Abstract
Virions of infectious pancreatic necrosis virus (IPNV) were completely disintegrated upon dialysis against salt-free buffers. Direct visualization of such preparations by electron microscopy revealed 5.0- to 6.5-nm-thick entangled filaments. By using a specific colloidal gold immunolabeling technique, these structures were shown to contain the viral protein VP3. Isolation by sucrose gradient centrifugation of the filaments, followed by serological analysis, demonstrated that the entire VP3 content of the virion was recovered together with the radiolabeled genomic material forming the unique threadlike ribonucleoprotein complexes. In a sensitive blotting assay, the outer capsid component of IPNV, i.e., the major structural protein VP2, was shown to specifically bind lectins recognizing sugar moieties of N-acetylgalactosamine, mannose, and fucose. Three established metabolic inhibitors of N-linked glycosylation did not prevent addition of sugar residues to virions, and enzymatic deglycosylation of isolated virions using N-glycosidase failed to remove sugar residues of VP2 recognized by lectins. However, gentle alkaline β elimination clearly reduced the ability of lectins to recognize VP2. These results suggest that the glycosylation of VP2 is of the O-linked type when IPNV is propagated in RTG-2 cells.
Infectious pancreatic necrosis virus (IPNV) is an agent of contagious fish disease causing high mortality among salmonid eggs and fingerlings (46). As for all members of the family Birnaviridae, the naked capsid encloses the two segments of double-stranded RNA (10, 33). Segment B (2.3 × 106 Da) encodes the RNA-dependent RNA polymerase VP1 (94 kDa) (11). The larger segment (segment A, 2.5 × 106 Da) contains two open reading frames. The short one encodes a 17-kDa polypeptide identified only in infected cells and not in purified virions (25). The long open reading frame encodes a 106-kDa polyprotein which is cotranslationally cleaved by a viral protease that is contained within the polyprotein (designated NS or VP4, 29 kDa) into pVP2 (62 kDa) and VP3 (31 kDa) (12, 24, 29). pVP2 is further processed during virus maturation into VP2 (54 kDa), which is the polypeptide constituting the icosahedral capsid (9). The glycosylation of this protein is still a matter of controversy. Estay et al. (16) were able to show the existence of N-linked oligosaccharides present on VP2 of virions propagated in CHSE-214 cells. On the contrary, Perez et al. (37) could not detect any carbohydrates on the viral proteins expressed in the same cell line. In this study, we have propagated IPNV in RTG-2 cells, and we found it worthwhile to investigate if any glycosylation of the virus occurs in this cell line, since it is possible that different cell lines give rise to different types of glycosylation, quantitatively and/or qualitatively (6, 23). Better knowledge of glycosylation is important for future studies on the tropism of this virus.
The second major structural protein, VP3, is known to be absent in empty capsids and is therefore referred to as an internal protein (8), but no reports regarding its function and exact localization inside the virion of IPNV have been published. In this investigation, we disrupted virions gently by incubation in a low-ionic-strength buffer and studied the released components by serological methods and electron microscopy (EM).
Analysis of disintegrated virions by EM.
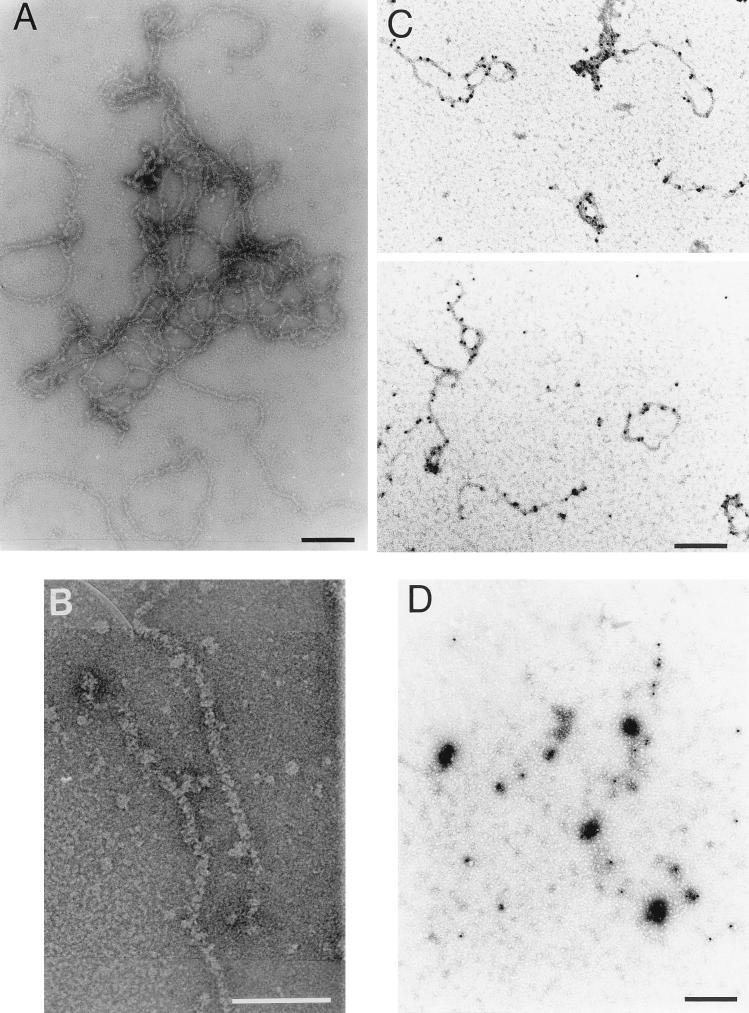
Highly purified IPNV (4), serotype VR-299 (14), was dialyzed for 2 days at room temperature against a low-ionic-strength buffer (5 mM Tris-HCl [pH 8.1], 0.2 mM EDTA) in order to gently disintegrate the virions. The result, as assessed by negative staining with 2% uranyl acetate and visualization by EM, was total disintegration of the viral capsids and quantitative and highly reproducible release of aggregated filamentous structures (Fig. 1A). Aggregation of the filaments prevented any measurement of strand length or estimation of the number of aggregated segments. Upon higher magnification, smaller entities covering the filaments were visible, thereby creating components with a thickness of 5.0 to 6.5 nm (Fig. 1B). Their appearance indicated that the structures might correspond to double-stranded RNA (2 nm) covered by repeated units of a nucleic acid-binding protein. By using the present preparation technique, we were not able to detect condensed complexes with a shape fitting an empty capsid, and we could not observe icosahedral structures similar to the tentative cores described by Dobos et al. (9). However, it is quite possible that VP3-RNA cores under other conditions may be released as more condensed and subvirion-like structures that would fit into the cavities seen in some virus preparations (9). The results of our investigation indicate that the core structure of IPNV displays similarities to the established cores of adenoviruses, where highly basic protein VII is closely and quantitatively associated with the DNA and is believed to neutralize the negative charges along the sugar-phosphate backbone of the entire genome (38, 39). Such adenovirus cores, released from virions, have been visualized by EM and reveal an appearance similar to that of the ribonucleoprotein complexes released from disrupted IPNV (34).
FIG. 1.
Subviral components of IPNV. Visualization by EM of filamentous structures released from virions dialyzed against a low-ionic-strength buffer (A) and an enlargement of a section of a tentative ribonucleoprotein complex (B) are shown. The horizontal bars indicate 100 (A) and 50 (B) nm. Subviral components labeled with colloidal-gold-conjugated MAbs against VP3 (C) or VP2 (D) and stained with 2% uranyl acetate before examination by EM are shown. Under the present conditions, the specimens in panels C and D were stained positively. Bars in panels C and D, 200 nm.
To identify the viral proteins of the released filaments, disintegrated virions were adsorbed to carbon-coated copper grids that were subsequently placed face down for 10 min on top of 30-μl droplets of suspensions of mouse immunoglobulin G (IgG) monoclonal antibodies (MAbs) against VP3 or VP2 (both produced in this laboratory [15]; Fig. 2A, inset) or total mouse IgG, all bound to protein A colloidal gold (10 nm; Sigma Chemical Co., St. Louis, Mo.), and diluted in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.02% sodium azide. After being washed three times in PBS, the specimens were stained with 2% uranyl acetate. With this preparation technique, using 1% BSA to quench the grid and to avoid unspecific protein-protein binding, uranyl acetate functions as a positive stain of the specimen (3, 34). The highly specific binding of MAbs against VP3 to the filaments strongly suggests that VP3 was a component of the structures released from disrupted virions (Fig. 1C). The aggregation of these structures into larger and more contracted complexes was most likely due to excessive amounts of unconjugated antibodies that were still present. Neither MAbs against VP2 (Fig. 1D) nor total mouse IgG (data not shown) caused any labeling or aggregation of the filaments, and consequently, these structures were almost undetectable in the quenching film of BSA covering the grid.
FIG. 2.
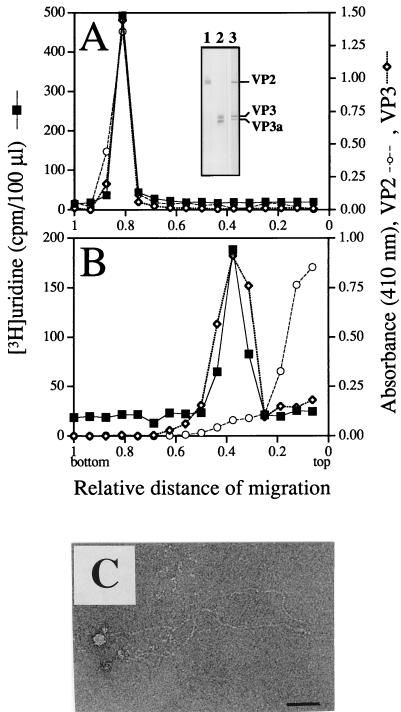
Rate zonal sedimentation of intact and disintegrated virions. IPNV stored in TNE (A) and IPNV dialyzed against low-ionic-strength buffer (B) were sedimented by sucrose gradient centrifugation. The RNA was measured as trichloroacetic acid-precipitable, [3H]uridine-labeled material, and viral proteins VP2 and VP3 were identified by MAbs in an ELISA and detected as A410. The specificities of the MAbs are shown in an immunoblot (inset in panel A). IPNV-infected RTG-2 cells were run in lanes 1 and 2, and purified IPNV was run in lane 3. Proteins in lane 1 were displayed by a MAb against VP2 (14/2d, also recognizing pVP2), those in lane 2 by a MAb against VP3 (1/2f-3), and the proteins in lane 3 were revealed by an anti-IPNV specific serum. Material of the fraction from the sucrose gradient sedimentation of disintegrated virions containing the highest concentrations of VP3 and RNA was observed by negative staining and EM (C). Bar, 50 nm.
Rate sedimentation analysis of disintegrated virions.
The existence of RNA in the filamentous structures observed by EM was investigated by rate zonal centrifugation of [3H]uridine-labeled disintegrated and intact virions. Runs were made in sucrose gradients consisting of 2 × 1.8 ml of 10 to 25% (wt/vol) sucrose in TNE buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) layered on top of 800-μl cushions of CsCl at 50% relative saturation and 20% (wt/vol) sucrose in TNE buffer. Samples of 150 to 300 μl were applied on top of the gradients, and centrifugations were carried out at 23,000 rpm (49,500 × g) for 6 h in an SW50.1 rotor at 10°C. The tubes were punctured, and fractions were collected dropwise from the bottom. Screening for virus-specific proteins was done by enzyme-linked immunosorbent assay (ELISA) using MAbs against VP2 and VP3. Radiolabeled RNA was quantified as trichloroacetic acid-precipitable material in a liquid scintillation spectrometer. Viral components released from the purified virions upon disintegration in the low-ionic-strength buffer were recovered in two major regions (Fig. 2B). The faster-migrating material consisted of the comigrating total detectable VP3 and RNA contents of the virus, whereas the released VP2 quantitatively remained on top of the gradient in a nonsedimentable form (Fig. 2B). In a parallel centrifugation, intact virions sedimented onto the high-density cushion at the bottom of the tube (Fig. 2A). The strong signal of the MAbs against VP3 is noteworthy, bearing in mind the suggested inner location of this polypeptide. The recognition of VP3 could be due to possible disruption of intact virions upon attachment to the plastic surface of the ELISA plates. Another possibility is that some part of VP3 is exposed on the surface of the virion since MAbs against VP3 have been shown to contain neutralizing capacity (44) and also to recognize VP3 on purified IPNV in ELISAs, as well as in immunodot assays (5). Furthermore, antibodies against VP3 from total anti-IPNV serum have been demonstrated to adsorb to intact virions (36).
Material recovered from fractions of the rate sedimentation analysis of disintegrated virions, containing cosedimenting VP3 and RNA, was visualized by EM, which unequivocally demonstrated the existence of filaments. These filaments appeared to be less aggregated and thinner (3 to 4 nm) (Fig. 2C) than prior to the rate zonal sedimentation (Fig. 1A). However, since no VP2 was detectable in the condensed form of the filaments by labeling with antibody-gold conjugates, and also since VP3 was still quantitatively associated with the radiolabeled RNA after sucrose gradient centrifugation of disrupted virions, it is most plausible that the decrease in thickness was simply due to relaxation of the more condensed form caused by the environment of the higher ionic strength of the sucrose gradients.
Our results suggest that VP3 is intimately associated with the segments of viral RNA. The diameter of the filaments, as observed in freshly disintegrated particles, implies that VP3 is in a low-molecular-weight form attached to the entire RNA segments. VP3 displays a pI of 6.6, which is the highest value among the proteins of IPNV (16), and of the last 43 carboxy-terminal amino acid residues, 25% consist of the basic amino acids arginine and lysine (12), and it is logical to believe that this part of the protein is associated with the viral genome. For infectious bursal disease virus, another member of the Birnaviridae family with biophysical and biochemical characteristics similar to those of IPNV, it has been proposed that VP3 may be a constituent of the capsid with the highly basic carboxy-terminal end associated with the RNA genome (2, 21). From our investigation, it is not possible to ascertain if VP3 is a part of the capsid, and as mentioned above, VP3 is not a constituent of empty capsids of IPNV (8). During the low-ionic-strength dialysis of virions, the capsids were totally disintegrated and no VP2 could be detected in the filaments with gold-labeled antibodies. This indicates that the binding of VP2 to VP3, if there is any, is weaker than the VP3-RNA association under the present conditions.
Lectin-binding properties of IPNV polypeptides.
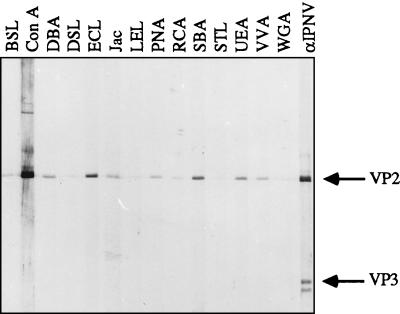
To detect any carbohydrates present on the proteins of highly purified virions, we used 14 different lectins (Table 1) in a sensitive lectin-blotting assay. The possibility of contaminating proteins being copurified with virions was minimized by an additional step of purification in which virions were briefly treated with 0.3% Tween 20 and reisolated by isodensity centrifugation. Subsequently, the viral polypeptides were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (26) and transferred to nitrocellulose membranes. To increase the sensitivity and specificity of the assay, the membranes were blocked with 2% polyvinylpyrrolidone (molecular weight, 44,000; BDH Laboratory Supplies, Poole, England) in PBS–0.05% Tween 20 overnight at 4°C (1). The membranes were mounted in a Mini-protean II multiscreen device (Bio-Rad Laboratories, Richmond, Calif.) and biotinylated lectins (Lectin Screening Kits I and III; Vector Laboratories, Inc., Burlingame, Calif.) with a variety of sugar specificities (Table 1) were added to the channels at a concentration of 2 μg/ml in 2% polyvinylpyrrolidone–PBS–0.05% Tween 20. Lectins recognizing any glycoprotein upon incubation for 2 h at 22°C were detected by incubation of the carefully washed membranes with avidin conjugated with alkaline phosphatase and developed by standard procedures. None of the lectins was able to bind at the 30,000-Mr position corresponding to VP3, and this protein can therefore be considered an internal negative control. At the 54,000-Mr position of VP2, several lectins bound with different specificities (Fig. 3; Table 1), suggesting the existence of oligosaccharides on VP2. Although the binding capacity differed from lectin to lectin, it was clear that lectins preferentially recognizing N-acetylgalactosamine (GalNAc), e.g., Dolichos biflorus agglutinin, jacalin, peanut agglutinin, soybean agglutinin (SBA), and Vicia villosa agglutinin, bound significantly stronger than lectins with their highest specificity toward N-acetylglucosamine (GlcNAc), e.g., Bandeiraea simplicifolia lectin, Datura stramonium lectin, Lycopersicon esculentum lectin, Solanum tuberosum lectin, and wheat germ agglutinin (WGA) (41; Vector Laboratories). This is interesting because GalNAc is the most-studied saccharide bound to the amino acid serine or threonine by O linkage and GlcNAc is the most abundant sugar in the core of N-linked oligosaccharides. Consequently, the binding pattern of the lectins implies O-linked glycosylation of VP2. Erythrina cristagalli lectin (ECL) is one of the lectins giving a strong signal at the position of VP2, and although ECL has its highest binding activity toward galactosyl (β-1,4)-linked N-acetylglucosamine, this lectin requires a galactose constituent in all bindings, again supporting the former general binding pattern (Vector Laboratories). Other sugars indicated to be present on VP2 were fucose and mannose, binding Ulex europaeus agglutinin (UEA) and concanavalin A (ConA), respectively (Fig. 3; Table 1). In an earlier study, the presence of mannose in IPNV replicated in CHSE-214 cells was detected with the lectin ConA (16). Furthermore, radiolabeled mannose is incorporated into the virus particles if added 7 h postinfection (p.i.) (16). However, if labeled mannose is added in the final stage of virus replication (at 16 h p.i.) (27), no labeling is found in the virions (37). This lack of labeling could be due to the fact that glycosylation of proteins is a posttranslational modification carried out before the final assembly of the virions. In a control experiment, we therefore had [3H]mannose (20 μCi/ml, 28.0 Ci/mmol) present in the culture medium from 1 h p.i. until harvest 70 h later. Virus was isolated and purified upon the occurrence of a total cytopathic effect on the cells, and the viral polypeptides were separated by SDS-PAGE. Quantification of radioactivity upon CuCl2 staining and alkaline hydrolysis of gel pieces containing the polypeptide bands revealed that 92% of the radioactivity was confined to the polypeptide of VP2 and 8% was in the region of polypeptides VP3 and VP3a, thus clearly demonstrating the presence of mannose residues associated with VP2 in virus propagated in RTG-2 cells. Furthermore, the label distribution also showed that mannose was not significantly metabolically degraded and reused in protein synthesis, although the label was allowed to be present for an excessive period of time.
TABLE 1.
Carbohydrate specificities of lectinsa
| Lectina | Carbohydrate specificityb |
|---|---|
| BSL | α- or β-linked GlcNAc |
| ConA | α-linked mannose |
| DBA | α-linked GalNAc |
| DSL | (β-1,4)-linked GlcNAc |
| ECL | Galactosyl (β-1,4)-linked GlcNAc |
| Jac | O-linked galactosyl (β-1,3)-linked GalNAc |
| LEL | GlcNAc |
| PNA | Galactosyl (β-1,3)-linked GalNAc |
| RCA | Galactose |
| SBA | GalNAc |
| STL | GlcNAc |
| UEA | α-linked fucose |
| VVA | α- or β-linked GalNAc |
| WGA | GlcNAc |
BSL, Bandeiraea simplicifolia lectin II; DBA, Dolichos biflorus agglutinin; DSL, Datura stramonium lectin; Jac, jacalin; LEL, Lycopersicon esculentum lectin; PNA, peanut agglutinin; RCA, Ricinus communis agglutinin; STL, Solanum tuberosum lectin; VVA, Vicia villosa agglutinin. The other abbreviations are defined in the text.
Carbohydrate specificities of lectins according to the manufacturer (Vector Laboratories) and Sharon and Lis (41). Only sugars with the highest specificities are listed, although some of the lectins can bind other carbohydrates but to a lesser extent.
FIG. 3.
Lectin blot of virion polypeptides. IPNV was purified by an additional step of isodensity centrifugation after detergent treatment. Upon SDS-PAGE of the recovered virus and transblotting of the polypeptides to a nitrocellulose membrane, carbohydrates were monitored with 14 different lectins. The total amount of virion proteins subjected to SDS-PAGE was 67 ng per channel (42). The specificities of the different lectins are explained in Table 1. The positions of the viral proteins were detected with a polyclonal serum against IPNV (αIPNV). For definitions of abbreviations, see the text and Table 1, footnote a.
Metabolic inhibition of glycosylation.
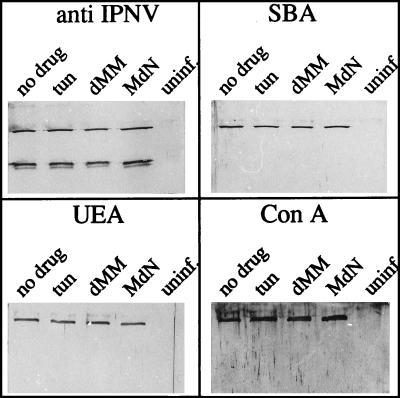
The biosynthetic events leading to virion glycosylation were investigated in the presence of three selected drugs that inhibit identified steps during the process of N-linked glycosylation. It would have been desirable to employ a drug with a documented effect only on O-linked glycosylation in this study, but no such drug is available. At 1 h p.i., the following drugs were added to separate cell culture flasks: 5- and 10-μg/ml tunicamycin (Sigma), a transferase inhibitor blocking the first step in the dolichol pathway; 3 mM deoxymannojirimycin (Calbiochem-Novabiochem Corp., La Jolla, Calif.), a mannose analogue inhibiting mannosidase; and 4 mM N-methyl-1-deoxynojirimycin (Sigma), an inhibitor of α-glucosidase (13, 18, 40). Upon the occurrence of an apparent cytopathic effect (3 days p.i.), the virus was purified by NaCl-polyethylene glycol precipitation (10), and the total yield was determined quantitatively as A260. Equal amounts of virus were analyzed by SDS-PAGE and subsequent transblotting of the polypeptides. For practical reasons, purification by precipitation was selected since several and different virus pools with low virus content were processed separately and at the same time. The extent of glycosylation, revealed as lectin binding, was monitored with the lectins SBA, ConA, and UEA, which recognize GalNAc, mannose, and fucose, respectively. These three sugars are found at different positions in common oligosaccharides. GalNAc is frequently found attached to the hydroxyl group of serine or threonine by O linkage, mannose is detected mostly in inner cores of N-linked oligosaccharides, and fucose is a terminal sugar (6, 31). Figure 4 shows that no quantitative differences in the carbohydrate contents of VP2 newly synthesized in the presence of any of the drugs could be detected by the selected lectins, suggesting a non-N-linked mechanism of glycosylation. These results do not agree with the results obtained by Estay et al. (16), who found a decrease of VP2-linked carbohydrates in the presence of tunicamycin at 2 μg/ml when it was added to CHSE-214 cells at 4 h p.i. In the present investigation, the highest concentration of tunicamycin was 10 μg/ml, and even though this concentration was shown to be toxic to the cells, reducing viability by 50% after 48 h of cell growth in the presence of the drug (data not shown) and, accordingly, resulting in a decreased virus yield, the relative amount of glycosylated VP2 was constant. The finding that the replication of IPNV per se is insensitive to tunicamycin, which has been reported by both Estay et al. (16) and Perez et al. (37), supports this observation.
FIG. 4.
Effects of inhibitors of glycosylation. Virus-infected cells were treated with the following glycosylation inhibitors added at 1 h p.i.: 10-μg/ml tunicamycin (tun), 3 mM deoxymannojirimycin (dMM), and 4 mM N-methyl-1-deoxynojirimycin (MdN). The two controls were virus replicated in the absence of any drug (no drug) and mock-infected cells treated in the same way as the infected cells, without any drug (uninf.). The latter material was recovered from positions in a CsCl gradient corresponding to the viral bands in gradients separating virus-containing material. To each SDS-PAGE lane, 0.30 μg of purified virion polypeptides (42) was applied, and three different lectins were selected for the detection of lectin-binding proteins (SBA, UEA, and ConA, recognizing GalNAc, fucose, and mannose, respectively). A polyclonal serum against IPNV was used for detection of viral proteins.
The inhibitory effect of tunicamycin was verified by growing RTG-2 cells for 48 h in the presence of the drug and [3H]mannose. The level of mannose incorporation, measured as specific radioactivity in counts per minute per microgram of protein, was reduced to 82% when the commonly used concentration of tunicamycin (2 μg/ml) (28, 30, 35) was added and to 0.8% in the presence of the excessive tunicamycin concentration of 10 μg/ml, thus demonstrating the anticipated effect of the drug.
Deglycosylation of VP2.
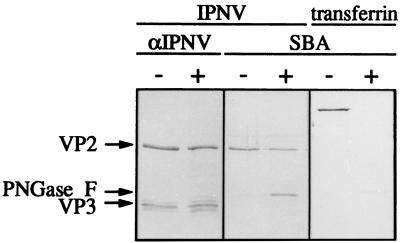
The enzyme N-glycosidase F (Boehringer GmbH, Mannheim, Germany) cleaves all types of asparagine-linked carbohydrates (43) and was used to further analyze the lectin-binding properties of the virus. IPNV was purified by an additional centrifugation step as described above and quantified spectrophotometrically as described by Smith et al. (42) to yield data on total protein content. The lyophilized virus was reconstituted in phosphate buffer (0.1 M sodium phosphate [pH 7.2], 10 mM EDTA, 0.02% sodium azide) containing 0.5% SDS and 5% 2-mercaptoethanol. The calculated amount of IPNV protein in each sample was 3 μg, and the concentration was 0.25 μg/μl. The samples were denatured by boiling prior to reduction of the SDS concentration to 0.1% by adding additional phosphate buffer, and to further prevent inactivation of the enzyme by SDS, octyl-glucoside was added to a concentration of 1% (20). Finally, the samples were incubated with N-glycosidase F (30 U/ml) overnight at 37°C. Evaluation of the enzymatic reactions by SDS-PAGE, followed by lectin blotting, showed that even a high ratio of N-glycosidase F to virion proteins under extreme conditions did not affect the assessed levels of VP2 glycosylation (Fig. 5). Faint bands of viral proteins migrating faster than VP2 after being subjected to N-glycosidase F treatment were unveiled with the anti-IPNV serum. This indicated a low level of proteolytic degradation rather than deglycosylation, since the increase in the rate of migration is too large to simply mirror a loss of sugar residues. The enzyme itself is glycosylated and is therefore visualized in the lectin blot as a band corresponding to an Mr of 34,000. The proper function of the enzyme was demonstrated by complete deglycosylation of human transferrin regarding carbohydrates recognized by SBA (Fig. 5). Transferrin possesses two N-linked complex carbohydrate chains with an inner core of GlcNAc and mannose and outer chains containing GlcNAc, galactose, and sialic acid (7).
FIG. 5.
Enzymatic deglycosylation of purified IPNV. Virions of IPNV were treated with N-glycosidase F (+) or left untreated (−). Transferrin treated in the same way was used as a control. Separation of the polypeptides by SDS-PAGE was followed by transblotting and detection with a polyclonal serum against IPNV (αIPNV) and SBA. VP2 and VP3 indicate the positions of the viral polypeptides, whereas PNGase F indicates the position of N-glycosidase F, which is also recognized by SBA.
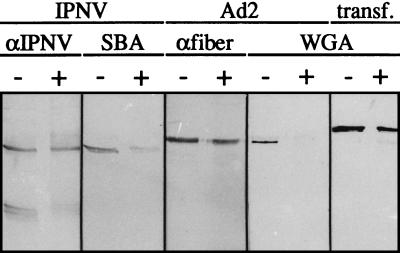
The inability of N-glycosidase F to remove the carbohydrates from VP2 further strengthened the indication that the linkage between the carbohydrates and the polypeptide was of an O-linked type. To show this, an effort was made to find a suitable assay for the exclusion of O-linked carbohydrates. A commercially available O-glycosidase, O-glycopeptide endo-d-galactosyl-N-acetyl-α-galactosamino hydrolase (EC 3.2.1.97; Boehringer), recognizing the disaccharide Galβ3GalNAc and the trisaccharide Fucα2Galβ3GalNAc (23, 45), was applied, but even with excessive concentrations of the enzyme and after prolonged periods of incubation, the optimal conditions for this enzyme were not found (data not shown). We therefore selected a pure chemical deglycosylation assay utilizing the sensitivity of the linkage between glycans and serine or threonine toward alkaline β elimination (23, 31). Conditions were optimized, and the lyophilized virus was subsequently reconstituted with 5 mM NaOH and 1 M NaBH4 and incubated for 20 h at 37°C in a toluene atmosphere (17). By using this mild treatment, degradation of the proteins was minimized. The reaction was stopped by pH neutralization with HCl and cooling of the samples at 4°C. The amount of carbohydrates still present was revealed by lectin blotting using IPNV incubated with PBS as a control. Any protein degradation or increase in the migration rate as a consequence of the treatment was detected with anti-IPNV serum. As expected, the N-linked carbohydrates of transferrin used as a negative control were unaffected by this treatment, whereas the positive control, the fiber of human adenovirus type 2 (Ad2), was totally deglycosylated (Fig. 6). This virus contains GlcNAc, which is strongly recognized by WGA. However, the linkage to the protein moiety is not, as expected, of the N-linked type but is established as a so-called dynamic O linkage (19, 22).
FIG. 6.
Detection of polypeptides and carbohydrates after alkaline β elimination. Lyophilized samples of IPNV, human Ad2, and transferrin (transf.) were incubated at 37°C for 20 h in 5 mM NaOH with 1 M NaBH4 (+) or in PBS (−) as described in the text. Analysis by SDS–13% PAGE was followed by transblotting and detection with antiserum (αIPNV and αfiber are antibodies against the glycosylated 62-kDa fiber protein of Ad2) or with lectins (SBA and WGA). The amount of IPNV polypeptides applied to each lane was 0.37 μg, and those of the controls Ad2 and transf. were 3.3 and 0.5 μg, respectively.
Although the amount of VP2, as detected by anti-IPNV serum, was constant upon β elimination, the carbohydrate content of VP2 was clearly reduced (Fig. 6), indicating that the VP2 of virions propagated in RTG-2 cells possessed O-linked oligosaccharides. Removal of the established small quantity of GlcNAc residues of the Ad2 fiber, three to four molecules per fiber molecule (32), revealed a minimal effect on the migration rate on SDS–13% PAGE. Likewise, the migration of VP2 was unaffected upon β elimination (Fig. 6), also suggesting small quantities of carbohydrates.
It was not in the scope of this investigation to determine the exact residue composition of the oligosaccharides; however, the lectin assay implies the presence of GalNAc, mannose, and fucose. In recent years, a large number of novel carbohydrate structures, monosaccharide constituents as well as new linkages between carbohydrates and the peptide backbones, have been discovered. For N-linked carbohydrates, the most abundant amino acid sequence required is NXS/T, and IPNV has four of these sequences in VP2 that theoretically could be suitable for glycosylation (8). However, our results indicate that the glycosylation of VP2 is O linked when virions are propagated in RTG-2 cells, and for O-linked carbohydrates, it is the linkage between GalNAc and serine or threonine that is best known, and any further amino acid requirements in a specific motif to establish this linkage are not known.
Acknowledgments
We are indebted to Blanka Boberg for expert technical assistance and to Lars-Olof Hedén for careful reading of the manuscript.
This investigation was supported by the Swedish Natural Science Research Council and the Crafoord Foundation, Lund.
REFERENCES
- 1.Bartles J R, Hubbard A L. 125I-wheat germ agglutinin blotting: increased sensitivity with polyvinylpyrrolidone quenching and periodate oxidation/reductive phenylamination. Anal Biochem. 1984;140:284–292. doi: 10.1016/0003-2697(84)90166-0. [DOI] [PubMed] [Google Scholar]
- 2.Böttcher B, Kieselev N, Stel’Mashchuk V, Perevozchikova N, Borisov A, Crowther R. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J Virol. 1997;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D, Westphal M, Burlingham B, Winterhoff U, Doerfler W. Structure and composition of the adenovirus type 2 core. J Virol. 1975;16:366–387. doi: 10.1128/jvi.16.2.366-387.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsson A, Kuznar J, Varga M, Everitt E. Purification of infectious pancreatic necrosis virus by anion exchange chromatography increases the specific infectivity. J Virol Methods. 1994;47:27–36. doi: 10.1016/0166-0934(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Caswell-Reno P, Reno P W, Nicholson B L. Monoclonal antibodies to infectious pancreatic necrosis virus: analysis of viral epitopes and comparison of different isolates. J Gen Virol. 1986;67:2193–2206. doi: 10.1099/0022-1317-67-10-2193. [DOI] [PubMed] [Google Scholar]
- 6.Datema R, Olofsson S, Romero P A. Inhibitors of protein glycosylation and glycoprotein processing in viral systems. Pharmacol Ther. 1987;33:221–286. doi: 10.1016/0163-7258(87)90066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong G, van Dijk J P, van Eijk H G. The biology of transferrin. Clin Chim Acta. 1990;190:1–46. doi: 10.1016/0009-8981(90)90278-z. [DOI] [PubMed] [Google Scholar]
- 8.Dobos P. The molecular biology of infectious pancreatic necrosis virus (IPNV) Annu Rev Fish Dis. 1995;5:25–54. [Google Scholar]
- 9.Dobos P, Hallett R, Kells D T C, Sorensen O, Rowe D. Biophysical studies of infectious pancreatic necrosis virus. J Virol. 1977;22:150–159. doi: 10.1128/jvi.22.1.150-159.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobos P, Hill B J, Hallett R, Kells D T C, Becht H, Teninges D. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J Virol. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan R, Mason C, Nagy E, Leong J, Dobos P. Sequence analysis of infectious pancreatic necrosis virus genome segment B and its encoded VP1 protein: a putative RNA-dependent RNA polymerase lacking the gly-asp-asp motif. Virology. 1991;181:541–552. doi: 10.1016/0042-6822(91)90887-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan R, Nagy E, Krell P J, Dobos P. Synthesis of the infectious pancreatic necrosis virus polyprotein, detection of a virus-encoded protease, and fine structure mapping of genome segment A coding regions. J Virol. 1987;61:3655–3664. doi: 10.1128/jvi.61.12.3655-3664.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbein A D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- 14.Espinoza E, Farias G, Soler M, Kuznar J. Identity between infectious pancreatic necrosis virus VR-299 and a Chilean isolate. Intervirology. 1985;24:58–60. doi: 10.1159/000149619. [DOI] [PubMed] [Google Scholar]
- 15.Espinoza J C, Everitt E, Vargas J, Soler M, Kuznar J. Reactividad de dos anticuerpos monoclonales preparados contra el virus de la necrosis pancreatica infecciosa. Acta Microbiol. 1993;4:23–32. [Google Scholar]
- 16.Estay A, Farias G, Soler M, Kuznar J. Further analysis on the structural proteins of infectious pancreatic necrosis virus. Virus Res. 1990;15:85–95. doi: 10.1016/0168-1702(90)90015-4. [DOI] [PubMed] [Google Scholar]
- 17.Florman H, Wassarman P. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell. 1985;41:313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuhrmann U, Bause E, Legler G, Ploegh H. Novel mannosidase inhibitor blocking conversion of high mannose to complex oligosaccharides. Nature. 1984;307:755–758. doi: 10.1038/307755a0. [DOI] [PubMed] [Google Scholar]
- 19.Hart G W. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 20.Haselbeck A, Hösel W. Studies on the effect of the incubation conditions, various detergents and protein concentration on the enzymatic activity of N-glycosidase F (glycopeptidase F) and endoglycosidase F. Top Biochem (Boehringer Mannheim GmbH) 1988;8:1–4. [Google Scholar]
- 21.Hudson P J, McKern N M, Power B E, Azad A A. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res. 1986;14:5001–5012. doi: 10.1093/nar/14.12.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishibashi M, Maizel J V. The polypeptides of adenovirus. VI. Early and late glycopolypeptides. Virology. 1974;58:345–361. doi: 10.1016/0042-6822(74)90070-1. [DOI] [PubMed] [Google Scholar]
- 23.Lis H, Sharon N. Protein glycosylation. Structural and functional aspects. Eur J Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald R, Dobos P. Identification of the proteins encoded by each segment of infectious pancreatic necrosis virus. Virology. 1981;114:414–422. doi: 10.1016/0042-6822(81)90222-1. [DOI] [PubMed] [Google Scholar]
- 25.Magyar G, Dobos P. Evidence for the detection of the infectious pancreatic necrosis virus polyprotein and the 17-kDa polypeptide in infected cells and of the NS protease in purified virus. Virology. 1994;204:580–589. doi: 10.1006/viro.1994.1572. [DOI] [PubMed] [Google Scholar]
- 26.Maizel J V. Polyacrylamide gel electrophoresis of viral proteins. In: Maramorosch K, Koprowski H, editors. Methods of virology. Vol. 5. New York, N.Y: Academic Press, Inc.; 1971. pp. 179–246. [Google Scholar]
- 27.Malsberger R G, Cerini C P. Multiplication of infectious pancreatic necrosis virus. Ann N Y Acad Sci. 1965;126:320–327. doi: 10.1111/j.1749-6632.1965.tb14283.x. [DOI] [PubMed] [Google Scholar]
- 28.McDowell W, Romero P A, Datema R, Schwarz R T. Glucose trimming and mannose trimming affect different phases of the maturation of Sindbis virus in infected BHK cells. Virology. 1987;161:37–44. doi: 10.1016/0042-6822(87)90168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertens P, Dobos P. Messenger RNA of infectious pancreatic necrosis virus is polycistronic. Nature. 1982;297:243–246. doi: 10.1038/297243a0. [DOI] [PubMed] [Google Scholar]
- 30.Mirazimi A, von Bonsdorff C, Svensson L. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology. 1996;217:554–563. doi: 10.1006/viro.1996.0150. [DOI] [PubMed] [Google Scholar]
- 31.Montreuil J, Bouquelet S, Debray H, Fournet B, Spik G, Strecker G. Glycoproteins. In: Chaplin M, Kennedy J, editors. Carbohydrate analysis: a practical approach. Oxford, England: IRL Press; 1986. pp. 143–204. [Google Scholar]
- 32.Mullis K, Haltiwanger G, Hart G, Marchase R, Engler J. Relative accessibility of N-acetylglucosamine in trimers of the adenovirus type 2 and 5 fiber proteins. J Virol. 1990;64:5317–5323. doi: 10.1128/jvi.64.11.5317-5323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Arch Virol. 1995;1995(Suppl. 10):240–244. [Google Scholar]
- 34.Nermut M V. The architecture of adenoviruses: recent views and problems. Arch Virol. 1980;64:175–196. doi: 10.1007/BF01322699. [DOI] [PubMed] [Google Scholar]
- 35.Olofsson S, Milla M, Hirschberg C, de Clercq E, Datema R. Inhibition of terminal N- and O-glycosylation specific for herpesvirus-infected cells: mechanism of an inhibitor of sugar nucleotide transport across Golgi membranes. Virology. 1988;166:440–450. doi: 10.1016/0042-6822(88)90515-6. [DOI] [PubMed] [Google Scholar]
- 36.Park J, Gajin J. Identification of VP3 as an important neutralising epitope from DRT strain, a Korean isolate of infectious pancreatic necrosis virus (IPNV) Fish Shellfish Imm. 1996;6:207–219. [Google Scholar]
- 37.Perez L, Chiou P, Leong J. The structural proteins of infectious pancreatic necrosis virus are not glycosylated. J Virol. 1996;70:7247–7249. doi: 10.1128/jvi.70.10.7247-7249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prage L, Pettersson U. Structural proteins of adenoviruses. VII. Purification and properties of an arginine-rich core protein from adenovirus type 2 and type 3. Virology. 1971;45:364–373. doi: 10.1016/0042-6822(71)90337-0. [DOI] [PubMed] [Google Scholar]
- 39.Prage L, Pettersson U, Höglund S, Lonberg-Holm K, Philipson L. Structural proteins of adenovirus. IV. Sequential degradation of the adenovirus type 2 virion. Virology. 1970;42:341–358. doi: 10.1016/0042-6822(70)90278-3. [DOI] [PubMed] [Google Scholar]
- 40.Romero P A, Datema R, Schwartz R T. N-methyl-1-deoxynojirimycin, a novel inhibitor of glycoprotein processing and its effect on fowl plaque virus maturation. Virology. 1983;130:238–242. doi: 10.1016/0042-6822(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 41.Sharon N, Lis H. Lectins. London, England: Chapman & Hall; 1989. [Google Scholar]
- 42.Smith R, Zweerink H, Joklik W. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 43.Tarentino A, Gómez C, Plummer T. Deglycosylation of aspargine-linked glycans by peptide: N-glycosidase F. Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 44.Tarrab E, Berthiaume L, Heppell J, Arella M, Lecomte J. Antigenic characterization of serogroup ‘A’ of infectious pancreatic necrosis virus with three panels of monoclonal antibodies. J Gen Virol. 1993;74:2025–2030. doi: 10.1099/0022-1317-74-9-2025. [DOI] [PubMed] [Google Scholar]
- 45.Umemoto J, Bhavanandan V, Davidson E. Purification and properties of an endo-α-N-acetyl-d-galactosaminidase from Diplococcus pneumoniae. J Biol Chem. 1977;252:8609–8614. [PubMed] [Google Scholar]
- 46.Wolf K, Snieszko S, Dunbar C, Pyle E. Virus nature of infectious pancreatic necrosis in trout. Proc Soc Exp Biol Med. 1960;104:105–108. doi: 10.3181/00379727-104-25743. [DOI] [PubMed] [Google Scholar]