Abstract
Pathogenic variants in the alpha-synuclein (SNCA) gene cause familial forms of Parkinson’s disease (PD). Here, we describe generation of six isogenic controls from iPS cell lines derived from two PD disease patients carrying the SNCAp.A53T variant. The controls were created using CRISPR/Cas9 technology and are available for use by the PD research community to study A53T-related synucleinopathies.
Resource utility
This unique set of lines will be an important resource to study the effects of the SNCA p.A53T variant in cell lines at an endogenous level. Differentiating varying types of neurons from these iPSC lines could help understand the role of cell type in selective vulnerability related to Parkinson’s disease.
Resource Details
Parkinson’s disease (PD) is a prevalent neurodegenerative disease characterised in part by the loss of dopaminergic neurons in substantia nigra as well as widespread pathology as the disease progresses. The motor symptomatology of the disease is associated with tremor, bradykinesia, gait and postural instability with cognitive symptoms arising later in disease (Langston, 2006).
Histopathologically, PD is typically characterised by the deposition of aggregated α-synuclein protein within surviving neurons, identified as Lewy bodies and Lewy neurites (Goedert, Jakes and Spillantini, 2017). The gene encoding α-synuclein protein is SNCA which is associated with familial and sporadic PD (Reed et al., 2019). More specifically the first genetic variant found to segregate with PD in an autosomal dominant manner was a single base substitution, SNCA p.A53T (Polymeropoulos et al., 1997).
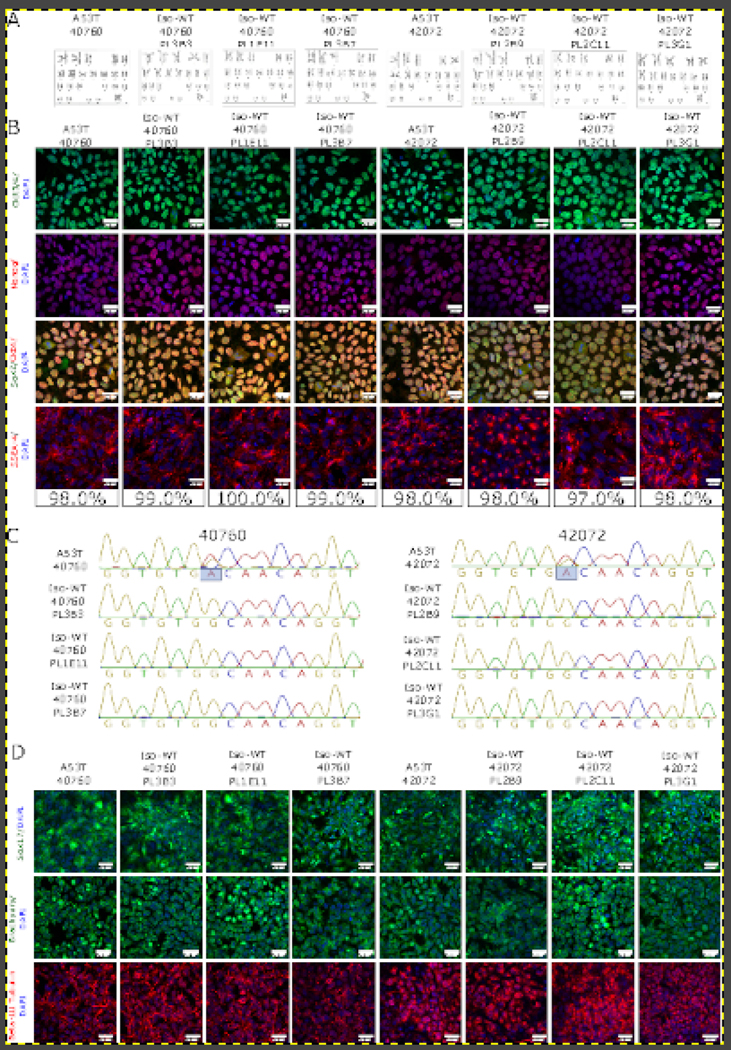
Here, we created six isogenic control lines (3 clones/line) for two iPSC lines carrying SNCA p.A53T from two unrelated PD patients (Table 1). We used Cas9 ribonucleoprotein (RNP) complexes with synthetic guide RNAs (crRNA/tracrRNA) to revert both p.A53T lines to WT using single stranded donor oligos (ssDO) (Table 3). After single cell cloning, two to three 96 well plates were picked per line and analysed by Sanger sequencing. Clones carrying homozygous WT nucleotides without off-target mutation in the amplicon sequence were expanded and re-sequenced to confirm target sequence (Figure 1C). Pluripotency of the lines was validated by expression of OCT4, SOX2, and NANOG by immunocytochemistry (ICC) (Figure 1B). Quantitative assessment showed that more than 98% of cells were positive for SSEA-4 in all iPSC lines (Figure 1B). All iPSC lines have a normal female karyotype (46, XX) without any obvious aberrations (Figure 1A). Short tandem repeat (STR) analysis of 16 genomic loci confirmed that the two parental iPSC lines were distinct and all reverted iPSC lines were identical to the appropriate original parent line (Supplementary File 1). The differentiation potential of the iPSC lines was confirmed by targeted differentiation into cells of all three germ layers. Additionally, ICC confirmed the expression of SOX17 (endoderm); Brachyury (mesoderm); and β-III Tubulin (ectoderm) (Figure 1D). All generated iPSC lines were free of mycoplasma contamination (Supplementary Figure 1).
Table 1:
Summary of lines
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| NIAi002-A | A53T 40760 | Female | N/A | N/A | SNCA p.A53T heterozygous variant | Parkinson disease |
| NIAi002-A-1 | Iso-WT 40760 PL1E11 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
| NIAi002-A-2 | Iso-WT 40760 PL3B3 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
| NIAi002-A-3 | Iso-WT 40760 PL3B7 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
| NIAi003-A | A53T 42072 | Female | N/A | N/A | SNCA p.A53T heterozygous variant | Parkinson disease |
| NIAi003-A-1 | Iso-WT 40760 PL2B9 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
| NIAi003-A-2 | Iso-WT 40760 PL2C11 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
| NIAi003-A-3 | Iso-WT 40760 PL3G1 | Female | N/A | N/A | WT SNCA gene sequence | N/A |
Table 3:
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:200 | Millipore, Cat# MAB4423, RRID:AB_11213224 |
| Pluripotency Markers | PE anti-human SSEA-4 | 1:200 | BioLegend Cat# 330406, RRID:AB_1089206 |
| Pluripotency Markers | Mouse anti-OCT3/4 | 1:200 | Santa Cruz Biotechnology Cat# sc5279, RRID: AB_628051 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:200 | Abcam Cat #ab19557, RRID:N/A |
| Pluripotency Markers | Mouse anti-Nanog | 1:200 | Millipore Cat# MABD24, RRID:AB_11203826 |
| Differentiation Markers | Goat anti-SOX17 | 1:200 | R and D Systems Cat# AF1924, RRID:AB_355060 |
| Differentiation Markers | Goat anti-Brachyury | 1:200 | R and D Systems Cat# AF2085, RRID:AB_2200235 |
| Differentiation Markers | Chicken anti-beta-III Tubulin | 1:200 | Novus Cat# NB100–1672, RRID:AB_522025 |
| Secondary antibodies | Donkey anti-mouse Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific Cat# A21202, RRID:AB_141607 |
| Secondary antibodies | Donkey anti-mouse Alexa Fluor 568 | 1:500 | Thermo Fisher Scientific Cat# A10037, RRID:AB_2534013 |
| Secondary antibodies | Donkey anti-rabbit Alexa Fluor 568 | 1:500 | Thermo Fisher Scientific Cat# A10042, RRID:AB_2534017 |
| Secondary antibodies | Donkey anti-goat Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific Cat# A11055, RRID:AB_2534102 |
| Secondary antibodies | Goat anti-chicken Alexa Fluor 568 | 1:500 | Thermo Fisher Scientific Cat# A11041, RRID:AB_2534098 |
| Primers | |||
| Target | Forward/Reverse primer (5′−3′) | ||
| Targeted variant analysis/sequencing | A53T | CTAGCTAATCAGCAATTTAAGGCTA/GCTC AGTGATTGTTTTACAATTTCA | |
| CRISPR reagents | |||
| Targeted mutation | crRNA used for Cas9 editing | Donor oligo sequence | |
| A53T | GTGGTGCATGGTGTGACAAC agg | AAAACTAGCTAATCAGCAATTTAAGGCTAGCTT GAGACTTATGTCTTGAATTTGTTTTTGTAGGCT CCAAAACCAAGGAGGGAGTGGTGCATGGTGT GGCAACAGGTAAGCTCCATTGTGCTTATATCC |
|
| AAAGATGATATTTAAAGTATCTAGTGATTAGTG TGGCCCAGTATTCAAGATTCCTATGAAATTGTAAAAC |
|||
Figure 1:
Generation of SNCA A53T isogenic controls. (A) Karyotype analysis of iPS unedited lines and isogenic clones (WT = wild type, clone ID above each corresponding graph). (B) Pluripotency analysis of selected lines. Top panel - OCT 3/4 (green), second panel - NANOG (red), third panel - Sox2 (green) and Oct4 (red), and fourth panel - SSEA-4 (red). The latter allowed for quantitative percentage estimation of pluripotent cells. All lines were counterstained with nuclear stain DAPI (blue). Scale bars = 20 μm. (C) Sequencing chromatograms of SNCA A53T lines and isogenic clones. (D) ICC analysis of differentiation potential. Endodermal layer (top panel SOX17, green); mesodermal layer (second panel, Brachuyry, green) and ectodermal layer (third panel, β-III Tubulin, red). All cells were counterstained with the nuclear dye DAPI (blue) and scale bars = 20 μm.
In conclusion, we successfully generated three isogenic control lines for each of two iPSC lines carrying SNCA p.A53T to support studies of PD in human cellular models.
Materials and Methods
Growth, propagation and morphology of iPSC lines.
PPMI40760 and PPMI42072 cell lines and isogenic clones derived from these lines were grown in Gibco StemFlex media (Fisher Scientific, cat #A3349401). 10uM Rock inhibitor (STEMCELL, Cat # 72304) was used for splitting and thawing. iPSC clones were cryopreserved in Synth-A-Freeze Cryopreservation media (Thermo Scientific, Cat #A1254201).
Genome editing of iPSC lines using ribonucleoprotein.
RNP complex formation:
Alt-R CRISPR-Cas9 guide RNA (crRNA) were custom designed using the IDT website https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM. Alt-R CRISPR-Cas9 crRNA for A53T SNCA targeted region is shown in Table 3. Alt-R CRISPR-Cas9 crRNA and Alt-R CRISPR-Cas9 tracrRNA (IDT, cat #1072533) were resuspended in nuclease-free duplex buffer (IDT, cat #11010301) at 200 μM. 5 μl of 200 μM crRNA and 5 μl of 200 μM tracrRNA were mixed together, heated at 95°C for 5 minutes and then cooled to RT. Lastly, 1.7 μl (104 pmol) of Alt-R S.p. Cas9 nuclease (IDT, cat # 1081058) was mixed together with 1.2 μl (120 pmol) Alt-R CRISPR-Cas9 crRNA/tracrRNA duplex and 2.1 μl of 1xPBS solution and incubated 30 minutes at RT.
ssDO preparation:
ssDO with conversion of A53T to WT (see Table 3) were synthetized by IDT and resuspended in DPBS at a concentration of 100 pmol/ μl.
Nucleofection:
iPSCs were dissociated into single cells using TrypLE (Fisher Scientific, cat #A12605036) and counted. 8×105 cells were then pelleted at 1000 rpm for 3 minutes and subsequently gently resuspended in 100μl of P3 Primary Cell Solution from P3 Primary Cell 4D-Nucleofector X Kit L (Lonza, #V4XP-3024). Immediately prior to nucleofection, 2 μl (100 pmol/μl) of ssDO (Table 3) was added to 5μl of pre-assembled Cas9/RNP complex. 100μl of iPS cells, resuspended in P3 primary cell solution, was then combined with the Cas9/RNP complex and ssDO; thoroughly mixed and transferred to the 100 μl nucleofector cuvette (Lonza; #V4XP-3024). Immediately upon transfer, cells were transfected using the ‘Primary Cell P3’ program and ‘CA-137’ pulse code on Lonza Nucleofection machine. iPSCs were then carefully transferred using a Lonza disposable Pasteur pipette into one well of a Matrigel-coated 6-well plate containing 3 mL StemFlex media with 10 μM Rock inhibitors. Cells were cultured in a 32C/5% CO2 incubator for two days and then transferred to a 37 C incubator. Edited iPSC pools were expanded and cryopreserved.
Generation of clones from single cell
Expanded pools were dissociated with TrypLE, counted and plated on 10cm Matrigel-coated dishes containing Stemflex media with 10 μM Rock inhibitor for two days at 10×103 and 15×103 density. Subsequently, the media was changed without Rock inhibitor for 5–7 days. Single cell colonies were expanded to 250–500 μM diameter and picked using a 100 μl pipet tip under a Bioimager microscope. Individual colonies were transferred to Matrigel-coated 96-well plates. Two to three 96 well plates were picked per line and expanded until 70–80% confluency. Each expanded plate was split into two plates: one third of the cells were transferred onto a new 96 well plate for further propagation and two thirds were used for sequencing analysis.
Sequencing
Plates with collected cells were centrifuged at 3,000 rpm for 5 minutes and resuspended in 30 μl of water. Cells were heated at 95°C for 10 minutes and 2 μl of cells were used for PCR with A53T region specific primers listed in Table 3. PCR was carried out using Terra polymerase (Takara Bio). Each PCR product was sequenced using forward and reverse primers with Applied Biosystems BigDye terminator v3.1 sequencing chemistry according to manufacturer’s instructions. The products were cleaned using Agencourt CleanSEQ reagent (Beckman Coulter), run on a 3730xl DNA analyzer (Applied Biosystems, Hitachi) and analysed with Sequencher software. Positive clones were expanded and re-sequenced.
Karyotyping and STR analysis
Karyotyping and STR analysis were performed by WiCell Research Institute.
Mycoplasma detection.
Mycoplasma test was performed using PCR (ATCC, cat # 30–1012K).
Pluripotency assessment
iPSC clones were grown on Matrigel-coated coverslips, fixed with 4% PFA, and stained with pluripotency markers listed in Table 3. Images were taken on a Zeiss 880 confocal microscope. SSEA4 quantifications were performed by PE-SSEA4 antibody (Table 3). At least 50 cells per sample were counted.
Differentiation potential
iPSC clones were differentiated into three layers according to STEMdiff™ Trilineage Differentiation Kit (StemCell, Cat# 05230), fixed in 4% PFA, and stained with differentiation markers listed in Table 3. Images were taken on a Zeiss 880 confocal microscope.
Supplementary Material
Supplementary File 1: STR analysis
Supplementary Figure 1: Mycoplasma test
Table 2:
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | All iPSC lines are morphologically normal | Not shown; available from authors. |
| Phenotype | Qualitative analysis | Confirmed expression of pluripotency markers: OCT4, NANOG, and SOX2 | Figure 1, panel B |
| Quantitative analysis | Assessed % of positive cells for pluripotency cell surface marker, SSEA-4. All lines are more than 98% positive. | Figure 1 panel B | |
| Genotype | Karyotype (G-banding) and resolution | 1. 46XX, Resolution 425–450 2. 46XX, Resolution 425–525 3. 46XX, Resolution 400–425 4. 46XX, Resolution 425–450 5. 46XX, Resolution 375–475 6. 46XX, Resolution 400–450 7. 46XX, Resolution 400–450 8. 46XX, Resolution 450–475 |
Figure 1 panel A |
| Identity | Microsatellite PCR (mPCR) OR STR analysis |
STR analysis | Supplementary data 1 |
| 16 specific loci were tested. All isogenic lines matched 100%. | Submitted in archive with journal | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | 1. A53T 40760 heterozygous | Figure 1 panel C |
| 2. Iso-WT 40760 PL3B3 3. Iso-WT 40760 PL1E11 4. Iso-WT 40760 PL3B7 5. A53T 42072 heterozygous 5. Iso-WT 42072 PL2B9 6. Iso-WT 42072 PL2C11 7. Iso-WT 42072 PL3G1 |
|||
| Southern Blot OR WGS | Not performed | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing was done by RT-PCR. All clones-negative | Supplementary figure 1 |
| Differentiation potential | e.g. Embryoid body formation OR Teratoma formation OR Scorecard OR Directed differentiation |
The STEMdiff™ Trilineage Differentiation Kit (StemCell) was used to test differentiation potential. We confirmed the expression of endoderm (SOX-17), mesoderm (Brachyury) and ectoderm ( -Tubulin III) markers in all clones. |
Figure 1 panel D |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | N/A |
| HLA tissue typing | Not performed | N/A |
Resource Table:
| Unique stem cell lines identifier | NIAi002-A NIAi002-A-1 NIAi002-A-2 NIAi002-A-3 NIAi003-A NIAi003-A-1 NIAi003-A-2 NIAi003-A-3 |
|
| |
| Alternative names of stem cell lines | A53T SNCA 40760 (NIAi002-A) Iso-WT SNCA 40760 PL1E11 (NIAi002-A-1) Iso-WT SNCA 40760 PL3B3 (NIAi002-A-2) Iso-WT SNCA 40760 PL3B7 (NIAi002-A-3) A53T SNCA 42720 (NIAi003-A) Iso-WT SNCA 42720 PL2B9 (NIAi003-A-1) Iso-WT SNCA 42072 PL2C11 (NIAi003-A-2) Iso-WT SNCA 42072 PL3G1 (NIAi003-A-3) |
|
| |
| Institution | National Institutes of Health, National Institute on Aging |
|
| |
| Contact information of distributor | Mark R Cookson: cookson@mail.nih.gov |
|
| |
| Type of cell lines | iPSC |
|
| |
| Origin | Human |
| Cell Source | The Michael J. Fox Foundation for Parkinson’s Research (MJFF), Parkinson’s Progression Markers Initiative (PPMI). |
|
| |
| Clonality | Clonal |
|
| |
| Method of reprogramming | Sendai virus |
|
| |
| Multiline rationale | Isogenic clones from two unrelated donors carrying the same pathogenic variant in a PD related gene |
|
| |
| Gene modification | Yes |
|
| |
| Type of modification | Revert SNCA p.A53T to WT sequence |
|
| |
| Associated disease | Parkinson’s disease |
|
| |
| Gene/locus | SNCA/PARK1 |
|
| |
| Method of modification | RNP CRISPR/Cas9 |
|
| |
| Name of transgene or resistance | N/A |
|
| |
| Inducible/constitutive system | N/A |
|
| |
| Date archived/stock date | N/A |
|
| |
| Cell line repository/bank | Indiana University https://hpscreg.eu/cell-line/NIAi002-A https://hpscreg.eu/cell-line/NIAi002-A-1 https://hpscreg.eu/cell-line/NIAi002-A-2 https://hpscreg.eu/cell-line/NIAi002-A-3 https://hpscreg.eu/cell-line/NIAi003-A https://hpscreg.eu/cell-line/NIAi003-A-1 https://hpscreg.eu/cell-line/NIAi003-A-2 https://hpscreg.eu/cell-line/NIAi003-A-3 |
|
| |
| Ethical approval | Original line obtained from MJFF. Ethical licence/Sponsor Protocol number: 001. |
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging and by the Michael J Fox Foundation for Parkinson’s Disease Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Langston JW. The Parkinson’s complex: parkinsonism is just the tip of the iceberg. Ann Neurol. 2006. Apr;59(4):591–6. doi: 10.1002/ana.20834. PMID: . [DOI] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Spillantini MG. The Synucleinopathies: Twenty Years On. J Parkinsons Dis. 2017;7(s1):S51–S69. doi: 10.3233/JPD-179005. PMID: ; PMCID: PMC5345650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed X, Bandrés-Ciga S, Blauwendraat C, Cookson MR. The role of monogenic genes in idiopathic Parkinson’s disease. Neurobiol Dis. 2019. Apr;124:230–239. doi: 10.1016/j.nbd.2018.11.012. Epub 2018 Nov 15. PMID: ; PMCID: PMC6363864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997. Jun 27;276(5321):2045–7. doi: 10.1126/science.276.5321.2045. PMID: . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: STR analysis
Supplementary Figure 1: Mycoplasma test