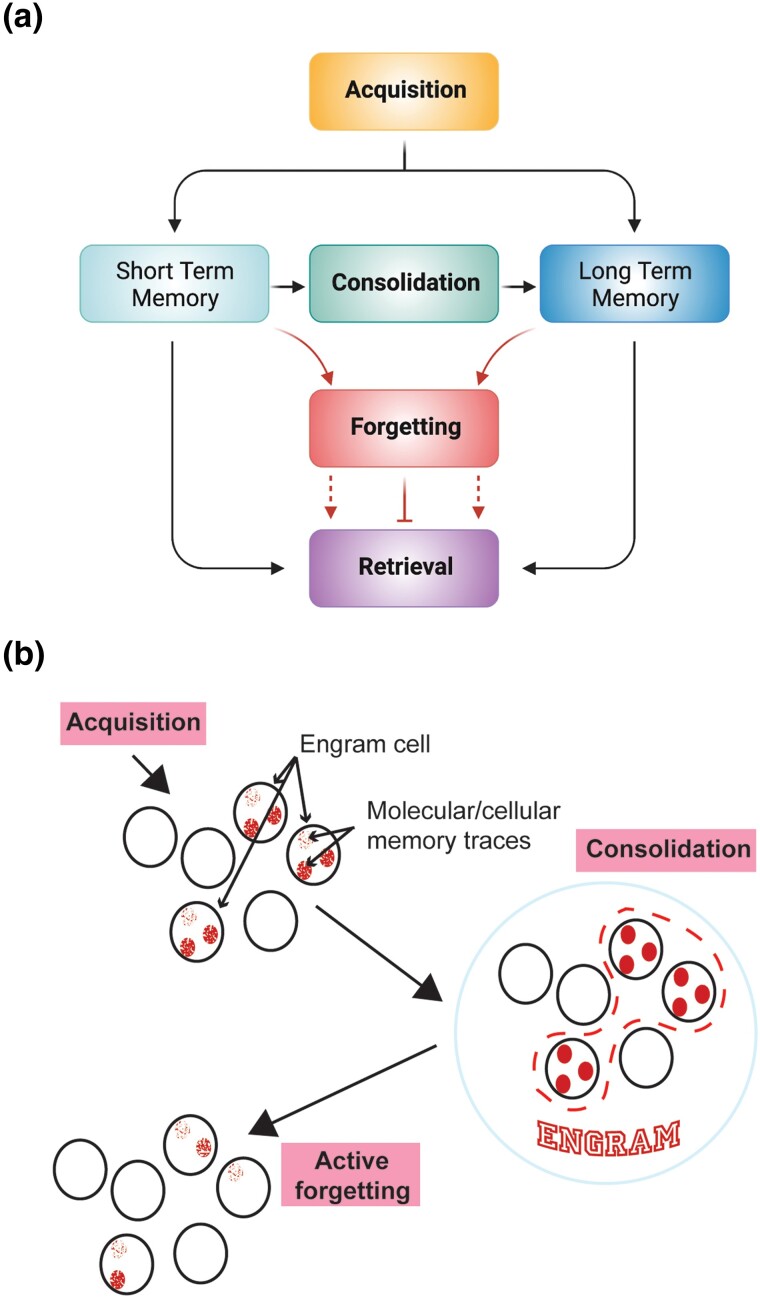
Five decades have lapsed since Quinn, Harris, and Benzer developed an associative, aversive, olfactory learning assay for adult Drosophila (Quinn et al. 1974). Since then, much has been learned about the mechanisms underlying memory formation in Drosophila and other organisms. Our current and broad understanding, offered as a brief overview for a perspective of the advances made in the last 10–12 years, is that the nervous system employs four different memory operations: acquisition, consolidation, forgetting, and retrieval (Fig. 1a). The acquisition of information, or learning, alters the physiological state of selected neurons, or engram cells, in ways that encode memory (Fig. 1b). These state changes, or molecular and cellular memory traces, can be any change in the activity of the cell that is induced by learning that becomes part of the neural code for that memory. After acquisition, memories can be stabilized by consolidation mechanisms. In Drosophila, two forms of consolidated memory are recognized: a protein synthesis-dependent (PSD) form and a form resistant to anesthesia. The collection of all molecular and cellular memory traces that are induced by learning across all neurons and circuits engaged by the acquisition event together comprise the overall memory engram that can guide behavior upon subsequent retrieval. Alternatively, memories can be permanently lost or transiently hidden from retrieval. The latter is transient forgetting.
Fig. 1.
a) The nervous system uses four operations for short- and long-term memory formation: acquisition, consolidation, forgetting, and retrieval. Acquisition is synonymous with “learning,” and represents the initial encoding of information. Consolidation refers to the processes involved in stabilizing memory over time. Forgetting involves mechanisms whereby memories can be erased or hidden from retrieval. Retrieval is simply the recollection, or recall, of existing memories. b) Cartoon illustrating the broad cellular and network events of memory formation. During acquisition, selected cells in the nervous system undergo molecular or biochemical changes that alter their physiological state. These selected cells are known as engram cells, and the molecular or biochemical changes within the engram cells are termed molecular or cellular memory traces. Consolidation mechanisms stabilize the cellular memory traces and the selected engram cells, together with their corresponding memory traces, represent the overall “engram” for a given memory. The activity of forgetting cells can erode the memory traces and cause memory failure. Modified with permission from Noyes et al. 2021 and Davis and Zhong 2017.
The initial assay developed by Quinn and colleagues was population-based, training groups of about 40 flies at a time and was partially operant in nature, requiring flies to learn an association between their phototactic behavior and the reinforcer of mild electric shock. The assay is discriminative, with learning and memory measured by the flies’ selective avoidance of an odor coupled to the reinforcer to a second odor uncoupled to the reinforcer. This assay marked the beginning of an intense research era of ∼25 years of behavioral genetics research, which focused on expanding the types of learning assays used to explore Drosophila memory formation, and identifying and characterizing single gene mutations that impair learning and memory (reviewed by Quinn and Greenspan 1984; Waddell and Quinn 2001; McGuire et al. 2005; Griffith and Ejima 2009; Pitman et al. 2009; Kahsai and Zars 2011;and Masek and Keene 2016). Other learning assays were developed and include aversive olfactory classical conditioning, appetitive olfactory conditioning, visual learning, taste learning, motor learning, spatial orientation learning, courtship learning, and others. Some of the assays like courtship learning, olfactory conditioning of the proboscis extension reflex, and spatial learning, employ single flies rather than small populations. Odor-shock learning assays for larvae were also developed in this period along with non-associative assays for habituation and sensitization. Learning assay development continues to this day, using more sophisticated equipment.
The identification and characterization of single gene mutations that impair learning and memory began shortly after behavioral assays were developed. Multiple ways were employed to create single gene mutants, including chemical mutagenesis, transposon mobilization, and enhancer detection (reviewed by Quinn and Greenspan 1984; Davis 1993; and Waddell and Quinn 2001). This led to a group of initial mutants that were studied behaviorally to detail the nature of the learning/memory impairment, molecularly to identify the gene product, and histologically using immunohistochemistry and RNA in situ hybridization to identify brain areas that express the gene product. A major conclusion that emerged from these studies is that some of the initially identified genes encode protein products involved in cAMP signaling (Davis 1993). This includes dunce, which encodes cAMP phosphodiesterase, rutabaga, which encodes a Ca2+-sensitive adenylyl cyclase, and DCO, which encodes the catalytic subunit of cAMP-dependent protein kinase A. Moreover, all three protein products were found to be preferentially expressed in mushroom body neurons (MBn), also known as Kenyon cells, with the rutabaga-encoded cyclase distributed primarily in the axons of these neurons. Heisenberg and colleagues (reviewed by Heisenberg 1989) had previously identified Drosophila brain structural mutants that disrupted both MBn anatomy and learning. These observations led to a dramatically increased focus on the MBn and a model for aversive olfactory classical conditioning which envisioned the axons of MBn as the principal sites for temporally integrating the conditioned stimulus (odor) and unconditioned stimulus (electric shock) (CS and US, respectively) employed to classically train flies. Prior anatomical studies of insects had shown that olfactory information (CS pathway) is sensed by olfactory receptor neurons on the antenna and this information is communicated to the MBn via the antennal lobe (AL) (Fig. 2 and Fig. 3a). The US pathway was hypothesized to be unidentified neuromodulatory cells that synapse on the axons of the MBn and activate the rutabaga-encoded adenylyl cyclase synergistically with calcium influx from the activation of the CS pathway (Fig. 2). This would initiate a signaling cascade of protein phosphorylation from protein kinase A, altering the physiology of the MBn to a state of “learning.” The Drosophila studies were not the first to suggest that MBn are central to olfactory learning in insects. Menzel and colleagues (reviewed by Menzel 2001) had previously described elegant studies, including localized cooling of honeybee MBn, to show their relevance to olfactory learning. Indeed, one must applaud the remarkable conceptual advances about memory formation made with this more intelligent insect (Menzel 2022). But the idea that MBn were a major site for olfactory memory formation was cemented by Drosophila behavioral genetic experiments on dunce, rutabaga, and DCO, which pointed to MBn as functional site for the products of several “learning genes.”
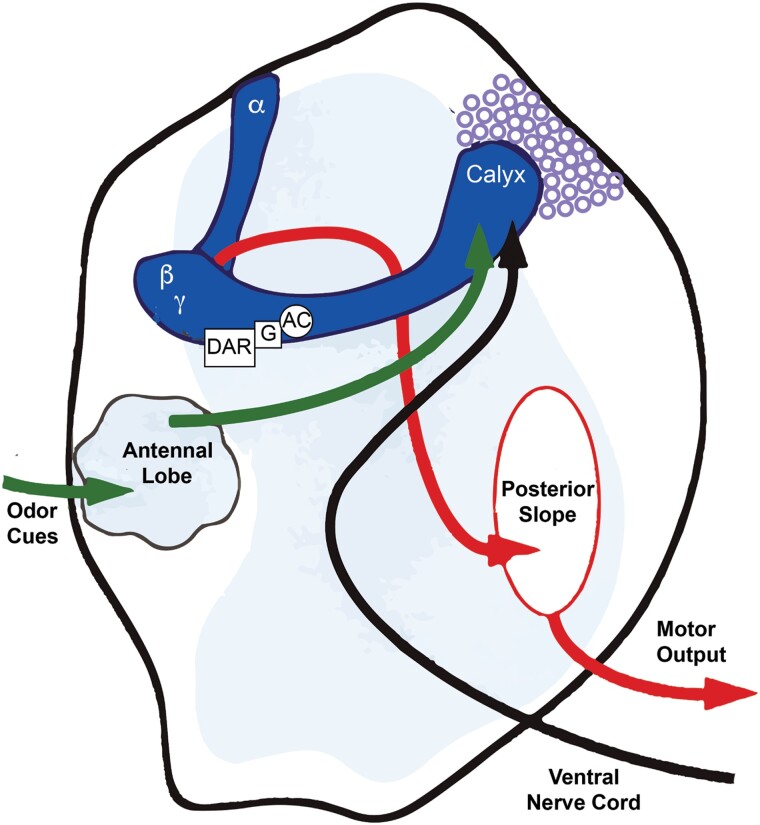
Fig. 2.
Early model for olfactory learning envisioning odor cues being communicated to the MBn via the antennal lobe, and aversive shock information being communicated to the MBn via an ascending pathway from the ventral nerve cord. The MB includes the cell bodies (purple circles), the dendritic region (calyx), and the axons distributed into neuropil regions called lobes (α,β,γ lobes). The α-lobes project dorsally, and the β and γ lobes project medially. The rutabaga-encoded adenylyl cyclase and its associated G-protein (G) was envisioned to be coupled to a neuromodulatory receptor, probably dopamine (DAR), to help integrate the CS and US signals. This would lead to altered behavior in response to the CS from altered motor output instructions to the thoracic ganglia. Reproduced with permission from Davis 1993.
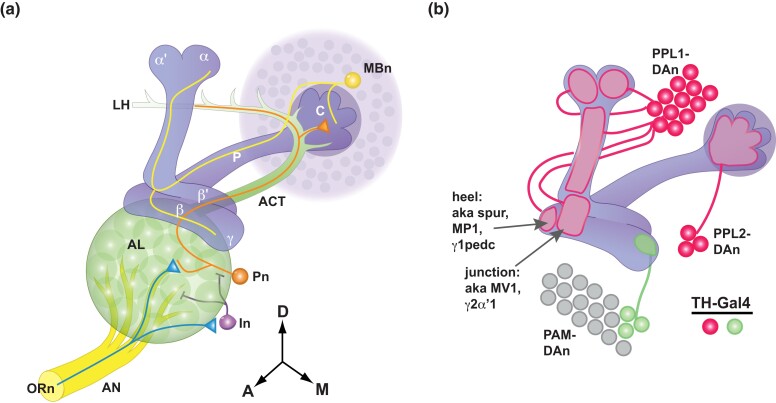
Fig. 3.
a). Structure of the olfactory nervous system. Olfactory information is communicated to the MBn from the olfactory receptor neurons (ORn) on the antennae, to the projection neurons (Pn) in the antennal lobe (AL), whose axons comprise the antennal cerebral tract (ACT), to the dendrites of the MBn in the calyx (C). The ACT continues beyond the calyx to the lateral horn (LH). The AL also contains GABAergic interneurons (In) that help shape information processing. The MBn extend their primary axons through the peduncle (P) to the vertical and horizontal lobes. The α/β MBn send axon branches into both the vertical (α) and horizontal (β) lobes. This arrangement is mirrored by the α′/β′ MBn (α′vertical and β′horizontal lobes). The axons of the γ MBn extend only into the horizontal γ lobe. Reproduced with permission from Davis 2011. b) Innervation pattern of the MB neuropil by DAn that are labeled by the TH-Gal4 driver. Three clusters of DAn—PPL1, PPL2, and PAM—send axon projections into the MB neuropil. The 12 PPL1-DAn innervate defined segments (compartments) of the vertical lobes, the junction area, and the heel. The junction and heel compartments have alternative names as indicated in the figure. PPL2-DAn innervate the calyx. Only a small number of PAM-DAn express the TH-Gal4 driver. Other PAM-DAn innervate compartments of the horizontal lobes. Modified with permission from Boto et al. 2019.
Three major developments led the field to combine behavioral genetics studies of learning and memory with systems neuroscience approaches. First, binary expression systems like the Gal4/UAS system and enhancements that provide for conditional expression were developed that allow researchers to direct gene expression to specific cells and at specific times (Brand and Perrimon 1993; McGuire et al. 2004).
Second, transgenes were developed that allow the experimenter to conditionally silence neurons of interest before or after learning. The first of these, Shibirets (Shits), was introduced by Waddell, Kitamoto, and Quinn (Waddell et al. 2000). The temperature-dependent expression of the Shits transgene in specific neurons allowed researchers to block synaptic transmission from the neurons in the living fly, providing insights into the role of the neurons in the processes of acquisition, consolidation, and retrieval. This, and other thermogenetic advances (trpA1, trpM8, etc.), were rapidly followed by optogenetic approaches used to activate or inhibit the activity of selected neurons in larvae (Schroll et al. 2006; Riemensperger et al. 2016). The third major development was engineering and utilizing fluorescent reporter transgenes, such as those that register increased synaptic transmission or Ca2+ influx, which allowed the visualization of neuronal activity in ex vivo brains or living flies under the microscope. Ng et al. (2002); Fiala et al. (2002); and Wang et al. (2003) first employed such transgenes to study the responses of antennal lobe neurons (ALn) to odors. Yu et al. (2004) and Riemensperger et al. (2005) employed them to study how neuronal responses are altered by olfactory learning, leading to the identification of the first cellular memory traces. Cellular memory traces, as indicated above, can include molecular, biophysical, or cellular changes induced by learning that subsequently alter the processing and response of the nervous system to the sensory information that is learned. There is currently widespread use of both thermo- and opto-genetics to query potential memory functions of specific neurons and functional optical imaging to identify and characterize altered neuronal response properties due to learning.
The role for neuromodulatory cells, and specifically dopaminergic (DAn) and octopaminergic neurons (OAn), respectively, as neurons communicating the US for aversive and appetitive classical conditioning, became a focus with the studies of Schwärzel, Heisenberg, and co-workers in 2003 (Schwaerzel et al. 2003). These researchers demonstrated that blocking synaptic release from DAn with Shibirets impaired aversive memory formation but not appetitive memory formation. Conversely, synaptic release from OAn was shown to be required for appetitive conditioning. This suggested that different neuromodulators are required for learning the valence of the same odor used as the CS. Although such blocking experiments were highly suggestive, capstone data was offered by showing that artificial activation of the neuromodulatory neurons in behaving Drosophila is sufficient to replace the US normally used for conditioning. Schroll et al. (2006) offered the first of these demonstrations by showing that optogenetic stimulation of DAn or OAn, respectively, is sufficient to replace the aversive or appetitive US employed for olfactory classical conditioning of larvae. Subsequent studies (Claridge-Chang et al. 2009; Aso et al. 2012) extended the observation for DAn to adult flies.
A more detailed anatomical picture of the structure of mushroom body (MB) and especially its dopaminergic innervation became a critical need to advance our understanding of learning and memory. The MBs of Drosophila are bilaterally symmetric structures consisting of ∼2000 intrinsic neurons (MBn) per brain hemisphere (Figs. 2 and 3a). The cell bodies are situated in the dorsal posterior aspect of the brain, and project dendrites into the calyx. The axons extend to the anterior region of the brain in a bundle known as the peduncle where they turn and give rise to axon collaterals in neuropil known as lobes, with axon branches projecting both dorsally and medially. Two studies, one based on the differential expression of immunohistochemical markers and the other on highlighting single cell clones, allowed the subdivision of the MBn and their axon projections into three broad classes (Crittenden et al. 1998; Lee et al. 1999). One class, known as α/β MBn, extend axon collaterals both dorsally and medially into the α and β lobes (Figs. 2 and 3a). A second class, known as α′/β′ MBn, also extend collaterals in both directions into the α′ and β′ lobes. A third class, the γ MBn, extend a single branch in the γ lobe. This subdivision offered the initial insight into the functional diversity of the MBn. Analysis of Gal4 expression lines revealed layers within these three classes for a total of seven types of MBn (Tanaka et al. 2008; Aso et al. 2014a). The MBn have a unique and interesting development, with the overall structure being derived from four neuroblasts, each of which contributes autonomously to all parts of the overall structure (Ito et al. 1997). Each neuroblast sequentially produces the three types of MBn, with the earliest neurons born between larval hatching through the mid-third larval instar projecting into to the γ lobe. Neurons born between the mid-third larval instar and puparium formation project into the α′/β′ lobes. MBn born after puparium formation project into the α/β lobes (Lee et al. 1999).
Mao and Davis (2009) offered initial insights into DAn innervation of the MB using anti-TH immunohistochemistry and analysis of single cell clones of the frequently used Gal4 driver line, TH-Gal4 (Fig. 3b). The anti-TH immunohistochemistry identified ∼282 positive cells that are distributed in clusters in the central brain. Single cell clones showed that three of the clusters project axons to the MB. PAM-DAn project to the horizontal lobes; PPL1-DAn project to the vertical lobes, junction area, the heel, and distal peduncle; and PPL2-DAn project to the calyx. Most importantly, the projections from single cell clones generated from the PPL1 cluster defined non-overlapping and separate axonal segments, or compartments, of the vertical lobes. This unique projection pattern strongly predicted the existence of functional differences among the DAn in the PPL1 cluster as well as the other DAn clusters, and functional differences between the compartments of the MBn axons. This segmental or compartment organization of the MB axons was echoed in future studies of the MB output neurons (MBOn) described below.
This summary of progress across the first ∼4 decades of Drosophila research on learning and memory serves as background for a focus on that occurring in the last 10–12 years. Additional details about the progress across the first 40 years can be found in the reviews cited above.
The connectome: insights and complexities
Excellent progress has been made in advancing our knowledge of brain circuits and neuron:neuron connections involved in memory formation for both larvae and adult Drosophila. The groundwork for subsequent massive electron microscopic efforts and artificial intelligence approaches was established using the larval brain by Cardona et al. (2010) by matching reconstructed microcircuitry from serial transmission electron microscopic images into the anatomical framework established from light microscopy. For the MB and its associated circuits, we define the MBn, with their cell bodies in the dorsal, posterior brain, along with the dendritic processes in the calyx and axonal processes in the peduncle and lobes (Figs. 2 and 3a), as MB intrinsic neurons. Some researchers also include neurons that appear from current information to be connected pre- and post-synaptically only to the MBn as MB intrinsic neurons [e.g. dorsal paired medial neurons (DPMn), anterior paired lateral neurons (APLn)], Here, for simplicity, we define all neurons that connect to the MBn as MB extrinsic neurons.
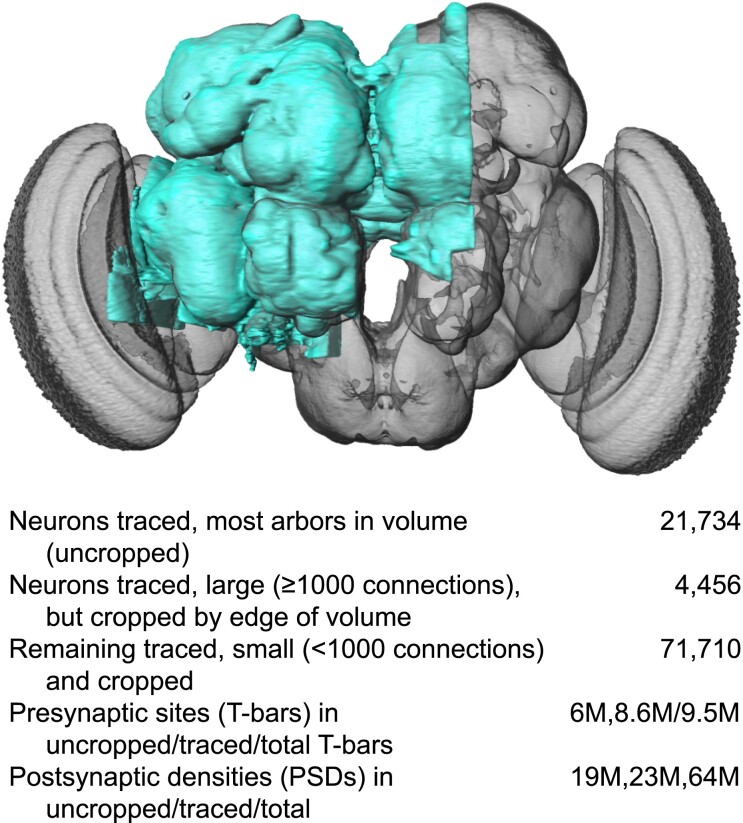
Scheffer et al. (2020) provided an examination of a major portion of the adult central brain that illustrates the precision and detail of its cells and connections. The accumulated data include the circuits, cell types, and synapses, referred to as the “connectome”. They detailed ∼25,000 central brain neurons along with ∼20 million connections using serial section electron microscopy and machine learning algorithms to reconstruct the central brain from individual images. Each neuron is identified by name and putative cell type along with its known structural connections. This information is publicly available using a web interface (https://neuprint.janelia.org). The NeuPrint browser interface allows the user to query connectivity, partners, connection strengths, and morphologies of all specified neurons, which aids in identifying upstream and downstream partners in the circuits of interest. Figure 4 illustrates the region of the adult brain connectome produced from these efforts in a frontal perspective along with a data table indicating the number of neurons, synapses, etc., found in the reconstruction.
Fig. 4.
Frontal image of the brain, with the connectome studied by Scheffer et al. 2020, in blue. Reproduced with permission from Scheffer et al. 2020.
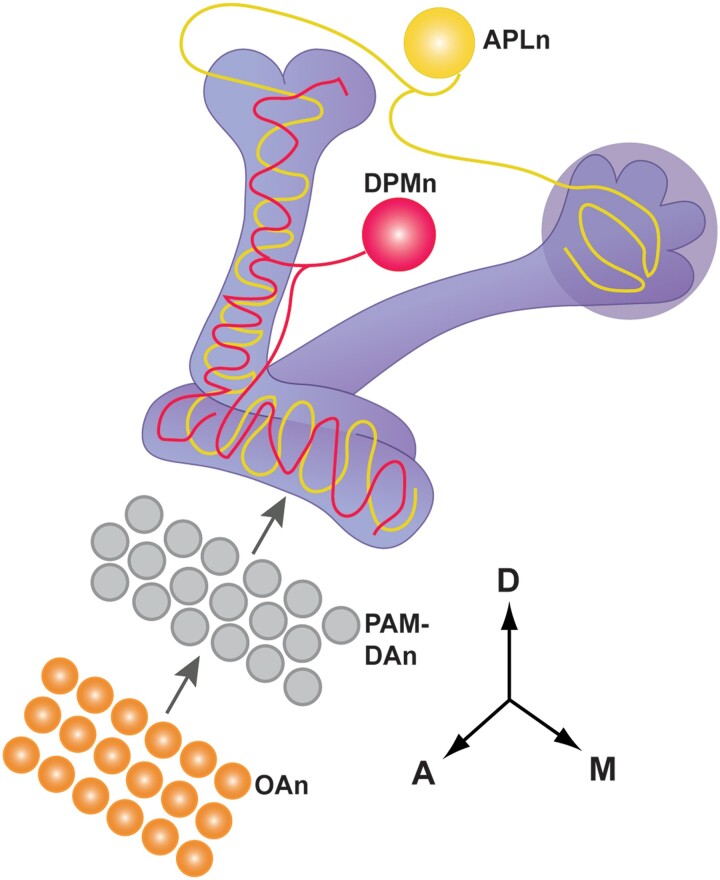
Takemura et al. (2017) employed these techniques and produced a connectome of the MB α-lobe and MB extrinsic neurons, building on the previously understood anatomy from light microscopic studies. The three major classes of MBn (α/β, α′/β′, and γ MBn) connect to four, well-studied types of extrinsic MBn (Fig. 5). The APLn are GABAergic with a single cell body located laterally in each hemisphere with axonal projections ramifying extensively in the vertical and horizontal lobes and calyx. These neurons negatively modulate olfactory associations by suppressing the strength of the CS pathway (Liu et al. 2007; Liu et al. 2009; Liu and Davis 2009). The DPMn, which are conspicuously absent in larvae, consist of a single cell body located medially in each brain hemisphere with neurites also ramifying throughout the vertical and horizontal lobes. Activity of the DPMn was originally reported to be required after acquisition, suggesting a role in the process of memory consolidation (Waddell et al. 2000, Yu et al. 2005; Keene et al. 2006; Lee et al. 2011; Haynes et al. 2015). A more recent study argued that it is also involved during acquisition, providing serotonergic feedback to MBn to constrain the time window needed for CS/US associations to occur (Zeng et al. 2023). DAn exist in multiple clusters in the brain with two clusters, PPL1 and PAM, responsible for DA input onto the MBn axons in the vertical and horizontal lobes, respectively, as discussed above. The ∼22 different types of DAn tile the MB axons into 15 compartments, with individual DAn innervating a single or sometimes two, compartments (Mao and Davis, 2009; Aso et al. 2014a, 2014b; Li et al. 2020). Figure 5 illustrates another feature of the PAM-DAn. They are layered between OAn and the MBn axons, explaining the requirement for OAn in appetitive conditioning (Burke et al. 2012). Twenty-two different MBOn mirror the DAn on the postsynaptic side of the MB (Aso et al. 2014a; Li et al. 2020). They project dendrites into 15 defined and non-overlapping MBn compartments, with some compartments innervated by multiple MBOn, to receive processed information from the MBn and direct it to downstream targets for olfaction and olfactory learning (Fig. 6).
Fig. 5.
Figure showing other classes of neurons in addition to the DAn (fig. 3b) that innervate the MB neuropil. There is a single DPMn per hemisphere that broadly innervates the vertical and horizontal lobes. The single APLn per hemisphere also broadly innervates the lobes and the calyx. The layering of PAM-DAn between OAn and the MB horizontal lobe is depicted. Modified with permission from Guven-Ozkan and Davis (2014).
Fig. 6.
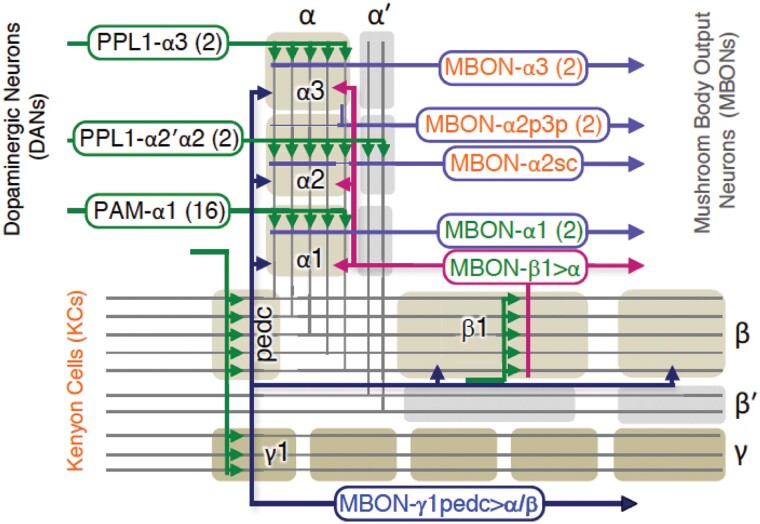
Subway map showing the compartment organization of MBn axons, illustrating the 3 major classes of MBn (α/β, α′/β′and γ MBn), compartment (gray and tan shading) specific input from PPL1 and PAM clusters of DAn, and compartment specific output from MBOn's. This figure focuses on the inputs and outputs from the vertical lobe compartments of α/β MBn, but a similar compartment and input (DAn) and output (MBOn) arrangement exists for the horizontal lobes (see Fig. 7b). Reproduced with permission from Takemura et al. 2017.
The connectome researchers extended this anatomical information about the MBn and MB neuropil in four major ways. First, they demonstrated that each MB axon makes synapses in each compartment, indicating that extrinsic neurons innervating each compartment probably have access to the same olfactory information carried by the MB axons. Second, they found that DAn axons synapse on both MBn axons and on MBOn. This observation complicates the simpler possibility that each MBOn receives only DAn-based information processed by the MBn axons in that compartment. Considering only synaptic connections and assuming that they are all functional, it appears that the input to each MBOn consists of information integrated (odor and DAn) by the MBn axons, and direct DAn input. Third, they found that APL axons innervate only MBn, and not the other cell types present in the neuropil, consistent with APL's role in suppressing the CS pathway either through MBn dendrites, axons, or both (Liu et al. 2009). The remaining connection patterns found extend this complicated picture: DPMn were found to be presynaptic to both MBn and DAn; and APLn, DPMn, and DAn receive reciprocal inputs from MBn. Fourth, the reconstructed presynaptic sites of MBn, DAn, APLn, and DPMn contain both dense core and clear synaptic vesicles, indicating that these neurons probably employ multiple neurotransmitters. It will be a large task to unravel the complexity of information processing in the various compartments of the MB neuropil given the reciprocal connections and the use of multiple neurotransmitters. Nevertheless, the impact of the work by Takemura et al. (2017) along with updates by Li et al (2020) and Otto et al (2020), and of Eichler et al (2017) on the larval brain (see below), will be felt in future years.
Functions of DAn and MBOn in appetitive and aversive olfactory classical conditioning
Despite the complexity of neuron:neuron interactions in the MB lobe neuropil, major advances have been made in understanding the role for these neurons and their circuits in aversive and appetitive olfactory classical conditioning. The unique pattern of DAn innervation of the MB lobes prompted studies to characterize the role of each type in olfactory classical conditioning. Similarly, the discovery of multiple types of MBOn prompted questions about the role of each MBOn in this type of learning. These studies have been combined into a model that envisions subsets of DAn providing the positive or negative valence of the US that becomes associated with the odor CS carried by the MBn, with the combined output of the MBOn driving approach or avoidance behavior by the fly. Many experimental techniques in the Drosophila research toolkit were used to produce this model, including neuron-type specific expression of transgenes; chemo-, thermo-, and optogenetic activation or inhibition of specific neuron types; electrophysiological recordings; and functional cellular imaging of neuronal activity using transgene-expressed sensors of calcium influx and neurotransmitter release.
As previously indicated (Fig. 2 and 3a), the neurons that form the initial parts in the CS pathway include ORn expressed primarily on the antennae, interneurons that process olfactory information at the first synaptic station, and projection neurons (Pn) that communicate olfactory information to the MBn dendrites in the calyx. The first step in learning associations about specific odors is olfactory discrimination. We focus here on MB intrinsic and extrinsic neurons beginning with the fact that different odors are represented by the sparse activation of subsets of MBn. The lateral horn (Fig. 3a) also receives olfactory information directly from the Pn. These neurons, in general, are thought to influence primarily innate odor preferences, while the path through the MBn provides for experience-dependent olfactory discrimination and learning (Heimbeck et al. 2001; Masse et al. 2009; Amin and Lin 2019; Dolan et al. 2018; Dolan et al. 2019).
The sparse, distributed, and accurate representation of different odors by the MBn population occurs from several different mechanisms. First, the activation of action potentials in the MBn requires the simultaneous input from ∼6 randomly chosen Pn, with only 5–10% of the MBn responding to any given odor (Honegger et al. 2011; Gruntman and Turner 2013). Second, sparseness is controlled, in part, by a negative feedback loop between MBn and the APLn (Lin et al. 2014). MBn activate APLn either directly or indirectly and the APLn provide GABAergic feedback onto the MBn through the resistance to dieldrin receptor (Liu et al. 2009). Notably, synaptic blockade of the APLn feedback loop decreases the sparseness of MBn odor responses. Third, neuromodulation by muscarinic axo-axonic connections in neighboring MBn axons functions to isolate axon activity, maintaining the integrity of odor representation by individual MBn axons in the bundled peduncle (Manoim et al. 2022).
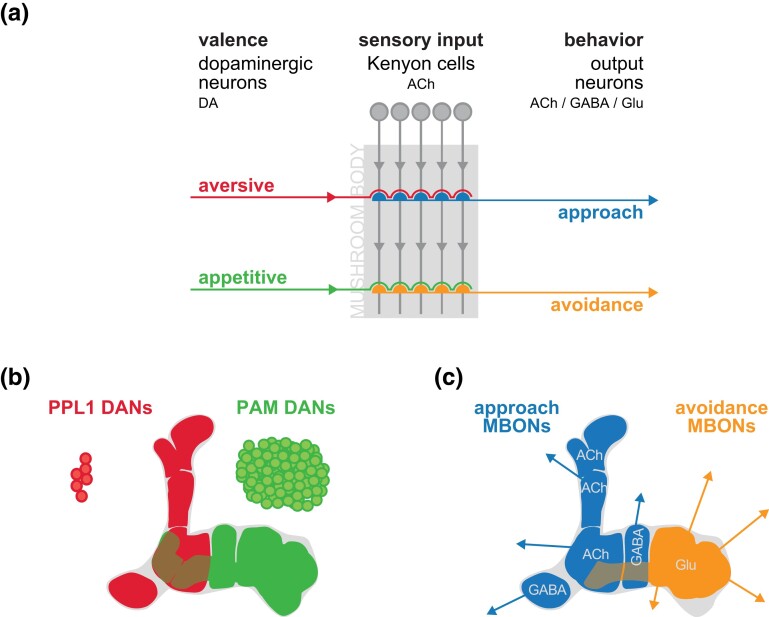
But what are the specific roles of the multiple types of DAn and MBOn that interact with MBn axons in the MB neuropil? A consensus model emerging from the contributions of multiple laboratories is that appetitive or aversive reinforcing DAn synapse onto the MBn carrying CS information. This interaction modifies synaptic communication to the MBOn to influence behavioral attraction or avoidance (Fig. 7) (Aso et al. 2014a, 2014b; Griffith 2014; Cohn et al. 2015; Hige et al. 2015; Owald and Waddell 2015; Aso and Rubin 2016; Kaun and Rothenfluh 2017; Cognigni et al. 2018; Amin and Lin, 2019). The 12 PPL1n DAn provide negative US valence, and the ∼100 PAM DAn provide positive valence to the MBn. In essence, the DAn input to the MB axons biases the valence of the odor CS with which it is paired by activating the dDA1 receptor (also known as DopR1) and initiating cAMP signaling pathways. This alters the MBn output to the MBOn, depressing the activity of these downstream neurons. Learning using an aversive US leads to CS avoidance by depressing the MBOn that drive approach behavior, whereas learning using an appetitive US leads to CS approach by depressing MBOn that drive avoidance behavior. Expressed in another way, DAn input assigns a valence—punishment or reward—to a given odor used as a CS during conditioning and represented by a subset of MBn axons. Processes in the MBn integrate the temporal coincidence of the CS and US and inhibit downstream MBOn that ultimately cause the fly to approach odors paired with appetitive stimuli or avoid odors paired with aversive stimuli. This model speaks to the role of DAn in the first step of memory formation—acquisition (Fig. 1). However, DAn have post-acquisition roles as well, highlighted by the ongoing activity of these neurons for active forgetting as discussed below. Additional studies and revised models (Ichinose et al. 2021; Adel and Griffith 2021) should help to disentangle the specific roles for DAn and MBOn across the four operations of memory (Fig. 1).
Fig. 7.
a) DAn in the PPL1 cluster provide aversive information to the MBn axons in the vertical lobe compartments, while DAn in the PAM cluster provide primarily appetitive information to the horizontal lobe compartments. This valence is integrated with odor information carried by the MBn axons and conveyed to the MBOn. Aversive signals decrease the activity of “approach” MBOn, while appetitive signals decrease the activity of “avoidance” MBOn. b) Color map showing the compartments innervated by PPL1- and PAM-DAn. c) Color map indicating the neurotransmitter phenotype of the compartment specific MBOn. Reproduced with permission from Cognigni et al., 2018.
The simple model ascribing valence to the binary input of two clusters of DAn and the behavioral response of attraction or avoidance to the balanced output of MBOn (Fig. 7) offers an explanation to some long-standing observations on olfactory classical conditioning in the fly. For instance, both appetitive and aversive conditioning require MBn expression of the dDA1 receptor, the rutabaga-encoded adenylyl cyclase, and downstream cAMP signaling (Davis 2005), and are thus independent of US valence. The model ascribes valence to independent channels of US input from the panel of DAn, and the behavioral response to distinct outputs from the panel of MBOn. Thus, the molecular signaling through the cAMP signaling system that occurs in the MBn during learning can be viewed as a universal mechanism for generating plasticity (Louis et al. 2018), with distinct behavioral responses attributed to distinct circuitry for valence. However, there are additional dimensions to the process of olfactory classical conditioning that need discussion in the context of the valence and behavioral action model.
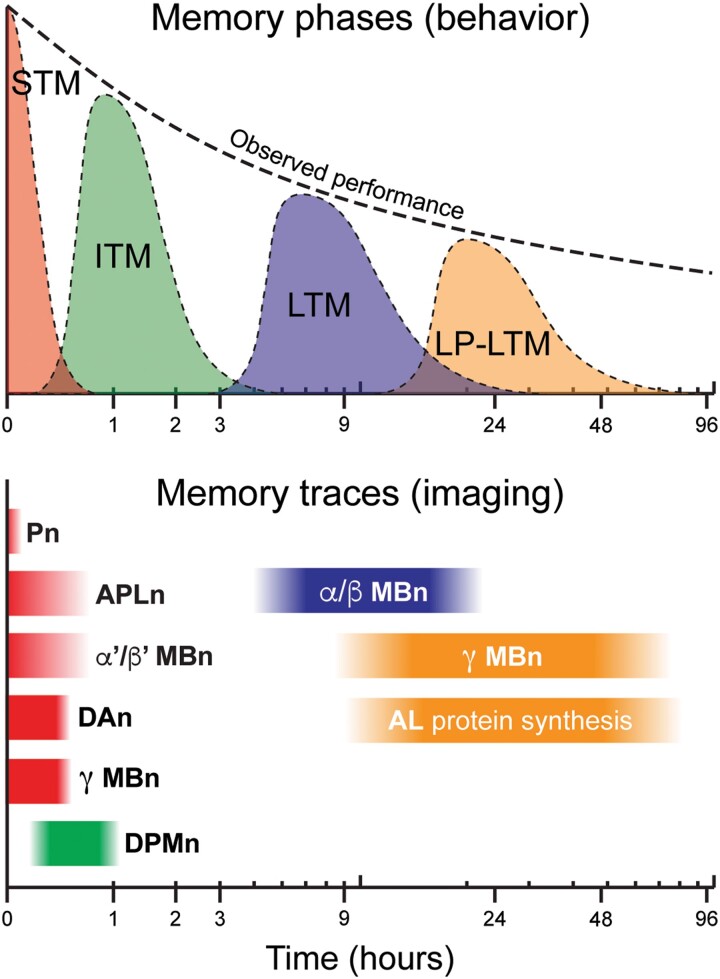
First, olfactory classical conditioning generates distinct temporal forms of memory, including short-term memory (STM), intermediate-term memory (ITM), and several forms of long-term memory (LTM; Fig. 8). The LTM forms include anesthesia-resistant memory (ARM), PSD-LTM, and late-phase LTM (LP-LTM). Current evidence from functional imaging experiments that detect the onset and persistence of cellular memory traces indicate these temporal forms are influenced by different populations of neurons in the olfactory pathway, with STM being influenced primarily by Pn, APLn, γ MBn (Boto et al. 2019), and α′/β′ MBn (Ueno et al. 2013; Zhang et al. 2019; Adel et al. 2022); ITM by DPMn; and forms of LTM by α/β and γ MBn, and ALn (early studies reviewed by Tomchik and Davis 2013). The relevance of the α/β MBn PSD-LTM cellular trace is supported by its abolishment in 26 different LTM mutants (Akalal et al. 2011). Short-term cellular memory traces also form in the α′3 MBOn (Zhang et al. 2019) and the γ2α′1 MBOn (Berry et al. 2018). More recent imaging approaches have advanced the technology to image calcium transients in individual presynaptic boutons (Bilz et al. 2020).
Fig. 8.
Memory phases and cellular memory traces. Behavioral memory expression is the combination of separate memory phases: short-term memory (STM), intermediate-term memory (ITM), long-term memory (LTM), and late-phase long-term memory (LP-LTM). Cellular memory traces have been found in Pn, APLn, α′/β′ MBn, DPMn, α/β MBn, γ MBn, DAn, and MBOn. Modified with permission from Tomchik and Davis 2013.
Neuronal blocking and spatial rescue experiments of Drosophila mutants are broadly aligned with the above summary, but with some minor differences (Tomchik and Davis 2013). First, STM in these experiments is heavily weighted towards γ MBn involvement, as rut expression in these cells rescues this temporal form, despite the multiple cellular memory traces assigned to this form. In addition, some research indicates that α/β MBn influence STM (McGuire et al. 2001; Dubnau et al. 2001), yet no memory trace for this form has been discovered in these neurons. These differences can easily be explained by the variable importance of individual cellular traces for behavioral memory and the limited set of fluorescent reporters (calcium, cAMP, etc.) for specific cellular functions that may underlie memory formation.
Second, not all DAn are equally potent in assigning valence and behavioral response patterns. Mao and Davis (2009) surveyed properties of the five types of PPL1 DAn and found distinct differences in response to electric shock, odor, and odor/shock pairing. This functional heterogeneity is echoed in subsequent studies of PAM DAn. For instance, PAM DAn that serve as water reinforcers for thirsty flies are distinct from those that provide reinforcement for nutritious sugars; and those that offer reinforcement for sweetness are distinct from those for nutrient value (Huetteroth et al. 2015). Sweetness is detected by OAn neurons, which synapse on a subpopulation of DAn that express the OA receptor, OAMB (Fig. 5). These DAn convey reinforcement for sweet taste and project to MBn axonal compartments that are different from those that convey nutrition. Such functional heterogeneity was predicted from the structural heterogeneity of the DAn (Mao and Davis 2009), with additional examples of functional heterogeneity being constantly discovered.
Similarly, the MBOn are not equally potent in driving approach or avoidance behavior and in regulating short-term appetitive and aversive olfactory memory expression. Photoactivation experiments using red-shifted CsChrimson revealed that activation of individual MBOn that innervate the horizontal lobes fall into two major classes (Aso et al. 2014a; Aso and Rubin 2016). The photoactivation of 10 such glutamatergic MBOn (Fig. 7c) led to avoidance behavior to red light, while the photoactivation of three such GABAergic MBOn led to attraction. Photoactivation of eight cholinergic MBOn that innervate the vertical lobes led to attraction to the light stimulus. In all cases, the magnitude of the effect varied due to the inherent potency of individual MBOn and/or the strength of the driver line used to express CsChrimson.
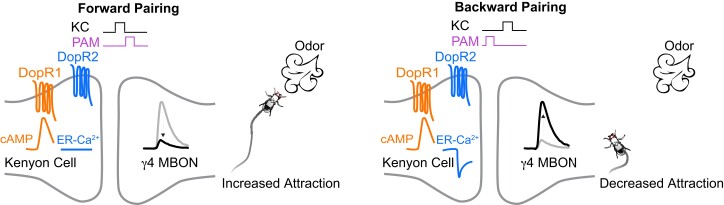
Third, what occurs inside of the MBn axons in response to DA input and how is synaptic strength of MBOn altered by conditioning? The axons of MBn express four distinct DA receptors—DopEcR, dDA1, DAMB, and D2R (Kondo et al. 2020)—with dDA1 and DAMB being the most extensively studied. The dDA1 receptor is coupled to Gαs leading to cAMP generation and protein kinase A activation. The DAMB receptor is coupled to Gαq leading to inositol phosphate (IP3) generation and IP3-dependent calcium efflux from the endoplasmic reticulum (Boto et al. 2014; Himmelreich et al. 2017; Handler et al. 2019). Using a novel conditioning paradigm which allows the investigator to rapidly change the temporal pattern of CS/US contingency from trial-to-trial, Handler et al. (2019) demonstrated that forward pairing with an appetitive odor and optogenetic activation of DAn leads to the anticipated change in walking behavior: increased approach with PAM activation and decreased approach with PPL1 activation. Backward pairing immediately after forward pairing reverses the change in behavior, indicating that flies can rapidly update their behavior on a trial-by-trial basis (Fig. 9). Other experiments showed that forward pairing decreases MBn > MBOn synaptic strength, and backward pairing reverses this change. Moreover, forward pairing activates cAMP accumulation and requires the dDA1 receptor, whereas backward pairing requires DAMB and leads to calcium release from the ER. Thus, these two DA receptors expressed in MBn axons oppose each other's effects in mediating approach vs avoidance behavior and MBn > MBOn synaptic strength. These observations may speak to the respective roles of the two receptors in behavioral acquisition and forgetting, respectively (see below).
Fig. 9.
Cartoon illustrating that forward pairing of an appetitive odor with PAM-DAn stimulation leads dDA1 receptor activation and cAMP accumulation, a decreased response to the studied MBOn, and behavioral attraction. Backwards pairing of the odor and PAM-DAn stimulation leads to DAMB activation and calcium release from the endoplasmic reticulum (ER), an increased response of the MBOn, and decreased attraction. Reproduced with permission from Handler et al. 2019.
Fourth, evidence has been presented indicating that some DAn are subject to feedback from their associated MBn or MBOn (Cervantes-Sandoval et al. 2017; Pavlowsky et al. 2018). Feedback mechanisms indicate a deeper complexity to the information processing not captured by the first-generation models (Adel and Griffith 2021).
Finally, an alternative model for long-term synaptic enhancement at Pn > MBn synapses has been presented that removes DA from conveying the US sensory input (Ueno et al. 2017; Saitoe et al. 2022). Rather, the data are consistent with a model in which olfactory information is transmitted to the MBn using nAChR activation and the aversive shock US sensory input transmitted to MBn via NMDA receptors from the ascending ventral nerve cord. DA release from pre-synaptic DAn is dependent on the coincidence of nAChR and NMDA receptor activation in the post-synaptic MBn, and a possible retrograde carbon monoxide signal from the MBn to the DAn. Similarly, an updated associative memory circuit model has been presented that addresses the major contradictions in the current model and incorporates the well-known concept of DA-based reward prediction error established from mammalian studies (Adel and Griffith 2021). Such models need to be embraced and tested, since the probability of any current model surviving the next decade of research is nil given the complexity of the system.
Calyx function in learning and memory
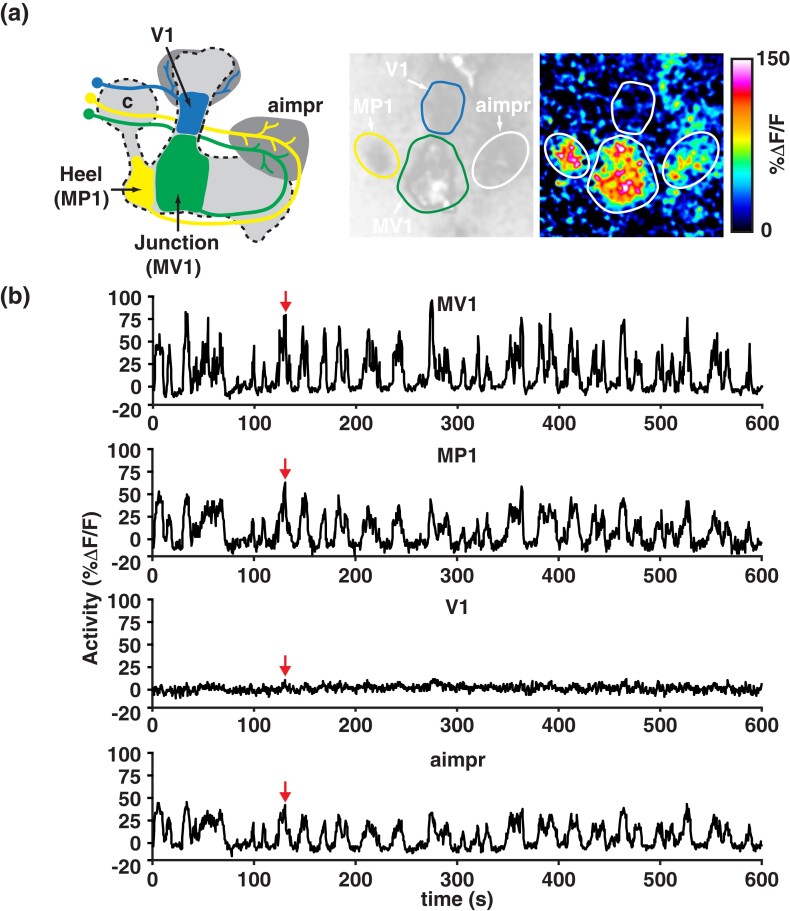
Most research, to date, has focused on the role of the MBn axons in the vertical and horizontal lobes and associated neurons in olfactory memory formation. This has led to very significant information gains about MBOn and DAn that innervate the MBn axons. However, a third cluster of DAn, named PPL2, innervates the calyx of the MB and a few additional brain regions (Fig. 3b). These neurons had no known role in memory formation until recently.
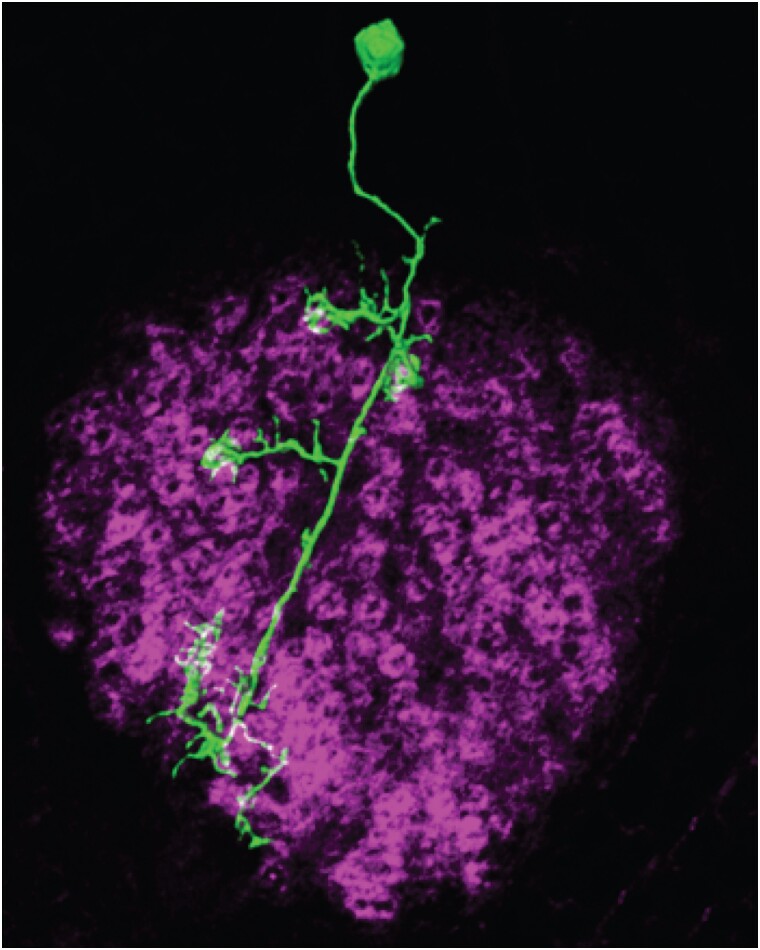
The calyx contains the dendrites of the MBn, a network of glial processes, GABAergic axon terminals from the APLn, and pre-synaptic elements from AL Pn (Leiss et al. 2009; Liu et al. 2009; Butcher et al. 2012; Li et al. 2020; Baltruschat et al. 2021). Most of the actin-rich, post-synaptic terminals of MBn form claw-like structures, with the claws from 11 MBn, on average, forming each of about 1,000 rings (or spheres in three dimensions) of actin-rich processes known as microglomeruli (Fig. 10). Pn branch as they enter the calyx to innervate the microglomeruli, with the center of the microglomerulus filled primarily by a single cholinergic Pn bouton along with smaller GABAergic terminals. This organization indicates that each Pn contacts several MBn, so that each MBn receives information from multiple Pn terminals located in different microglomeruli, and each microglomerulus receives GABAergic inhibition through synaptic connections onto MBn dendrites and the presynaptic terminals of Pn.
Fig. 10.
Dendritic projections from a single MBn (green) extending into the calyx, with claw-like dendritic terminals. The microglomeruli are visible as ring-like structures highlighted by actin staining (magenta). Reproduced with permission from Leiss et al. 2009.
Several advances were made in the last decade regarding the function of Pn and MBn dendrites in memory formation. Baltruschat et al. (2021) demonstrated that consolidation of appetitive olfactory memory is accompanied by structural changes in the microglomeruli delivering the CS to the post-synaptic MBn. Differential conditioning using two odors generates STM and LTM to the odor paired with the appetitive US and an increase in the number of microglomeruli that represent the conditioned odor after PSD-LTM formation. This observation suggests that new boutons are formed, or recruited, during the consolidation of appetitive LTM. Cell body responses should be a downstream effect of dendritic responses. Thus, the increased number of MBn cell bodies exhibiting a significant calcium response to the CS odor after aversive LTM may be a related observation (Delestro et al. 2020). Although the mechanism underlying the recruitment of additional microglomeruli into the representation of the CS odor for PSD-LTM remains unknown, an intriguing possibility is that this occurs as a downstream effect of the recruitment of additional AL glomeruli into the CS representation during STM formation (Yu et al. 2004).
Two studies were recently reported that are consistent with a model that DA input to the cell bodies and/or calyx of the MBn work to increase the salience of a learned odor, leaving valence to be registered by DA inputs into the lobes. Boto et al. (2019) investigated plasticity in the MB γ lobe using G-CaMP as a reporter after pairing odor with the stimulation of PPL1, PPL2, or both classes of DAn. They observed an increase in odor responses after forward but not backward pairing using the TH-gal4 driver—estimated to persist for ∼30 min, consistent with γ lobe involvement in STM described above. This plasticity was also observed upon imaging the calyx, suggesting that the lobe plasticity is a downstream effect of the dendritic plasticity. Although PPL2 neurons respond to odors, pairing odor with PPL2 stimulation failed to produce significant behavioral memory, unlike similar experiments with odor stimulation and PPL1 pairing. In addition, the PPL2-DAn had no effect on appetitive odor learning. Nevertheless, the odor/PPL2 pairing increased the response in the γ lobe MBn axons. These data suggest that PPL2-DAn enhance odor-evoked responses in the MBn but do not play a part in valence.
Qiao et al. (2022) brought electrophysiological analyses of the Pn > MBn synapse into play and discovered a timing-dependent facilitation in evoked excitatory post-synaptic potential that occurs with AL stimulation and depolarization of the MBn. The facilitation persists for >60 min and is observed only with Pn connections to γ MBn. It requires normal expression of Dop2R, a D2-like receptor, on the cell bodies and dendrites of γ MBn. Surprisingly, the DA input was attributed to the PPL1 cluster of DAn, which have no known direct connections to this region. The connection may be indirect. The authors hypothesize that the long-lasting facilitation offers a mechanism for increasing the salience of a learned odor, like the studies by Boto et al. (2019).
The above summary represents a simplified overview of calyx organization and known roles in memory formation. For instance, Christiansen et al. (2011) reported the existence of active presynaptic sites along the dendrites of γ and α/β MBn, but not α′/β′ MBn. These will probably prove to be important for memory formation. In addition, there are known morphological and functional subdivisions of the entire calyx region beyond the main calyx described above. These include dorsal, lateral, and ventral accessory calyces (d-, l-, vACA). Connectome analysis (Yagi et al. 2016; Li et al. 2020) indicates that the vACA and dACA receive predominantly visual input, and the lACA predominantly thermosensory information. Thus, the accessory calyces appear to be more dedicated to non-olfactory information. Bielopolski et al. (2019) reported that odor responses of γ MBn are limited by activation of muscarinic ACh receptors expressed on the dendrites, and that receptor function is required for effective conditioning. Andlauer et al. (2014) identified a novel synaptic protein, Drep-2, that is enriched at the MBn postsynaptic terminals in the calyx. Drep-2 is required for aversive STM and probably interacts with mGluR proteins for their signaling function. There remains much more to learn about the involvement of MBn dendrites and the calyx in olfactory memory formation, and how learning induced changes in the calyx are integrated with those in the lobes to produce behavioral memory.
Larval learning
Most of the progress in memory formation in the last decade has come by using adult flies. However, larvae were shown to learn environmental information soon after olfactory memory formation was demonstrated with adult flies (Aceves-Piña and Quinn 1979). The interest in larvae as a subject for learning assays was renewed by the development of robust odor-taste paradigms in the early 2000s (Gerber and Stocker 2007). Larvae provide both advantages and disadvantages over adult flies for research into the mechanisms of memory formation. The major advantages include their small size, translucent body wall, and simpler nervous system, which consists of ∼10,000 instead of ∼100,000 adult brain neurons (Thum and Gerber 2019). Given the complexity of the adult nervous system described earlier, even just within the MBn lobe neuropil, an order of magnitude reduction in the number of neurons offers a very significant advantage. A possible disadvantage is that they are continuously developing, so experimental results could vary by using larvae that differ in age by only a few hours or days. Nevertheless, the last decade has witnessed increased effort and progress in studying larval learning.
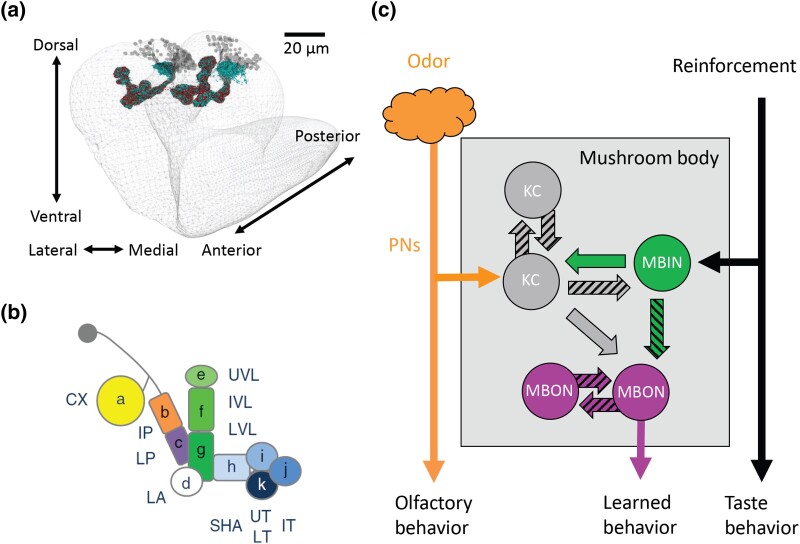
Many of the techniques used to study adult memory formation are used with larvae. In addition, the broad anatomical and functional features of the adult brain are found in the larval brain (Fig. 11a), with the notable exception of the visual system. For instance, the olfactory nervous system is conserved in structure and function, but in miniature (Eichler et al. 2017; Saumweber et al. 2018; Thum and Gerber 2019). The larval MB is considered the major center for olfactory memory formation. At 6 hr of age (1st instar), there are about 200 MBn vs 2,000 in the adult. As with the adult MB, the axons tracts in the lobes are compartmentalized, defined by the spatial segregation of connections with MB input neurons (MBIn) that are dopaminergic, octopaminergic, or of unknown nature, and by connections with MBOn (Fig. 11b). Each compartment is innervated by 1–3 MBIn and 1–5 MBOn. Similarly, there exists reciprocal connections between the GABAergic APLn and MBn in the larval calyx as in adult flies. The interneuron structural connections within the MB lobe neuropil are similarly complex (Fig. 11c), with connections between MBn > MBIn, MBn > MBOn, MBn > MBn, MBn > APLn, MBIn > MBn, and MBIn > MBOn. Most MBn > MBn synapses are axo-axonic and located in the peduncle and lobes. Multiple connections exist between the MBOn of different compartments. Additional complexity was recently added with the discovery that each MBOn sends information back to the dopaminergic MBIn via feedback neurons (Eschbach et al. 2020). Indeed, the MBIn typically receive more than half of their input from such feedback. One prominent motif discovered is the convergence of both excitatory and inhibitory connections from MBOn that sample opposite valence compartments, which may allow MBIn to integrate appetitive and aversive signals.
Fig. 11.
a) a reconstruction of the larval MB from 3-dimensional electron microscopy, showing the MBn cell bodies, the calyx, and the MB vertical and horizontal lobes. b) The MB lobes have both input neurons (MBIn) and output neurons (MBOn), organized as 11 compartments. c) The circuit organization is like adult flies, with Pn providing olfactory input to the MBn, the MBIn providing input to both the MBn and MBOn, and with reciprocal communication between the population of MBn and the population of MBOn. Reproduced with permission from Thum and Gerber 2019.
The function of the MBn as conveying olfactory information and integrating this information with other inputs is thus conserved between larvae and adult flies. In addition, the modulatory DA and OA neurons can drive aversive and appetitive olfactory conditioning with their optogenetic activation (Schroll et al. 2006), with DAn of the DL1 cluster innervating the vertical lobes and driving aversive conditioning, and DAn from the pPAM cluster innervating the horizontal lobes and driving appetitive conditioning (Eichler et al. 2017; Saumweber et al. 2018; Thum and Gerber 2019). It is important to recognize that the discovery of sufficiency was first made using larvae.
Larvae also display olfactory STM, LTM, and ARM like adult flies (Thum and Gerber 2019; Lesar et al. 2021). STM depends on the normal function of genes encoding classical cAMP signaling components (Davis 1993, 1996). Protein synthesis dependent LTM forms after prolonged or repeated olfactory training and requires cAMP response element-binding protein (CREB) function. ARM is resistant to cold-shock anesthesia and forms concurrently with STM but decays more slowly. As with adult flies, ARM requires the normal function of several genes including radish, protein kinase C, and bruchpilot.
Research on olfactory memory formation using larvae previously lagged that using the adult fly, with a few notable and recent discoveries. Nevertheless, the conservation of the olfactory nervous system but with reduced complexity, conditioning principles, available connectomics, and recent progress may slingshot the larvae ahead of the adult in producing new conceptual insights across the next decade of research (Thum and Gerber 2019).
Advances in behavioral genetics
Although research of Drosophila memory formation was born from behavioral genetic approaches, new tools and approaches that were subsequently introduced widened the research scope. Nevertheless, behavioral genetics research has continued to flourish across the last decade and produced new insights into how specific molecules mediate memory formation. Indeed, the power of the fly for memory research now lies in combining behavioral genetics and systems neuroscience approaches. This synthesis is necessary for obtaining a deep understanding of memory formation.
CREB function and PSD-LTM
STM and LTM occur after training due to cellular memory traces that form in multiple neuronal populations, or nodes, of the nervous system (Fig. 1 and 8). For the olfactory nervous system, the nodes can include the ALn, MBn, DAn, APLn, DPMn, MBOn, and others. In principle, it could be that STM is molecularly encoded in neurons at some nodes and LTM in others. The extreme alternative model is that both STM and LTM are encoded in all neurons and all nodes of the circuit. Several different advances were recently made that address this and other important issues concerning CREB and its role in PSD-LTM.
Importantly, four papers reported that CREB activity and/or protein synthesis is required in at least seven nodes to support olfactory PSD-LTM: the α/β and α′/β′ MBn (but not the γ MBn or DAn); the dorsal-anterior-lateral neurons; and the α3, γ3, γ3β′1, β′2mp MBOn (Chen et al. 2012; Hirano et al. 2013; Plaçais et al. 2013; Wu et al. 2017; Widmer et al. 2018). This is an important insight that can be contrasted with earlier thoughts that perhaps PSD-LTM is a dedicated function of a singular node, e.g. the MBn. In addition, this insight prompts the question of whether CREB signaling and/or protein synthesis works in similar ways in different nodes of the circuit. However, it is notable that the reports above examined a mix of aversive and appetitive PSD-LTM, and CREB may have distinct functions due to the type of conditioning used. In addition, Plaçais et al. (2013) made the interesting observation that α3 MBOn activity is required at retrieval but not before, so that CREB may participate in establishing retrieval machinery in this neuron, but in building consolidated synapses after conditioning in others. Moreover, Wu et al. (2017) attribute the effects of blocking protein synthesis in the γ3, γ3β′1, β′2mp MBOn to the role for Orb2-mediated local protein synthesis (see below), rather than to CREB nuclear signaling. An early study showed that PSD-LTM is also represented by new protein synthesis in AL Pn (Ashraf et al. 2006). In sum, the collective data indicate that PSD-LTM is associated with changes in protein synthesis across multiple nodes of the sensory system rather than in a single node.
Miyashita et al. (2018) turned their attention to another important, long-standing question of how the duration and spacing of training bouts dictates the probability of forming PSD-LTM after aversive, differential conditioning (Tully et al. 1994). In this conditioning paradigm, aversive olfactory PSD-LTM is formed by spaced training, in which flies are presented 5–10 sessions of conditioning separated by an optimal period of rest of ∼15 min. The critical importance of rest periods between multiple training epochs dates to similar studies using the sea snail, Aplysia (Carew and Kandel, 1973). An equal amount of conditioning without rest periods fails to generate PSD-LTM. Thus, a mystery has persisted regarding the molecular and cellular interactions that occur during rest periods between bouts of conditioning. However, in an important and recent twist, Zhao et al. (2019) found that LTM forms after a single training trial and in the presence of protein synthesis inhibitors if a copper shock grid, originally used only in tubes used for training, is included in the odor-choice distribution tubes during testing. Thus, the spaced training requirement appears to be specific to LTM of the odor cues, since the requirement is removed when the context used for testing is the same as that used for training.
Miashita and colleague discovered that rest periods are required to stably engage dynamic and reciprocal transcriptional cycling in the α/β MBn between CREB, whose expression is regulated by the transcription factor, AP-1; and c-Fos, a CREB-regulated component of the AP-1 transcription factor. The activity of both transcription factors is regulated by the protein kinase, ERK. ERK activity (pERK) decreases across a training cycle (Pagani et al. 2009), but then increases during a subsequent rest period, reaching a maximum at ∼8 min into the rest period. The increased pERK activity during the rest period activates CREB activity which in turn, transcriptionally enhances c-Fos. The result is transcriptional cycling between CREB and c-Fos, which is thought to define the neurons that encode PSD-LTM from the expression of downstream genes regulated by these transcription factors. The report also included data indicating that both dDA1 and rutabaga AC activity are required for rest period activation of ERK, and that memory recall requires reactivation of the cFos/CREB cycling α/β MBn. Awata et al. (2019) derived a circuit-based explanation for the spacing effect. Their results suggest that the activity of a subset of γMBn is decreased and paralleled by decreased pERK activity due to spaced conditioning, and this leads to decreased activity of a postsynaptic MBOn (MBOn-γ1pedc > α/β). The MBOn positively influences the activity of a downstream DAn, PPL1α′2α2, which in turn, synapses upon a subset of the α/β MBn to facilitate PSD-LTM. Complicated? Without a doubt. Has the mystery of the spaced training effect been solved? No. Nevertheless, there are now viable models backed by molecular, circuit, and behavioral data that can be further explored in search for a complete answer.
Lin et al. (2022) re-investigated the discrepant results previously reported that heat-shock-induced expression of a transgene encoding one CREBB isoform, CREBB-a, enhances PSD-LTM formation (Yin et al. 1995) or is without effect (Perazzona et al. 2004). The latter study discovered a nonsense mutation in the previously used transgenic construct and found no effect on PSD-LTM formation with the original construct or one with the nonsense mutation repaired. Lin et al. (2022) employed the corrected CREBB-a construct and confirmed the results showing no effect on PSD-LTM after heat-shock-induced overexpression. However, they report that PSD-LTM was formed after minimal training when the corrected construct was expressed in the α/β MBn using Gal4 drivers and attributed the early discrepant results due partly to variable and weak expression of heat shock promoters. They also found that 5-HT1A receptor expression generated by weak training protocols inhibits PSD-LTM formation and strong training promotes CREBB expression that overcomes this inhibition. These observations make three important conclusions. First, the corrected CREBB-a construct can promote PSD-LTM when expressed specifically in the α/β MBn. Second, PSD-LTM as mediated by CREBB occurs at least in part, through MBn involvement, reproducing earlier results (Yu et al. 2006; Hirano et al. 2013; Widmer et al. 2018). Third, there exist both molecular suppressors and enhancers of PSD-LTM. It remains difficult to explain the PSD-LTM-enhancing effects of the nonsense-mutation carrying transgene originally reported.
Further insights into the regulation of CREBB and PSD-LTM were made by Lee et al. (2018). They focused on protein kinase genes that are expressed in the MBn. Beginning with 27 putative kinase-encoding genes, they found that 12 are expressed in the MBn and used RNAi knockdown to investigate their functional importance. They identified a new, evolutionary-conserved serine/threonine protein kinase gene, named Meng-Po, that impaired 3 hr and PSD-LTM aversive memory but not acquisition. Overexpressing Meng-Po enhanced 24 hr PSD-LTM and the activity of CREBB. Although Meng-Po influences CREBB activity, it is not a substrate of Meng-Po. Rather Meng-Po influences the levels of CREBB by promoting the translation or stabilization of CREBB. Meng-Po, like CREB, is phosphorylated by PKA and this phosphorylation enhances its activity. Thus, the two kinases, PKA and Meng-Po, work together to regulate CREBB activity and PSD-LTM. It remains unknown how the activities of Meng-Po and PKA are integrated with the activity of pERK to perhaps initiate the transcriptional cycling reported by Miyashita et al. (2018).
Orb2 and PSD-LTM
Several advances were recently made concerning the role of Orb2 proteins and PSD-LTM consolidation. Orb2 encodes isoforms of the cytoplasmic polyadenylation element binding (CPEB) protein subfamily-II, which are candidate proteins for the synaptic tag that marks specific synapses to be selectively modified for enduring memory. Like the selection of a few cells to become engram cells within a much larger population (Fig. 1b), conditioning probably engages many synaptic elements of which only a fraction is reorganized to provide long-lasting, local PSD changes in synaptic strength. The CPEB proteins bind to polyadenylation elements in synaptically localized mRNAs and help trigger their polyadenylation and translation. Members of the CPEB-II family have been found to function in synaptic plasticity and LTM.
Two reports published in 2012 extended this hypothesis and crystallized the fact that the formation of multimers through the polyglutamine domains is critical for LTM persistence (Krüttner et al. 2012; Majumdar et al. 2012). Orb2 encodes two protein isoforms of primary interest, Orb2A and Orb2B, that are broadly expressed in the adult brain and localized synaptically. They both contain a glutamine-rich (Q-domain) and RNA-binding domain. The Q-domain is dispensable in both isoforms for STM formation, but both protein isoforms are required for PSD-LTM. For PSD-LTM, Orb2A requires only the Q-domain and Orb2B only the RNA binding domain, indicating separable functions. Orb2A is expressed at very low levels in the adult brain relative to Orb2B but together, and with neural activity they can form oligomeric and filamentous assemblies. The Orb2A Q-domain and the N-terminal 8 amino acids before the Q-domain are particularly important in seeding the formation of these self-assembling, heat and detergent stable assemblies that take on the appearance of amyloid fibrils (Hervas et al. 2020). A point mutation within the 8 N-terminal amino acids [Orb2A-F5Y] was identified that has a 5× reduced propensity to oligomerize. LTM persists for 1, but not 2 days, using flies that express this mutant form, suggesting that fibril formation is critical for the persistence of PSD-LTM. Interestingly, the Orb2A transcript expressed in the adult nervous system retains an unspliced intron that attenuates protein expression. Behavioral conditioning in at least two paradigms, including appetitive classical conditioning, facilitates Orb2A splicing, protein expression, and LTM (Gill et al. 2017). Thus, experience-dependent intron removal serves as a gate for LTM expression.
Other reports investigated the neuromodulatory pathway that leads to acquisition and consolidation of courtship conditioning memory involving Orb2 (Keleman et al. 2012; Krüttner et al. 2015). Courtship conditioning is a robust form of memory whereby attempts of a naïve male to court an unreceptive mated female leads to the suppression of subsequent courtship behavior. The activity of as few as 2–6 DAn, named aSP13 and located in the PAM cluster, were found to be required for the acquisition and consolidation of LTM of courtship conditioning. The role in LTM acquisition was demonstrated by blocking the aSP13 DAn concurrent with STM conditioning. The role in LTM consolidation was demonstrated by artificially activating or blocking the neurons at a remarkably long period after the STM conditioning trials (8–11 hr later). The dDA1 receptor expression in γ MBn is required to consolidate STM into PSD-LTM. Consistent with prior results, PSD-LTM was formed and consolidated only when the Q-domain of synaptically located Orb2A in the MBn and the RNA binding domain of Orb2B remained intact.
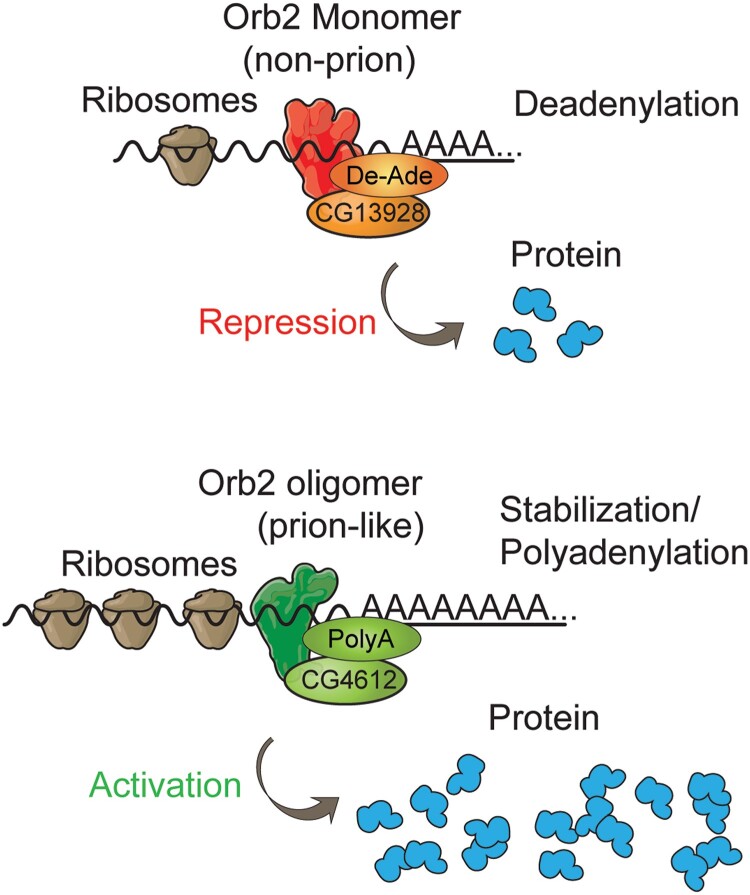
Using a combination of in vivo and in vitro biochemical studies, Khan et al. (2015) contributed an extraordinarily deep study of Orb2 focused on the role of monomeric vs oligomeric forms in mRNA binding, stability, and translatability, for LTM formation. They identified sequences in the 3′ UTR mRNA of a model gene known to be involved in LTM that are involved in recruiting Orb2, and subsequently showed that mutations of these 3′ UTR sequences increased the translation of an attached reporter gene in a system where only Orb2 monomers were present. This indicated that monomers of Orb2 inhibit translation. When oligomeric Orb2 was present, translation increased. Mutations in the Orb2 binding site of the mRNA reduced the monomer-dependent decrease and abolished the oligomer-dependent increase in translation. The enhanced translation required the Q-domain of Orb2A but not the RNA binding domain, along with Orb2B, indicating that increased translation occurs through oligomerization of Orb2A to Orb2B. What effect does monomeric and oligomeric Orb2 have on mRNA structure? Orb2A monomer promotes deadenylation of the poly(A) tail and destabilizes the mRNA, accounting for inhibited translation. Oligomeric Orb2 stabilizes mRNA, at least in part, by elongating the poly(A) tail. These effects occur through Orb2-interacting protein complexes identified from pull-down experiments. Translational repressors that promote deadenylation were identified including the novel protein, CG13928. And proteins that enhance polyadenylation and translation, including CG4612, were identified as interacting with Orb2 oligomers. A final connection was made between CG4612 and LTM: RNAi knockdown of this protein in the γ MBn inhibits LTM formation. These studies support the attractive model that Orb2 monomers interact with proteins that cause deadenylation and subsequent translation repression, and oligomers forming between Orb2A and Orb2B recruit proteins that increase polyadenylation and protein translation (Fig. 12) to facilitate LTM formation and stability. Thus, oligomeric Orb2 that forms in the γ MBn axons represents a long-term, biochemical memory trace (Li et al. 2016).
Fig. 12.
Model for the role of Orb2 monomers and oligomers in mRNA stability and protein translation. Reproduced with permission from Khan et al. 2015.
There are other notable and recent contributions made towards understanding the role of individual genes in Drosophila learning and memory that do not lend themselves to the topics discussed. A sampling of these is listed in Table 1 along with a short summary of the advances made.
Table 1.
A sampling of genes recently identified using behavioral genetics and their reported involvement in memory formation.
| Report | Summary |
|---|---|
| Wu et al., 2011 | APLn and DPMn form Gap junctions within the MB neuropil that are required for robust olfactory STM. |
| Miyashita et al., 2012 | Flies defective for the Mg2+ block of the NMDA receptor are defective in olfactory PSD-LTM and CREB activation. |
| Tan et al., 2013 | Wnt signaling through armadillo, arrow, and Wingless in MBn is required for olfactory PSD-LTM. |
| Zhang and Roman, 2013 | Go signaling in the γ MBn flies is required for decreased synaptic release elicited by the CS- odor relative to the CS+ odor and normal differential conditioning. |
| Masek et al., 2015 | Single fly proboscis extension assays reveal that aversive taste memory require two groups of PPL1 DAn and their corresponding MBOn that innervate the α MBn axons. |
| Niewalda et al, 2015 | Synapsin levels in the MBn influence the strength of both punishment and relief-learning. |
| Hirano et al., 2016 | Describes PSD-LTM maintenance as requiring the activity of transcription factors CREB/CBP, followed by CREB/CRTC, and then Bx. |
| Liu et al., 2016a, 2016b | Gap junctions are required in α/β and α′/β′ MBn for normal visual learning and memory. |
| Drago and Davis 2016 | Mitochondrial calcium uniporter activity is required during development for normal adult olfactory STM. |
| Scholz-Kornehl and Schwärzel, 2016 | The dopamine D2 receptors are required for normal ARM in diverse neuron types. |
| Scheunemann et al., 2018 | The dunce-encoded cAMP phosphodiesterase is inhibited during LTM formation in a pair of serotonergic neurons that activate DAn. |
| Yamazaki et al., 2018 | MBn encode valence like DAn, with two subpopulations of γMBn, termed γCRE-p and γCRE-n, for aversive and appetitive cues, respectively. |
| APLn are suppressed at acquisition from pre-synaptic PPL1 DAn. The suppression, needed for efficient learning, is mediated by APLn-expressed DopR2 receptors. | |
| Jacob and Waddell, 2020 | Demonstrates that aversive spaced conditioning can generate both an aversive memory for the CS+ odor and a safety-memory for the CS- odor. |
| Lin et al., 2021 | PSD-LTM is enhanced upon over-expressing the activating form of CREB, CREBA, in the DALn. |
| Yamagata et al., 2021 | APLn provide inhibitory input into the presynaptic terminals of PAM DAn through the GABA-B-R3 receptor. The APLn input through the receptor regulates the salience of rewarding DA inputs to the MBn and odor CS specificity. |
| Zhao et al., 2021 | PSD-LTM is formed after a single, aversive, olfactory differential conditioning trial if the CS+ and CS- are tested against a 3rd novel odor, rather than between themselves. |
Memory suppressor molecules and genes
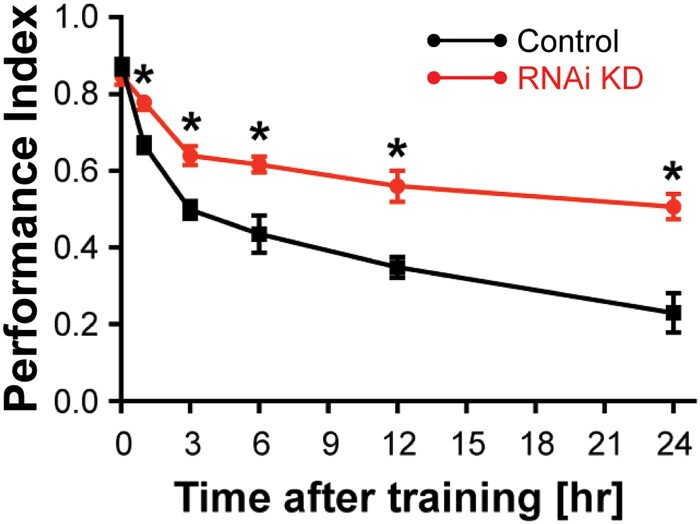
Although early research of Drosophila memory formation focused on identifying mutants that impair memory formation, more recent strategies have sought gene expression impairments that increase memory formation (Fig. 13). Such genes are termed memory suppressor genes since the function of the unimpaired gene is to limit memory formation. The number and diversity of memory suppressor genes is surprising, with almost 100 currently identified in Drosophila and mammalian systems (Noyes et al. 2021). This fact has led to the realization that the brain is designed not only with molecular and cellular processes to form memories, but also with molecules and cellular processes that suppress the formation and retention of memory.
Fig. 13.
An example of enhanced memory. A forgetting curve for the memory suppressor gene, scribble, showing reduced rate of forgetting with RNAi knockdown. Reproduced with permission from Cervantes-Sandoval et al. 2016).
Memory suppressor molecules and processes are revealed by genetic or pharmacological insults that lead to enhanced memory expression. The recent focused and systematic research on memory suppressor genes is important for two reasons. First, memory suppressors identify the control points in neurophysiological processes that limit memory formation. Second, memory suppressors may be relevant for understanding enhanced and compromised memory that occurs in the human population. Savant syndrome, for example, is characterized by remarkable memory capacity, often occurring in individuals with autism spectrum disorder (Treffert 2009). Conversely, post-traumatic stress disorder presents as maladaptive, abnormally strong memories—potentially arising from the dysregulation of memory suppressor genes and/or their products. Similarly, some types of intellectual disability could arise from overly efficient memory suppressor systems, rather than inefficient processes that promote memory.
Although memory suppressors were recognized more than 20 years ago (Abel et al. 1998), there were few systematic studies of memory suppressors until 8 years ago (Walkinshaw et al. 2015). New experimental tools for Drosophila became available, especially libraries of RNA interfering transgenes, that allow the investigator to behaviorally screen through thousands of different genes to identify those that increase memory. Such screens and subsequent gene analyses have helped in understanding memory suppressors and their mechanisms. One major conclusion drawn so far is that there are many different neurophysiological processes that function at least in part, to suppress memory formation.
For instance, the initial RNAi screen reported by Walkinshaw et al. (2015) identified RNAi transgenes to several dozen genes that increase memory at 3 hr after aversive olfactory classical conditioning. This included genes that had been characterized for their roles in other aspects of Drosophila biology along with genes of unknown function. Several of these genes were then studied in detail, including solute carrier DmSLC22a (Gai et al. 2016), scribble (Cervantes-Sandoval et al. 2016), stromalin (Phan et al. 2019), ras85D (Noyes et al. 2020), and sickie (Zhang et al. 2022). DmSLC22A is a member of the organic cation transporter family and appears to function by helping to terminate acetylcholine neurotransmission at the Pn > MBn synapse. Scribble is a pre- and post-synaptic scaffolding protein that interacts physically with Cofilin, Pak3, and Rac1. Its memory suppressor function maps to α/β, γ MBn, and DAn. Stromalin is a protein component of the cohesion complex and limits the pool size of synaptic vesicles in DAn that innervate the heel of the MB lobes. Interestingly, normal Stromalin expression is required during a critical developmental period of the 3rd larval instar for its memory suppressing function in adult flies. Ras85D is a small GTPase required for normal Ras/Raf/Rock signaling in the α/β and γ MBn. Sickie is a member of the AAA+ ATPase domain containing proteins, interacts physically with the pre-synaptic protein, Bruchpilot, and is required in the DAn that innervate the heel of the MB lobes for its memory suppressing function. Thus, insults to many different molecular processes can produce elevated memory. In retrospect, this is not surprising given the complexity of the brain and that it employs many different strategies and mechanisms to form memories.
A second major conclusion about memory suppressors from such studies is that they can function at most, if not all, of the operations involved in memory formation. In principle, memory suppressors could act at any of the four major operations involved in memory formation: acquisition, consolidation, forgetting, and retrieval (Fig. 1a). Behavioral assays have been designed to help pinpoint the operation influenced by memory suppressor genes and their products (Noyes et al. 2021). DmSLC22A functions during acquisition, consistent with its role in limiting Pn > MBn neurotransmission. Stromalin also has a role in acquisition by limiting the synaptic vesicle pool in DAn. Ras85D functions as a switch for the consolidation of LTM. Scribble and Sickie function in the process of forgetting, which is described in more detail in the next section.
Many other memory suppressors are now known, and include molecules involved in GABAergic inhibition, transcriptional regulation, cAMP signaling and CREB-regulated gene expression, regulation of protein abundance such as small non-coding RNAs, constraining protein synthesis, and other processes. Additional details can be found in two recent reviews of memory suppressors (Noyes et al. 2021; Noyes and Davis, 2022). Understanding the genetic constraints on memory offers a promising area for future research.
Active forgetting
Among the four operations that underlie memory formation (Fig. 1a), acquisition and consolidation have been studied mechanistically in many different organisms for more than 5 decades. The origin for understanding the mechanisms underlying acquisition might be traced to the discovery of long-term potentiation by Bliss and Lomo in 1973 (reviewed by Nicoll, 2017). One might consider the origin of mechanistic studies of consolidation as the experiments performed in the 1960s by Flexner and Flexner, Agranoff, and others, showing the disrupting effect of protein synthesis inhibitors on LTM (reviewed by Hernandez and Abel 2008). Retrieval, or recall, remains largely unexplored mechanistically but the current information suggests that recall occurs from internal or external stimuli that activate the same neural circuits and associated cellular memory traces that form during acquisition and consolidation (Fig. 1b; Frankland et al. 2019). Research on the mechanisms of forgetting has been very fruitful across the last decade, with Drosophila providing the pioneering understanding of the circuitry and molecular biology for forgetting (Davis and Zhong 2017). The term “active forgetting,” refers to mechanisms that are stimulated to cause forgetting, as opposed to “passive forgetting,” which may occur due to the natural turnover of proteins involved in memory maintenance.
The seminal study that put a biochemical handle on active forgetting belongs to Shuai, Zhong, and coworkers (Shuai et al. 2010). They demonstrated using olfactory classical conditioning of adult flies that expressing a dominant negative form of Rac1 in the MBn produced a slowly decaying memory, while expressing a constitutively active form accelerated memory loss. Rac1 regulates the forgetting “labile memory,” or anesthesia-sensitive memory (ASM), without having effects on ARM, one form of consolidated memory. Rac1 has many different cellular functions but is critically important for regulating the dynamics of the actin cytoskeleton, at least in part, by regulating the phosphorylation state of Cofilin, a potent actin depolymerizing molecule. Flies expressing a dephosphorylated and persistently active mutant of Cofilin exhibit the same, slowly decaying memory as Rac1 dominant negative flies. Other players identified in the Rac1 → Cofilin pathway include Pak3 and LIMK. Thus, the Rac1 pathway and regulation of the actin cytoskeleton in MBn emerged as initial biochemical pathway that produces active forgetting. Rac1 has been shown to have a conserved function in forgetting a motor learning and object recognition memory in mice (Hayashi-Takagi et al. 2015; Liu et al. 2016b).
A cellular and circuit entrée to active forgetting that eventually merged with the Rac1 → Cofilin biochemical pathway came from Berry, Davis, and collaborators (Berry et al. 2012). They discovered from functional cellular imaging experiments that the calcium activity in the axon terminals of DAn innervating the junction and heel areas of the MB lobes exhibit modest, but chronic or “ongoing” activity, in naïve flies compared to the V1 region which was silent (Fig. 14). Zhang et al. (2022) subsequently reproduced and extended these data using a sensor for the neurotransmitter DA. Surprisingly, the activity does not change after conditioning, remaining constant for at least 45 min afterwards (Zhang et al. 2022), although DAn activity is decreased during sleep and increased with locomotor activity (Berry et al. 2015). Most importantly, stimulating the DAn after behavioral conditioning increases forgetting, and inhibiting the neurons after conditioning decreases forgetting. These data led to the model that the heel and junction DAn chronically release DA to slowly erase memory and cellular memory traces formed in MBn axons and associated neurons like the downstream MBOn (Davis and Zhong 2017).
Fig. 14.
Calcium activity measured with an early version of G-CaMP in the DAn axon processes that innervate the heel, junction, and V1 regions of the MB neuropil. a) Cartoon of the PPL1 DAn that innervate these regions (blue, yellow, and green). Grayscale and pseudocolor images of the regions imaged at the peak of calcium fluctuations. b) Time-based traces illustrating the chronic, ongoing fluctuations in calcium in the DAn terminals innervating the heel and junction regions of the MB lobes. Reproduced with permission from Berry et al. 2012.
This model predicts that a DA receptor involved in forgetting processes should be expressed on the MBn axons. Berry et al. (2012) found that the DAMB DA receptor fits this prediction, with DAMB mutant flies exhibited markedly reduced forgetting. Other studies of the memory suppressor gene, scribble, made the connection to the Rac1 → Cofilin pathway in the MBn (Cervantes-Sandoval et al. 2016). As a scaffolding protein, Scribble forms a signaling complex by binding to Rac1, Pak3, and Cofilin to mediate forgetting. The Scribble signaling complex is genetically downstream of DA inputs into the MBn mediated by DAMB, and upstream of Rac1, established from epistasis experiments. Together, these studies identify a neural circuit from the DAn, which in this context functions as “forgetting cells”, to the MBn (engram cells) expressing the DAMB receptor. This activates downstream biochemical signaling through the Scribble scaffolding complex of Rac1/Pak3/Cofilin to mediate a reorganization of the actin cytoskeleton for forgetting (Fig. 15). Other notable studies have added features to this model or provided information that supports it. The DAMB receptor was found to be biochemically coupled to Gαq, and RNAi knockdown of Gαq inhibits forgetting (Himmelreich et al. 2017). This indicates that DAMB signals through this G-protein to cause active forgetting. Sickie (Zhang et al. 2022) is required for the normal release of DA from the heel DAn through its interaction with Bruchpilot, so that insults to its function decrease release of the DA forgetting signal.
Fig. 15.
Model for DA-based forgetting of olfactory memory. In this model, the chronic and slow release of DA is envisioned to slowly degrade memory traces in MBn or other associated neurons, through the DAMB receptor coupled to Gαq, the Scribble scaffolding complex and Rac1 → Cofilin components, and the actin cytoskeleton. Reproduced with permission from Davis and Zhong 2017.
Recently, Aso et al. (2019) provided an interesting set of data that extends the model illustrated in Fig. 15. They found that some DAn, specifically the DAn that innervates the heel region of the MB axons, release nitric oxide (NO) in addition to DA. Released NO spurs signaling in the post-synaptic MBn by activating guanylyl cyclase, which was previously identified as a memory suppressor (Walkinshaw et al. 2015). Interestingly, the behavioral effects of NO are slower and require more potent stimuli than DA. Most interestingly, NO signaling in the MBn for its behavioral effects requires Scribble, the same scaffolding protein required for DA-based forgetting. Thus, the combined data offer an updated model in which the post-acquisition effects of DA and NO, released by the forgetting-DAn, initiate distinct signaling pathways in the MBn that converge on Scribble to degrade behavioral forgetting. Given the differences in stimulus strength and time course for these two neurotransmitters, it may be that NO is brought into action only when more robust forgetting is required.
A critical question posed from the above studies is whether forgetting is transient or permanent. This question is difficult to answer with complete confidence using behavioral assays because a failure to observe memory recovery can always be attributed to an inadequate lapse of time prior to testing. However, one can gain some insight into the issue by determining whether cellular memory traces are eliminated or retained upon activating a forgetting signal, with the assumption that the cellular memory trace monitored is critical to behavioral performance. Berry et al. (2018) approached the question in this way. They found that a cellular memory trace forms in junction MBOn that is registered by decreased fluorescence from G-CaMP in response to the conditioned odor after training. Blocking experiments indicated that the junction MBOn activity is required for robust olfactory memory performance, consistent with the idea that the cellular memory trace is relevant for behavioral memory expression. Most importantly, activating the junction DAn optogenetically or in other ways eliminated the expression of this cellular memory trace, consistent with the conclusion that this DAn pathway can produce a permanent loss of memory.
This result poses several critical questions that need to be addressed in future studies regarding the number and diversity of cellular memory traces that represent a behavioral memory and form with learning in a neural circuit. Given the number of cellular memory traces discovered so far, it seems likely that dozens and perhaps hundreds of different biochemical and cellular memory traces form in a circuit upon learning. Are all such cellular memory traces equivalent in their importance for behavioral memory, or are there kingpin memory traces, with others providing a supporting role so that they can be removed with minimal effects on memory? A related question is how many cellular memory traces can be lost in a circuit without influencing behavioral expression? Just one, or many? Answers to such questions are critical to understand the process of active forgetting more fully.
Several other recent advances of active forgetting that extend the paradigm described above are notable. First, Zhang et al. (2018) discovered that the expression of a Raf-GOF (gain of function) transgene in the γ MBn enhances the stability of labile memory through its activation of MAPK, which classifies Raf a memory promoting gene. Combining Raf-GOF with Rac1-DN expression stabilized memory beyond that conferred by each individual transgene, consistent with a model that they function in memory stability through different pathways. A search for downstream effectors of the Raf/MAPK pathway identified spaghetti squash (sqh), which encodes the regulatory light chain of non-muscle myosin II, a regulator of the actin cytoskeleton. Behavioral experiments employing transgenes expressing modified sqh coding regions including a constitutively active Sqh were consistent with this protein being an effector of the Raf/MAPK pathway. Second, other neurons beyond heel and junction DAn are involved in forgetting. A search for these by seeking bidirectional effects on memory stability from neuronal activation vs inhibition experiments identified DAn PAM-β′1, which innervates the MB β′1 compartment; and MBON-γ4 > γ1γ2, which receives information from the γ4 compartment and distributes it to γ1γ2 (Shuai et al. 2015). Third, small G-proteins other than Rac1 are involved in intrinsic forgetting. The small G-protein, Cdc42, a member the Rho GTPase family that includes Rac1, mediates the forgetting of ARM (Zhang et al. 2016). Manipulating Cdc42 activity has no effect on the formation of ARM, but the decay of ARM is accelerated with expression of a constitutively active form of Cdc42 and prolonged with expression of a dominant negative form of Cdc42 in the MBn. Manipulating Cdc42 activity has no effect on the formation or decay of ASM. These advances magnify the complexity of active forgetting mechanisms in Drosophila, involving many different molecules, signaling pathways, and neural circuits after only a decade or so of research. Clearly, much more remains to be learned and decades of additional research will be required, like for acquisition and consolidation.
The advances described above on intrinsic forgetting are provisionally classed as permanent forgetting, based on observing the erasure of associated cellular memory traces. However, forgetting can also be transient, which involves the temporary inability to retrieve memory. Sabandal et al. (2021) recently reported the first neurobiological-based mechanism for transient forgetting, which, remarkably, also involves DA, the DAMB receptor, and the MBn, but different PPL1 DAn from those involved in active forgetting.
They employed spaced conditioning to generate PSD-LTM of aversive olfactory memory and showed that this memory persists for at least 14 days. Using three types of interfering stimuli—strong airflow, electric shock, or intense blue light—they found that memory retrieval was blocked when the interfering stimuli were presented 1 min prior to the retrieval test at 72 hr after conditioning, but not when the interfering stimuli were presented 1 hr prior to the retrieval test. This indicates that the retrieval block was temporary, providing an assay system for interference-based, transient forgetting. They subsequently asked whether any of the PPL1 DAn were involved in transient forgetting using neuronal stimulation and blocking experiments. These experiments indicated that brief stimulation of PPL1α2α′2 DAn, which innervates the “stalk” region of the vertical lobes, mimicked the effects of interfering stimuli in causing transient forgetting. The disruption of memory retrieval induced by the PPL1α2α′2 DAn predicts the existence of a DA receptor expressed on the MBn to propagate the transient forgetting signal. Interestingly, normal expression of DAMB, the same DA receptor involved in permanent forgetting, was found to be required for transient forgetting produced by external sensory stimuli. Finally, in strategic ways that parallel those used to study active forgetting, the researchers searched for a relevant cellular memory trace and discovered one that forms with spaced training in the MBOn that receives information from the MB α2α′2 compartment, namely MBOn-α2sc. This cellular memory trace was found to be dependent on normal protein synthesis and is registered by increase G-CaMP fluorescence in response to the CS. Most importantly, the trace was not altered by activating the transient forgetting signal from PPL1α2α′2 DAn, indicating that unlike active forgetting that is permanent, a relevant cellular memory trace persists across the period of transient forgetting. Thus, the behavioral and functional imaging data reveal that PPL1-α2α′2 DAn, working through the DAMB receptor expressed in the α2α′2 MBn axonal compartment, mediates the transient forgetting of PSD-LTM. This effect occurs without altering a cellular memory trace in the postsynaptic MBOn-α2sc. This research performed with the fly represents the first insight into the neurobiological mechanisms for transient forgetting. Our current understanding of the mechanisms for permanent and transient forgetting is depicted in Fig. 16.
Fig. 16.
Model comparing “permanent vs” transient forgetting. Two forms of forgetting include permanent (red) and transient (orange) forgetting. Permanent forgetting involves a PPL1 DAn that synapses onto the γ2α′1-MBn (junction) compartment (red). The slow, ongoing DAn activity after learning is transduced by the Gq-coupled, DAMB receptor. This forgetting signal mobilises the Scribble scaffolding complex and recruits Rac1, PAK3 and Cofilin to erode labile, nonconsolidated memory. The cellular memory traces formed and stored in the following neuron, γ2α′1-MBOn, are also eroded. This process can be exacerbated by enhanced sensory stimulation, or repressed by sleep/rest (Berry et al. 2015). Transient forgetting incorporates a different PPL1 DAn that synapses onto the α2α′2-MBn compartment (orange). This forgetting signal, transduced by DAMB, temporarily impairs the expression of consolidated, PSD-LTM. A cellular memory trace stored in α2sc-MBOn is not abolished after activating the forgetting pathway. This process can be triggered by interfering or distracting stimuli to transiently block the retrieval of PSD-LTM. Reproduced with permission from Sabandal et al. 2021.
We have learned from these studies that the brain is designed to forget, and this begs the question of why. Why should the brain be designed with molecular and circuit mechanisms for forgetting? This design runs against the human notion that maximum learning is good and forgetting is bad. But there are good reasons and concepts that explain it. First, biological processes generally operate with separate and dedicated pathways for synthesis and degradation. At least some forms of forgetting, as we currently understand them, follow this design, with the intracellular signaling for acquisition (Gαs) being distinct from forgetting (Gαq). Second, biological systems generally operate within pre-set limits and when events bump them away from their normal operating range, homeostatic mechanisms are engaged to return the system to the proper state. From this perspective, forgetting may be the default state of the brain to help cleanse it from the enormous amount of information assimilated daily, operating chronically at a low level to slowly remove each newly acquired memory. Consolidation processes would override forgetting, allowing relevant memories to remain and irrelevant ones to be removed. Third, it is intuitive that the brain should be designed with mechanisms to remove irrelevant memories to help organize and manage important memories more efficiency. Lastly, forgetting increases fitness by eliminating undesirable decisions, such as continuing to seek food in the same location after resources are depleted. Forgetting offers the flexibility in memory for this.
Conclusions and perspectives
The fly has offered itself as an extremely attractive system to uncover the mysteries of learning and memory. From its beginnings using behavioral genetic approaches, to the inclusion of functional systems neuroscience approaches, to a recent focus on connectomics, and to the discoveries of mechanisms of forgetting, the fly has pioneered the evolution of concepts and principles for encoding, stabilizing, and forgetting information. This will continue across the next decade with its unique and powerful toolset to extract new concepts.
Although the progress reviewed above is very broad, there remain important areas of learning and memory research with the fly that are not covered in detail due to space constraints. For instance, significant progress has been made in using the fly as a model for memory disorders, probing the effects of metabolism on learning and memory processes, analyzing state dependent effects on memory formation, and revealing the mechanisms for non-associative learning and higher order processes such as extinction and second order conditioning. Literature searches will lead the reader to reports documenting advances in these and other areas of research.
Acknowledgements
I want to acknowledge all the scientists studying Drosophila memory formation for their passion and contributions to this important area of neuroscience. I also want to thank Drs. Mohammed Adel, Jacob Berry, Bertram Gerber, Leslie Griffith, Junjiro Horiuchi, Nathanial Noyes, Minoru Saitoe, Kausik Si, Seth Tomchik, and two anonymous reviewers for their comments and suggestions on the manuscript.
Funding
Research on Drosophila memory formation in the author's laboratory is currently supported by NIH grant R35NS097224.
Literature cited
- Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279(5349):338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- Aceves-Piña EO, Quinn WG. Learning in normal and mutant Drosophila larvae. Science. 1979;206(4414):93–96. doi: 10.1126/science.206.4414.93. [DOI] [PubMed] [Google Scholar]
- Adel M, Chen N, Zhang Y, Reed ML, Quasney C, Griffith LC. Pairing-dependent plasticity in a dissected fly brain is input-specific and requires synaptic CaMKII enrichment and nighttime sleep. J Neurosci. 2022;42(21):4297–4310. doi: 10.1523/JNEUROSCI.0144-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adel M, Griffith LC. The role of dopamine in associative learning in Drosophila: an updated unified model. Neurosci Bull. 2021;37(6):831–852. doi: 10.1007/s12264-021-00665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalal DB, Yu D, Davis RL. The long-term memory trace formed in the Drosophila α/β mushroom body neurons is abolished in long-term memory mutants. J Neurosci. 2011;31(15):5643–5647. doi: 10.1523/JNEUROSCI.3190-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin H, Lin AC. Neuronal mechanisms underlying innate and learned olfactory processing in Drosophila. Curr Opin Insect Sci. 2019;36:9–17. doi: 10.1016/j.cois.2019.06.003. [DOI] [PubMed] [Google Scholar]
- Andlauer TF, Scholz-Kornehl S, Tian R, Kirchner M, Babikir HA, Depner H, Loll B, Quentin C, Gupta VK, Holt MG, et al. Drep-2 is a novel synaptic protein important for learning and memory. Elife. 2014;3:e03895. doi: 10.7554/eLife.03895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014a;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Herb A, Ogueta M, Siwanowicz I, Templier T, Friedrich AB, Ito K, Scholz H, Tanimoto H. Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 2012;8(7):e1002768. doi: 10.1371/journal.pgen.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Ray RP, Long X, Bushey D, Cichewicz K, Ngo T-T, Sharp B, Christoforou C, Hu A, Lemire AL, et al. Nitric oxide acts as a cotransmitter in a subset of dopaminergic neurons to diversify memory dynamics. Elife. 2019;8:e49257. doi: 10.7554/eLife.49257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Rubin GM. Dopaminergic neurons write and update memories with cell-type-specific rules. Elife. 2016;5:e16135. doi: 10.7554/eLife.16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais P-Y, Robie AA, Yamagata N, Schnaitmann C, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife. 2014b;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata H, Takakura M, Kimura Y, Iwata I, Masuda T, Hirano Y. The neural circuit linking mushroom body parallel circuits induces memory consolidation in Drosophila. Proc Natl Acad Sci U S A. 2019;116(32):16080–16085. doi: 10.1073/pnas.1901292116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltruschat L, Prisco L, Ranft P, Lauritzen JS, Fiala A, Bock DD, Tavosanis G. Circuit reorganization in the Drosophila mushroom body calyx accompanies memory consolidation. Cell Rep. 2021;34(11):108871. doi: 10.1016/j.celrep.2021.108871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell. 2015;161(7):1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, Davis RL. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74(3):530–542. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Phan A, Davis RL. Dopamine neurons mediate learning and forgetting through bidirectional modulation of a memory trace. Cell Rep. 2018;25(3):651–662.e5. doi: 10.1016/j.celrep.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielopolski N, Amin H, Apostolopoulou AA, Rozenfeld E, Lerner H, Huetteroth W, Lin AC, Parnas M, et al. Inhibitory muscarinic acetylcholine receptors enhance aversive olfactory learning in adult Drosophila. Elife. 2019;8:e48264. doi: 10.7554/eLife.48264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilz F, Geurten BRH, Hancock CE, Widmann A, Fiala A. Visualization of a distributed synaptic memory code in the Drosophila brain. Neuron. 2020;106(6):963–976.e4. doi: 10.1016/j.neuron.2020.03.010. [DOI] [PubMed] [Google Scholar]
- Boto T, Louis T, Jindachomthong K, Jalink K, Tomchik SM. Dopaminergic modulation of cAMP drives nonlinear plasticity across the Drosophila mushroom body lobes. Curr Biol. 2014;24(8):822–831. doi: 10.1016/j.cub.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto T, Stahl A, Zhang X, Louis T, Tomchik SM. Independent contributions of discrete dopaminergic circuits to cellular plasticity, memory strength, and valence in Drosophila. Cell Rep. 2019;27(7):2014–2021.e2. doi: 10.1016/j.celrep.2019.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492(7429):433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher NJ, Friedrich AB, Lu Z, Tanimoto H, Meinertzhagen IA. Different classes of input and output neurons reveal new features in microglomeruli of the adult Drosophila mushroom body calyx. J Comp Neurol. 2012;520(10):2185–2201. doi: 10.1002/cne.23037. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, Hartenstein V. An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 2010;8(10):e1000502. doi: 10.1371/journal.pbio.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew TJ, Kandel ER. Acquisition and retention of long-term habituation in Aplysia: correlation of behavioral and cellular processes. Science. 1973;182(4117):1158–1160. doi: 10.1126/science.182.4117.1158. [DOI] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Chakraborty M, MacMullen C, Davis RL. Scribble scaffolds a signalosome for active forgetting. Neuron. 2016;90(6):1230–1242. doi: 10.1016/j.neuron.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Phan A, Chakraborty M, Davis RL. Reciprocal synapses between mushroom body and dopamine neurons form a positive feedback loop required for learning. Elife. 2017;6:e23789. doi: 10.7554/eLife.23789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, Chiang AS. et al. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335(6069):678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- Christiansen F, Zube C, Andlauer TF, Wichmann C, Fouquet W, Owald D, Mertel S, Leiss F, Tavosanis G, Farca Luna AJ, et al. Presynapses in Kenyon cell dendrites in the mushroom body calyx of Drosophila. J Neurosci. 2011;31(26):9696–9707. doi: 10.1523/JNEUROSCI.6542-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, Miesenböck G, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139(2):405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P, Felsenberg J, Waddell S. Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila. Curr Opin Neurobiol. 2018;49:51–58. doi: 10.1016/j.conb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P, Felsenberg J, Waddell S. Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila. Curr Opin Neurobiol. 2018;49:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R, Morantte I, Ruta V. Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell. 2015;163(7):1742–1755. doi: 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;5(1–2):38–51. doi: 10.1101/lm.5.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Mushroom bodies and Drosophila learning. Neuron. 1993;11(1):1–14. doi: 10.1016/0896-6273(93)90266-T. [DOI] [PubMed] [Google Scholar]
- Davis RL. Physiology and biochemistry of Drosophila learning mutants. Physiol Rev. 1996;76(2):299–317. doi: 10.1152/physrev.1996.76.2.299. [DOI] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70(1):8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Zhong Y. The biology of forgetting—A perspective. Neuron. 2017;95(3):490–503. doi: 10.1016/j.neuron.2017.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delestro F, Scheunemann L, Pedrazzani M, Tchenio P, Preat T, Genovesio Auguste. In vivo large-scale analysis of Drosophila neuronal calcium traces by automated tracking of single somata. Sci Rep. 2020;10(1):7153. doi: 10.1038/s41598-020-64060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MJ, Belliart-Guérin G, Bates AS, Frechter S, Lampin-Saint-Amaux A, Aso Y, Roberts RJV, Schlegel P, Wong A, Hammad A, et al. Communication from learned to innate olfactory processing centers is required for memory retrieval in Drosophila. Neuron. 2018;100(3):651–668.e8. doi: 10.1016/j.neuron.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MJ, Frechter S, Bates AS, Dan C, Huoviala P, Roberts RJ, Schlegel P, Dhawan S, Tabano R, Dionne H, et al. Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife. 2019;8:e43079. doi: 10.7554/eLife.43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago I, Davis RL. Inhibiting the mitochondrial calcium uniporter during development impairs memory in adult Drosophila. Cell Rep. 2016;16(10):2763–2776. doi: 10.1016/j.celrep.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411(6836):476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM., Saumweber T, Huser A, Eschbach C, Gerber B, et al. The complete connectome of a learning and memory centre in an insect brain. Nature. 2017;548(7666):175–182. doi: 10.1038/nature23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach C, Fushiki A, Winding M, Schneider-Mizell CM, Shao M, Arruda R, Eichler K, Valdes-Aleman J, Ohyama T, Thum AS, et al. Recurrent architecture for adaptive regulation of learning in the insect brain. Nat Neurosci. 2020;23(4):544–555. doi: 10.1038/s41593-020-0607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A, Spall T, Diegelmann S, Eisermann B, Sachse S, Devaud JM, Buchner E, Galizia CG. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr Biol. 2002;12(21):1877–1884. doi: 10.1016/S0960-9822(02)01239-3. [DOI] [PubMed] [Google Scholar]
- Frankland PW, Josselyn SA, Köhler S. The neurobiological foundation of memory retrieval. Nat Neurosci. 2019;22(10):1576–1585. doi: 10.1038/s41593-019-0493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Y, Liu Z, Cervantes-Sandoval I, Davis RL. Drosophila SLC22A transporter is a memory suppressor gene that influences cholinergic neurotransmission to the mushroom bodies. Neuron. 2016;90(3):581–595. doi: 10.1016/j.neuron.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32(1):65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- Gill J, Park Y, McGinnis JP, Perez-Sanchez C, Blanchette M, Si K. Regulated intron removal integrates motivational state and experience. Cell. 2017;169(5):836–848.e15. doi: 10.1016/j.cell.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC. A big picture of a small brain. Elife. 2014;3:e05580. doi: 10.7554/eLife.05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC, Ejima A. Courtship learning in Drosophila melanogaster: diverse plasticity of a reproductive behavior. Learn Mem. 2009;16(12):743–750. doi: 10.1101/lm.956309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E, Turner GC. Integration of the olfactory code across dendritic claws of single mushroom body neurons. Nat Neurosci. 2013;16(12):1821–1829. doi: 10.1038/nn.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21(10):519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler A, Graham TGW, Cohn R, Morantte I, Siliciano AF, Zeng J, Li Y, Ruta V. Distinct dopamine receptor pathways underlie the temporal sensitivity of associative learning. Cell. 2019;178(1):60–75.e19. doi: 10.1016/j.cell.2019.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, Kuhlman B, Hahn KM, Kasai H. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525(7569):333–338. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes PR, Christmann BL, Griffith LC. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife. 2015;4:e03868. doi: 10.7554/eLife.03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimbeck G, Bugnon V, Gendre N, Keller A, Stocker RF. A central neural circuit for experience-independent olfactory and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98(26):15336–15341. doi: 10.1073/pnas.011314898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Genetic approach to learning and memory (mnemogenetics) in Drosophila melanogaster. Fundamentals of memory formation: Neuronal plasticity and brain function progress in Zoology. 1989;37:3–44. [Google Scholar]
- Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89(3):293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas R, Rau MJ, Park Y, Zhang W, Murzin AG, et al. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science. 2020;367(6483):1230–1234. doi: 10.1126/science.aba3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T, Aso Y, Modi MN, Rubin GM, Turner GC. Heterosynaptic plasticity underlies aversive olfactory learning in Drosophila. Neuron. 2015;88(5):985–998. doi: 10.1016/j.neuron.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich S, Masuho I, Berry JA, MacMullen C, Skamangas NK, Martemyanov KA, Davis RL. Dopamine receptor DAMB signals via Gq to mediate forgetting in Drosophila. Cell Rep. 2017;21(8):2074–2081. doi: 10.1016/j.celrep.2017.10.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Ihara K, Masuda T, Yamamoto T, Iwata I, Takahashi A, Awata H, Nakamura N, Takakura M, Suzuki Y. et al. Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies. Nat Commun. 2016;7:13471. doi: 10.1038/ncomms13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Masuda T, Naganos S, Matsuno M, Ueno K, Miyashita T, Horiuchi J, Saitoe M. Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science. 2013;339(6118):443–446. doi: 10.1126/science.1227170. [DOI] [PubMed] [Google Scholar]
- Honegger KS, Campbell RA, Turner GC. Cellular-resolution population imaging reveals robust sparse coding in the Drosophila mushroom body. J Neurosci. 2011;31(33):11772–11785. doi: 10.1523/JNEUROSCI.1099-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huetteroth W, Perisse E, Lin S, Klappenbach M, Burke C, Waddell S. Sweet taste and nutrient value subdivide rewarding dopaminergic neurons in Drosophila. Curr Biol. 2015;25(6):751–758. doi: 10.1016/j.cub.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose T, Kanno M, Wu H, Yamagata N, Sun H, Abe A, Tanimoto H. Mushroom body output differentiates memory processes and distinct memory-guided behaviors. Curr Biol. 2021;31(6):1294–1302.e4. doi: 10.1016/j.cub.2020.12.032. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jacob PF, Waddell S. Spaced training forms complementary long-term memories of opposite valence in Drosophila. Neuron. 2020;106(6):977–991.e4. doi: 10.1016/j.neuron.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahsai L, Zars T. Learning and memory in Drosophila: behavior, genetics, and neural systems. Int Rev Neurobiol. 2011;99:139–167. doi: 10.1016/B978-0-12-387003-2.00006-9. [DOI] [PubMed] [Google Scholar]
- Kaun KR, Rothenfluh A. Dopaminergic rules of engagement for memory in Drosophila. Curr Opin Neurobiol. 2017;43:56–62. doi: 10.1016/j.conb.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Krashes MJ, Leung B, Bernard JA, Waddell S. Drosophila Dorsal paired medial neurons provide a general mechanism for memory consolidation. Curr Biol. 2006;16(15):1524–1530. doi: 10.1016/j.cub.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Keleman K, Vrontou E, Krüttner S, Yu JY, Kurtovic-Kozaric A, Dickson BJ. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489(7414):145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- Khan MR, Li L, Pérez-Sánchez C, Saraf A, Florens L, Slaughter BD, Unruh JR, Si K. Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell. 2015;163(6):1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Takahashi T, Yamagata N, Imanishi Y, Katow H, Hiramatsu S, Lynn K, Abe A, Kumaraswamy A, Tanimoto H. Neurochemical organization of the Drosophila brain visualized by endogenously tagged neurotransmitter receptors. Cell Rep. 2020;30(1):284–297.e5. doi: 10.1016/j.celrep.2019.12.018. [DOI] [PubMed] [Google Scholar]
- Krüttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. Drosophila CPEB Orb2A mediates memory independent of its RNA-binding domain. Neuron. 2012;76(2):383–395. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüttner S, Traunmüller L, Dag U, Jandrasits K, Stepien B, Iyer N, Fradkin LG, Noordermeer JN, Mensh BD, Keleman K. Synaptic Orb2A bridges memory acquisition and late memory consolidation in Drosophila. Cell Rep. 2015;11(12):1953–1965. doi: 10.1016/j.celrep.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126(18):4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee PT, Lin HW, Chang YH, Fu TF, Dubnau J, Hirsh J, Lee T, Chiang AS. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci U S A. 2011;108(33):13794–13799. doi: 10.1073/pnas.1019483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Lin G, Lin WW, Diao F, White BH, Bellen HJ. A kinase-dependent feedforward loop affects CREBB stability and long term memory formation. Elife. 2018;7:e33007. doi: 10.7554/eLife.33007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiss F, Groh C, Butcher NJ, Meinertzhagen IA, Tavosanis G. Synaptic organization in the adult Drosophila mushroom body calyx. J Comp Neurol. 2009;517(6):808–824. doi: 10.1002/cne.22184. [DOI] [PubMed] [Google Scholar]
- Lesar A, Tahir J, Wolk J, Gershow M. Switch-like and persistent memory formation in individual Drosophila larvae. Elife. 2021;10:e70317. doi: 10.7554/eLife.70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lindsey JW, Marin EC, Otto N, Dreher M, Dempsey G, Stark I, Bates AS, Pleijzier MW, Schlegel P, et al. The connectome of the adult Drosophila mushroom body provides insights into function. Elife. 2020;9:e62576. doi: 10.7554/eLife.62576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sanchez CP, Slaughter BD, Zhao Y, Khan MR, Unruh JR, Rubinstein B, Si K. A putative biochemical engram of long-term memory. Curr Biol. 2016;26(23):3143–3156. doi: 10.1016/j.cub.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Bygrave AM, de Calignon A, Lee T, Miesenböck G. Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat Neurosci. 2014;17(4):559–568. doi: 10.1038/nn.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HW, Chen CC, de Belle JS, Tully T, Chiang AS. CREBA and CREBB in two identified neurons gate long-term memory formation in Drosophila. Proc Natl Acad Sci U S A. 2021:118(37):e2100624118. doi: 10.1073/pnas.2100624118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HW, Chen CC, Jhang RY, Chen L, de Belle JS, Tully T, Chiang AS. CREBB repression of protein synthesis in mushroom body gates long-term memory formation in Drosophila. Proc Natl Acad Sci U S A. 2022;119(50):e2211308119. doi: 10.1073/pnas.2211308119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Buchanan ME, Han KA, Davis RL. The GABAA receptor RDL suppresses the conditioned stimulus pathway for olfactory learning. J Neurosci. 2009;29(5):1573–1579. doi: 10.1523/JNEUROSCI.4763-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Davis RL. The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat Neurosci. 2009;12(1):53–59. doi: 10.1038/nn.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du S, Lv L, Lei B, Shi W, Tang Y, Wang L, Zhong Y. Hippocampal activation of Rac1 regulates the forgetting of object recognition memory. Curr Biol. 2016b;26(17):2351–2357. doi: 10.1016/j.cub.2016.06.056. [DOI] [PubMed] [Google Scholar]
- Liu X, Krause WC, Davis RL. GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron. 2007;56(6):1090–1102. doi: 10.1016/j.neuron.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yang X, Tian J, Gao Z, Wang M, Li Y, Guo A. Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila. Elife. 2016a;5:e13238 doi: 10.7554/eLife.13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis T, Stahl A, Boto T, Tomchik SM. Cyclic AMP-dependent plasticity underlies rapid changes in odor coding associated with reward learning. Proc Natl Acad Sci U S A. 2018;115(3):E448–E457. doi: 10.1073/pnas.1709037115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148(3):515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Manoim JE, Davidson AM, Weiss S, Hige T, Parnas M. Lateral axonal modulation is required for stimulus-specific olfactory conditioning in Drosophila. Curr Biol. 2022;32(20):4438–4450.e5. doi: 10.1016/j.cub.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Keene AC. Gustatory processing and taste memory in Drosophila. J Neurogenet. 2016;30(2):112–121. doi: 10.1080/01677063.2016.1185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Worden K, Aso Y, Rubin GM, Keene AC. A dopamine-modulated neural circuit regulating aversive taste memory in Drosophila. Curr Biol. 2015;25(11):1535–1541. doi: 10.1016/j.cub.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse NY, Turner GC, Jefferis GS. Olfactory information processing in Drosophila. Curr Biol. 2009;19(16):R700–R713. doi: 10.1016/j.cub.2009.06.026. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog Neurobiol. 2005;76(5):328–347. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le and R PT, Davis L. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293(5533):1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20(8):384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Menzel R. Searching for the memory trace in a mini-brain, the honeybee. Learn Mem. 2001;8(2):53–62. doi: 10.1101/lm.38801. [DOI] [PubMed] [Google Scholar]
- Menzel R. In search for the retrievable memory trace in an insect brain. Front Syst Neurosci. 2022;16:876376. doi: 10.3389/fnsys.2022.876376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Kikuchi E, Horiuchi J, Saitoe M. Long-term memory engram cells are established by c-Fos/CREB transcriptional cycling. Cell Rep. 2018;25(10):2716–2728.e3. doi: 10.1016/j.celrep.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Oda Y, Horiuchi J, Yin JC, Morimoto T, Saitoe M. Mg(2+) block of Drosophila NMDA receptors is required for long-term memory formation and CREB-dependent gene expression. Neuron. 2012;74(5):887–898. doi: 10.1016/j.neuron.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenböck G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36(3):463–474. doi: 10.1016/S0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Nicoll RA. A brief history of long-term potentiation. Neuron. 2017;93(2):281–290. doi: 10.1016/j.neuron.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Niewalda T, Michels B, Jungnickel R, Diegelmann S, Kleber J, Kähne T, Gerber B. Synapsin determines memory strength after punishment- and relief-learning. J Neurosci. 2015;35(19):7487–7502. doi: 10.1523/JNEUROSCI.4454-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes NC, Davis RL. Genetics and molecular biology of memory suppression. Neuroscientist. 2022;10738584221138527. doi: 10.1177/10738584221138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes NC, Phan A, Davis RL. Memory suppressor genes: modulating acquisition, consolidation, and forgetting. Neuron. 2021;109(20):3211–3227. doi: 10.1016/j.neuron.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes NC, Walkinshaw E, Davis RL. Ras acts as a molecular switch between two forms of consolidated memory in Drosophila. Proc Natl Acad Sci U S A. 2020;117(4):2133–2139. doi: 10.1073/pnas.1819925117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto N, Pleijzier MW, Morgan IC, Edmondson-Stait AJ, Heinz KJ, Stark I, Dempsey G, Ito M, Kapoor I, Hsu J, et al. Input connectivity reveals additional heterogeneity of dopaminergic reinforcement in Drosophila. Curr Biol. 2020;30(16):3200–3211.e8. doi: 10.1016/j.cub.2020.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D, Waddell S. Olfactory learning skews mushroom body output pathways to steer behavioral choice in Drosophila. Curr Opin Neurobiol. 2015;35:178–184. doi: 10.1016/j.conb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani MR, Oishi K, Gelb BD, Zhong Y. The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell. 2009;139(1):186–198. doi: 10.1016/j.cell.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A, Schor J, Plaçais PY, Preat T. A GABAergic feedback shapes dopaminergic input on the Drosophila mushroom body to promote appetitive long-term memory. Curr Biol. 2018;28(11):1783–1793.e4. doi: 10.1016/j.cub.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24(40):8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A, Thomas CI, Chakraborty M, Berry JA, Kamasawa N, Davis RL. Stromalin constrains memory acquisition by developmentally limiting synaptic vesicle pool size. Neuron. 2019;101(1):103–118.e5. doi: 10.1016/j.neuron.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, DasGupta S, Krashes MJ, Leung B, Perrat PN, Waddell S. There are many ways to train a fly. Fly (Austin). 2009;3(1):3–9. doi: 10.4161/fly.3.1.7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaçais PY, Trannoy S, Friedrich AB, Tanimoto H, Preat T. Two pairs of mushroom body efferent neurons are required for appetitive long-term memory retrieval in Drosophila. Cell Rep. 2013;5(3):769–780. doi: 10.1016/j.celrep.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Qiao J, Yang S, Geng H, Yung WH, Ke Y. Input-timing-dependent plasticity at incoming synapses of the mushroom body facilitates olfactory learning in Drosophila. Curr Biol. 2022;32(22):4869–4880.e4. doi: 10.1016/j.cub.2022.09.054. [DOI] [PubMed] [Google Scholar]
- Quinn WG, Greenspan RJ. Learning and courtship in Drosophila: two stories with mutants. Annu Rev Neurosci. 1984;7(1):67–93. doi: 10.1146/annurev.ne.07.030184.000435. [DOI] [PubMed] [Google Scholar]
- Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1974;71(3):708–712. doi: 10.1073/pnas.71.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Kittel RJ, Fiala A. Optogenetics in Drosophila neuroscience. Methods Mol Biol. 2016;1408:167–175. doi: 10.1007/978-1-4939-3512-3_11. [DOI] [PubMed] [Google Scholar]
- Riemensperger T, Völler T, Stock P, Buchner E, Fiala A. Punishment prediction by dopaminergic neurons in Drosophila. Curr Biol. 2005;15(21):1953–1960. doi: 10.1016/j.cub.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Sabandal JM, Berry JA, Davis RL. Dopamine-based mechanism for transient forgetting. Nature. 2021;591(7850):426–430. doi: 10.1038/s41586-020-03154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoe M, Naganos S, Miyashita T, Matsuno M, Ueno K. A non-canonical on-demand dopaminergic transmission underlying olfactory aversive learning. Neurosci Res. 2022;178:1–9. doi: 10.1016/j.neures.2021.12.008. [DOI] [PubMed] [Google Scholar]
- Saumweber T, Rohwedder A, Schleyer M, Eichler K, Chen YC, Aso Y, Cardona A, Eschbach C, Kobler O, Voigt A, et al. Functional architecture of reward learning in mushroom body extrinsic neurons of larval Drosophila. Nat Commun. 2018;9(1):1104. doi: 10.1038/s41467-018-03130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. A connectome and analysis of the adult Drosophila central brain. Elife. 2020;9:e57443. doi: 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheunemann L, Plaçais PY, Dromard Y, Schwärzel M, Preat T. Dunce phosphodiesterase acts as a checkpoint for Drosophila long-term memory in a pair of serotonergic neurons. Neuron. 2018;98(2):350–365.e5. doi: 10.1016/j.neuron.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz-Kornehl S, Schwärzel M. Circuit analysis of a Drosophila dopamine type 2 receptor that supports anesthesia-resistant memory. J Neurosci. 2016;36(30):7936–7945. doi: 10.1523/JNEUROSCI.4475-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Völler T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16(17):1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23(33):10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai Y, Hirokawa A, Ai Y, Zhang M, Li W, Zhong Y. Dissecting neural pathways for forgetting in Drosophila olfactory aversive memory. Proc Natl Acad Sci U S A. 2015;112(48):E6663–E6672. doi: 10.1073/pnas.1512792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai Y, Lu B, Hu Y, Wang L, Sun K, Zhong Y. Forgetting is regulated through Rac activity in Drosophila. Cell. 2010;140(4):579–589. doi: 10.1016/j.cell.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Aso Y, Hige T, Wong A, Lu Z, Xu CS, Rivlin PK, Hess H, Zhao T, Parag T, et al. A connectome of a learning and memory center in the adult Drosophila brain. Elife. 2017;6:e26975. doi: 10.7554/eLife.26975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Yu D, Busto GU, Wilson C, Davis RL. Wnt signaling is required for long-term memory formation. Cell Rep. 2013;4(6):1082–1089. doi: 10.1016/j.celrep.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508(5):711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Thum AS, Gerber B. Connectomics and function of a memory network: the mushroom body of larval Drosophila. Curr Opin Neurobiol. 2019;54:146–154. doi: 10.1016/j.conb.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Tomchik S, Davis RL. Drosophila memory research through four eras: genetic, molecular biology, neuroanatomy, and systems neuroscience. Invertebrate Learning and Memory; 2013. Elsevier, London. doi: 10.1016/B978-0-12-415823-8.00027-7. [DOI] [Google Scholar]
- Treffert DA. The savant syndrome: an extraordinary condition. A synopsis: past, present, future. Philos Trans R Soc Lond B Biol Sci. 2009;364(1522):1351–1357. doi: 10.1098/rstb.2008.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79(1):35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Ueno K, Naganos S, Hirano Y, Horiuchi J, Saitoe M. Long-term enhancement of synaptic transmission between antennal lobe and mushroom body in cultured Drosophila brain. J Physiol. 2013;591(1):287–302. doi: 10.1113/jphysiol.2012.242909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Suzuki E, Naganos S, Ofusa K, Horiuchi J, Saitoe M. Coincident postsynaptic activity gates presynaptic dopamine release to induce plasticity in Drosophila mushroom bodies. Elife. 2017;6:e21076. doi: 10.7554/eLife.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103(5):805–813. doi: 10.1016/S0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Waddell S, Quinn WG. Flies, genes, and learning. Annu Rev Neurosci. 2001;24(1):1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, Davis RL. Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics. 2015;199(4):1173–1182. doi: 10.1534/genetics.114.173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112(2):271–282. doi: 10.1016/S0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Widmer YF, Fritsch C, Jungo MM, Almeida S, Egger B, Sprecher SG. Multiple neurons encode CrebB dependent appetitive long-term memory in the mushroom body circuit. Elife. 2018;7:e39196. doi: 10.7554/eLife.39196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Shih MF, Lai JS, Yang HT, Turner GC, Chen L, Chiang AS. Heterotypic gap junctions between two neurons in the Drosophila brain are critical for memory. Curr Biol. 2011;21(10):848–854. doi: 10.1016/j.cub.2011.02.041. [DOI] [PubMed] [Google Scholar]
- Wu JK, Tai CY, Feng KL, Chen SL, Chen CC, Chiang AS. Long-term memory requires sequential protein synthesis in three subsets of mushroom body output neurons in Drosophila. Sci Rep. 2017;7(1):7112. doi: 10.1038/s41598-017-07600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Mabuchi Y, Mizunami M, Tanaka NK. Convergence of multimodal sensory pathways to the mushroom body calyx in Drosophila melanogaster. Sci Rep. 2016;6:29481. doi: 10.1038/srep29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, Ezaki T, Takahashi T, Wu H, Tanimoto H. Presynaptic inhibition of dopamine neurons controls optimistic bias. Elife. 2021;10:e64907. doi: 10.7554/eLife.64907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Hiroi M, Abe T, Shimizu K, Minami-Ohtsubo M, Maeyama Y, Horiuchi J, Tabata T. Two parallel pathways assign opposing odor valences during Drosophila memory formation. Cell Rep. 2018;22(9):2346–2358. doi: 10.1016/j.celrep.2018.02.012. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81(1):107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52(5):845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123(5):945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Yu D, Ponomarev A, Davis RL. Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron. 2004;42(3):437–449. doi: 10.1016/S0896-6273(04)00217-X. [DOI] [PubMed] [Google Scholar]
- Zeng J, Li X, Zhang R, Lv M, Wang Y, Tan K, Xia X, Wan J, Jing M, Zhang X, et al. Local 5-HT signaling bi-directionally regulates the coincidence time window for associative learning. Neuron. 2023;111(7):1118–1135.e5. doi: 10.1016/j.neuron.2022.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li Q, Wang L, Liu ZJ, Zhong Y. Cdc42-dependent forgetting regulates repetition effect in prolonging memory retention. Cell Rep. 2016;16(3):817–825. doi: 10.1016/j.celrep.2016.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li Q, Wang L, Liu ZJ, Zhong Y. Active protection: learning-activated Raf/MAPK activity protects labile memory from Rac1-independent forgetting. Neuron. 2018;98(1):142–155.e4. doi: 10.1016/j.neuron.2018.02.025. [DOI] [PubMed] [Google Scholar]
- Zhang X, Noyes NC, Zeng J, Li Y, Davis RL. Aversive training induces both presynaptic and postsynaptic suppression in Drosophila. J Neurosci. 2019;39(46):9164–9172. doi: 10.1523/JNEUROSCI.1420-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Roman G. Presynaptic inhibition of gamma lobe neurons is required for olfactory learning in Drosophila. Curr Biol. 2013;23(24):2519–2527. doi: 10.1016/j.cub.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Zhang X, Sabandal JM, Tsaprailis G, Davis RL. Active forgetting requires Sickie function in a dedicated dopamine circuit in Drosophila. Proc Natl Acad Sci U S A. 2022;119(38):e2204229119. doi: 10.1073/pnas.2204229119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Sun J, Li Q, Zhong Y. Differential conditioning produces merged long-term memory in Drosophila. Elife. 2021;10:e66499. doi: 10.7554/eLife.66499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Sun J, Zhang X, Mo H, Niu Y, Li Q, Wang L, Zhong Y. Long-term memory is formed immediately without the need for protein synthesis-dependent consolidation in Drosophila. Nat Commun. 2019;10(1):4550. doi: 10.1038/s41467-019-12436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]