Abstract
Microglia transform in response to changes in sensory or neural activity, such as sensory deprivation. However, little is known about how specific frequencies of neural activity, or brain rhythms, affect microglia and cytokine signaling. Using visual noninvasive flickering sensory stimulation (flicker) to induce electrical neural activity at 40 hertz, within the gamma band, and 20 hertz, within the beta band, we found that these brain rhythms differentially affect microglial morphology and cytokine expression in healthy animals. Flicker induced expression of certain cytokines independently of microglia, including interleukin-10 and macrophage colony-stimulating factor. We hypothesized that nuclear factor κB (NF-κB) plays a causal role in frequency-specific cytokine and microglial responses because this pathway is activated by synaptic activity and regulates cytokines. After flicker, phospho–NF-κB colabeled with neurons more than microglia. Inhibition of NF-κB signaling down-regulated flicker-induced cytokine expression and attenuated flicker-induced changes in microglial morphology. These results reveal a mechanism through which brain rhythms affect brain function by altering microglial morphology and cytokines via NF-κB.
Frequency-specific sensory stimulation regulates cytokines, microglia, and microglia-independent cytokine expression via NF-κB.
INTRODUCTION
Brain rhythms are found in many brain structures and result from coordinated electrical neural activity at specific frequencies in response to task demands and environmental stimuli (1–3). Gamma frequency neural activity (~30 to 100 Hz) increases during attention and sensory processing, whereas beta frequency (~12.5 to 30 Hz) is associated with active concentration and suppression of motor behaviors (2, 4). Furthermore, neural activity is modulated rhythmically in response to rhythmic sensory stimuli, and this entrainment enhances sensory processing, including attention (5). While brain rhythms and neural entrainment to sensory stimuli are well known to reflect and influence neuronal electrical function, little is known about how different frequencies of neural activity affect other brain functions, such as immune signaling (6). Our recent work has shown that gamma frequency (40 Hz) noninvasive flickering sensory stimulation (flicker) recruits microglia to engulf pathogens in mouse models of Alzheimer’s disease, revealing that this specific frequency of neural activity alters microglia, the primary immune cells of the brain in the presence of Alzheimer’s pathology (7–11). However, microglial responses vary with pathological context, and, therefore, it is unclear whether these effects of gamma flicker on microglia generalize to healthy brains (12). Furthermore, the mechanisms by which gamma frequency activity alters microglia and immune signaling pathways remain unknown.
Brain immune function is typically ascribed to microglia, the primary phagocytes in the brain, because of their roles in engulfing pathogens and secreting both cytokines and related extracellular signals that support neuronal health (12–14). Microglia are known to respond to changes in sensory and neural activity, such as sensory deprivation or seizures (15–17). In the healthy adult brain, microglial processes dynamically shift to survey surrounding synapses, acquiring elongated phenotypes to scan their environment and monitor neuronal activity (e.g., hyper-ramified). Microglia rapidly change their phenotype through intermediate stages to shortened processes and enlarged soma when detecting pathogens (e.g., hypo-ramified) and evolve into ameboid phenotypes that can lack processes altogether (18). Nevertheless, little is known about how specific patterns of neural activity, beyond gross increases or decreases, affect microglia and cytokines in healthy individuals. While microglia are a commonly accepted source of cytokines, neurons are known to secrete specific cytokines, such as fractalkine (CX3CL1) (19–22). Although the mechanisms that govern neuronally sourced cytokines have received limited attention, cytokine expression is controlled by canonical phospho-protein signaling pathways, which are present in both microglia and neurons, including the nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling cascades (23–26). NF-κB is activated in neurons by synaptic activity, and we have shown that 40 Hz flicker increases activation of this pathway (10). Because of its regulation by synaptic activity and its role in cytokine expression, the NF-κB pathway could be a key mechanism by which brain rhythms regulate microglial responses (27–30).
Sensory experiences can induce frequency-specific brain rhythms, such as gamma, and neural activity is modulated by rhythmic sensory stimuli. Thus, it is important to understand how specific frequencies of neural activity affect cytokines and microglia. Cytokine and microglial responses to specific frequencies of neural activity may play a role in how the brain adapts to the environment. Flicker stimulation, which precisely induces specific frequencies of neural activity, allows us to noninvasively test the effects of different frequencies of neural activity without significantly increasing or decreasing mean firing rates, which has more commonly been investigated (9). We and others have shown that simple flickering lights and sounds, similar to a fast strobe light and beeping, drive specific frequencies of neural activity in primary sensory areas at the flicker frequency of the stimulus (7–9, 31, 32) and beyond (33, 34). Thus, flicker stimulation noninvasively induces specific frequencies of neural activity to test how this activity affects microglia and cytokines without significantly changing average neural firing rates (8, 9). Previously, we have shown that 20 and 40 Hz visual flicker led to different patterns of cytokine protein expression in healthy mice, but the effects of specific flicker frequencies on microglia in healthy mice and the causal biomolecular mechanisms by which neural activity causes these changes are unknown.
To elucidate how brain rhythms modulate microglia and cytokines, we tested the hypothesis that flicker differentially affects microglia and cytokines via NF-κB, which is a canonical phospho-protein signaling pathway. First, we established frequency-specific effects of flicker on microglia and cytokine-related transcription in healthy mice. We used 20 and 40 Hz flickering stimuli because they drive frequencies of neural activity within common, naturally occurring brain rhythms, and we previously found that specific frequencies have distinct effects on cytokine protein levels. We found that 1 hour of 40 or 20 Hz flicker was sufficient to differentially transform microglial morphology, consistent with different functional roles, and to alter transcription of genes that control cytokine expression (12). We next investigated the cellular source of flicker-induced cytokine expression because cytokines can both cause microglial changes or arise from microglia. We found that 40 Hz flicker in microglia-depleted mice increases tissue levels of the cytokines macrophage colony-stimulating factor (M-CSF), which recruits microglia, and interleukin-10 (IL-10), which is anti-inflammatory, indicating that these cytokines are expressed from a different cellular source. Because NF-κB and MAPK pathways regulate synapses and immune function, we tested the hypothesis that these pathways are necessary for flicker-induced cytokine expression and microglial changes. We found that inhibition of either NF-κB or MAPK signaling pathways before flicker exposure down-regulated expression of cytokines, while inhibition of NF-κB only prevented microglial branching responses to 40 Hz flicker. Together, these results show that specific frequencies of flicker elicit a distinct effect on microglia and cytokine transcription. Furthermore, these findings reveal an NF-κB–mediated signaling mechanism by which 40 Hz flicker activity affects cytokines and microglia. Collectively, our data thus define a previously unknown pathway linking flicker stimulation, brain rhythms, and brain immune signaling.
RESULTS
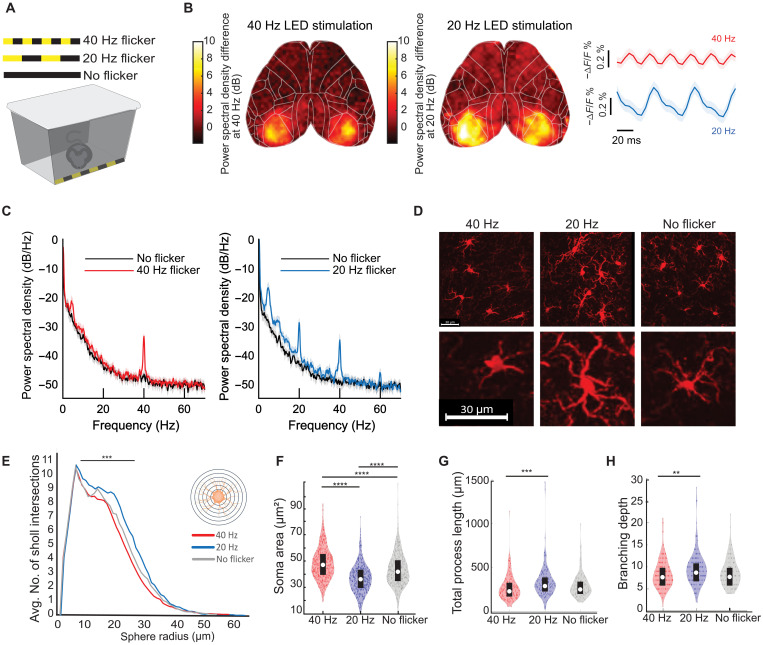
Visual flicker elicits corresponding frequency-specific activity in primary visual cortex
We first established that visual flicker induces neural activity at the specific frequency of flicker across all of the visual cortex. While prior work has shown that visual flicker induces neural activity at the specific frequency of flicker in the visual cortex using local field potential recordings (35), these recordings do not assess neural activity on a large spatial scale across cortex or even across all primary visual cortex. Thus, we began the current study by asking whether both 20 and 40 Hz flicker stimuli would respectively drive 20 and 40 Hz neural activity across visual cortex. To test this, we performed wide-field voltage imaging at 200 frames/s during 20 or 40 Hz flicker with a fast voltage indicator, JEDI-1P-Kv, that can detect high-frequency neural activity (35). With this approach, we detected fast fluctuations in voltage within neurons. We observed strong neural responses in the frequency of the visual flicker throughout bilateral primary visual cortex at both 20 and 40 Hz (Fig. 1B and fig. S1). During 20 Hz flicker, the voltage increases and decreases over 50 ms, consistent with a 20 Hz signal, while during 40 Hz flicker, the voltage increases and decreases over 25 ms, consistent with a 40 Hz signal (Fig. 1B and fig. S1). Twenty-hertz flicker produces a peak in the power spectral density at 20 Hz, while 40 Hz flicker produces a peak at 40 Hz. Note that peaks at multiples of the stimulus frequency at 40 and 60 Hz during 20 Hz flicker are expected harmonics of 20 Hz due to the fact that the neural response is not a perfect sinusoid and represent the difference between the real signal and a perfect sinusoid. A peak at harmonic frequencies in the power spectral density does not mean that there is a corresponding periodic signal in time domain, as evidenced by the clearly slower oscillations during 20 Hz flicker. Thus, 20 Hz flicker induces 20 Hz neural activity, and 40 Hz flicker induced 40 Hz neural activity.
Fig. 1. Visual flicker affects microglial morphology in a frequency-specific manner in wild-type mice.
(A) Light-emitting diode (LED)–lined stimulation box for mice. (B) Representative spatial heatmap of power differences at 40 Hz (left) or 20 Hz (right) during visual flicker relative to no flicker from wide-field voltage imaging of JEDI-1P-Kv, shared scale bar for visualization. Right: Mean and 95% confidence interval (CI) of voltage imaging traces. n = 6 mice, 10 trials per mouse. (C) Mean and 95% CI power spectral density of voltage imaging signal in primary visual cortex (2 by 2 pixels) during 40 Hz visual flicker (left, red) or 20 Hz flicker (right, blue) and baseline (black). Peaks at 40 and 60 Hz are expected harmonics of 20 Hz due to the fact that the neural response is not a perfect sinusoid. (D) Example Iba-1+ microglia (red). Scale bars, 30 μm. (E) Sholl analysis of microglia after 40 Hz flicker (red), 20 Hz flicker (blue), or no flicker (gray) shows significant differences between groups (linear mixed model with repeated measures). (F) Soma area differed after 40 Hz (n = 6 mice, 459 microglia), 20 Hz (n = 5 mice, 398 microglia), or no flicker (n = 5 mice, 388 microglia) [F(12,1242) = 98.63, P = 1.75 ×10−40; t tests: 40 Hz versus 20 Hz, P = 9.56 × 10−10; 40 Hz versus no flicker, P = 1.04 ×10−8; 20 Hz versus no flicker, P = 9.56 ×10−10]. (G) Total process length differed after 40 or 20 Hz flicker [F(2,443) = 6.8925, P = 0.0011; t tests: 40 Hz versus 20 Hz, P = 0.0007; 40 Hz versus no flicker, P = 0.432; 20 Hz versus no flicker, P = 0.0622]. (H) Full branching depth differed after 40 or 20 Hz flicker [F(2,443) = 4.789, P = 0.0088; t tests: 40 Hz versus 20 Hz, P = 0.0069; 40 Hz versus no flicker, P = 0.7235; 20 Hz versus no flicker, P = 0.0915]. For (F) to (H), one-way analysis of variance (ANOVA) and post hoc t tests with Tukey’s post hoc test correction. Box plots inside violin plots indicate median and quartiles, and dots indicate individual microglia. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Visual 20 and 40 Hz flicker stimuli elicit distinct microglial morphological responses in healthy mice
Having established that 20 and 40 Hz flicker induce different brain rhythms throughout primary visual cortex, we next asked whether 20 and 40 Hz stimulation led to different microglial morphologies that correspond to different microglial functions (18, 36). We hypothesized that 40 Hz flicker would induce a transition to hypo-ramified microglial morphology, as observed during synaptic engulfment and other microglia activity, while 20 Hz flicker would induce microglia with hyper-ramified morphology that occurs during surveillance or homeostatic processes (13, 37). We compared 40 Hz flicker to 20 Hz flicker to best identify the effects of the specific frequency of periodic activity rather than the general effects due to any stimulus that turns on and off. We compared 40 to 20 Hz instead of random stimulation to compare two periodic stimuli and the related neural activity and because we have found that random stimulation exhibits variable effects on cytokines (10). We expected that while 20 and 40 Hz would elicit different microglial responses, these effects should be more subtle than those due to injury or disease, instead reflecting a range of microglial morphology present in the healthy brain (36, 38–41). To test this hypothesis, we analyzed microglial morphology after the mice were exposed to 1 hour of flicker at either 20 Hz flicker, 40 Hz flicker, or no flicker (constant light) (Fig. 1, A and C). We found that the 40 Hz flicker elicited a significant increase in soma area compared to 20 Hz, whereas 20 Hz elicited significantly increased process length and branching compared to 40 Hz (Fig. 1, E to G; fig. S2; and table S1). However, these changes were not associated with significant differences in Iba-1 (encoded by Aif1) transcription (P = 0.6 unadjusted, DESeq2). Thus, 40 Hz stimulation leads to more ameboidal morphology with fewer ramifications consistent with engulfing microglial phenotypes, in line with ours and others' prior studies (9, 18, 36), and 20 Hz stimulation results in a more ramified morphology with longer processes consistent with surveillance phenotypes.
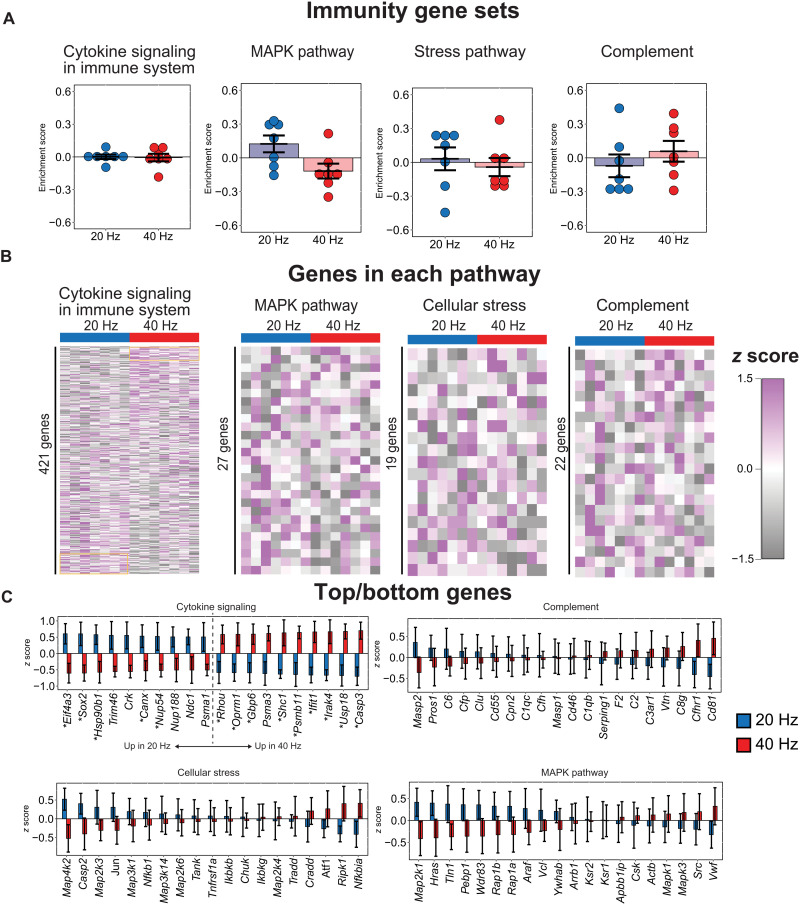
RNA sequencing analysis reveals differences in cytokine-related transcription factors between 20 and 40 Hz flickers
To identify potential molecular mechanisms of these different microglial morphological responses to brain rhythms, we asked which immune-related gene pathways also differentially respond to 20 and 40 Hz flickers. To broadly examine changes in expression of several potentially microglia-related pathways with relatively high sequencing depth, we used bulk RNA sequencing (RNA-seq) to quantify gene expression in the visual cortices of mice exposed to 20 or 40 Hz combined audio/visual flicker, which is expected to have similar effects in the visual cortex as visual-only flicker (10). We then used a gene set variation analysis (GSVA) with the C2 canonical signaling pathways gene sets database (MSigDB, Broad) (42). We found that different frequencies of flicker differentially affect transcription factors that control cytokine expression with a more nuanced effect than injury or disease. We did not identify clear changes in signaling pathways that have previously been shown to affect microglial responses to neural activity, including norepinephrine and chemoreceptor P2Y12 (fig. S3). We also did not identify any significantly enriched gene sets between 20 and 40 Hz flicker related to immunity, including MAPK pathway, cytokine signaling, stress response, or complement cascade (Fig. 2A and data S1). However, these cytokine-related immune pathways are made up of broad lists of more than 50 genes, and a more detailed examination revealed significant differences in expression of transcription factors that control cytokine expression, such as Ifit1 and Irak4 (Fig. 2, B and C) (43–45). While the 20 and 40 Hz flickers induced significant differences in cytokine expression, neither produced coordinated expression patterns indicative of broad inflammation that is often found in lipopolysaccharide models, for example (46). Given that these changes in cytokine expression are associated with changes in microglial morphology and occur in healthy brains, they suggest an overall frequency-specific modulation of cytokines that is distinct from that due to injury or disease.
Fig. 2. Audio/visual flicker for 1 hour at 20 Hz or 40 Hz significantly modulates cytokine-related transcription factors.
(A) GSVA did not identify significant enrichment of immune-related gene sets after 20 Hz (blue) or 40 Hz (red) flicker (means ± SEM, statistical testing via gene set permutation analysis, and dots indicate individual animals). (B) Heatmap representation of all genes within each pathway reveals that 20 Hz versus 40 Hz flicker drive distinct patterns of gene expression within the “cytokine signaling” pathways but no clear patterns in other pathways (rows are z-scored, each row is a gene, and each column is an animal). (C) Many of the top and bottom 10 genes within the cytokine signaling pathway are each significantly differentially regulated (top left, DESeq2, *P < 0.05 unadjusted). Top/bottom 10 genes for “complement” (top right), “cellular stress” (bottom left), and “MAPK pathway” (bottom right) signaling show no significant differences.
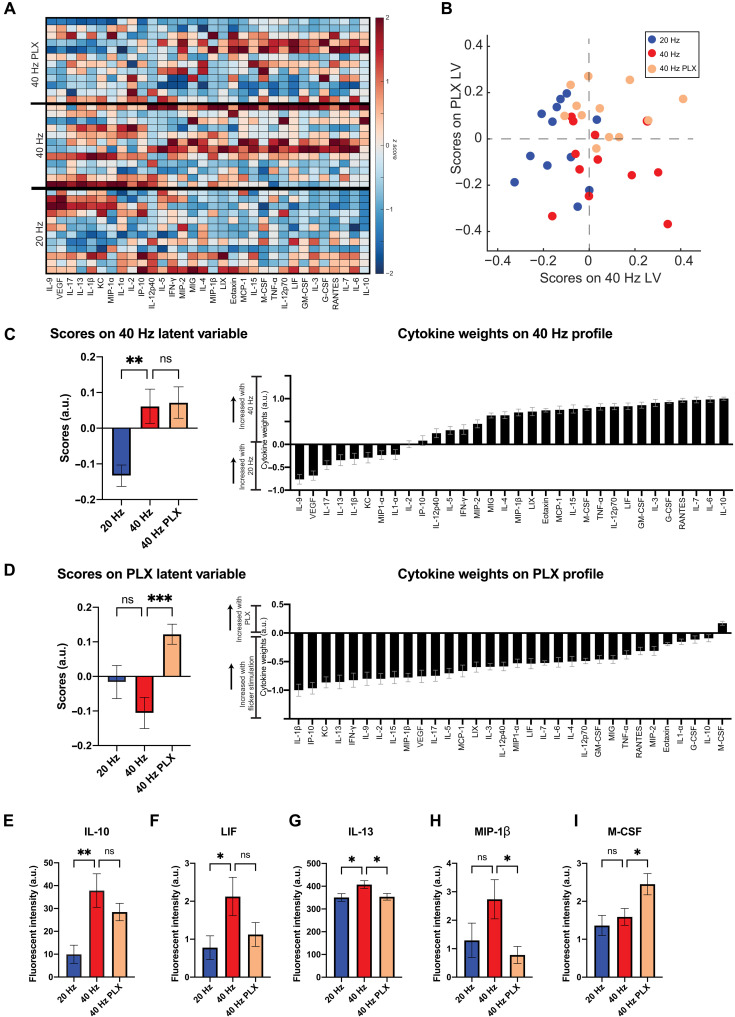
Flicker stimulation induces microglia-dependent and -independent cytokines
Because cytokines both signal to and are secreted by microglia, we determined the necessity of microglia for flicker-induced cytokine expression by examining protein expression changes after depleting microglia. We compared 40 Hz effects to those of 20 Hz because these stimuli differentially affected cytokines and microglia and because 20 Hz flicker controls for key aspects of the flickering stimulus, including light exposure, stimuli turning on and off repeatedly, and any salience associated with flickering stimuli. We depleted microglia using PLX3397 (PLX) (290 parts per million in the chow; see Materials and Methods and fig. S4), exposed mice to visual flicker, and then used a Luminex multiplexed immunoassay to quantify protein expression of 32 cytokines in the visual cortex, including many that previously showed differential expression in response to 40 and 20 Hz flicker in our prior work (Fig. 3 and data S3) (10). We initially compared protein expression in control animals fed a regular diet versus PLX diet, showing that expression of some cytokines persisted in microglia-depleted mice (fig. S5). We accounted for the multidimensional nature of the data using a discriminant partial least-squares regression [partial least-squares discriminant analysis (PLSDA)], as we have done previously (10). We identified two latent variables to separate flicker frequency condition [latent variable 1 (LV1)] and intact versus depletion of microglia (LV2). LV1 separated 40 Hz flickered animals to the right and 20 Hz flickered animals to the left along LV1 (Fig. 3B). In addition, microglia-depleted animals (40 Hz PLX) separated toward the top from microglial-intact animals toward the bottom along LV2. We evaluated potential overfitting of the model using a permutation analysis, wherein sample labels were permuted 1000 times, and distances between group centroids were compared against true group labels. This analysis revealed that true group assignments were better than random (40 Hz versus 20 Hz, P = 0.316; 40 Hz versus 40 Hz PLX, P = 0.20; 40 Hz PLX versus 20 Hz, P = 0.107).
Fig. 3. Forty-hertz flicker up-regulates expression of cytokines M-CSF and IL-10 in microglia-depleted mice.
(A) Cytokine expression in visual cortices of mice fed PLX and exposed to 1 hour of 40 Hz flicker (top “40 Hz PLX”) or control diet then exposed to 40 Hz (center “40 Hz”) or 20 Hz (bottom “20 Hz”) flicker. Cytokines (columns) are ordered by their LV1 weights. Color indicates z-scored expression levels. Each row is an animal. (B) PLSDA identified LV1, the axis that separated 40 Hz (red and beige)– and 20 Hz (blue)–exposed animals. LV2 separated animals treated with PLX and 40 Hz flicker (beige) from control and 40 Hz flicker exposure (red). (C) Left: LV1 (40 Hz LV) scores were significantly different between groups [means ± SEM; F(2,33) = 7.747, P = 0.0017] with significant differences between control diet and 40 Hz versus 20 Hz flicker (P = 0.0043, Dunnett’s multiple comparisons). Right: Weighted profile of cytokines in the 40 Hz profile based on which cytokines best correlated with separation of 40 Hz (positive) versus 20 Hz flicker control (negative) (means ± SD, a leave-one-out cross-validation). (D) As in (C) for LV2 (PLX LV). Scores were significantly different between groups [means ± SEM; F(2,33) = 7.630, P = 0.0019] with significant differences between animals exposed to 40 Hz flicker and pretreated with PLX diet or control diet (P = 0.0010, Dunnett’s multiple comparisons). (E) IL-10 expression differed across the three groups [F(2,33) = 7.195, P = 0.0026], with a significant increase in 40 Hz versus 20 Hz control (IL-10, P = 0.0014). (F) LIF expression differed between 40 and 20 Hz control groups (P = 0.0362). (G) IL-13 expression differed across groups [F(2,33) = 3.840, P = 0.0317] with significant differences between 20 and 40 Hz flicker (P = 0.0345) and between 40 Hz flicker control and PLX pretreated animals (P = 0.0479). (H) MIP-1β expression differed across groups [F(2,33) = 3.315, P = 0.0488] with significant differences between 40 Hz control and 40 Hz with PLX diet (P = 0.0339). (I) M-CSF expression differed across groups [F(2,33) = 4.959, P = 0.0131) with significant differences between 40 and 40 Hz plus PLX diet (P = 0.0445). (E) to (I) show means ± SEM; one-way ANOVA throughout. * P < 0.05, ** P < 0.005, *** P < 0.0025, ns, not significant. a.u., arbitrary units.
We identified which cytokines had decreased or intact expression levels when microglia were depleted. First, we determined the profile of cytokines in LV1 (Fig. 3C) that were associated with 40 Hz (positive) or 20 Hz (negative) and the profile of cytokines in LV2 (Fig. 3D) that were associated with microglial depletion (PLX, positive) versus microglial intact mice (negative). Top correlates from LV1, including IL-10, which has anti-inflammatory functions, were not significantly changed by microglial depletion, showing that it is microglia independent (Fig. 3E). In contrast, the top correlate from LV2 was M-CSF (CSF1, the agonist for CSF1R, which is antagonized by PLX3397), which was increased by microglial depletion and is known to recruit microglia, showing that its expression levels were microglia dependent, but lead to increased rather than decreased expression (Fig. 3I). Some cytokines including IL-13, an anti-inflammatory cytokine, and macrophage inflammatory protein–1β (MIP-1β), a chemokine, were microglia dependent, consistent with the known role of microglia in cytokine expression (47). Unexpectedly, flicker-induced expression of IL-10 and M-CSF is independent of microglia or increased in the absence of microglia. Because microglia are commonly thought of as the primary source of cytokines, we sought further evidence of cytokine expression in the absence of microglia. Accordingly, we assessed cytokine levels after microglia depletion alone in animals that were not exposed to flicker. Consistent with our hypothesis that cytokines can be expressed in the absence of microglia, we find that some cytokines are elevated following microglia depletion without flicker exposure. Because we have previously established that 40 Hz flicker elevates cytokines including M-CSF and IL-10, these results, combined with our prior findings, support our hypothesis that these cytokines can arise from nonmicroglia cells (10). Thus, flicker stimulation increased the expression of both microglia-dependent and -independent cytokines.
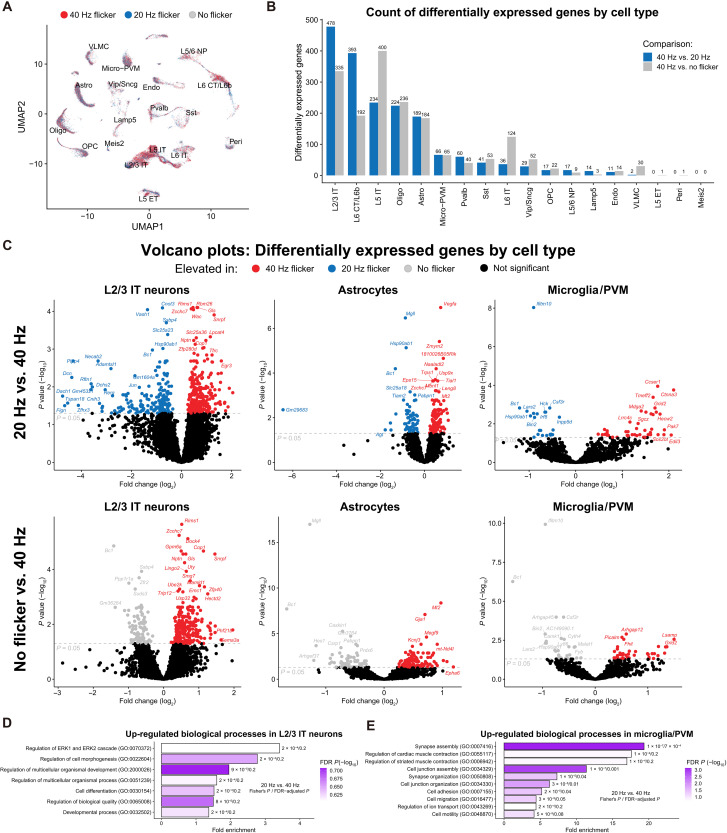
Single-nuclear RNA-seq identifies most of differentially expressed genes in neurons
Since we found that flicker stimulation induces both microglia-dependent and -independent cytokine expression, we next used single-nuclear RNA-seq (snRNA-seq) to profile transcriptional changes within each cell type in the visual cortices of mice exposed to no flicker, 20 Hz, or 40 Hz visual stimulation for 1 hour (n = 4 per group). More than 5000 nuclei were analyzed per sample, and Azimuth (see Materials and Methods) was used to identify multiple neuronal populations (layer L2/3 glutamatergic and layer L5 glutamatergic), astrocytes, oligodendrocytes, and microglia/perivascular macrophages (PVMs), among other cell types (Fig. 4A and fig. S5A). Although 20 and 40 Hz are modestly different stimuli, differential gene expression analysis revealed that 40 Hz flicker had pronounced effects on L2/3, L6, and L5 neuronal populations, with fewer differentially expressed genes (DEGs) in oligodendrocytes, astrocytes, and microglia/PVMs compared to 40 Hz and no flicker (Fig. 4B and data S2). Comparing 40 to 20 Hz in L2/3 glutamatergic neurons revealed that 40 Hz flicker increased genes associated with neurotransmission and synaptic plasticity (e.g., Gls, Gria2, and Rims1) as well as cell signaling and transcription pathways, e.g., genes related to NF-κB and/or MAPK (e.g., Ptn, Wnk1, and Egr3). Many of these genes were also up-regulated in 40 Hz compared to no flicker. In astrocytes, 40 Hz versus 20 Hz showed increased expression of growth factors and receptors (e.g., Vegfa and Eps15), cell signaling (e.g., Prkd1 and Ppp2r5a), and scaffolding proteins (e.g., Abi1, Faf1, and Gab2). Last, 40 Hz compared to 20 Hz in microglia showed increased expression of membrane receptors (e.g., Grid2, Lrrc4c, Pcdh7, and Cldn11) and cytoskeletal genes (e.g., Sgcz). Gene ontology (GO) analysis of genes up-regulated in 40 Hz versus 20 Hz flicker revealed enriched GO terms associated with extracellular signal–regulated kinase 1 (ERK1)/ERK2 cell signaling and cellular morphogenesis in L2/3 neurons (Fig. 4D) but no enriched terms in astrocytes. In microglia/PVMs, we found enrichment of terms associated with cell organization, cell migration, and cell motility (Fig. 4E), transcriptional changes that are consistent with our observed morphological changes (Fig. 1). Collectively, these data show that 40 Hz flicker elicits multiple changes in the visual cortex across diverse cell types and biological functions. Moreover, enrichment of GO terms for cell signaling at this 1-hour time point at the transcript level is consistent with our prior findings that 40 Hz flicker robustly activates NF-κB and MAPK signaling in the visual cortex after 15 min (10).
Fig. 4. snRNA-seq analysis reveals DEGs in multiple cell types after flicker.
(A) Uniform Manifold Approximation and Projection (UMAP) projection of single-cell data from all 12 samples reveals separation into clusters by cell type (reference mapping and cell type labeling via Azimuth) with approximately even representation across stimulation conditions. (B) Number of DEGs by cell type from pseudo-bulk data of 40 Hz, 20 Hz, or no flicker stimulation [n = 4 per stimulation group; P < 0.05, false discovery rate (FDR)–adjusted Wald test]. (C) Volcano plots of DEGs from pseudo-bulk data from layer 2/3 intratelencephalon-projecting (L2/3 IT) neurons (left), astrocytes (center), and microglia/PVM (right). Differential expression analysis was conducted for 40 Hz versus 20 Hz flicker stimulation (top row) and 40 Hz versus no flicker stimulation (bottom row) (FDR-adjusted Wald test). (D) GO analysis of DEGs identified significantly enriched biological processes in L2/3 IT neurons and (E) microglia/PVMs (PANTHER 17.0 overrepresentation test, FDR-adjusted Fisher’s exact P < 0.25).
Phosphorylated NF-κB colocalizes with neurons following flicker
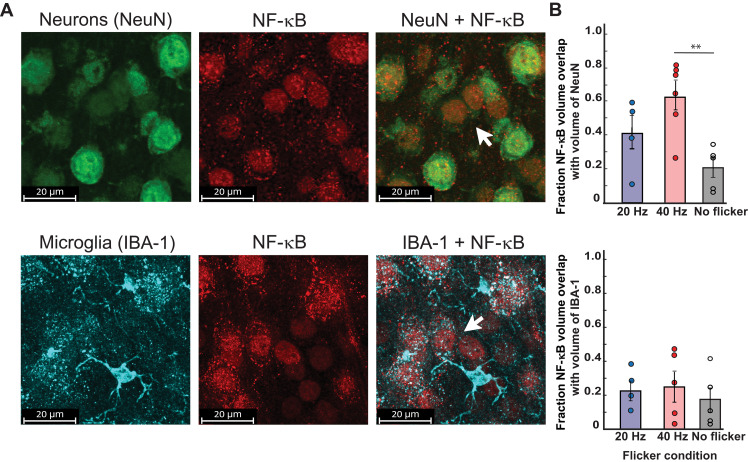
Having previously found increased phosphorylation of the NF-κB pathway in visual cortex after 15 min of 40 Hz flicker (10) and that NF-κB gene sets were enriched in neuronal populations, we next hypothesized that phosphorylated NF-κB (pNF-κB) would localize to neurons after 40 Hz flicker stimulation, indicating NF-κB activation within neurons. We examined colocalization of pNF-κB and neurons or microglia after 15 min of 40 Hz flicker because phosphorylation within the NF-κB pathway is inherently transient, and we previously found the most robust activation of the pathway after 15 min of 40 Hz flicker. To address this question, we colabeled pNF-κB with cell markers for neurons (NeuN, neuronal nuclear protein) and microglia (Iba-1) (10). After 40 Hz flicker stimulation, pNF-κB colocalized with most NeuN+ labeling and little Iba-1 labeling. Specifically, 43% (SEM = 0.064) of pNF-κB signal colocalized with neurons compared to 22% (SEM = 0.04) of microglia [F(1,26) = 8.95, P = 0.006] (Fig. 5). These findings indicate that pNF-κB signaling occurs within neurons after flicker. There was a significant effect of flicker condition [F(2,26) = 5.56, P = 0.01]; however, Tukey’s post hoc tests revealed that the effect of flicker condition was due to significant differences between the flicker conditions 40 Hz versus no flicker for NeuN overlap (P = 0.008).
Fig. 5. pNF-κB colocalizes with neurons after exposure to 40 Hz flicker.
(A) Top: Representative images show colocalization of pNF-κB volume (red) with neuronal marker, NeuN, (green) volume after 15 min of 40 Hz flicker exposure. Scale bars, 20 μm. Bottom: Representative images labeling microglia marker Iba-1 (cyan) show low colocalization of pNF-κB (red) volume within microglia volume after 15 min of 40 Hz flicker exposure. Scale bars, 20 μm. (B) Percent of total pNF-κB volume colocalized with cells (top, neurons; bottom, microglia) across flicker frequencies. ANOVA results show that pNF-κB was highly colocalized with neurons (mean = 43% across all experimental groups; SEM = 0.064) compared to microglia (mean = 22%; SEM = 0.04) [F(1,26) = 8.95, P = 0.006], with a significant effect of flicker condition [F(2,26) = 5.56, P = 0.01). Tukey’s post hoc tests revealed that the effect of flicker condition was due to significant differences between flicker conditions 40 Hz versus no flicker for NeuN (P = 0.008) compared to 40 Hz versus 20 Hz (P = 0.17) or 20 Hz versus no flicker (P = 0.28).**P < 0.01.
Gamma flicker–induced microglial changes are mediated by NF-κB
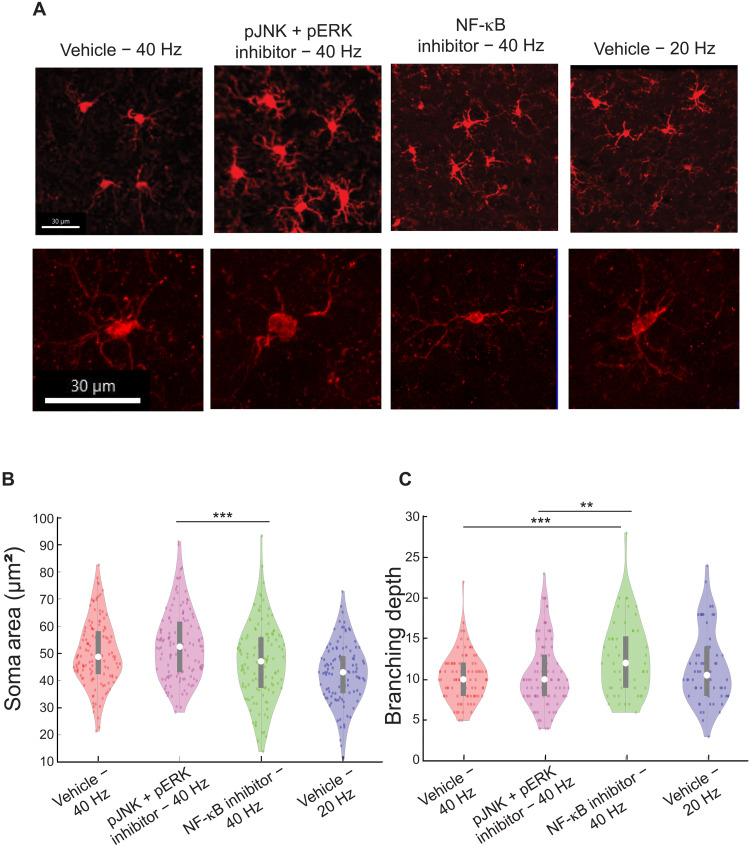
Having found that pNF-κB colabeled with neurons and that microglia-modulating cytokine expression was stimulated by 40 Hz flicker in microglia-depleted mice, we next reasoned that inhibition of predominantly neuronal pNF-κB would inhibit changes in microglial morphology. To test this, we examined inhibition of two pathways previously shown to be altered by flicker (10). We expanded our approach to examining both NF-κB, which localized to neurons, and the MAPK pathway because we previously found that phosphorylation of both pathways are altered by 40 Hz flicker, both are known to respond to synaptic activity (10, 48, 49), and our snRNA-seq identified genes and GO terms associated with general signaling that could encompass both pathways. As above, we compared microglial morphology in mice exposed to 40 Hz and vehicle, 40 Hz and MAPK inhibitors, or 40 Hz and NF-κB inhibitors. We compared 40 Hz flicker and vehicle or MAPK and NF-κB inhibitors to 20 Hz flicker because these stimuli isolate the effects of different frequencies of stimulation while controlling for nonspecific effects of flickering stimuli. Furthermore, these stimuli elicit different microglial morphologies with 40 Hz flicker eliciting microglia to shift toward an ameboid-like shape, with increased soma area and shorter processes, while 20 Hz flicker elicited microglia to shift toward a phenotype where processes are more branched and elongated. Mice were injected intraperitoneally with small-molecule inhibitors of NF-κB [IMD0354 (10 mg/kg) plus curcumin (100 mg/kg)], which inhibits phosphorylation of NF-κB and its translocation into the nucleus or MAPK [MAPK/MAPK kinase (MEK) inhibitor SL327 (100 mg/kg) plus MAPK/c-Jun N-terminal kinase (JNK) inhibitor SP600125 (15 mg/kg)] pathways 1 hour before exposing animals to flicker using multiple inhibitors for each pathway because of the known compensatory effects within each pathway. Following exposure to 40 Hz flicker, we found that microglia soma area was significantly smaller after NF-κB inhibition than MAPK inhibition (pJNK + pERK inhibitor) but was not different from the 40 Hz and vehicle control, suggesting that these inhibitors have distinct but mild effects on soma size [F(2,370) = 7.5909, P = 0.0006; Tukey’s post hoc test: vehicle + 40 Hz versus MAPK inhibitor + 40 Hz, P = 0.1757; MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0795; MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0003; Tukey’s post hoc test corrected for multiple comparisons; Fig. 6C]. Assessing branching depth, NF-κB inhibition increased process branching when compared to both the 40 Hz vehicle group and MAPK inhibitor [F(2,229) = 7.3418, P = 0.0008; post hoc t tests: vehicle + 40 Hz versus MAPK inhibitor + 40 Hz, P = 0.7765; MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0006; MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0051; Tukey’s post hoc test corrected for multiple comparisons; Fig. 6D]. Because we found that NF-κB affects microglia processes, and we previously showed that 40 and 20 Hz flickers have different effects on microglia processes, we then aimed to better understand how 40 Hz and no flicker differentially affect microglia processes. To do so, we performed an experiment in which animals underwent either 40 Hz flicker or no flicker after vehicle injection or 40 Hz or no flicker after NF-κB inhibitor (fig. S6). Forty hertz flicker led to smaller convex hull volume but more branches than no flicker, suggesting more, but shorter, process segments (fig. S6, B and C). Forty hertz flicker led to more short-process segments than no flicker, and NF-κB inhibitor abolished this effect, leading to longer segments (fig. S6, E and F). Together, these results show that 40 Hz flicker exposure leads to more but shorter branches and that NF-κB is required for 40 Hz flicker–induced changes in microglia branching but not increases in cell body diameter. NF-κB inhibition with 40 Hz flicker appears to shift microglia into a more ramified state similar to that elicited by 20 Hz flicker (i.e., longer branching depth and process length) rather than the hypo-ramified pattern we observed following exposure to 40 Hz flicker, indicating NF-κB’s role as a mediator in frequency-specific effects on microglia morphology.
Fig. 6. Forty-hertz flicker–induced microglia changes are mediated by phospho-signaling pathways.
(A) Example 20× images show clusters of representative Iba-1+ microglia and single representative microglia (bottom) from each experimental condition [vehicle + 40 Hz, MAPK inhibitor (pJNK + pERK inhibitor) + 40 Hz, NF-κB inhibitor + 40 Hz, and vehicle + 20 Hz] taken at 40× magnification. Scale bars, 30 μm. (B) Soma area of Iba-1+ microglia differed between groups [F(3,475) = 14.8797, P = 2.84 ×10−9; (post hoc t tests: MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0003; Tukey’s post hoc test corrected for multiple comparisons] and (C) full branching depth of Iba-1+ microglia differed between groups [F(2,229) = 7.3419, P = 0.0008; post hoc t tests: vehicle + 40 Hz versus MAPK inhibitor + 40 Hz, P = 0.7765; MAPK inhibitor +40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0007; MAPK inhibitor + 40 Hz versus NF-κB inhibitor + 40 Hz, P = 0.0051; Tukey’s post hoc test corrected for multiple comparisons]. For (B and C) violin plots, F and P values were generated from one-way, two-tailed, unpaired ANOVA test. Differences between groups were found from Tukey’s post hoc multiple-comparisons test. Box plots inside violin plots indicate median and quartiles, and dots indicate individual microglia. **P < 0.01 and ***P < 0.001.
Cytokine expression is down-regulated by MAPK and NF-κB inhibitors
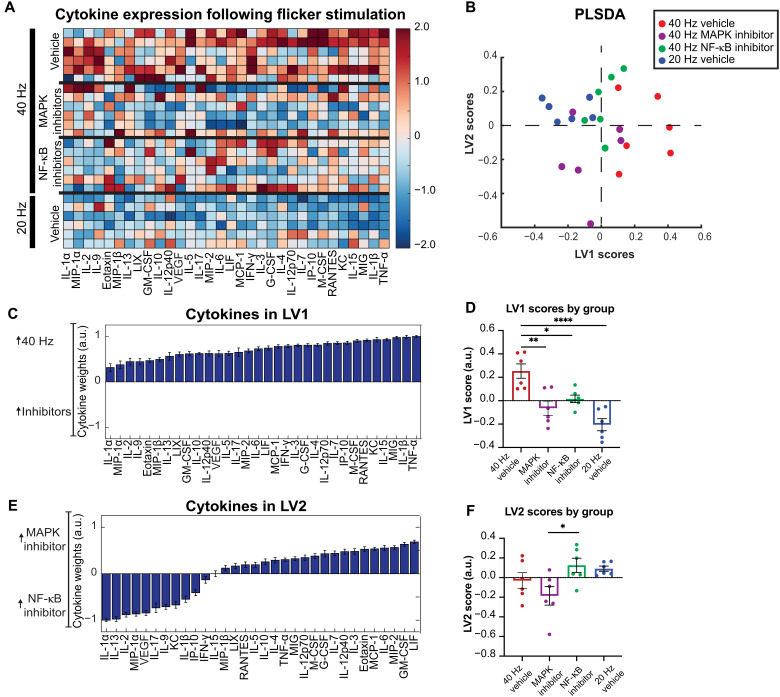
Having found that the NF-κB and MAPK signaling pathways mediate flicker-induced changes in microglial morphology, we concluded by asking whether inhibition of these pathways affects cytokine expression after 40 Hz flicker. To test this, we quantified 32 cytokines using Luminex analysis in mice exposed to 40 Hz and MAPK inhibitors, 40 Hz and NF-κB inhibitors, 40 Hz and vehicle, or 20 Hz and vehicle. PLSDA revealed that 40 Hz samples separated to the right, while 20 or 40 Hz with either MAPK or NF-κB separated to the left along LV1 (Fig. 7B and data S3). LV1 (Fig. 7, C and D) consisted of a profile of cytokines associated with 40 Hz stimulation, including tumor necrosis factor–α (TNF-α), IL-1β, monokine induced gamma interferon (MIG), IL-15, and keratinocyte chemoattractant (KC), all of which have proinflammatory properties (24, 49, 50), indicating that inhibition of each pathway suppresses the proinflammatory effects of 40 Hz compared to 20 Hz. The PLSDA also revealed that MAPK inhibition separates toward the bottom and NF-κB separates toward the top, along LV2, indicating that each inhibitor has distinct effects on cytokine expression (Fig. 7E). As above, the model was evaluated for potential overfitting using a permutation analysis with sample labels permuted 1000 times, and distances between group centroids were compared against true group labels. This analysis revealed that true group assignments were better than random (Pperm = 1.0 for 40 Hz versus 20 Hz, Pperm = 0.128 for MAPK inhibitor versus 40 Hz, Pperm = 0.417 for NF-κB inhibitor versus 40 Hz, Pperm = 0.227 for MAPK inhibitor versus NF-κB inhibitor, and Pperm = 0.67 for NF-κB inhibitor versus 20 Hz). Several of the cytokines we found that were expressed after microglia depletion were decreased following inhibition of NF-κB and MAPK, including IL-10, M-CSF, and leukemia inhibitory factor (LIF) (fig. S7), indicating that expression of these cytokines is both independent of microglia and dependent on these phospho-signaling pathways.
Fig. 7. Forty-hertz flicker–induced cytokine protein expression is mediated by MAPK and NF-κB pathways.
(A) Cytokine expression in visual cortices of mice administered either vehicle (top and bottom), MAPK inhibitors (top center), or NF-κB inhibitors (bottom center) phospho-signaling inhibitors and then exposed to 1 hour of 40 Hz (top) or 20 Hz (bottom) visual flicker. Each row represents one animal (n = 6). Cytokines (columns) are arranged in the order of their weights on the LV1 in (D). Color indicates z-scored expression levels for each cytokine. (B) PLSDA identified LV1, the axis that separated vehicle + 40 Hz flicker–exposed animals (red) to the right and all other groups to the left (vehicle + 20 Hz flicker, blue; MAPK inhibited + 40 Hz flicker, purple; NF-κB inhibited + 40 Hz flicker, green). Dots indicate individual animals for all graphs in this figure. LV2 separated the two inhibitor types (NF-κB inhibited, green; MAPK inhibited, purple). (C) The weighted profile of cytokines that make up LV1 based on which cytokines best correlated with separation of 40 Hz (positive) versus other groups (means ± SD from a leave-one-out cross-validation). (D) LV1 scores were significantly different for the 40 Hz group compared with the comparison groups [means ± SEM; F(3,20) = 13.28, P < 0.0001, one-way ANOVA]. (E) The weighted profile of cytokines that make up LV2 based on which cytokines best correlated with separation of the two inhibited groups MAPK inhibitor (positive) and NF-κB inhibitor (negative) (means ± SD from a leave-one-out cross-validation). (F) LV2 scores were significantly different between the four groups [means ± SEM; F(3,20) = 3.689, P = 0.0291, one-way ANOVA], with post hoc analysis revealing a significant difference between animals injected with the MAPK versus NF-κB inhibitors (P = 0.0415). *P < 0.05, **P < 0.01, ***P < 0.001.
In sum, our data show that different frequencies of flicker elicit different brain rhythms with divergent microglial and cytokine responses. Furthermore, our results show that much of the 40 Hz flicker–induced expression of cytokines arises from microglia, although not all, and that changes in microglia and cytokine expression depend on pNF-κB. Our results are thus consistent with a model in which 40 Hz flicker induces phosphorylation of NF-κB that, in turn, leads to expression of cytokines that affect microglial phenotypes. The initial effect on microglia may result from microglia-independent cytokines, such as M-CSF and IL-10, which arise from neuronal pNF-κB (Fig. 8). This, in turn, could initiate further release of cytokines from microglia. Our results point to previously unknown mechanisms of brain neuron-microglia communication in response to flicker and flicker-induced brain rhythms.
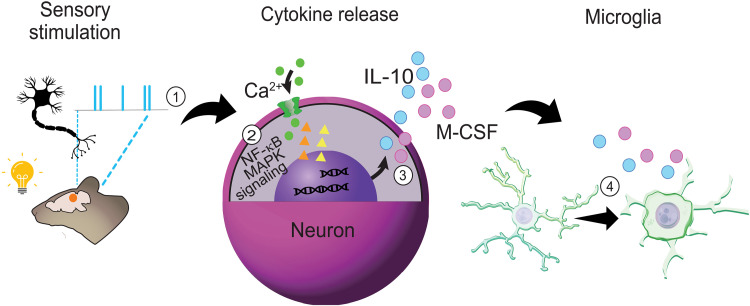
Fig. 8. Proposed mechanisms by which 40 Hz flicker changes microglia and cytokine signaling.
(1) Visual flicker entrains neurons in mouse visual cortex, (2) activating MAPK and NF-κB pathway signaling in the neuron to regulate distinct (3) cytokines, and, subsequently, (4) microglial morphology. (2) pNF-κB labeling was highly colocalized with neurons, more so than microglia, and was present even in the absence of microglia, suggesting that neurons play a key role in the modulation of the neuroimmune environment via NF-κB signaling. (3) Forty-hertz flicker increased the expression of several key cytokines including M-CSF, known to recruit microglia, and IL-10, a powerful anti-inflammatory, even in the absence of microglia. (4) Frequency-specific changes were evident in microglia morphology accompanying the altered expression of anti-inflammatory cytokines: 20 Hz flicker elicited a surveillance morphology phenotype, whereas 40 Hz flicker elicited an engulfment/amoeboid morphology phenotype, suggesting that specific frequencies play an important role in regulating brain immunity. When the NF-κB pathway was inhibited, 40 Hz flicker did not fully induce this ameboid-like state in microglia. Furthermore, cytokine expression following flicker was mediated by NF-κB phospho-signaling, suggesting that NF-κB signaling mediates flicker’s effects on microglial cell morphology and cytokine signaling.
DISCUSSION
In the current study, we found that flicker-induced brain rhythms have frequency-specific effects on microglia and cytokines via activation of NF-κB. We found that 1 hour of visual stimulation at specific frequencies is sufficient to alter microglial morphology into different phenotypes consistent with performing disparate functions (36). Forty-hertz flicker induced 40 Hz neural activity, microglia morphology associated with engulfment, and an altered profile of cytokine expression. In contrast, 20 Hz flicker induced 20 Hz neural activity, microglia surveillance or homeostatic phenotypes, and a different pattern of cytokine expression compared to 40 Hz. Furthermore, we found transcriptional changes in microglia/PVM consistent with these morphological changes. Through microglia depletion, we identified 40 Hz flicker–induced cytokines that were microglia independent, including M-CSF, a microglial chemokine and colony-stimulating factor, and IL-10, an anti-inflammatory cytokine, which may, in turn, induce microglial responses. We then investigated the role of NF-κB and MAPK signaling in mediating flicker-induced microglia and cytokine changes because these pathways are both regulated by cytokine expression and respond to synaptic activity (27–30). pNF-κB was highly localized within neurons compared to microglia. Inhibition of NF-κB or MAPK signaling revealed that 40 Hz flicker–induced cytokines are mediated in part by NF-κB and MAPK signaling, while 40 Hz flicker–induced changes in microglial ramification were mediated by NF-κB signaling only. Together, these results show that different flicker frequencies and flicker-induced brain rhythms elicit distinct microglia and cytokine responses mediated by the NF-κB signaling pathway. These studies reveal previously unrecognized effects of different brain rhythms in microglia and cytokines and important mediators underlying how brain rhythms and flicker stimulation rapidly elicit changes in brain signaling and cytokines.
Interactions between neurons, microglia, and cytokines support neuron health and survival and are key to normal brain functions, such as development and learning (13, 14, 16, 51). Moreover, microglial dysfunction plays a key role in multiple diseases, for example, by overly pruning synapses in anxiety and depression disorders or failing to clear pathogenic proteins that accumulate in neurodegenerative diseases (12). While prior studies have shown that broad increases or decreases in neural activity, such as seizure activity or silencing, alter microglia, little work has shown how specific patterns of neural activity affect microglia (14, 22, 52, 53). This is especially important to understand because extensive research has shown that different frequencies of neural activity occur naturally and are associated with different brain functions (54). Our own recent work has shown that 40 Hz frequency neural activity recruits microglia into an engulfing state in mouse models of Alzheimer’s disease (7–9). However, prior studies have primarily considered pathological states and 40 Hz (gamma) activity. Whether these effects generalize and indicate that brain rhythms beyond gamma control microglial responses in normal adult brains was unknown. We previously showed that 40 Hz flicker alters cytokine protein expression in healthy mice (10), which is consistent with our current findings that 40 Hz flicker alters transcription factors in control of cytokine expression (Fig. 3). Here, we also found that flicker-induced neural activity has distinct frequency-specific effects on microglia in healthy mice. We compared different frequencies of flicker to best identify the effects of the specific frequency of rhythmic activity rather than the general effects due to any stimulus that turns on and off. In particular, we compared 40 Hz flicker to 20 Hz flicker based on our earlier findings that 20 Hz flicker induces 20 Hz neural activity without changing mean firing or behavior and that 20 Hz flicker leads to lower cytokine expression. Because microglial morphology changes as microglia shift between surveillance and engulfing states, we examined microglial morphology histologically after 40 and 20 Hz flicker (13, 37). We found that 1 hour of 40 or 20 Hz flicker led to significant differences in microglial morphology in visual cortex. Specifically, after 20 Hz flicker, microglia have longer, more branched processes closer to the cell soma with smaller cell bodies, changes that are consistent with surveillance functions, while 40 Hz had the opposite effects (13, 37). The microglial morphological changes in response to 40 Hz are similar to those we observed in prior studies in animal models with amyloid β pathology, revealing that microglial responses to 40 Hz flicker are not specific to Alzheimer’s disease pathology. Microglial morphology with shortened processes and enlarged soma are typically observed during development or under pathological conditions when microglia engulf synapses or pathogens. Thus, we found a nonpathological stimulus that induces this microglial morphology in adulthood in healthy brains. The current study was conducted in healthy mice in the absence of pathology, showing that the effect of flicker on brain immunity is a general process that may play a role in normal brain function.
We have found that NF-κB and MAPK phospho-signaling pathways mediate the effects of flicker-induced gamma on cytokines and microglia. Prior work studying the effects of neural activity on microglia has typically not examined NF-κB and MAPK phospho-protein signaling pathways (52, 53, 55). Thus, this study reveals previously unidentified mechanisms of neural activity–cytokine-microglia interactions. While the MAPK and NF-κB pathways and downstream cytokine signaling represent canonical immune-regulatory pathways, they work in concert with other diverse pro- and anti-inflammatory pathways. These pathways are parallel to, or mediate the functions of, other diverse immune-regulatory mechanisms, including Janus kinase/signal transducers and activators of transcription (56), phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (57), the complement system (58), sphingosine-1-phosphate (59), and scavenger receptors (60), among many others. Moreover, microglia express purinergic receptors, including P2Y12, which can evoke migration and activation (61). Although we do not rule out the importance of any of these pathways, we did not interrogate them in the current study because of our prior findings that 40 Hz flicker stimulates rapid MAPK and NF-κB pathway activity within 5 min. Furthermore, our unbiased gene RNA-seq screen and pathway analysis (fig. S3) did not implicate these alternative mechanisms.
We found that flicker induced the expression of a few cytokines even when microglia were depleted. While microglia are commonly thought of as the primary source of cytokines, prior evidence shows that neurons also release cytokines (19–22). Furthermore, we found that some cytokines are microglia dependent. Unexpectedly, however, some flicker-induced cytokines were microglia independent, including IL-10 and M-CSF (Fig. 3). M-CSF promotes microglia survival, proliferation, and phagocytosis, and IL-10 has anti-inflammatory functions (62–67). Therefore, these results establish that a subset of flicker-induced cytokines do not arise from microglia. Because 40 Hz flicker–induced cytokine expression requires NF-κB and some of these cytokines are microglia independent, we wondered in which cell type NF-κB is activated. We found that pNF-κB was highly colocalized with neurons following 40 Hz flicker. These results show that neurons are abundant sources of pNF-κB, which leads to cytokine expression and microglia recruitment in healthy adult mice, and uncover a role for NF-κB in gamma frequency effects on microglia and cytokine signaling.
The present study has several limitations that suggest avenues of future research. First, we used small-molecule MAPK (JNK + ERK) and NF-κB signaling inhibitors to define the roles of these pathways in mediating cytokine expression in response to 40 Hz flicker. We selected these inhibitors because they inhibit cell signaling across multiple cell types, but this leaves open the question of which cells play a causal role in the observed cytokine-suppressive effects. We believe that this concern is partially abrogated because we quantified cytokines 1 hour after the start of stimulation, and, thus, secondary effects through other cell types would be unlikely to have sufficient time to translate into protein changes or would have minimal effects (68). Moreover, we have shown that phospho–NF-κB colocalizes with neurons, meaning that the main effects of these phospho-signaling inhibitors may be in neurons. Furthermore, our snRNA-seq analysis identified changes in genes associated with cellular signaling in L2/3 neurons but primarily changes associated cytoskeletal reorganization in microglia. Nevertheless, there remains the possibility that NF-κB from nonneuronal cells is playing a role in flicker-induced cytokine and microglial responses. Future cell type–specific knockout studies will confirm whether neuronal signaling is indeed responsible for flicker-induced cytokine expression. Future studies will also investigate the role of other glia, such as astrocytes, in NF-κB signaling and cytokine expression. In addition, it is possible that the NF-κB inhibitor acts by inhibiting NF-κB in microglia; thus, flicker could induce NF-κB in microglia to alter microglia and the production of cytokines. Second, we used the small-molecule CSF1R inhibitor PLX3397 to define the role of microglia in mediating cytokine expression in response to 40 Hz flicker because it effectively depletes microglia without the need for transgenic animals or induction of Cre recombination using tamoxifen, which is immunomodulatory (69). However, it has recently been shown that CSF1R inhibitors can have direct effects on neurons in addition to their well-known effects on peripheral monocytes (70). We are encouraged that several of our most important up-regulated cytokines (e.g., M-CSF and IL-10) in response to 40 Hz flicker are preserved in CSF1R-treated/microglia-depleted mice. We therefore conclude that these cytokines are secreted independent of microglia. Flicker-induced cytokine expression changes after microglia depletion could result from multiple cell types, including neurons or astrocytes. DEGs and GO terms in L2/3 neurons associated with cellular signaling suggest that neurons are likely an important source of flicker-induced extracellular signals, such as cytokines. Future studies will verify the roles of microglia using Cre-Lox–mediated microglial depletion. Furthermore, brain rhythms are diverse not only with different frequencies of activity but also different circuit mechanisms and relationships to sensory stimuli. Thus, further work is needed to fully elaborate how the diverse array of brain rhythms affect microglia and cytokines. Last, we conducted the present work in males to be consistent with prior studies, but the effects of brain rhythms on microglia and cytokine responses in females need to be established.
In total, we reveal a causal link between flicker, brain rhythms, microglial state, and the secretion of cytokines via the NF-κB pathway. On the basis of our combined results, we propose a model in which specific frequencies of neural activity, including gamma, increase activation of NF-κB phospho-signaling in neurons, leading to neuronal cytokine release, which, in turn, alters microglial state. This model is in line with prior work showing that NF-κB phospho-signaling in neurons is regulated by synaptic activity (27–30). Thus, our data reveal mechanistic bases for frequency-specific neuron-to-microglia signaling elicited by visual flicker and brain rhythms. This neuronal regulation of microglia and cytokine responses may play a role in plasticity since gamma oscillations have been shown to increase during learning and memory or in response to injury (71–73). Furthermore, because we find that flicker effects generalize beyond the context of Alzheimer’s pathology, this sensory-induced and brain rhythm–induced regulation of microglia and cytokine responses could be harnessed to mitigate pathological neuroimmune activity present in multiple diseases.
MATERIALS AND METHODS
Mice
The Georgia Tech Institutional Animal Care and Use Committee or the Emory Institutional Animal Care and Use Committee approved all animal work performed in this study. For experiments assessing microglia, cytokines, and phospho-signaling, adult (2- to 3-month-old) male C57BL/6J mice were obtained from the Jackson Laboratory. Mice were pair-housed and acclimated to the vivarium for at least 5 days before experiments began. Food and water were provided ad libitum. Experiments were performed during the light cycle and conducted at different times of day and with varying order of stimulus presentation to control for circadian effects. Animals were randomly assigned to flicker exposure groups, and experimenters were blind to flicker exposure conditions during analysis for all experiments. For wide-field imaging experiments, EMX1-Cre male mice were bred in-house to allow neonatal injection of the adeno-associated virus (AAV) vectors on postnatal day 0 (P0) (described below). Mice for imaging experiments were housed on reversed light cycle (12-hour light and 12-hour dark), and imaging experiments were performed during the dark cycle. Mice started to receive water restriction 5 days before the experiments and were habituated to the head-fixation setup.
Voltage imaging of visual cortex
To achieve pan-cortical expression of voltage indicators and reference fluorescence for widefield imaging, P0 EMX1-Cre pups received intracerebroventricular injections bilaterally with 4 μl of AAV vectors (2 μl per hemisphere) (52). The injected solution consisted of AAV.php.eb-EF1a-DIO-JEDI-1P-Kv2.1-WPRE (3 × 1012 viral genomes per ml or vg/ml) and AAV9-hSyn-mCherry (9 × 1012 vg/ml). A 10-μl Nanofil syringe and a 34-gauge beveled needle (World Precision Instrument) were used to load and inject the vector mixture. The injection site was identified using two-fifth of the distance between the lambda sutures to each eye with a targeting depth of 3 mm.
Mice (3 months old) that expressed voltage sensor, JEDI-1P-Kv, and reference fluorescence, mCherry, were implanted with an imaging window using the clear skull preparation (52, 55) and headplate and were allowed to recover for 7 days after surgery. Wide-field voltage imaging was conducted using a high-speed complementary metal-oxide semiconductor imaging system (MiCAM ULTIMA, SciMedia Ltd.) at 200 frames/s. During voltage imaging experiments, a light barrier was placed around the imaging window to prevent visual flicker from contaminating imaging signals. Animals were imaged during 20 and 40 Hz visual flickers for 10 trials in which each trial consisted of 10-s baseline (no flicker) and 10-s flicker stimulation. Light-emitting diodes (LEDs) were used to apply visual flicker with intensity approximately 400 lux at the head of the animal.
Voltage imaging analysis
Signals from 100 by 100 pixels (10 mm by 10 mm) were binned to groups of 2 by 2 pixels yielding 2500 (50 by 50) traces. A preprocessing pipeline was used to subtract background fluorescence, correct photobleaching, and regress hemodynamics, motion artifact, and potential light contamination caused by visual flicker (35). The JEDI-1P-kv signals were low pass–filtered at 70 Hz. The reference mCherry signals were filtered at 0 to 1, 1 to 10, 10 to 19.5, 19.5 to 20.5, 20.5 to 38, and 38 to 42 Hz, respectively, to regress out the shared nonvoltage artifacts from the voltage signals using ordinary least-squares methods. The extracted voltage signals were used for further analyses. To identify the cortical areas that responded to visual flicker, we calculated pixel-wise power spectrum. Pixel-wise power at 20 or 40 Hz during either baseline or visual flicker was averaged, and the mean power during baseline was subtracted from that during visual flicker, n = 10 trials. The difference in power at the frequency of interest from binned pixels was used to generate spatial heatmaps. The heatmaps were resized to 100 by 100 pixels and masked with a cortical map adapted from Brain Explorer (Allen Institute for Brain Science, 2004) (74). The heatmaps were used to select a 2 by 2 pixel region of interest (ROI) in V1 that responded strongly to visual flicker. Voltage signal at the selected ROI from each animal was used to calculate the power spectrum from each trial, which was averaged to plot the mean power spectral density during baseline and stimulation, respectively, using MATLAB DSP System toolbox (dsp.SpectrumAnalyzer).
Visual stimulation exposure
Mice were habituated in a dark room in the laboratory for at least 1 hour before experimentation. At the start of experimentation, mice were transferred to enclosures for flicker exposure, which had three opaque sides and one clear side that faced a strip of LEDs. Animals were exposed to either LED lights flashing at 40 Hz frequency (50% duty cycle: 12.5-ms light on and 12.5-ms light off), 20 Hz frequency (50% duty cycle: 25-ms light on and 25-ms light off), or no flicker for either 15 min or 1 hour. LED intensity was about 400 lux at the head of the animal. Immediately after stimulation exposure, mice were anesthetized with isoflurane, and within 3 min, mice were decapitated, and brains were removed. The left hemisphere’s visual cortex was microdissected, placed in microcentrifuge tube, and flash-frozen using liquid nitrogen to be used for cytokine and phospho-protein assays. The right hemisphere was used for immunohistochemistry.
Phospho-protein inhibitors
The NF-κB pathway was inhibited using inhibitor of NF-κB kinase inhibitor (IMD0354 at 10 mg/kg) combined with curcumin (S1848 at 100 mg/kg). The MAPK pathway was inhibited using MEK inhibitor (SL327 at 100 mg/kg) and JNK inhibitor (SP600125 at 15 mg/kg). All drugs were solubilized in vehicle consisting of dimethyl sulfoxide, polyethylene glycol, polysorbate 20 (Tween 20), and sterile physiologic saline. Drugs were delivered via intraperitoneal injection 30 min before the start of 1 hour of flicker stimulation.
Microglia depletion
Microglia were depleted using pexidartinib (PLX3397, MedChemExpress) incorporated into open standard diet with 15 kcal% fat (Research Diets Inc.) at a dose of 290 mg/kg (fig. S4). A control group was fed an open standard diet with 15 kcal%. Mice were fed depletion or control diet for 3 weeks, and animal weight was monitored to ensure that weight gain or loss did not differ between groups. Microglia depletion was confirmed via histology.
Immunohistochemistry and microscopy
Following brain extraction, right hemispheres were drop-fixed into cold 4% paraformaldehyde for 24 hours, rinsed in 1× phosphate-buffered saline (PBS), and stored in PBS with 0.02% sodium azide (NaN3). Each hemisphere was rinsed and placed in 30% sucrose for 3 days and then frozen. Sagittal sections were obtained at 30 μm for phosphoprotein staining and 40 μm for microglia staining. Two sections per animal were rinsed three times for 10 min each in 1× PBS on a shaker, blocked for 1 hour in blocking buffer (0.3% Triton X-100, 7.5% normal goat/donkey serum, and 1× PBS), incubated overnight at 4°C in primary antibody (anti–pNF-κB, anti-NeuN, and anti–Iba-1), and diluted in blocking buffer at antibody combination and concentrations shown in table S2. Sections were then washed in 1× PBS three times for 10 min each and incubated on shaker at room temperature for 2 hours in secondary antibody solution (goat anti-rabbit 555 and goat anti-mouse 488) diluted in blocking buffer with antibody combinations and dilutions shown in table S2. Sections were then washed in 1× PBS three times for 10 min each, and nuclei were stained with 4′,6-diamidino-2-phenylindole for 1 min. Sections were mounted using VECTASHIELD, and Z-stack images of visual cortex were taken on a Nikon Crest spinning disk confocal microscope with a 40× objective or a Zeiss LSM700 with a 20× objective for glial imaging and reconstruction and pNF-κB colocalization, respectively. For pNF-κB colocalization, nonspecific background staining was reduced by increasing the salt concentration (NaCl) in the blocking buffer from 0.3 to 0.5 M (75).
Cytokine and phospho-protein assays
For signaling and cytokine analysis, the visual cortex was thawed on ice and lysed using Bio-Plex lysis buffer (Bio-Rad). After lysing, samples were centrifuged at 4°C for 10 min at 13,000 rpm. Protein concentrations in each sample were determined using a Pierce bicinchoninic acid protein assay (Thermo Fisher Scientific). Total protein concentrations were normalized in each sample using Bio-Plex lysis buffer (Bio-Rad), and 6 μg was loaded for cytokine analysis. Cytokine analysis was conducted using the Milliplex Mouse MAP Mouse Cytokine/Chemokine Magnetic Bead Panel 32-Plex Kit [Eotaxin, granulocyte CSF (G-CSF), granulocyte M-CSF (GM-CSF), interferon-γ (IFN-γ), IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17, interferon gamma-induced protein 10 (IP-10), KC, leukemia inhibitory factor (LIF), lipopolysaccharide-induced CXC chemokine (LIX), monocyte chemoattractant protein-1, M-CSF, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, TNF-α, and vascular endothelial growth factor (VEGF)]. All kits were read on a MAGPIX system (Luminex, Austin, TX).
Morphology and colocalization analysis
Bitplane Imaris 9.7.1 (Oxford Instruments, Concord, MA) was used to assess morphology of Iba-1+ microglia and colocalization of pNF-κB with both NeuN+ neurons and Iba-1+ microglia while blinded to treatment groups. Surface reconstruction parameters were optimized across samples and examined by eye while blinded and before any hypothesis testing. To assess microglia morphology, only microglia whose processes were completely within the bounds of the image were included in analysis. Z-stack images of Iba-1+–stained visual cortex tissue were modified using Gaussian filtering and cropped to standard dimensions (2301 μm by 3301 μm by 101 μm). Within Imaris, microglia were automatically reconstructed as surfaces using standardized thresholds pertaining to diameter of clustered signal and level of surface detail. Irrelevant surfaces were filtered by number of voxels and total area. Surfaces were then manually corrected on the basis of visual inspection. Surfaces that belonged to the same cell were grouped together as a single surface. Following reconstruction, microglia surfaces were filtered again to remove any remaining surfaces not part of a reconstructed cell. Surfaces were transformed into processes based on standardized thresholds including diameter, gaps between surfaces, and surface detail, and process properties were quantified. We measured soma area, total process length, branching depth, and number of branches at each Sholl radii from the soma (fig. S7).
To assess colocalization of stained pNF-κB with both NeuN+ neurons and Iba-1+ microglia, Z-stack images of stained visual cortex tissue were modified using baseline subtraction and gamma correction filtering and cropped to standard dimensions (2400 μm by 2400 μm by 120 μm). Within Imaris, cells (either neurons or microglia) were reconstructed as surfaces using standardized thresholds pertaining to diameter of clustered signal and level of surface detail. pNF-κB staining was reconstructed as surfaces using a different set of thresholds. Using both sets of surfaces, all pNF-κB staining was eliminated except for the surfaces that were located a maximum distance of 5 μm from the reconstructed cells. Volumetric percent of total pNF-κB compared to overlap of pNF-κB staining with NeuN or Iba-1 cell stain was used to determine whether pNF-κB colocalized to a cell type.
Partial least-squares discriminant analysis
Data were z-scored within each group, and a PLSDA was performed in MATLAB (MathWorks) using the algorithm by C. Nunes (MathWorks File Exchange). To identify LVs that best separated conditions, an orthogonal rotation in the plane of the first two LVs (LV1-LV2 plane) was performed. Error bars for LV1 figures represent the mean and SDs after iteratively excluding single samples (a leave-one-out cross-validation) and recalculating the PLSDA 1000 times. To test different flicker conditions and durations, cytokine experiments were performed over several cohorts, and samples were normalized by removing batch effects. We performed a batch effects analysis (combat package in R v 3.5.0) to remove any between-experiment variability before conducting the PLSDA. Last, for analyzing the effect of 40 Hz flicker following microglia depletion, the data from three cohorts were combined by performing a batch-effects analysis (combat package in R version 3.2.0) to remove any between-experiment variability before conducting the PLSDA.
Bulk RNA-seq
mRNA were quantified from male, 4- to 6-month-old wild-type mice exposed to either 20 or 40 Hz audio/visual flickering for 1 hour (n = 8). RNA was isolated from visual cortices using a QIAGEN RNeasy kit (217804, QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Libraries were sequenced for paired-end mRNA using the NovaSeq 6000 sequencing system to obtain a sequencing depth of 30 to 40 million reads per sample. Before sequencing, quality control was performed using a bioanalyzer for RNA integrity number greater than seven. The NEBNext Poly(A) mRNA Magnetic Isolation Module and NEBNext Ultra II Directional RNA Library Prep Kit were used to prepare sequencing libraries. RNA alignment was performed by Molecular Research Laboratory, and data were validated for fastq integrity and quality. Specifically, each sample was run on four lanes and merged, followed by using DNAstar ArrayStar. Reads were mapped using the mouse reference genome GRCm39. For read assignment, the threshold was set at 20 base pairs and 80% of the bases matching within each read. Duplicated reads were eliminated, and genes with less than 20 total raw counts were removed from analysis. All gene counts were normalized using the DESeq2 package in R. RNA-seq data are deposited on the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession number GSE225842.
GSVA and statistical analysis of gene expression data
To study holistic gene changes within different pathways relevant to brain immunity and neurotransmission function, GSVA analysis was conducted. GSVA is an unsupervised enrichment algorithm, which identifies variations of pathway activity by defining enrichment score for gene sets, each containing a set of genes with shared cellular function. For GSVA gene set reference and symbols, the Molecular Signatures Database C2 (v7) gene sets (MSigDB) were used. To compare statistical differences between groups for each pathway of interest, enrichment scores for each gene set between groups were computed by measuring the true differences in means versus the differences computed by a random distribution obtained by permuting the gene labels 1000 times. False discovery rate (FDR)–adjusted P values were computed for detection of differences between two groups, and significant level was set at an FDR of <0.25. All data are presented as means ± SEM. Nonparametric Wilcoxon rank sum test was used to determine the group differences (40 Hz versus 20 Hz) in individual genes within each pathway. P < 0.05 was considered statistically significant. All analyses were performed in R using the stats package v.3.6.2.
Single-nuclear RNA sequencing
Twelve male wild-type mice aged 2 months were randomly assigned to three groups: 1 hour each of ambient light, 20 Hz visual stimulation, or 40 Hz of visual stimulation. Immediately after stimulation, right visual cortex was removed and flash-frozen in liquid nitrogen. Nuclei extraction, 10x Chromium Next GEM Single Cell 3′ GEM v3.1 library preparation (5000 cells), and Illumina sequencing with 250 M paired-end reads were conducted by Admera Health. Count matrices were calculated from two lanes per sample using 10x Genomics Cell Ranger 7.0.1 using 10x Genomics Cloud Analysis with introns included (76). We removed cells containing fewer than 200 features, more than 3000 features, or greater than 20% mitochondrial RNA to account for droplets, multiple cells, or dying cells, respectively. Count matrices were log-normalized in R using the Seurat package function NormalizeData() through which feature counts were divided by the total counts per cell, multiplied by a scale factor of 10,000, and then naturally log-transformed by ln(1 + x) (77–80). The resulting data for each sample were uploaded to the Azimuth web app (National Institutes of Health Human Biomolecular Atlas Project) for cell type assignment by reference-based mapping against the Yao et. al. (81) mouse primary motor cortex cell atlas. snRNA-seq data are deposited un the NCBI Gene Expression Omnibus under accession number GSE226822.
Pseudo-bulk data were prepared by aggregating counts of each gene across all cells of each Azimuth-assigned subclass label to produce a cell type–specific count matrix of 12 samples and 32,285 features. Each count matrix was normalized across samples using median of ratios method and analyzed for DEGs using DESeq2 (82). DEGs were defined as having a Wald test FDR-adjusted P value less than 0.05.
GO was conducted separately for L2/3 neuron, astrocytes, and microglia data using PANTHER overrepresentation test on PANTHER 17.0, available on the GO Resource. We used the mm10 GO biological process complete annotation set with Fisher’s exact test and FDR-corrected P values. The test was applied using a query set of genes significantly elevated after 40 Hz stimulation (Wald test FDR-adjusted P value less than 0.05; n = 249 genes in L2/3 IT, n = 112 genes in astrocytes, and 40 genes in microglia) and a background of all nonzero features within each cell type, which pass DESeq2-independent filtering of the mean of normalized counts (9465 genes in L2/3 IT, 5571 genes in astrocytes, and 4751 genes in microglia) (82).
Statistical analysis
For morphology and colocalization analyses, hypothesis testing was performed between the four groups using one-way, two-tailed, and unpaired analyses of variance (ANOVAs) assuming equal variance, with an α value of 0.05. Differences between groups were found from post hoc multiple-comparisons tests with a Tukey’s post hoc test correction. We used the linear mixed model procedure in SPSS 26 (IBM) for Sholl analyses after binning data by radii, with maximum-likelihood estimation of fixed effects for the frequency and intercept, including random effects for individual differences, repeated measures for radius and glial cell, and identity covariance structure. For multiplex analysis, either a one-way ANOVA (more than two groups) or two-tailed unpaired t test (two groups) was used to determine whether there was a significant LV1 separation between groups. The top correlated cytokines and/or phospho-proteins of LV1 were isolated, and an ANOVA or two-tailed unpaired t test was performed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA) to determine statistical significance between groups. These tests were followed by a post hoc Dunnett’s multiple comparisons test to determine differences between specific groups or a Tukey’s multiple comparisons test to compare differences in phosphorylation levels across time. To further confirm significant differences between groups using PLSDA, a permutation analysis was conducted, which randomly assigned animals into experimental groups and ran the PLSDA based on these shuffled values 1000 times. The mean for each permuted value was then compared as the square root of the difference between the sums of squares on the LV1 and LV2 axis of PLSDA. For each test, true group assignment showed Ppermute < 0.05 compared to the randomly permuted distribution, further confirming the validity of our data. To compare individual cytokine expression in the microglia depletion and flicker experiment, a one-way ANOVA was used, following by multiple comparisons using the Tukey method.
Acknowledgments
We thank the Wood and Singer laboratory for critical feedback and technical assistance on this work.
Funding: This work was supported by National Institutes of Health grant R01-NS-109226 (to A.C.S.), National Institutes of Health grant R01-NS-109226-01S1 (to K.M.G.), National Institutes of Health grant R01-NS-111470 (to D.J.), National Institutes of Health grant R01-AG-075820 (to L.B.W.), National Institutes of Health Cell and Tissue Engineering Biotechnology Training Grant (T32-GM008433), The Coins for Alzheimer’s Research Trust Fund (to L.B.W. and A.C.S.), Packard Award in Science and Engineering (to A.C.S.), friends and alumni of Georgia Tech (to A.C.S.), and NSF CAREER 1944053 (to L.B.W.).
Author contributions: Conceptualization: K.M.G., Y.W., D.J., L.B.W., and A.C.S. Methodology: K.M.G., A.Pr., A.S., C.H., S.B., Y.W., M.C.G., D.J., L.B.W., and A.C.S. Investigation: K.M.G., A.Pr., A.S., C.H., S.B., A.Py., Y.W., E.S., M.C.G., T.C.F., D.J., L.B.W., and A.C.S. Visualization: K.M.G., A.Pr., A.S., C.H., S.B., A.Py., Y.W., M.C.G., D.J., L.B.W., and A.C.S. Supervision: D.J., L.B.W., and A.C.S. Writing—original draft: A.Pr., A.S., L.B.W., and A.C.S. Writing—review and editing: A.Pr., A.S., C.H., S.B., A.Py., M.C.G., T.C.F., L.B.W., and A.C.S.
Competing interests: A.C.S. owns shares of Cognito Therapeutics. Her conflicts are managed by Georgia Institute of Technology. A.C.S., K.M.G., and L.B.W. are inventors of “Systems and methods for driving neural activity to control brain signaling and gene expression,” U.S. patent application no. 16/979,226, filed 09 September 2020. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data and code are available via Zenodo at DOI: 10.5281/zenodo.8121569 and via GitHub at https://github.com/singerlabgt/PrichardGarza_ScienceAdvances/ and https://github.com/JaegerLab/VisualFlickerVoltageImaging for custom code, NCBI GEO for transcriptomic data (NCBI Gene Expression Omnibus accession number GSE225842 and GSE226822), and NeuroMorpho.Org for histological data (identified by paper DOI).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 and S2
Legend for movie S1
Legends for data S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movie S1
Data S1 to S3
REFERENCES AND NOTES
- 1.M. G. Frank, Brain rhythms, in Encyclopedia of Neuroscience, M. D. Binder, N. Hirokawa, U. Windhorst, Eds. (Springer, 2009), pp. 482–483. [Google Scholar]
- 2.G. Buzsaki, X. J. Wang, Mechanisms of gamma oscillations. Annu. Rev. Neurosci. 35, 203–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. Buzsáki, A. Draguhn, Neuronal oscillations in cortical networks. Science 304, 1926–1929 (2004). [DOI] [PubMed] [Google Scholar]
- 4.C. Adaikkan, L.-H. Tsai, Gamma entrainment: Impact on neurocircuits, glia, and therapeutic opportunities. Trends Neurosci. 43, 24–41 (2020). [DOI] [PubMed] [Google Scholar]
- 5.P. Lakatos, G. Karmos, A. D. Mehta, I. Ulbert, C. E. Schroeder, Entrainment of neuronal oscillations as a mechanism of attentional selection. Science 320, 110–113 (2008). [DOI] [PubMed] [Google Scholar]
- 6.A. Ferro, Y. S. S. Auguste, L. Cheadle, Microglia, cytokines, and neural activity: Unexpected interactions in brain development and function. Front. Immunol. 12, 703527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. C. Singer, A. J. Martorell, J. M. Douglas, F. Abdurrob, M. K. Attokaren, J. Tipton, H. Mathys, C. Adaikkan, L. H. Tsai, Noninvasive 40-Hz light flicker to recruit microglia and reduce amyloid beta load. Nat. Protoc. 13, 1850–1868 (2018). [DOI] [PubMed] [Google Scholar]
- 8.H. F. F. Iaccarino, A. C. C. Singer, A. J. J. Martorell, A. Rudenko, F. Gao, T. Z. Z. Gillingham, H. Mathys, J. Seo, O. Kritskiy, F. Abdurrob, C. Adaikkan, R. G. G. Canter, R. Rueda, L.-H. H. Tsai, E. N. Brown, E. S. Boyden, L.-H. H. Tsai, Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. J. Martorell, A. L. Paulson, H.-J. Suk, F. Abdurrob, G. T. Drummond, W. Guan, J. Z. Young, D. N.-W. Kim, O. Kritskiy, S. J. Barker, V. Mangena, S. M. Prince, E. N. Brown, K. Chung, E. S. Boyden, A. C. Singer, L.-H. Tsai, Multi-sensory gamma stimulation ameliorates Alzheimer’s-associated pathology and improves cognition. Cell 177, 256–271.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. M. Garza, L. Zhang, B. Borron, L. B. Wood, A. C. Singer, Gamma visual stimulation induces a neuroimmune signaling profile distinct from acute neuroinflammation. J. Neurosci. 40, 1211–1225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Q. He, K. M. Colon-Motas, A. F. Pybus, L. Piendel, J. K. Seppa, M. L. Walker, C. M. Manzanares, D. Qiu, S. Miocinovic, L. B. Wood, A. I. Levey, J. J. Lah, A. C. Singer, A feasibility trial of gamma sensory flicker for patients with prodromal Alzheimer’s disease. Alzheimers Dement 7, e12178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. W. Salter, B. Stevens, Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 13.M. È. Tremblay, B. Stevens, A. Sierra, H. Wake, A. Bessis, A. Nimmerjahn, The role of microglia in the healthy brain. J. Neurosci. 31, 16064–16069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Ferro, Y. S. S. Auguste, L. Cheadle, Microglia, mytokines, and neural Activity: Unexpected interactions in brain development and function. Front. Immunol. 12, 703527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.W. S. Chung, C. A. Welsh, B. A. Barres, B. Stevens, Do glia drive synaptic and cognitive impairment in disease? Nat. Neurosci. 18, 1539–1545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y. Wu, L. Dissing-Olesen, B. A. MacVicar, B. Stevens, Microglia: Dynamic mediators of synapse development and plasticity. Trends Immunol. 36, 605–613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Hong, L. Dissing-Olesen, B. Stevens, New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 36, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Z. Szepesi, O. Manouchehrian, S. Bachiller, T. Deierborg, Bidirectional microglia–neuron communication in health and disease. Front. Cell. Neurosci. 12, 323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. du Yan, H. Zhu, J. Fu, S. F. Yan, A. Roher, W. W. Tourtellotte, T. Rajavashisth, X. Chen, G. C. Godman, D. Stern, A. M. Schmidt, Amyloid-beta peptide-receptor for advanced glycation endproduct interaction elicits neuronal expression of macrophage-colony stimulating factor: A proinflammatory pathway in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 94, 5296–5301 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. L. Tchelingerian, L. Vignais, C. Jacque, Tnf alpha gene expression is induced in neurones after a hippocampal lesion. Neuroreport 5, 585–588 (1994). [DOI] [PubMed] [Google Scholar]
- 21.S. Suzuki, K. Tanaka, E. Nagata, D. Ito, T. Dembo, Y. Fukuuchi, Cerebral neurons express interleukin-6 after transient forebrain ischemia in gerbils. Neurosci. Lett. 262, 117–120 (1999). [DOI] [PubMed] [Google Scholar]
- 22.G. Gunner, L. Cheadle, K. M. Johnson, P. Ayata, A. Badimon, E. Mondo, M. A. Nagy, L. Liu, S. M. Bemiller, K.-W. Kim, S. A. Lira, B. T. Lamb, A. R. Tapper, R. M. Ransohoff, M. E. Greenberg, A. Schaefer, D. P. Schafer, Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat. Neurosci. 22, 1075–1088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. Kaltschmidt, B. Kaltschmidt, H. Neumann, H. Wekerle, P. A. Baeuerle, Constitutive NF-kB activity in neurons. Mol. Cell. Biol. 14, 3981–3992 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. H. Shih, C. Y. Wang, C. M. Yang, NF-kappaB signaling pathways in neurological inflammation: A mini review. Front. Mol. Neurosci. 8, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Y. B. Lee, J. W. Schrader, S. U. Kim, p38 map kinase regulates TNF-α production in human astrocytes and microglia by multiple mechanisms. Cytokine 12, 874–880 (2000). [DOI] [PubMed] [Google Scholar]
- 26.A. D. Bachstetter, L. J. van Eldik, The p38 map kinase family as regulators of proinflammatory cytokine production in degenerative diseases of the CNS. Aging Dis. 1, 199–211 (2010). [PMC free article] [PubMed] [Google Scholar]
- 27.L. B. Rosen, D. D. Ginty, M. J. Weber, M. E. Greenberg, Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron 12, 1207–1221 (1994). [DOI] [PubMed] [Google Scholar]
- 28.R. S. Fiore, T. H. Murphy, J. S. Sanghera, S. L. Pelech, J. M. Baraban, Activation of p42 mitogen-activated protein kinase by glutamate receptor stimulation in rat primary cortical cultures. J. Neurochem. 61, 1626–1633 (1993). [DOI] [PubMed] [Google Scholar]
- 29.M. K. Meffert, J. M. Chang, B. J. Wiltgen, M. S. Fanselow, D. Baltimore, NF-κB functions in synaptic signaling and behavior. Nat. Neurosci. 6, 1072–1078 (2003). [DOI] [PubMed] [Google Scholar]
- 30.L. A. J. O’Neill, C. Kaltschmidt, NF-kB: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 20, 252–258 (1997). [DOI] [PubMed] [Google Scholar]
- 31.M. Jones, B. McDermott, B. L. Oliveira, A. O’Brien, D. Coogan, M. Lang, N. Moriarty, E. Dowd, L. Quinlan, B. McGinley, E. Dunne, D. Newell, E. Porter, M. A. Elahi, M. O’Halloran, A. Shahzad, Gamma band light stimulation in human case studies: Groundwork for potential Alzheimer’s disease treatment. J. Alzheimers Dis. 70, 171–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Adaikkan, S. J. Middleton, A. Marco, P. C. Pao, H. Mathys, D. N. Kim, F. Gao, J. Z. Young, H. J. Suk, E. S. Boyden, T. J. McHugh, L. H. Tsai, Gamma entrainment binds higher-order brain regions and offers neuroprotection. Neuron 102, 929–943.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Tian, X. Qin, Y. Wang, Y. Shi, X. Yang, 40 Hz light flicker promotes learning and memory via long term depression in wild-type mice. J. Alzheimers Dis. 84, 983–993 (2021). [DOI] [PubMed] [Google Scholar]
- 34.L. Zheng, M. Yu, R. Lin, Y. Wang, Z. Zhuo, N. Cheng, M. Wang, Y. Tang, L. Wang, S. T. Hou, Rhythmic light flicker rescues hippocampal low gamma and protects ischemic neurons by enhancing presynaptic plasticity. Nat. Commun. 11, 3012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.X. Lu, Y. Wang, Z. Liu, Y. Gou, D. Jaeger, F. St-Pierre, Detecting rapid pan-cortical voltage dynamics in vivo with a brighter and faster voltage indicator. bioRxiv 10.1101/2022.08.29.505018 [Preprint]. 31 August 2022. 10.1101/2022.08.29.505018. [DOI]
- 36.B. Spittau, Aging microglia—Phenotypes, functions and implications for age-related neurodegenerative diseases. Front. Aging Neurosci. 9, 194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. C. Savage, M. Carrier, M. È. Tremblay, Morphology of microglia across contexts of health and disease. Methods Mol. Biol. 2034, 13–26 (2019). [DOI] [PubMed] [Google Scholar]
- 38.S. R. Subramaniam, H. J. Federoff, Targeting microglial activation states as a therapeutic avenue in Parkinson’s disease. Front. Aging Neurosci. 9, 176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. Zhang, J. Zhang, Z. You, Switching of the microglial activation phenotype is a possible treatment for depression disorder. Front. Cell. Neurosci. 12, 306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.L. Du, Y. Zhang, Y. Chen, J. Zhu, Y. Yang, H. L. Zhang, Role of microglia in neurological disorders and their potentials as a therapeutic target. Mol. Neurobiol. 54, 7567–7584 (2017). [DOI] [PubMed] [Google Scholar]
- 41.M. E. Tremblay, Microglial functional alteration and increased diversity in the challenged brain: Insights into novel targets for intervention. Brain Behav. Immun. Health. 16, 100301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. Hänzelmann, R. Castelo, J. Guinney, GSVA: Gene set variation analysis for microarray and RNA-seq data. 14, 7 (2013). [DOI] [PMC free article] [PubMed]
- 43.G. Pattabiraman, M. Murphy, F. Agliano, K. Karlinsey, A. E. Medvedev, IRAK4 activity controls immune responses to intracellular bacteria Listeria monocytogenes and Mycobacterium smegmatis. J. Leukoc. Biol. 104, 811–820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L. Cushing, A. Winkler, S. A. Jelinsky, K. Lee, W. Korver, R. Hawtin, V. R. Rao, X. M. Fleming, L. L. Lin, IRAK4 kinase activity controls Toll-like receptor-induced inflammation through the transcription factor IRF5 in primary human monocytes. J. Biol. Chem. 292, 18689–18698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. S. Diamond, M. Farzan, The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. Seemann, F. Zohles, A. Lupp, Comprehensive comparison of three different animal models for systemic inflammation. J. Biomed. Sci. 24, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.U. K. Hanisch, Microglia as a source and target of cytokines. Glia 40, 140–155 (2002). [DOI] [PubMed] [Google Scholar]
- 48.M. S. Hayden, S. Ghosh, Regulation of NF-κB by TNF family cytokines. Semin. Immunol. 26, 253–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.P. Zhang, M. Martin, S. M. Michalek, J. Katz, Role of mitogen-activated protein kinases and NF-κB in the regulation of proinflammatory and anti-inflammatory cytokines by Porphyromonas gingivalis Hemagglutinin B. Infect. Immun. 73, 3990–3998 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.M. D. Turner, B. Nedjai, T. Hurst, D. J. Pennington, Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 1843, 2563–2582 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Q. Li, B. A. Barres, Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18, 225–242 (2018). [DOI] [PubMed] [Google Scholar]
- 52.A. Badimon, H. J. Strasburger, P. Ayata, X. Chen, A. Nair, A. Ikegami, P. Hwang, A. T. Chan, S. M. Graves, J. O. Uweru, C. Ledderose, M. G. Kutlu, M. A. Wheeler, A. Kahan, M. Ishikawa, Y. C. Wang, Y. H. E. Loh, J. X. Jiang, D. J. Surmeier, S. C. Robson, W. G. Junger, R. Sebra, E. S. Calipari, P. J. Kenny, U. B. Eyo, M. Colonna, F. J. Quintana, H. Wake, V. Gradinaru, A. Schaefer, Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. P. Schafer, E. K. Lehrman, A. G. Kautzman, R. Koyama, A. R. Mardinly, R. Yamasaki, R. M. Ransohoff, M. E. Greenberg, B. A. Barres, B. Stevens, Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.G. Buzsaki, Rhythms of the Brain (Oxford University Press, 2006). [Google Scholar]
- 55.R. C. Paolicelli, G. Bolasco, F. Pagani, L. Maggi, M. Scianni, P. Panzanelli, M. Giustetto, T. A. Ferreira, E. Guiducci, L. Dumas, D. Ragozzino, C. T. Gross, Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 56.O. S. Kim, E. J. Park, E.-H. Joe, I. Jou, JAK-STAT signaling mediates gangliosides-induced inflammatory responses in brain microglial cells. 277, 40594–40601 (2002). [DOI] [PubMed]
- 57.H. Y. Huang, H. F. Chang, M. J. Tsai, J. S. Chen, M. J. Wang, 6-Mercaptopurine attenuates tumor necrosis factor-α production in microglia through Nur77-mediated transrepression and PI3K/Akt/mTOR signaling-mediated translational regulation. J. Neuroinflammation 13, 78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Rotshenker, Microglia and macrophage activation and the regulation of complement-receptor-3 (CR3/MAC-1)-mediated myelin phagocytosis in injury and disease. J. Mol. Neurosci. 21, 65–72 (2003). [DOI] [PubMed] [Google Scholar]
- 59.I. Karunakaran, S. Alam, S. Jayagopi, S. J. Frohberger, J. N. Hansen, J. Kuehlwein, B. V. Hölbling, B. Schumak, M. P. Hübner, M. H. Gräler, A. Halle, G. van Echten-Deckert, Neural sphingosine 1-phosphate accumulation activates microglia and links impaired autophagy and inflammation. Glia 67, 1859–1872 (2019). [DOI] [PubMed] [Google Scholar]
- 60.J. Husemann, J. D. Loike, R. Anankov, M. Febbraio, S. C. Silverstein, Scavenger receptors in neurobiology and neuropathology: Their role on microglia and other cells of the nervous system. Glia 40, 195–205 (2002). [DOI] [PubMed] [Google Scholar]
- 61.S. E. Haynes, G. Hollopeter, G. Yang, D. Kurpius, M. E. Dailey, W. B. Gan, D. Julius, The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Y. Imai, S. Kohsaka, Intracellular signaling in M-CSF-induced microglia activation: Role of Iba1. Glia 40, 164–174 (2002). [DOI] [PubMed] [Google Scholar]
- 63.M. R. P. Elmore, A. R. Najafi, M. A. Koike, N. N. Dagher, E. E. Spangenberg, R. A. Rice, M. Kitazawa, B. Matusow, H. Nguyen, B. L. West, K. N. Green, Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.V. Chitu, E. R. Stanley, Colony-stimulating factor-1 in immunity and inflammation. Curr. Opin. Immunol. 18, 39–48 (2006). [DOI] [PubMed] [Google Scholar]
- 65.A. Shemer, I. Scheyltjens, G. R. Frumer, J. S. Kim, J. Grozovski, S. Ayanaw, B. Dassa, H. van Hove, L. Chappell-Maor, S. Boura-Halfon, D. Leshkowitz, W. Mueller, N. Maggio, K. Movahedi, S. Jung, Interleukin-10 prevents pathological microglia hyperactivation following peripheral endotoxin challenge. Immunity 53, 1033–1049.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 66.M. Saraiva, A. O’Garra, The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 (2010). [DOI] [PubMed] [Google Scholar]
- 67.D. F. Fiorentino, A. Zlotnik, T. R. Mosmann, M. Howard, A. O’Garra, IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 147, 3815–3822 (1991). [PubMed] [Google Scholar]
- 68.D. Tweedie, H. K. Karnati, R. Mullins, C. G. Pick, B. J. Hoffer, E. J. Goetzl, D. Kapogiannis, N. H. Greig, Time-dependent cytokine and chemokine changes in mouse cerebral cortex following a mild traumatic brain injury. eLife 9, e55827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. Behjati, M. H. Frank, The effects of tamoxifen on immunity. 16, 3076–3080 (2009). [DOI] [PMC free article] [PubMed]
- 70.J. Luo, F. Elwood, M. Britschgi, S. Villeda, H. Zhang, Z. Ding, L. Zhu, H. Alabsi, R. Getachew, R. Narasimhan, R. Wabl, N. Fainberg, M. L. James, G. Wong, J. Relton, S. S. Gambhir, J. W. Pollard, T. Wyss-Coray, Colony-stimulating factor 1 receptor (CSF1R) signaling in injured neurons facilitates protection and survival. J. Exp. Med. 210, 157–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. Pevzner, A. Izadi, D. J. Lee, K. Shahlaie, G. G. Gurkoff, Making waves in the brain: What are oscillations, and why modulating them makes sense for brain injury. Front. Syst. Neurosci. 10, 30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.H. Kaltiainen, L. Helle, M. Liljeström, H. Renvall, N. Forss, Theta-band oscillations as an indicator of mild traumatic brain injury. Brain Topogr. 31, 1037–1046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.M. X. Huang, C. W. Huang, D. L. Harrington, S. Nichols, A. Robb-Swan, A. Angeles-Quinto, L. Le, C. Rimmele, A. Drake, T. Song, J. W. Huang, R. Clifford, Z. Ji, C. K. Cheng, I. Lerman, K. A. Yurgil, R. R. Lee, D. G. Baker, Marked increases in resting-state MEG gamma-band activity in combat-related mild traumatic brain injury. Cereb. Cortex 30, 283–295 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Q. Wang, S. L. Ding, Y. Li, J. Royall, D. Feng, P. Lesnar, N. Graddis, M. Naeemi, B. Facer, A. Ho, T. Dolbeare, B. Blanchard, N. Dee, W. Wakeman, K. E. Hirokawa, A. Szafer, S. M. Sunkin, S. W. Oh, A. Bernard, J. W. Phillips, M. Hawrylycz, C. Koch, H. Zeng, J. A. Harris, L. Ng, The Allen mouse brain common coordinate framework: A 3D reference atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.J. A. Ramos-Vara, Technical aspects of immunohistochemistry. Vet. Pathol. 42, 405–426 (2005). [DOI] [PubMed] [Google Scholar]
- 76.G. X. Y. Zheng, J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, T. D. Wheeler, G. P. McDermott, J. Zhu, M. T. Gregory, J. Shuga, L. Montesclaros, J. G. Underwood, D. A. Masquelier, S. Y. Nishimura, M. Schnall-Levin, P. W. Wyatt, C. M. Hindson, R. Bharadwaj, A. Wong, K. D. Ness, L. W. Beppu, H. J. Deeg, C. McFarland, K. R. Loeb, W. J. Valente, N. G. Ericson, E. A. Stevens, J. P. Radich, T. S. Mikkelsen, B. J. Hindson, J. H. Bielas, Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck, Y. Hao, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. Butler, P. Hoffman, P. Smibert, E. Papalexi, R. Satija, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.R. Satija, J. A. Farrell, D. Gennert, A. F. Schier, A. Regev, Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Z. Yao, H. Liu, F. Xie, S. Fischer, R. S. Adkins, A. I. Aldridge, S. A. Ament, A. Bartlett, M. M. Behrens, K. Van den Berge, D. Bertagnolli, H. R. de Bézieux, T. Biancalani, A. S. Booeshaghi, H. C. Bravo, T. Casper, C. Colantuoni, J. Crabtree, H. Creasy, K. Crichton, M. Crow, N. Dee, E. L. Dougherty, W. I. Doyle, S. Dudoit, R. Fang, V. Felix, O. Fong, M. Giglio, J. Goldy, M. Hawrylycz, B. R. Herb, R. Hertzano, X. Hou, Q. Hu, J. Kancherla, M. Kroll, K. Lathia, Y. E. Li, J. D. Lucero, C. Luo, A. Mahurkar, D. McMillen, N. M. Nadaf, J. R. Nery, T. N. Nguyen, S. Y. Niu, V. Ntranos, J. Orvis, J. K. Osteen, T. Pham, A. Pinto-Duarte, O. Poirion, S. Preissl, E. Purdom, C. Rimorin, D. Risso, A. C. Rivkin, K. Smith, K. Street, J. Sulc, V. Svensson, M. Tieu, A. Torkelson, H. Tung, E. D. Vaishnav, C. R. Vanderburg, C. van Velthoven, X. Wang, O. R. White, Z. J. Huang, P. V. Kharchenko, L. Pachter, J. Ngai, A. Regev, B. Tasic, J. D. Welch, J. Gillis, E. Z. Macosko, B. Ren, J. R. Ecker, H. Zeng, E. A. Mukamel, A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 and S2
Legend for movie S1
Legends for data S1 to S3
Movie S1
Data S1 to S3