Abstract
Equipping multiple functionalities on adoptive effector cells is essential to overcome the complex immunological barriers in solid tumors for superior antitumor efficacy. However, current cell engineering technologies cannot endow these functionalities to cells within a single step because of the different spatial distributions of targets in one cell. Here, we present a core-shell anti-phagocytosis-blocking repolarization-resistant membrane-fusogenic liposome (ARMFUL) to achieve one-step multiplexing cell engineering for multifunctional cell construction. Through fusing with the M1 macrophage membrane, ARMFUL inserts an anti-CD47 (aCD47)–modified lipid shell onto the surface and simultaneously delivers colony-stimulating factor 1 receptor inhibitor BLZ945-loaded core into the cytoplasm. The surface-presenting aCD47 boosts macrophage's phagocytosis against the tumor by blocking CD47. The cytoplasm-located BLZ945 prompts its polarization resistance to M2 phenotype in the immunosuppressive microenvironment via inactivating the intracellular M2 polarization signaling pathway. This ARMFUL provides a versatile cell engineering platform to customize multimodal cellular functions for enhanced adoptive cell therapy.
A membrane-fusion-mediated cell-engineering technique is presented to construct multifunctional adoptive cells for solid tumor.
INTRODUCTION
Adoptive cell therapy (ACT) involves the ex vivo engineering and reinfusion of immune effector cells, such as T cells, macrophages, and natural killer cells, with the aim of enabling them to recognize and eliminate tumor cells for cancer treatment. The in vitro engineering of effector cells is a crucial step for ACT, as it regulates cellular functions and behaviors to overcome specific immunological barriers in vivo, ultimately leading to therapeutic efficacy (1, 2). Now, cell engineering can be achieved using a diverse toolbox of bioengineering methodologies to intervene intercellular or extracellular molecular targets for functionality endowment, such as gene editing (3, 4), metabolic engineering (5–8), drug/protein regulation (9), polymeric layer/patch coating (10, 11), chemical/enzymatic modification (12, 13), lipid/antibody/synthetic DNA anchoring (1–16), and nanoparticle hybridization (17–22). Despite the progress in customizing ACTs for certain subsets of hematologic malignancies [e.g., chimeric antigen receptor (CAR)–engineered T cells] (23), these technologies are far from satisfactory because of the poor therapeutic efficacy in solid tumors (24). This failure can be attributed to the resistance of multiple immunological barriers in solid tumors toward the back-fused effector cells (25–32). To address this, multiple functionalities should be equipped on effector cells by engineering two or more types of cellular targets (29–32). Unfortunately, because of the different spatial distributions of targets in the cell, the construction of multifunctional effector cells requires a tedious step-by-step engineering process, inevitably adding cost and complexity (33–35). Therefore, a refined cell engineering technology, which allows the synchronous engineering of multiple targets at different subcellular levels within a single process, is urgently needed for enhanced ACT against tumors.
Membrane fusion is a fundamental biological process in natural organisms that enables the blending of membrane compositions and the mixing of inner cytosols between cells (36–39). Inspired by this natural process, membrane-fusogenic liposomes, which contain a membrane-fusogenic lipid layer and a cargo-loaded inner core, are developed to mimic this fusion interaction with cell membranes (39, 40). As a result, their lipid shells are inserted into cell membranes while the interior payloads are released into the cell cytosol at the same time, enabling them as a promising system for membrane engineering and cytoplastic delivery (40–44). Therefore, we hypothesize that membrane-fusogenic liposome is an optimal tool for cell engineering to simultaneously remodel multiple targets on/in cell membranes and cytosols, resulting in the efficient construction of multifunctional effector cells for ACT.
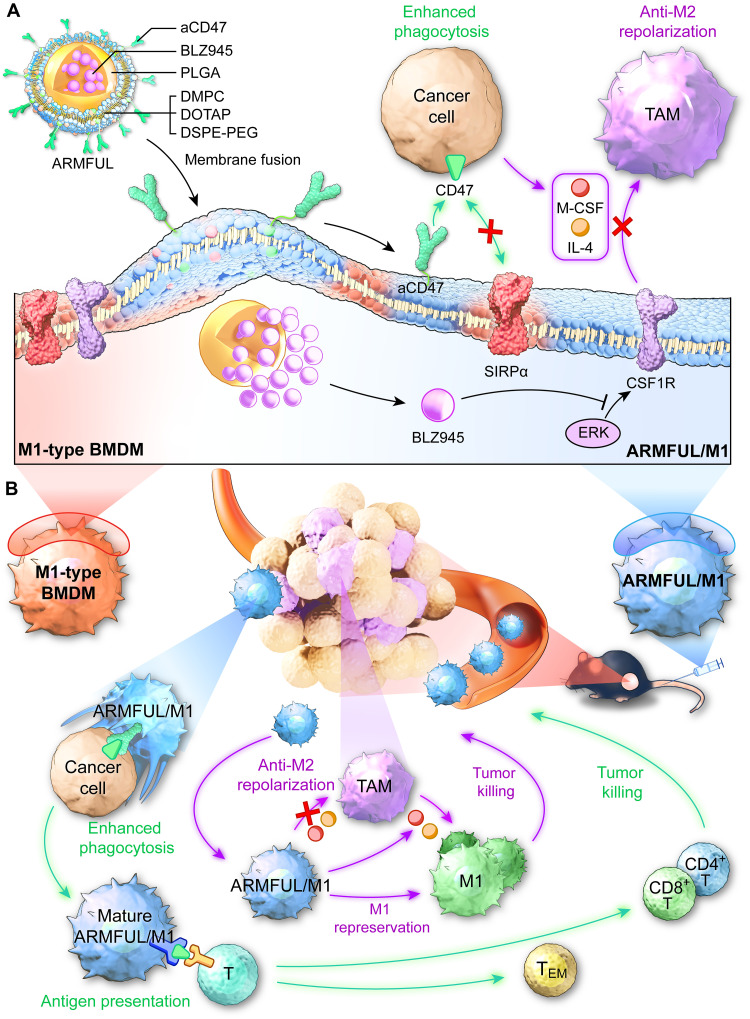
Here, we used membrane-fusogenic liposomes to engineer M1 phenotype macrophages with multiple functionalities for effective ACT against solid tumors in one step. Current clinical trials have shown that adoptive transfer of nonengineered M1 macrophages for tumor treatment is hindered by two key factors: (i) antiphagocytic molecules (e.g., CD47) overexpressed on tumor cell surfaces, which resist phagocytosis, and (ii) cytokines [e.g., macrophage colony-stimulating factor (M-CSF), interleukin-4 (IL-4), and IL-13] in the tumor environment, which could easily repolarize M1 macrophages to tumor-promoting M2 phenotypes (11, 17, 28, 45–47). To overcome these barriers, M1 macrophages should be designed with dual capacities of both efficiently phagocytosing tumor cells and robustly maintaining antitumor M1 phenotype in the tumor environment. To this end, we first synthesized an anti-phagocytosis-blocking repolarization-resistant membrane-fusogenic liposome (ARMFUL) with a core-shell structure, in which the CSF1 receptor (CSF1R) inhibitor BLZ945 was loaded into the poly(lactic-co-glycolic acid) (PLGA)–based polymeric core, and anti-CD47 (aCD47) was conjugated onto a fusogenic lipid shell surface (Fig. 1A). This ARMFUL was then fused with an M1 macrophage cell membrane to not only insert aCD47-modified lipid shells on the surfaces directly but also release the BLZ945-loaded core into the cytoplasm, forming ARMFUL-engineered M1 macrophages (ARMFUL/M1) for back-transfer (Fig. 1A). The release of BLZ945 in the cytoplasm effectively inhibited the intracellular tyrosine kinase of CSF1R to inactivate subsequent M2 polarization–related signaling pathway, maintaining the antitumor M1 phenotype of ARMFUL/M1 in the tumor environment after adoptive transfer (48–50). Meanwhile, the presentation of aCD47 on the ARMFUL/M1 surface ensured the effective phagocytosis of tumor cells under the blockade of antiphagocytotic CD47 (48, 51). Eventually, this multifunctional ARMFUL/M1 can effectively inhibit B16F10 tumor growth in mice, activate T cell–mediated immunity to suppress distant tumors, and impede tumor metastasis (Fig. 1B). Therefore, this ARMFUL presents a versatile tool for the synchronous engineering of adoptive cells through both intracellular and extracellular pathways, providing a platform to achieve the multimodal customization of cellular functions and behaviors for enhanced ACT against tumors.
Fig. 1. Schematic illustration of in vitro engineering of M1 macrophages using membrane-fusogenic liposomes in a membrane fusion manner for enhanced ACT against tumors.
(A) M1 macrophages were engineered with ARMFUL in a membrane fusion manner. ARMFUL has a core-shell structure, in which the CSF1R inhibitor BLZ945 was loaded in the PLGA-based polymeric core, and aCD47 was conjugated on the fusogenic lipid shell surface. This ARMFUL can fuse with M1 macrophage cell membrane to simultaneously insert aCD47-modified lipid shells on the surfaces directly and release the BLZ945-loaded core into the cytoplasm, formulating ARMFUL/M1 for back-transfer. The surface-presenting aCD47 endows macrophages with enhanced phagocytic ability toward tumors through blocking antiphagocytosis CD47 molecules on tumors. Meanwhile, the cytoplasm-located BLZ945 effectively inactivates intracellular tyrosine kinase of CSF1R and subsequent M2 polarization signaling pathway to allow M1 macrophages to resist the polarization to tumor-promoting M2 phenotype in immunosuppressing microenvironment for durable therapeutic effect. (B) ARMFUL/M1 macrophages could remodel the tumor microenvironment, activate T cell cytotoxicity, and induce systemic immunological memory to synergistically inhibit tumor growth after adoptive transfer. DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; DSPE-PEG-Mal, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000]; TAM, tumor-associated macrophage; BMDM, bone marrow–derived macrophage; ERK, extracellular signal–regulated kinase; TEM, effector memory T cell; SIRPα, signal regulatory protein α.
RESULTS
Construction and characterization of ARMFUL/M1
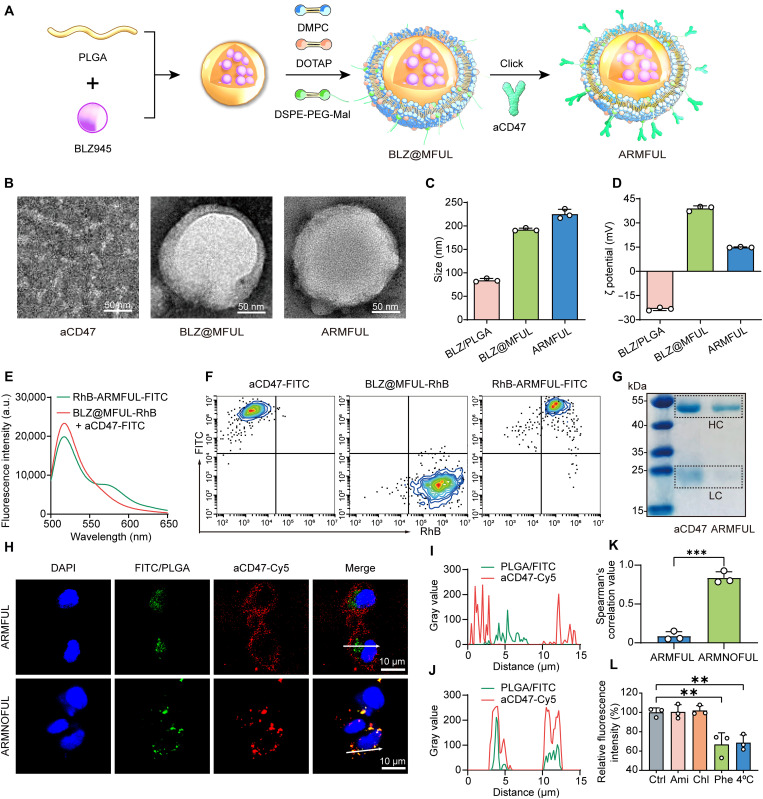
Before the construction of ARMFUL/M1, ARMFUL was first prepared through a three-step process (Fig. 2A). First, the PLGA polymeric core loaded with BLZ945 (BLZ/PLGA) was synthesized via a nanoprecipitation method. Then, the BLZ/PLGA was coated with a fusogenic lipid shell containing 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG-Mal) at a molar ratio of 76.2/20/3.8 using a film hydration method to form BLZ@MFUL (40–44). The transmission electron microscope (TEM) image of BLZ@MFUL showed an obvious lipid layer (Fig. 2B and fig. S1A). Meanwhile, the average hydrodynamic diameter increased from 84.3 nm of BLZ/PLGA to 192.2 nm of BLZ@MFUL (Fig. 2C), and the zeta potential changed from −23.3 mV of BLZ/PLGA to 39 mV of BLZ@MFUL (Fig. 2D). These results collectively demonstrated the successful coating of lipid shell to form BLZ@MFUL with a core-shell structure. In addition, the encapsulated BLZ945 can be completely released from PLGA cores in the acidic environment (fig. S3A).
Fig. 2. Construction of ARMFUL/M1.
(A) Schematic illustration of the preparation of ARMFUL. (B) TEM images of aCD47, BLZ@MFUL, and ARMFUL. (C and D) Size distribution (C) and zeta potential (D) of BLZ/PLGA, BLZ@MFUL, and ARMFUL. (E) Fluorescence intensity curves of RhB-ARMFUL-FITC and the mixture of BLZ@MFUL-RhB and aCD47-FITC excited at the wavelength of 488 nm. (F) Fluorescence-activated cell sorting (FACS) profiles showed the conjugation of aCD47 to BLZ@MFUL. (G) Gel electrophoresis showing the heavy chain (HC) and light chain (LC) released from free aCD47 and ARMFUL. The gel was stained with Coomassie Brilliant Blue. (H) Confocal laser scanning microscopy (CLSM) images of M1-type BMDMs after incubation with FITC@ARMFUL-Cy5 and FITC@ARMNOFUL-Cy5. Blue fluorescence, nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI); green fluorescence, fluorescein isothiocyanate (FITC)/PLGA core; red fluorescence, Cy5-labeled aCD47. (I and J) Gray values of both green (FITC/PLGA) and red (aCD47-Cy5) channels along with the corresponding white arrows in the “Merge” CLSM images in (H) analyzed using ImageJ software. (K) Spearman’s correlation value of green and red fluorescence in the CLSM images of FITC@ARMFUL-Cy5–treated and FITC@ARMNOFUL-Cy5–treated groups. (L) Effects of various inhibitors on the membrane fusion of FITC@ARMFUL-Cy5 in BMDMs. Data represent means ± SD; n = 3 biologically independent samples. Statistical significance in (K) and (L) was calculated by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison tests: **P < 0.01, and ***P < 0.001. a.u., arbitrary units.
Last, aCD47 was conjugated onto the surface of the BLZ@MFUL via a facile maleimide-thiol Michael addition reaction to obtain ARMFUL nanoparticles (52). The conjugation of aCD47 was confirmed by fluorescence resonance energy transfer (FRET)–based and fluorescence-activated cell sorting (FACS)–based assays. For these experiments, the DSPE-PEG-rhodamine B (RhB)–labeled BLZ@MFUL (BLZ@MFUL-RhB) was coupled with fluorescein isothiocyanate (FITC)–labeled aCD47 (aCD47-FITC) to form RhB-ARMFUL-FITC using the same synthetic procedure of ARMFULs. Figure 2E shows that the solution of RhB-ARMFUL-FITC emitted fluorescence at a peak of 563 nm (RhB emission) when excited at 488 nm (FITC excitation), whereas no similar phenomenon was observed in the mixture of an equal amount of aCD47-FITC and BLZ@MFUL-RhB. This FRET occurrence suggested the close distance (<10 nm) between the donor (FITC) and the acceptor (RhB) (53–55), substantiating the successful conjugation of aCD47 to the lipid layer. Moreover, the FACS profile of RhB-ARMFUL-FITC exhibited the location of its population in the RhB+FITC+ region while that of BLZ@MFUL-RhB and aCD47-FITC were in the RhB+FITC− and RhB−FITC+ regions, respectively (Fig. 2F). This result forcefully suggested the colocalization of these two labeling dyes in one particle, indicating the successful conjugation of aCD47 to BLZ@MFUL. Consequently, the hydrodynamic diameter of ARMFUL (225.3 nm) increased slightly compared with BLZ@MFUL (192.2 nm), and a great change in the zeta potential of ARMFUL was observed mainly because of the charge neutralization of aCD47 (Fig. 2, C and D). TEM images exhibited a rough surface on ARMFUL compared with BLZ@MFUL because of the conjugation of aCD47 on the surface (Fig. 2B). Gel electrophoresis displayed similar bands of heavy chains and light chains of aCD47 cleaved from ARMFULs with those from free aCD47, suggesting its structural integrity after conjugation (Fig. 2G and fig. S3B).
Construction of ARMFUL/M1 via membrane fusion interaction
Such ARMFULs could fuse with the macrophage cell membrane to engineer both membranous and cytoplasmic targets to construct bifunctional ARMFUL/M1. To evaluate this membrane fusion–mediated engineering of M1 macrophages, the core of ARMFULs and aCD47 were labeled with FITC and cyanine5 (Cy5), respectively, followed by formulating the dual-labeled ARMFULs (FITC@ARMFUL-Cy5) as the same preparation of ARMFULs. Similarly, contrastive dual-labeled aCD47-decorated repolarization-resistant membrane-nonfusogenic liposomes (FITC@ARMNOFUL-Cy5) were also prepared through the same protocol using DMPC and DSPE-PEG-Mal at the molar ratio of 96.2:3.8. These two types of liposomes were both incubated with M1 phenotype bone marrow–derived macrophages (BMDMs) isolated from mice, followed by confocal laser scanning microscopy (CLSM) observation. As shown in Fig. 2H, the fluorescence signals of aCD47-Cy5 (red) were mostly presented on the cell membrane, while those of FITC/PLGA (green) were dispersed in the cytoplasm of FITC@ARMFUL-Cy5–treated BMDMs. Comparatively, the fluorescence signals of aCD47-Cy5 and FITC/PLGA were almost completely colocalized within BMDMs after incubation with FITC@ARMNOFUL-Cy5 (Fig. 2H). Fluorescence intensity profiles and Spearman’s correlation values of green (FITC/PLGA) and red (aCD47-Cy5) channels were also in accordance with these observations (Fig. 2, I to K). The Western blot analysis of aCD47 in macrophage membrane proteins revealed a distinct band solely presented in the ARMFUL-engineered group, indicating successful anchoring of aCD47 onto the cell membrane post-ARMFUL engineering (fig. S3E). Collectively, these phenomena revealed that the ARMFULs could interact with BMDMs via a membrane fusion manner to insert the aCD47-modified lipid shell into the cell membrane and deliver the FITC/PLGA core into the cytoplasm concurrently. To further validate this membrane fusion interaction with M1 macrophages, we incubated the FITC@ARMFUL-Cy5 with BMDMs in the presence of several well-known cell uptake inhibitors, including amiloride (Ami) for inhibiting micropinocytosis, chlorpromazine (Chl) for clathrin-dependent endocytosis, and a peptide analog, Z-Phe-Phe-Phe-OH (Phe), as fusion inhibitor (56, 57). Only Phe and low-temperature (4°C) pretreatments caused a notable decrease of ARMFUL uptake by BMDMs, indicating an energy-dependent membrane fusion–mediated cell internalization manner (Fig. 2L), which was further confirmed by CLSM images (fig. S2A). Moreover, this intracellular delivery mediated by membrane fusion can circumvent endocytosis and lysosomal degradation, as evidenced by the absence of colocalization between FITC/PLGA cores of ARMFUL and lysosomes in CLSM images (fig. S2B).
On the basis of this membrane fusion effect, we constructed ARMFUL/M1 by coculturing ARMFULs with M1-type BMDMs with an optimized incubation time (4 hours) and concentration (100 μg/ml) without noticeable toxicity (figs. S3C and S7A). The ARMFUL can effectively penetrate macrophages, while the BLZ945 remains stable within the cells for at least 72 hours (figs. S1, D to F, and S3D). Contrastive M1 macrophages engineered with other interventions, including MIX (PLGA@MFUL + BLZ + aCD47), PLGA@MFUL-aCD47, BLZ@MFUL, and ARMNOFUL, were also prepared using the same method for further experiments (fig. S1). These engineering modifications did not affect the activation of pattern recognition receptors or the metabolic profile in macrophages (fig. S7, B to E). This ARMFUL is completely competent for the synchronous engineering of adoptive cells through both intracellular and extracellular pathways. As a comparison, the traditional liposomes with endocytosis-dependent cell internalization (ARMNOFUL) can only transport the whole nanoparticles including the cargo-loaded core and shell into the intercellular environment.
In vitro evaluation of the functionalities of ARMFUL/M1
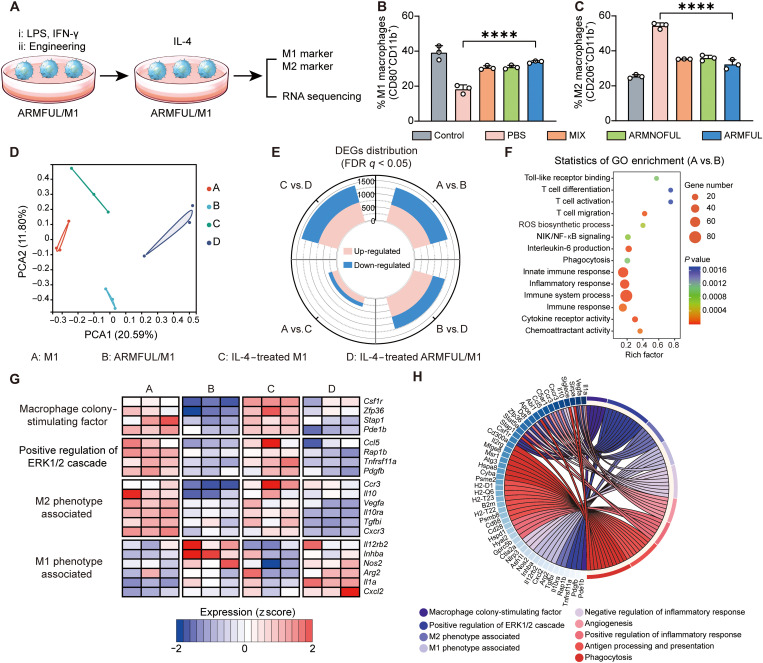
We first tested the anti-M2 polarization capacity of ARMFUL/M1 macrophages by detecting the surface levels of M1/M2-related biomarkers after treatment with the medium containing M2 polarizing cytokine, IL-4 (Fig. 3A). As shown in Fig. 3B and fig. S4A, IL-4 treatment caused a remarkable decrease of the percentage of M1 biomarker, CD80, on the surface of nonengineered M1 macrophages to 18.2% compared with that without IL-4 treatment (39.1%), but only led to a slight down-regulation of that on ARMFUL/M1 to 33.9%. Meanwhile, IL-4 treatment also caused distinct up-regulation of the surface level of M2 biomarker, CD206, on M1 macrophages engineered with phosphate-buffered saline (PBS), but less elevation of that on ARMFUL/M1 (32.25%) compared with control (25.34%) (Fig. 3C and fig. S4B). In addition, no obvious difference in M1 or M2 phenotype between M1 macrophages engineered with these nanoparticle groups before treatment with IL-4 was observed (fig. S4, C to F). Treatment of engineered macrophages with tumor cell–conditioned medium showed a similar result (fig. S4, G to J). These results illustrated that the nonengineered M1 macrophages are easily switched to M2 phenotype under the tumor microenvironment, but ARMFUL/M1 could resist this M2 polarization, indicating its satisfactory anti-M2 polarization capacity after ARMFUL engineering. To comprehensively investigate the M1 phenotype–maintaining effect of ARMFULs, we performed RNA sequencing analysis of native M1 macrophages and ARMFUL/M1 with/without IL-4 treatment. Both the principal components analysis (PCA) and Venn diagram indicated the substantial discrepancy of the transcriptome landscapes among the four groups (Fig. 3D and fig. S5A). The distribution of differentially expressed genes (DEGs) between different groups was visualized as a stacked rose chart (Fig. 3E). To further uncover the differences between native and ARMFUL/M1, DEGs in these two groups were submitted to gene ontology (GO) enrichment analysis, indicating ARMFUL engineering attached M1 macrophages with the enhanced immunological functions (Fig. 3F). Furthermore, two heatmaps and a Circos plot were presented to depict the detailed expression patterns of 53 selected DEGs among different groups (Fig. 3, G and H, and fig. S5, B to N). Consistent with the literature, BLZ945 in ARMFULs could down-regulate the expression of M-CSF and extracellular signal–regulated kinase 1/2 cascade genes to inhibit M2 polarization, consequently maintaining antitumorigenic M1 phenotype in IL-4–containing environment (48, 49). Moreover, ARMFUL engineering also slightly improved other antitumor biological functions of macrophages (e.g., M1 phenotype, positive regulation of inflammatory response, antigen processing and presentation, and phagocytosis) and dropped protumor biological functions (e.g., M2 phenotype, negative regulation of inflammatory response, and angiogenesis) substantially (Fig. 3G and fig. S5B). All the aforementioned bioinformatic results suggested that ARMFUL/M1 could resist M2 polarization by inhibiting CSF1R and subsequent M2 polarization signaling pathway to promote antitumor immunity.
Fig. 3. In vitro evaluation of anti-M2 polarization capacity of ARMFUL/M1.
(A) Schematic representation of the antipolarization assay. BMDMs are first polarized by lipopolysaccharide (LPS) and interferon-γ (IFN-γ), subsequently engineered with ARMFULs, and finally treated with IL-4. The cells are collected for the detection of M1/M2 markers and RNA sequencing. (B and C) The percent of M1 (CD80+CD11b+) (B) and M2 (CD206+CD11b+) (C) populations in engineered macrophages after treatment with IL-4, respectively. (D) PCA score plot of the expressed genes in M1 macrophages and ARMFUL/M1 with/without IL-4 treatment (n = 3 per group). (E) Landscape of DEG distribution between different groups. (F) GO enrichment analysis of DEGs between M1 and ARMFUL/M1 macrophages. (G) Expression of selected genes related to M-CSF, M2 phenotype, and M1 phenotype in M1 macrophages and ARMFUL/M1 with/without IL-4 treatment (n = 3 per group). (H) Circos plot depiction of the subordination between the selected genes and their enriched pathways. Data represent means ± SD; n = 3 biologically independent samples. Statistical significance in (B) and (C) was calculated by one-way ANOVA with Dunnett’s multiple comparison tests: ****P < 0.0005. FDR, false discovery rate.
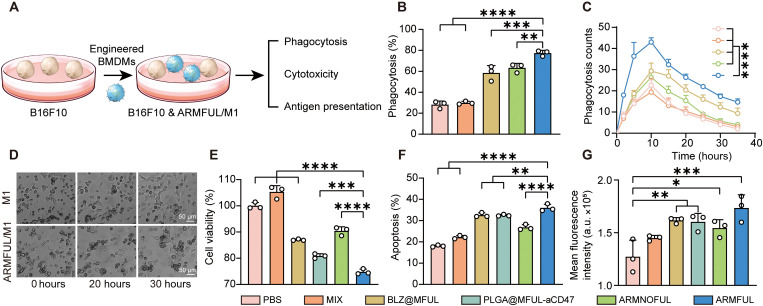
Next, to access the enhanced phagocytosis ability, engineered M1 macrophages and B16F10 cells were labeled with DSPE-PEG-Cy5 and carboxyfluorescein diacetate succinimidyl ester (CFSE), respectively, followed by coculturing and detecting Cy5+CFSE+ cell populations using FACS (Fig. 4A). Figure 4B and fig. S6A showed that ARMFUL/M1 exhibited 2.7-fold of B16F10 cell phagocytosis to nonengineered macrophages. The phagocytosis process was also recorded by a live-cell dynamic imaging and analysis system. As shown in movies S1 and S2, in the coculture, engineered M1 macrophages (cells with a round shape) were recognizing, chasing, and nibbling sole cancer cells (cells with fusiform). Quantification of phagocytosis cell clusters, a single cancer cell attacked by macrophages, showed a peak at 10 hours (Fig. 4, C and D, and fig. S6B). During this process, the phagocytosis cell clusters in the cocultures of ARMFUL/M1 and cancer cells maintained a higher level than that in the other groups. Collectively, the enhanced tumor phagocytosis ability of ARMFUL/M1 was confirmed, which should be attributed to the blockage of CD47 by aCD47 presented on the cell membrane.
Fig. 4. In vitro evaluation of antitumor efficacy of ARMFUL/M1.
(A) Schematic representation of the phagocytosis, cytotoxicity, and antigen presentation assay. Engineered M1 phenotype BMDMs were cocultured with B16F10/B16F10-OVA cells to evaluate the ability of phagocytosis, cytotoxicity, and antigen presentation. (B) Percentage of macrophages phagocyting B16F10 cancer cells after coculturing. (C) Phagocytosis cell clusters quantified in the images from live-cell dynamic imaging and analysis system. (D) Images of macrophages phagocytosing cancer cells at time points of 0, 20, and 30 hours. (E and F) The antitumor ability of macrophages evaluated by measuring cell viability (E) and apoptosis (F) of B16F10 cancer cells. (G) Antigen presentation profiles of macrophages exposed to B16F10-OVA cancer cells. Data represent means ± SD; n = 3 biologically independent samples. Statistical significance in (B), (E), (F), and (G) was calculated by one-way ANOVA, while (C) was calculated via two-way ANOVA, both with Dunnett’s multiple comparison tests: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0005.
This enhanced phagocytosis by engineered macrophages will cause the promotion of cancer cell death, as demonstrated by the detection of cell viability and apoptosis of cancer cells after coincubation. Figure 4E revealed that ARMFUL/M1 treatment resulted in remarkably lower cell viability than other contrastive nanoparticle-engineered macrophages. Meanwhile, the percent of apoptotic cancer cells cocultured with ARMFUL/M1 (36.0%) was relatively higher than macrophages engineered with PBS (18.1%), MIX (22.2%), BLZ@MFUL (32.6%), PLGA@MFUL-aCD47 (32.5%), and ARMNOFUL (26.8%) (Fig. 4F and fig. S6C). These results collectively confirmed the remarkably enhanced antitumor capacity of macrophages after ARMFUL engineering, which was superior to the endocytosis-based ARMNOFUL-engineering approach. Because the enhanced tumor phagocytosis can lead to the presentation of tumor antigens on the macrophage surface for downstream adaptive immune activation, we tested their antigen-specific major histocompatibility complex II (MHCII) protein expressions after coculturing with B16F10 tumor cells expressing ovalbumin (B16F10-OVA). Compared with natural M1 macrophages and those engineered with other nanoparticles, ARMFUL/M1 presented significantly higher OVA fragments onto the MHCII (Fig. 4G). The results substantiated the enhanced antigen-presenting ability of ARMFUL/M1, which is essential to activate the subsequent T cell–based adaptive antitumor immunity for a synergetic antitumor response. Overall, these experiments confirmed that ARMFUL engineering brought both excellent anti-M2 polarization and enhanced tumor phagocytosis ability to natural M1 macrophages owing to its membrane fusion effect, leading to a reinforced antitumor ability and even antigen-presenting capacity to potentially activate the adaptive antitumor immunity.
In vivo anti-M2 polarization capacity of ARMFUL/M1
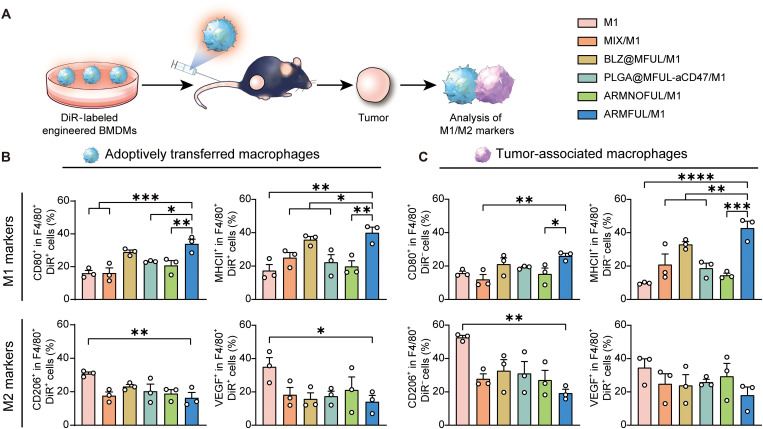
Whereafter, we sought to test the anti-M2 polarization capacity of ARMFUL/M1 in a tumor immunosuppressive environment on a B16F10 melanoma-bearing mouse model. To distinguish the injected engineering macrophages from native tumor-associated macrophages (TAMs), engineered M1 macrophages were stained with 1,1-dioctadecyl-3,3,3,3-tetramethylindotricarbocyaine iodide (DiR), followed by intravenous administration into mouse model twice at day 0 and day 1. On day 2, mice were euthanized to collect tumors for examining the infiltration of the injected M1 macrophages in the tumor and their M1/M2 phenotype (Fig. 5A). The CLSM images of tumor tissue slices showed that DiR-labeled macrophages could effectively infiltrate into tumors (fig. S8). As expected, the back-fused ARMFUL/M1 (F4/80+DiR+ populations) exhibited a sustained up-regulation of CD80 and MHCII on their surfaces within the tumor microenvironment compared to unmodified or MIX-, BLZ@MFUL-, PLGA@MFUL-aCD47–, and ARMNOFUL-engineered macrophages (Fig. 5B and fig. S9). Meanwhile, the M2 biomarkers CD206 and vascular endothelial growth factor (VEGF) in adoptively transferred ARMFUL/M1 cells maintained a relatively lower level than macrophages without engineering or engineered with MIX, BLZ@MFUL, PLGA@MFUL-aCD47, and ARMNOFUL. These results were consistent with that of the evaluation of the anti-M2 polarization capacity of ARMFUL/M1 in vitro (Fig. 5, C and D), demonstrating that ARMFUL engineering could maintain the M1 phenotype of macrophages in the immunosuppressive microenvironment of solid tumors. As the M1-type macrophages can secrete proinflammatory cytokines to modulate the immunosuppressive tumor microenvironment, we also evaluated the phenotype of TAMs (F4/80+DiR− population) in tumors after the adoptive transfer of these engineered macrophages. Figure 5C and fig. S10 showed that the TAMs of mice treated with ARMFUL/M1 macrophages exhibited a trend of repolarization toward M1 phenotypes, as evidenced by notably higher expressions of CD80 and MHCII. Correspondingly, the relative levels of CD206 and VEGF on TAMs in tumors treated with ARMFUL/M1 were significantly lower than those with all the other groups. In any case, these results indicated that ARMFUL/M1 could not only retain their own antitumorigenic M1 phenotype in solid tumors but also repolarize aboriginal protumorigenic TAMs toward the M1 phenotype to effectively remodel the tumor microenvironment. Collectively, the adoptive transfer of ARMFUL/M1 could be a potential therapy for effective cancer immunotherapy.
Fig. 5. In vivo evaluation of anti-M2 polarization ability of ARMFUL/M1.
(A) Schematic representation of the anti-M2 polarization experiment in vivo. BMDMs were engineered with liposomes and labeled by DiR, subsequently administrated into the mice burdened with B16F10 tumors. Then, the tumors are collected to analyze M1/M2 markers of adoptively transferred macrophages and autologous TAMs. (B and C) Expression of M1 (CD80 and MHCII) and M2 (CD206 and VEGF) markers on exogenous adoptively transferred macrophages (F4/80+DiR+) (B) and endogenous TAMs (F4/80+DiR−) (C) in tumor tissues after injection. Data are presented as means ± SD; n = 3 biologically independent mice. Statistical significance in (B) and (C) were calculated by one-way ANOVA with Dunnett’s multiple comparison tests: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0005.
Antitumor efficacy of ARMFUL/M1 in B16F10 tumor-bearing mice
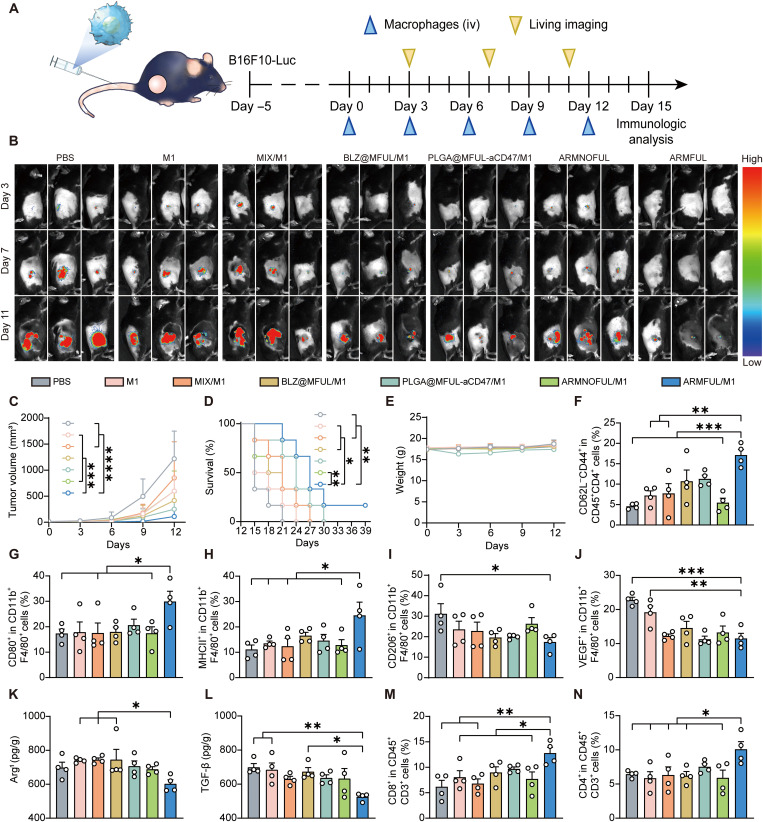
Inspired by this excellent anti-M2 polarization capacity in vivo, we next evaluated the therapeutic efficacy of ARMFUL/M1 on C57BL/6 mice burdened with B16F10-luciferase (B16F10-Luc) tumors (Fig. 6A). As shown in Fig. 6C and fig. S11, ARMFUL/M1 treatment led to a 90.9% suppression of tumor growth compared with natural M1 macrophages (50.9%) and those engineered with MIX (30.2%), BLZ@MFUL (65.8%), PLGA@MFUL-aCD47 (79.5%), and ARMNOFUL (50.9%) at day 12. Noninvasive Fluc bioluminescence imaging monitoring validated this result (Fig. 6B). Further immunofluorescence staining observation also showed the largest necrotic areas, the lowest expression of Ki67 and CD31, as well as the highest expression of terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) in tumor tissue sections of mice treated with ARMFUL/M1 (fig. S12). These analyses indicated that ARMFUL/M1 therapy could effectively impede tumor proliferation and cause remarkable cell apoptosis, which was coincident with the tumor progression observation. As a result, the mice that received ARMFUL/M1 macrophage administration showed an improved survival rate than the other groups, and notably, one ARMFUL/M1-treated mouse had a complete elimination of tumor (Fig. 6D). Furthermore, these engineered macrophages resulted in no obvious changes in body weight and serum biochemical index, as well as unnoticeable pathological damage in hematoxylin and eosin (H&E) staining of major organs, eliminating safety concerns (Fig. 6E and figs. S13 and 14).
Fig. 6. Antitumor efficacy of ARMFUL/M1 in vivo.
(A) Schematic representation of the antitumor effect in B16F10-Luc tumor-bearing mice. C57BL/6 mice were inoculated with B16F10-Luc cells, received intravenous (iv) injections of engineered BMDMs, and were imaged at the schedule after oncogenesis. (B) In vivo bioluminescence imaging of tumor growth of mice burdened with B16F10-Luc cells (n = 3 per group). (C) Tumor growth curves of mice burdened with B16F10-Luc cells after intravenous injection of PBS, M1 macrophages, and engineered M1 macrophages (n = 6 per group). (D and E) Survival plot (D) and weight change (E) of mice burdened with B16F10-Luc cells after treatment (n = 6 per group). (F) The percentage of TEM cells (CD62−CD44+) in the spleen. (G to J) The percentage of M1-type macrophages [(G) CD80+; (H) MHCII+] and M2 macrophages [(I) CD206+; (J) VEGF+] in isolated tumor tissues after treatment (n = 4 per group). (K and L) Cytokine levels in isolated tumor tissues after treatment (n = 4 per group). (M and N) The percentage of CD8+ T cells and CD4+ T cells in isolated tumor tissues after treatment (n = 4 per group). Data represent means ± SD. Statistical significance in (C) and (E) was calculated by two-way ANOVA with Turkey’s multiple comparison test, while (F) to (N) was calculated via one-way ANOVA with Dunnett’s multiple comparison tests, and log-rank (Mantel-Cox) test for (D): *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0005. TGF-β, transforming growth factor–β.
To investigate the therapeutic mechanism of engineered M1 macrophages, we analyzed the infiltrated immune cells and cytokines in the tumor microenvironment. As shown in Fig. 6 (G and H) and fig. S16, the percentage of M1 phenotype macrophages (CD80+ and MHCII+ populations) in tumors from ARMFUL/M1 macrophage-treated mice (30.0 and 24.6%) was higher than that treated with M1 macrophages without engineering (17.9 and 13.5%) or engineered with MIX (17.5 and 12.4%), BLZ@MFUL (18.0 and 16.5%), PLGA@MFUL-aCD47 (20.6 and 14.6%), and ARMNOFUL (17.4 and 12.9%). Oppositely, the percentage of TAMs (CD206+ or VEGF+ populations) showed a decreased level compared with the other treatment groups (Fig. 6, I and J, and fig. S16). This revealed that ARMFUL/M1 macrophages could reverse the intratumoral immune homeostasis from the M2 macrophage-enriched protumorigenic microenvironment to an M1 macrophage-dependent antitumorigenic type. With this remodeling of macrophage phenotype in the tumor microenvironment, the immunosuppressive cytokines, arginase-1 (Arg1) and transforming growth factor–β (TGF-β), showed a reduced level in tumors from ARMFUL/M1-treated mice (Fig. 6, K and L). As M1 phenotype macrophages play a crucial role in antigen presentation to T cells and subsequent adaptive immune activation, we next analyzed the activation of tumor-infiltrating T cells and splenic effector memory T (TEM) cells. The results displayed a notably higher proportion of CD8+ and CD4+ T cells in tumors from mice injected with ARMFUL/M1 macrophages compared to M1 macrophages without engineering or engineered with MIX, BLZ@MFUL, PLGA@MFUL-aCD47, and ARMNOFUL (Fig. 6, M and N, and fig. S17). This confirms the efficacy of this therapy in activating T lymphocyte cytotoxicity to synergistically inhibit tumor growth. Meanwhile, the percentage of TEM cells (CD62L−CD44+ populations) in the spleen from mice with ARMFUL/M1 macrophages treatment was higher than that with the other groups, suggesting ARMFUL/M1 therapy–induced immune memory in vivo (Fig. 6F and fig. S15). To further demonstrate this result, we extracted CD8+ T cells in spleens from that one cured mouse in the ARMFUL/M1 treatment group and two same-age blank mouse donors, followed by coculturing with B16F10 cancer cells (fig. S18A). T cells from the cured mouse caused higher cytotoxicity to B16F10 cancer cells than those from blank donors (fig. S18B). Meanwhile, the cured mouse–derived T cells also expressed higher levels of cytokines, including tumor necrosis factor–α (TNF-α), interferon-γ (IFN-γ), perforin, and granzyme B, after coincubation, indicating mouse-obtained T cell–based immune memory after ARMFUL/M1 therapy (fig. S18, C to J). In total, the ARMFUL/M1 macrophages could remodel the tumor microenvironment, activate T cell cytotoxicity, and even induce systemic immunological memory to synergistically inhibit tumor growth and provide long-term protection.
Systemic antitumor effect of ARMFUL/M1
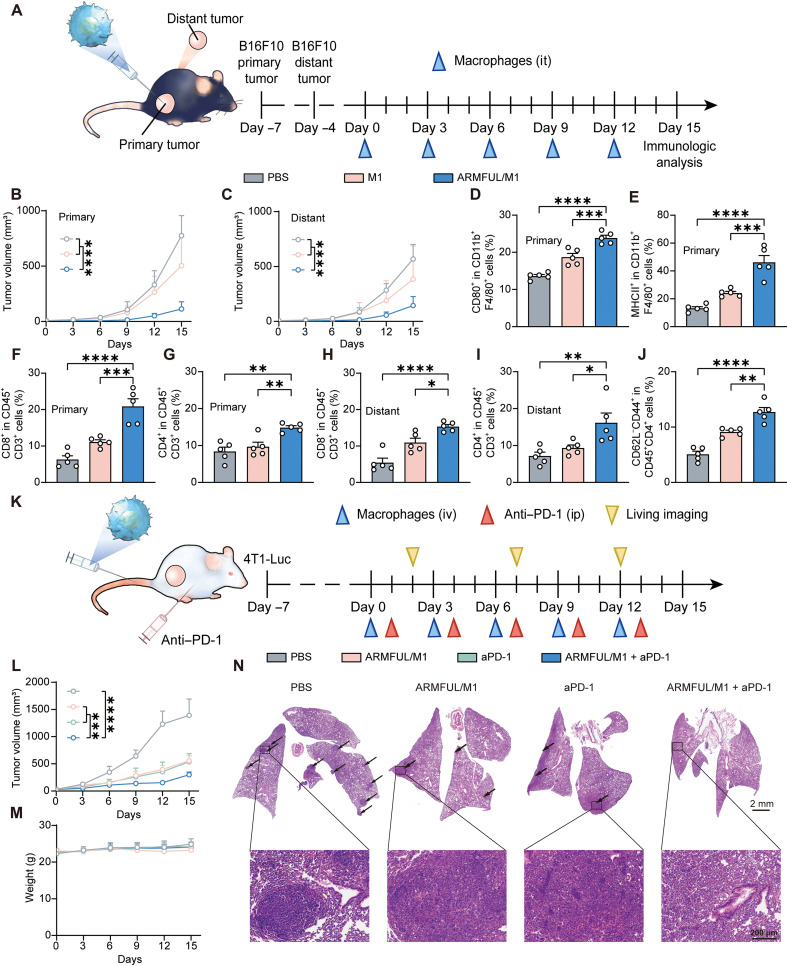
Inspired by the systemic T cell activation of ARMFUL/M1 therapy, we further explored whether this systemic T cell immunity could benefit abscopal tumor inhibition. To this end, we established a C57BL/6 mouse model bearing two B16F10 tumors in the right (the primary tumor) and left (distant tumor) flanks, followed by intratumoral injection of ARMFUL/M1 macrophages in the primary tumor (Fig. 7A). Tumor growth curves showed that ARMFUL/M1 macrophage treatment caused significant suppression of both primary and distant tumors (Fig. 7, B and C, and fig. S19). To investigate the antitumor mechanism, M1/M2 macrophages in both primary and distant tumors were analyzed. There was an increased percentage of M1 macrophages (CD80+ and MHCII+) (Fig. 7, D and E, and fig. S20, A and B) and a decreased percentage of M2 macrophages (CD206+ and VEGF+) in primary tumors (fig. S20, C to F). However, for distant tumors, M1 or M2 macrophage percentages have no distinct difference (fig. S21). To uncover the mechanism of distant tumor suppression, tumor-infiltrating T cells in both primary and distant tumors were also analyzed. As shown in Fig. 7 (F to I) and fig. S22 (A to D), both primary and distant tumors showed significantly higher infiltration of CD8+ and CD4+ activated T cells in ARMFUL/M1 macrophages groups, compared with natural M1 macrophages and the PBS group. These results strongly demonstrated that ARMFUL/M1 could remodel the primary tumor microenvironment and subsequently activate the immunity of systemic T cells to further inhibit distant tumor progression. Predictably, TEM cells in the spleen from ARMFUL/M1-treated mice also showed an elevation to induce the immunological memory (Fig. 7J and fig. S22E).
Fig. 7. Systemic antitumor effect of ARMFUL/M1 and their combination with aPD-1 treatment.
(A) Schematic representation of the systemic antitumor effect on B16F10 tumor-bearing mice. C57BL/6 mice were inoculated with B16F10 cells to form primary and distant tumors, followed by receiving an intratumor (it) injection of engineered BMDMs in primary tumors at the schedule. (B and C) Primary tumor (B) and distant tumor (C) growth curves of mice burdened with B16F10 cells after intratumoral injection of PBS, M1 macrophages, or ARMFUL/M1 (n = 6 per group). (D and E) The percentage of (D) CD80+ and MHCII+ (E) M1 macrophages in isolated primary tumor tissues after treatment (n = 5 per group). (F and G) The percentage of CD8+ (F) and CD4+ (G) T cells in isolated primary tumor tissues after treatment (n = 5 per group). (H and I) The percentage of CD8+ (H) and CD4+ (I) T cells in isolated primary tumor tissues after treatment (n = 5 per group). (J) The percentage of TEM cells (CD62−CD44+) in the spleen. (K) Schematic representation of the combination of ARMFUL/M1 with aPD-1 in 4T1-Luc tumor-bearing mice. BALB/c mice were inoculated with 4T1-Luc cells, followed by receiving intravenous injection of engineered BMDMs and intraperitoneal (ip) injection of aPD-1 at the schedule after oncogenesis. (L) Tumor growth curves of mice burdened with 4T1-Luc cells after intravenous injection of PBS, ARMFUL/M1, aPD-1, and ARMFUL/M1 + aPD-1 (n = 6 per group). (M) Weight changes of mice burdened with 4T1-Luc cells in different groups (n = 6 per group). (N) Representative H&E staining images of metastatic nodules in the lung after treatment. Data represent means ± SD. Statistical significance in (B), (C), (L), and (M) was calculated by two-way ANOVA, while (D) to (J) was calculated via one-way ANOVA, both with Dunnett’s multiple comparison tests: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0005.
Antitumor efficacy of ARMFUL/M1 combined with anti–programmed cell death protein 1 and their inhibition of cancer metastasis
The programmed death ligand 1 (PD-L1) expressed on cancer cells is also a key factor to impair the cytotoxicity of activated T cells and the antitumor responses of macrophages by interacting with programmed cell death protein 1 (PD-1) on these cell surfaces. Thus, we hypothesized that blocking PD-L1/PD-1 interactions using PD-1 antibody (aPD-1) can enhance ARMFUL/M1 macrophage-induced T cell–based immune response to improve their antitumor efficacy. To verify this, we established the breast cancer spontaneous lung metastasis BALB/c mouse model burdened with 4T1-luciferase (4T1-Luc) tumors and injected M1 macrophages intravenously and/or aPD-1 intraperitoneally into them (Fig. 7K). The tumor growth curve and bioluminescence imaging observation showed that both ARMFUL/M1 macrophages and aPD-1 treatments afforded significant antitumor efficacy compared with control, while their combination showed better suppression of tumor growth (Fig. 7L and fig. S23). The negligible change in the body weight of mice indicated the biosafety of this combinational therapy (Fig. 7M). In addition, we found that mice treated with either ARMFUL/M1 macrophages or aPD-1 had significantly fewer metastatic nodules in the lung than the control mice, and the combination of them almost completely inhibited lung metastasis (Fig. 7N and fig. S24). These results suggested that ARMFUL/M1 macrophage treatment can also work on the 4T1-Luc tumor-bearing BALB/c mouse model and decrease tumor metastasis to other organs. Meanwhile, the combination of this ARMFUL/M1 therapy with aPD-1 can further enhance antitumor efficacy.
DISCUSSION
In 2017, two cell-based products, i.e., tisagenlecleucel (Kymriah) and axicabtagene ciloleucel (Yescarta), were approved by the U.S. Food and Drug Administration for the treatment of certain B cell leukemia and lymphomas, respectively. However, these sole-target engineered cellular therapeutics exhibited limited antitumor efficacy on solid tumors (24). To this end, multifunctional engineering effector cells with the remolding of multiple intracellular or extracellular targets have been recently developed to overcome the more complex barriers in solid tumors for enhanced therapeutic efficacy (29, 30). The engineering of these targets for multifunctional adoptive cell construction always requires the integration of diverse cell engineering techniques, which makes the operation procedure costly and complicated, hindering their clinical translation. To address this, our study has outlined a facile multiplexing engineering technique that could synchronously remold multiple spatially inconsistent cellular targets in a one-step process via a membrane fusion manner. Through fusion with M1 phenotype macrophage membranes, our designed ARMFUL could simultaneously transport the BLZ-loaded core and aCD47-modified lipid shell to the cytoplasm and membrane, respectively. This spatial distribution of ARMFUL core and shell in the cell optimized the remolding efficiency of two engineering drugs according to their distinct intrinsic mechanisms, thereby improving both tumor phagocytic ability and anti-M2 polarization capacity of macrophages (Figs. 3 and 4). Therefore, our proposed membrane fusion–mediated multipath cell engineering approach is distinct from any currently used method, which could only aim at one target at one active site for remolding (11, 45). Armed with ARMFUL, the antitumor efficacy of dual-target engineered ARMFUL/M1 significantly outperformed those macrophages engineered with one target or two targets via traditional endocytosis pathway (Fig. 5). Moreover, this ARMFUL can be synthesized with an uncomplicated procedure, which makes them clinically translatable. It is expected that this membrane fusion–mediated cell engineering technique can be further combined with CAR-based cell therapies for a more extensive biomedical application.
In summary, we have developed a facile membrane fusion–mediated cell engineering technique using ARMFUL to synchronously engineer M1 phenotype macrophages with multiple functionalities in a simple step for ACT against solid tumors. To our knowledge, this is the first example of the utilization of the membrane fusion effect for cell engineering to construct multifunctional effector cells. Given the existence of various types of cell engineering reagents, this ARMFUL can be adapted to integrate other cell engineering reagent combinations to construct adoptive effector cells with diverse functionalities, providing a universal platform to customize cell behaviors and functions for improved antitumor efficacy. Furthermore, ARMFUL also shows the potential to remold a wider scope of immune effector cells used in the clinically tested ACT (such as T cells, natural killer cells, dendritic cells, and stem cells), serving as a versatile tool for the personalized design of effective cell-based immunotherapies.
MATERIALS AND METHODS
Materials
DMPC, DOTAP, DSPE-PEG-Mal, and PLGA (Mn = 50 kDa, lactic acid/glycolic acid = 75/25, mol/mol) were purchased from SunLipo NanoTech (Shanghai, China). 4′,6-Diamidino-2-phenylindole (DAPI) (catalog no.: 28718-90-3), FITC (catalog no.: 27072-45-3) (58), and sulfo-Cy5 succinimidyl ester (catalog no.: 146368-14-1) were obtained from Oukainasi (Beijing, China). Hexamethylene Ami was purchased from MedChemExpress (Shanghai, China). Chl was purchased from Macklin (Shanghai, China). Z-triphenylalanine (Phe) and TRIzol reagent were provided by GLPBIO (Shanghai, China). M-CSF protein (mouse recombinant, catalog no.: 51112-MNAH-20) and murine IFN-γ (catalog no.: Z02916-20) were purchased from Sino Biological (Beijing, China) and GenScript (Nanjing, China), respectively. Murine IL-4 (catalog no.: 214-14-5) and IL-13 (catalog no.: 210-13-2) were acquired from PeproTech (Suzhou, China). Lipopolysaccharide (LPS) was purchased from Sigma-Aldrich (Shanghai, China). Membrane protein extraction kit was acquired from Applygen (Beijing, China). Polyacrylamide gel electrophoresis (PAGE) gel (4 to 20%; catalog no.: P0468S) and enhanced chemiluminescence kit (#P0018AS) were purchased from Beyotime (Shanghai, China). Horseradish peroxidase (HRP)–conjugated AffiniPure goat anti-human IgG (SA00001) was obtained from Proteintech (Wuhan, China). Phycoerythrin (PE)/Cy7-conjugated anti-mouse CD45 (dilution 1:200; rat monoclonal, catalog no.: 157206), Allophycocyanin (APC)-conjugated anti-mouse F4/80 (dilution 1:200; mouse recombinant, catalog no.: 157306), Peridinin-chlorophyll-protein complex (PerCP)/Cy5.5-conjugated anti-mouse CD11b (dilution 1:200; rat monoclonal, catalog no.: 101228), PE-conjugated anti-mouse CD80 (dilution 1:200; Armenian Hamster monoclonal, 104707), FITC-conjugated anti-mouse CD206 (dilution 1:500; rat monoclonal, catalog no.: 141704), PE-conjugated anti-mouse CD309 (VEGF) (dilution 1:200; rat monoclonal, catalog no.: 121905), FITC-conjugated anti-mouse I-Ad (MHCII) (dilution 1:500; mouse monoclonal, catalog no.: 115005), PerCP/Cy5.5-conjugated anti-mouse CD3 (dilution 1:200; rat monoclonal, catalog no.: 100218), APC-conjugated anti-mouse CD4 (dilution 1:200; rat monoclonal, catalog no.: 100516), FITC-conjugated anti-mouse CD8 (dilution 1:500; rat monoclonal, catalog no.: 100705), FITC-conjugated anti-mouse CD44 (dilution 1:500; rat monoclonal, catalog no.: 103005), PE-conjugated anti-mouse CD62L (concentration: dilution 1:200; rat monoclonal, catalog no.: 104407), PE-conjugated anti-mouse TNF-α (dilution 1:200; rat monoclonal, catalog no.: 506305), FITC-conjugated anti-mouse IFN-γ (dilution 1:500; rat monoclonal, catalog no.: 505805), PE-conjugated anti-mouse perforin (dilution 1:200; rat monoclonal, catalog no.: 154305), FITC-conjugated anti-mouse granzyme B (dilution 1:200; mouse recombinant, catalog no.: 372205), PE-conjugated anti-mouse H-2Kb bound to SIINFEKL (dilution 1:200; mouse recombinant, catalog no.: 141603), PE anti-mouse CD282 [Toll-like receptor 2 (TLR2)] (dilution 1:200; rat monoclonal, catalog no.: 148603), PE anti-mouse CD284 (TLR4) (dilution 1:200; rat monoclonal, catalog no.: 145403), and PE anti-Nos2 [inducible nitric oxide synthase (iNOS)] (dilution 1:200; rat monoclonal, catalog no.: 696805) were purchased from BioLegend. Alexa Fluor 488 hypoxia-inducible factor 1α (HIF-1α) (dilution 1:200; rabbit monoclonal, catalog no.: ab208419) was obtained from Abcam. Annexin V–FITC/PI, Cell Counting Kit-8 (CCK-8), DiR, BCA Protein Quantification Kit, and d-Luciferin Potassium Salt D were obtained from Yeasen (Shanghai, China). Arg1- and TGF-β–specific enzyme-linked immunosorbent assay (ELISA) kits were purchased from Anoric (catalog nos.: TAEm-802 and TAEm-914, respectively, Tianjin, China). Dimethyl sulfoxide, N,N′-dimethylformamide (DMF), CHCl3, and other chemical reagents were at least analytical grade and used as received.
Instruments
Dynamic light scattering (DLS) and zeta potential studies were performed on a Litesizer 500 particle analyzer (Anton Paar, Austria). Transmission electron microscopy (Besselink, #197) measurements were performed on the high-solution TEM (FEI Tecnai G2 F30) operated at an acceleration voltage of 300 kV. Fluorescence spectra were recorded using a spectrofluorometer (RF-6000, Shimadzu, Japan). Ultraviolet absorption spectra were recorded by applying a microplate reader (Synergy H1, BioTek). Flow cytometry analysis was performed on a Beckman Coulter Cytoflex flow cytometer. CLSM images were captured on a Leica TSC SP8 confocal laser scanning microscope. Imaging macrophage phagocytosis was recorded through a live-cell dynamic imaging and analysis system (zenCELL owl). In vivo images of tumor growth were captured by an in vivo imaging system (AniView600, BLT Photon Technology).
Cell culture
The B16F10 melanoma cell line (obtained from the National Collection of Authenticated Cell Cultures), B16F10-Luc cell line (purchased from iCell Bioscience, Shanghai, China), and B16F10-OVA cell line (purchased from iCell Bioscience, Shanghai, China) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. The 4T1-Luc mouse breast tumor cell line (purchased from iCell Bioscience, Shanghai, China) was cultured in RPMI 1640 with 10% FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Murine BMDMs were extracted from the bone marrow of C57/BL6 or BALB/c mouse femurs according to the previously published protocol (19, 59) and cultured in DMEM growth medium containing M-CSF (20 ng/ml), 10% FBS, and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Fresh BMDM growth medium was changed on day 3, and mature BMDMs were collected on day 7. For M1 activation, mature BMDMs were incubated with fresh DMEM containing 10% FBS, LPS (100 ng/ml), and IFN-γ (25 ng/ml) for 24 hours.
Preparation of aCD47-decorated repolarization-resistant membrane-fusogenic liposomes
ARMFUL was prepared by a three-step process, including firstly encapsulating BLZ945 in PLGA polymeric cores, subsequently coating BLZ/PLGA with a fusogenic lipid shell to obtain BLZ@MFUL, and finally conjugating aCD47 on the BLZ@MFUL surface. First, the PLGA nanoparticles encapsulating the inhibitors of tyrosine kinase of CSF1R, BLZ945, were prepared using a nanoprecipitation method and used as the core of the ARMFUL. Briefly, PLGA and BLZ945 were dissolved in DMF with a concentration of 10 mg/ml, respectively. Subsequently, 20 μl BLZ and 80 μl PLGA solutions were mixed at the volume ratio of 1/4. Under the strong vibration of the vortex mixer, the mixture was rapidly added to 2 ml of distilled water. The solution was vibrated for another 5 min, followed by dialysis using distilled water for further use. BLZ@MFUL was then prepared by a film hydration method, in which the lipid films were hydrated using BLZ/PLGA nanoparticle solutions to obtain a core-shell structure. First, a stock solution of DMPC, DSPE-PEG-Mal, and DOTAP was prepared by dissolving them in CHCl3 with a concentration of 1 mg/ml, respectively. The lipid films were prepared from DMPC, DSPE-PEG-Mal, and DOTAP at the molar ratio of 76.2/3.8/20. Moreover, the organic solvent containing 667.2 μg of DMPC, 153.6 μg of DSPE-PEG-Mal, and 179.2 μg of DOTAP were mixed in a 25 ml round-bottom flask, followed by evaporating under the rotary evaporators to form a thin film on the bottle of the flask. After drying in the vacuum for 3 hours, the film was hydrated by rapidly mixing the above-obtained BLZ/PLGA nanoparticle solutions under ultrasonic vibration. The weight ratio of total lipids to BLZ/PLGA was fixed at 1/1. After 15 min of vibration, the nanoparticle solution was extruded through a 450 nm polycarbonate membrane, followed by freeze-drying to obtain the BLZ@MUFL. Last, aCD47 was conjugated on the surfaces of BLZ@MFUL to obtain ARMFUL. In addition, aCD47 (InVivoMab anti-CD47, Bio X Cell) was incubated with PBS supplemented with 1 mM dithiothreitol and 5 mM EDTA for 1 hour at room temperature for disulfide linkage reduction. The excess dithiothreitol was subsequently removed by buffer exchange using PD-10 desalting columns (Cytiva) with Sephadex G-25 (eluent: PBS). Afterward, the reduced aCD47 was mixed with BLZ@MFUL solution at the molar ratio of 1/4 (aCD47/DSPE-PEG-Mal). The mixture was incubated for 2 hours at room temperature with shaking, followed by overnight incubation at 4°C. Last, ARMFUL was separated from the free antibody using Sepharose CL-6B (Solarbio) beads on a gel filtration column with PBS as the mobile phase (52).
The contrastive liposomes were prepared using a similar protocol as described. The empty ARMFUL (PLGA@MFUL) and PLGA@MFUL-aCD47 (with aCD47 conjugation but without BLZ-945 loading) were synthesized using the same molar ratio of 76.2/3.8/20 (DMPC/DSPE-PEG-Mal/DOTAP), while ARMNOFUL was prepared using the molar ratio of 96.2/3.8 (DMPC/DSPE-PEG-Mal) to obtain BLZ@MNOFUL (42), followed by conjugating aCD47 on the surface.
Membrane fusion of ARMFUL
The membrane fusion mechanism of ARMFUL was examined by CLSM observation and FACS analysis. For these experiments, dually labeled ARMFUL (FITC@ARMFUL-Cy5) and ARMNOFUL (FITC@ARMNOFUL-Cy5), in which the PLGA core was loaded with FITC and aCD47 was labeled with Cy5, were prepared using the similar procedures of ARMFUL and ARMNOFUL, respectively. Briefly, 5 μl of FITC/DMF solution (10 mg/ml) was mixed with 95 μl of PLGA/DMF solution (10 mg/ml) in a tube, followed by rapidly adding 2 ml of distilled water. The mixture was then vibrated for another 5 min and dialyzed against distilled water to obtain the FITC/PLGA core nanoparticles. Next, the lipid components, which contain DMPC, DOTAP, and DSPE-PEG-Mal at a molar ratio of 76.2/20/3.8, were homogenized to form a thin film by mixing and vortically drying these solutions in a round-bottle flask. The vacuum-dried films were rapidly mixed with 2 ml of FITC/PLGA solution (~1 mg/ml) to form FITC@MFUL. Last, FITC@MFUL was conjugated with aCD47-Cy5 to obtain FITC@ARMFUL-Cy5 using dithiothreitol as the reductive reagent according to the same procedure of ARMFUL preparation. FITC@ARMNOFUL-Cy5 was prepared using a similar protocol at a molar ratio of 96.2/3.8 (DMPC/DSPE-PEG-Mal).
For CLSM observation, M1-type BMDMs were seeded in confocal dishes with approximately 80% confluence. FITC@ARMFUL-Cy5 or FITC@ARMNOFUL-Cy5 (100 μg/ml) was then added and cultured with cells for 2 hours. After incubation, cells were washed with PBS three times and fixed using 4% paraformaldehyde, while the nuclear was stained with DAPI. CLSM images were captured on a Leica CLSM with a 63-fold oil immersion objective using the excitation wavelengths of 405 nm for DAPI, 488 nm for FITC, and 663 nm for Cy5.
For membrane fusion mechanism validation, M1-type BMDMs were seeded in a confocal dish with approximately 80% confluence, followed by pretreating with DMEM containing inhibitors as follows: Ami (10 μg/ml), Chl (10 μg/ml), and Phe (50 μg/ml) at 37°C for 1 hour. The 4°C group or the PBS group was set as the control, and, therein, the 4°C group was treated with PBS at 4°C for 1 hour. After pretreatment, the cells of all groups were washed with PBS and further incubated with FITC@ARMFUL-Cy5 for 2 hours. Thereafter, cells were washed with PBS three times and fixed using 4% paraformaldehyde, while the nuclear was stained with DAPI. CLSM images were captured on a Leica CLSM with a 63-fold oil immersion objective using the excitation wavelengths of 405 nm for DAPI, 488 nm for FITC, and 663 nm for Cy5. The mean fluorescence for each group was analyzed using a flow cytometer.
For the percentage and half-life of liposomes in the macrophages, DiR-labeled liposomes were prepared by adding 40 μg of DiR to the lipid components when forming a thin film. Subsequently, the DiR-labeled liposomes were acquired according to the previously mentioned protocol. M1-type BMDMs were seeded in 10 cm dishes and incubated with DiR-labeled liposomes for 4 hours in a serum-free medium. After treatment, the cells of all groups were washed with PBS and cultured in a serum-containing medium. After treatment, the cells of all groups and supernatant were collected, and cells were washed with PBS for further use. The percentage of liposomes that successfully got into the macrophages was calculated by measuring the fluorescence intensity of the supernatant using a spectrofluorometer. For the half-life of liposomes, 2 × 105 collected cells were seeded in 24-well plates in a serum-containing medium. At the scheduled time, the cells were collected and washed with PBS and resuspended in 4% paraformaldehyde. The mean fluorescence intensity of the cells collected at different time points was determined by flow cytometer.
Preparation of ARMFUL/M1 phenotype BMDMs
First, mature BMDMs were activated to M1 macrophages. Afterward, BMDMs were collected, washed with PBS, and then engineered using ARMFUL. Typically, 1 × 106 cells were treated with 100 μg ARMFUL (containing 9.4 μg of BLZ945 and 58 μg of aCD47) dispersed in FBS-free DMEM at a fixed concentration of 100 μg/ml in 10 cm dishes for 4 hours. Other groups with the same dosage of BLZ945 and aCD47 were used for engineering. After incubation, engineered BMDMs were collected via a gentle blow and beat, followed by twice-repetitive centrifugation and washing with PBS to remove the free nanoparticles for further use.
Cell viability
Cell viability of engineered BMDMs was determined by CCK-8 assay. M1-type macrophages (2 × 104) were seeded in 96-well plates, with serum-containing medium allowed to adhere for 2 hours, followed by treatment with ARMFUL and other groups (MIX, BLZ@MFUL, PLGA@MFUL-aCD47, and ARMNOFUL) dispersed in serum-free medium. After treatment, the cells of all groups were washed with PBS and cultured in a serum-containing medium. At 24, 48, and 72 hours, the medium in the plates was removed, and 100 μl of CCK-8/DMEM [1/9 (v/v)] solution was added. After 3 hours of incubation, the absorbance was measured at the wavelength of 450 nm using a microplate reader, and cell viability was calculated.
Pattern recognition receptor activation and metabolism profile analysis
M1 polarized macrophages (2 × 105) were seeded in a 24-well plate and engineered by incubating with PBS, MIX, BLZ@MFUL, PLGA@MFUL-aCD47, ARMNOFUL, and ARMFUL for 4 hours in a serum-free medium. After treatment, the cells of all groups were washed with PBS and cultured in a serum-containing medium for 24 hours. Afterward, cells were collected and washed with PBS, resuspended in 4% paraformaldehyde containing 1% bovine serum albumin (BSA), and stained by APC-conjugated anti-mouse F4/80, PerCP/Cy5.5-conjugated anti-mouse CD11b, PE-conjugated anti-mouse TLR2, and PE-conjugated anti-mouse TLR4 for pattern recognition receptor activation analysis or PE-conjugated anti-mouse iNOS and FITC- conjugated anti-mouse HIF-1α for metabolism profile analysis using a flow cytometer.
In vitro anti-M2 polarization assay
M1 polarized macrophages (2 × 105) were seeded in a 24-well plate and engineered by incubating with PBS, MIX, ARMNOFUL, and ARMFUL for 4 hours in a serum-free medium. After treatment, the cells of all groups were washed with PBS and cultured in M2 polarization medium [DMEM containing 10% FBS and IL-4 (20 ng/ml)] or tumor cell conditioned medium (supernatant medium of B16F10 cells after incubation for 24 hours) for 24 hours. M1 polarized macrophages were set as the positive control. Afterward, cells were collected and washed with PBS, resuspended in 4% paraformaldehyde containing 1% BSA, and stained by APC-conjugated anti-mouse F4/80, PerCP/Cy5.5-conjugated anti-mouse CD11b, and PE-conjugated anti-mouse CD80 for M1 phenotype analysis or FITC-conjugated anti-mouse CD206 for M2 phenotype analysis using a flow cytometer.
Western blot analysis against aCD47
M1 polarized macrophages (1 × 106) were seeded in 6 cm dishes and engineered by incubating with PBS, MIX, ARMNOFUL, and ARMFUL for 4 hours in a serum-free medium. After treatment, the cells of all groups were collected and washed with PBS. Subsequently, the cell membrane protein of engineered M1 macrophages was extracted via the Membrane Protein Extraction Kit as the protocol. Then, the proteins were heated to 95°C for 20 min before ~15 μg of each sample was loaded onto a 4 to 20% PAGE gel (Beyotime) and treated for ~120 min (80 V). Sample proteins were then transferred to polyvinylidene difluoride (PVDF) membranes (0.45 μm) and subsequently blocked with buffer [tris-buffered saline with Tween 20 (TBST)] containing 5% BSA for 1 hour at room temperature. PVDF membranes were then bathed in a buffer containing HRP-conjugated IgG antibody for 2 hours at room temperature. Then, the membrane was washed with TBST and detected using an enhanced chemiluminescence kit.
In vitro phagocytosis assay
Phagocytosis was determined by coculturing Cy5-labeled macrophages and CFSE-labeled B16F10 tumor cells and subsequently detected using FACS. M1 polarized macrophages were engineered by incubating with PBS, MIX, BLZ@MFUL, ARMNOFUL, and ARMFUL for 4 hours in serum-free DMEM. Afterward, the cells of all groups were stained with DSPE-PEG-Cy5 (50 μg/ml) for 0.5 hours and washed with PBS three times. B16F10 tumor cells were stained with CFSE (Yeasen). B16F10 tumor cells were seeded in 12-well plates at 1 × 105 cells per well and allowed to adhere for 2 hours. Then, 2 × 105 macrophages were added and cocultured with the cancer cells for 4 hours at 37°C with a serum-containing medium. Afterward, cells were collected and washed with PBS and resuspended in 4% paraformaldehyde that contained 1% BSA. Phagocytosis was analyzed with a flow cytometer and calculated as the percentage of CFSE+ cells gated from Cy5+ macrophages.
Imaging macrophage phagocytosis
To visualize the phagocytosis, 2 × 104 B16F10 tumor cells were seeded in a 24-well plate and allowed to adhere for 2 hours. Then, 2 × 105 macrophages engineered with PBS, MIX, BLZ@MFUL, ARMNOFUL, and ARMFUL were added and cocultured at 37°C with a serum-containing medium respectively. The views of each well in the plate were recorded through a live-cell dynamic imaging and analysis system (zenCELL owl) every 10 min for 36 hours. In the recorded images, B16F10 tumor cells were fusiform, and macrophages were round, making them distinguishable. Observing the video formed by playing the images continuously, round macrophages were in groups attacking the single fusiform B16F10 tumor cell, and this cell cluster was regarded as a phagocytosis count. Phagocytosis cell cluster counts of all groups were quantified every 5 hours.
In vitro antitumor assay
Antitumor ability was evaluated by coculturing macrophages and B16F10 tumor cells followed by detecting cell viability and apoptosis. M1 polarized macrophages were engineered by incubating with PBS, MIX, BLZ@MFUL, PLGA@MFUL-aCD47, ARMNOFUL, and ARMFUL for 4 hours in serum-free DMEM beforehand. B16F10 tumor cells were seeded in 12-well plates (2 × 104 cells per well) or 48-well plates (1 × 104 cells per well) and allowed to adhere for 2 hours. Then, macrophages were added into plates (1 × 105 cells per well in a 12-well plate and 5 × 104 cells per well in a 48-well plate) and cocultured with tumor cells for 48 hours at 37°C with serum-containing medium. The group that was treated with PBS was set as the control. Afterward, the medium in the 48-well plate was removed, and 300 μl of CCK-8/DMEM [1/9 (v/v)] solution was added. After 3 hours of incubation, the absorbance was measured at the wavelength of 450 nm using a microplate reader, and cell viability was calculated. Meanwhile, cells in the 12-well plates were collected and washed with PBS for staining using the Annexin V–FITC/PI apoptosis detection kit (Yeasen) according to the manufacturer’s instructions.
Antigen presentation
B16F10-OVA tumor cells (2 × 104) were seeded in a 12-well plate and allowed to adhere for 2 hours. Then, 2 × 105 macrophages engineered with PBS, MIX, BLZ@MFUL, PLGA@MFUL-aCD47, ARMNOFUL, and ARMFUL were added and cocultured at 37°C with serum-containing medium for 3 days. Afterward, cells were collected and washed with PBS, resuspended in 4% paraformaldehyde containing 1% BSA, and stained by PE-conjugated anti-mouse H-2Kb bound to SIINFEKL and APC-conjugated anti-mouse F4/80. The antigen presentation capacity of macrophages was assessed with a flow cytometer and measured as the mean fluorescence intensity of PE within APC+ macrophages.
Transcriptome analysis
M1 phenotype BMDMs (2 × 106) were seeded in 6 cm dishes and engineered by incubating with PBS and ARMFUL for 4 hours in a serum-free medium. After treatment, the cells of all groups were washed with PBS and cultured in DMEM containing 10% FBS with/without IL-4 (20 ng/ml) for 48 hours, respectively. Then, the total RNA of cells was obtained by lysing cells using TRIzol. High-throughput sequencing and data analysis were performed by LC-Bio Technologies (Hangzhou, China).
In vivo anti-M2 polarization ability evaluation
All animal procedures were performed following the Guidelines for Care and Use of Laboratory Animals of Sun Yat-sen University and approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (protocol numbers: SYSU-IACUC-2022-000363 and SYSU-IACUC-2022-001491). Female C57BL/6 mice (8-week-old) were inoculated with 1 × 106 B16F10 cells in 100 μl of PBS by subcutaneous injection into the back of the right lower limb. Tumors were measured with calipers, and tumor volume was calculated according to the formula (1/2 × length × width2). When tumors reached a volume of approximately 200 mm3, mice intravenously received two dosages of macrophages engineered with different interventions on the 12th and 13th day after inoculation. Before injection, macrophages were engineered and stained with dye to distinguish them from endogenous macrophages. Briefly, M1 polarized BMDMs were engineered by incubating with PBS, MIX, BLZ@MFUL, PLGA@MFUL-aCD47, ARMNOFUL, or ARMFUL for 4 hours in serum-free DMEM beforehand, and acquired as previously. Then, the macrophages were collected and washed twice and stained with DiR (Yeasen) according to the manufacturer’s instructions. Each mouse was administrated with the same number of cells (i.e., 2 × 106 macrophages in 100 μl of PBS per mouse per injection). After 24 hours of the second injection, the mice were euthanized, and tumors were collected, crumbled, homogenized using a homogenizer, and fixed using 4% paraformaldehyde that contained 1% BSA. The tumor suspensions were separated into two tubes, followed by staining with APC-conjugated anti-mouse F4/80, PerCP/Cy5.5-conjugated anti-mouse CD11b, PE-conjugated anti-mouse CD80, FITC-conjugated anti-mouse CD206, PE-conjugated anti-mouse CD309, or FITC-conjugated anti-mouse I-Ad. After staining, the suspensions were analyzed using a flow cytometer for the expression of M1/M2 markers in adoptively transferred macrophages or TAMs.
In vivo antitumor efficacy
Female C57BL/6 mice (8-week-old) were inoculated with 1 × 106 B16F10-Luc cells in 100 μl of PBS by subcutaneous injection into the back of the right lower limb. Tumors were measured with calipers, and tumor volume was calculated according to the formula (1/2 × length × width2). The tumor-bearing mice were randomly allocated to the control and treatment groups on day 5 after implantation. The mice in the treatment groups received intravenous injections of 1 × 106 engineered macrophages in 100 μl of PBS per mouse every 3 days for 12 days. Macrophages were engineered and acquired before injection. Briefly, M1 polarized BMDMs were engineered by incubating with PBS, MIX, BLZ@MFUL, PLGA@MFUL-aCD47, ARMNOFUL, or ARMFUL for 4 hours in serum-free DMEM. Then, the macrophages were collected, washed twice, and suspended in PBS for injection. The mice in the control group were intravenously injected with 100 μl of PBS. The tumor size and mouse body weight were monitored. The mice were euthanized when their tumor sizes reach 2000 mm3 or when rapidly lost weight (15 to 20%).
To monitor tumor growth, the mice received in vivo imaging on days 3, 7, and 11 after the first injection. Briefly, mice were administrated with 200 μl of d-luciferin potassium salt bioluminescence substrate (15 mg/ml; Yeasen) in PBS and 200 μl of pentobarbital sodium (0.6 wt %) in PBS via intraperitoneal injection. Ten minutes after injection, mice were imaged under anesthesia using an in vivo imaging system (AniView600, BLT Photon Technology).
For cancer immunotherapy studies, mice were euthanized 14 days after the first injection, and the tumors were collected. One part of every tumor was used for FACS analysis and determination of cytokine expressions, and the other part was used for tissue section. For FACS and cytokine analysis, the tumors were crumbled, homogenized using a homogenizer, and diluted with PBS at a weight ratio of 1:10 (tumor:PBS). After centrifugation at 2500g for 10 min, the supernatant of the homogenates was collected for determination of cytokine expressions, while the cell sediment was fixed using 4% paraformaldehyde that contained 1% BSA to obtain tumor suspensions for FACS analysis. For FACS analysis, the tumor suspensions were stained with PE/Cy7-conjugated anti-mouse CD45, APC-conjugated anti-mouse F4/80, PerCP/Cy5.5-conjugated anti-mouse CD11b, PE-conjugated anti-mouse CD80, FITC-conjugated anti-mouse CD206, PE-conjugated anti-mouse CD309, and FITC-conjugated anti-mouse I-Ad for expression of M1/M2 markers. To investigate the T cells in the tumor, the suspensions were stained with PE/Cy7-conjugated anti-mouse CD45, Percy/Cy5.5-conjugated anti-mouse CD3, APC-conjugated anti-mouse CD4, and FITC-conjugated anti-mouse CD8. After staining, the suspensions were analyzed using a flow cytometer. For the determination of tumor-associated cytokine expressions (Arg1 and TGF-β), the tumor supernatant was detected using the ELISA kits (Anoric Biotechnology) according to the manufacturer’s protocol. For tissue staining, the tumors were fixed with 4% paraformaldehyde and sliced for a frozen section. The sections were stained with H&E, TUNEL, and fluorescently labeled antibodies of CD31 and Ki67, respectively. The fluorescently labeled slices were imaged using a Nikon A1R-SIMe confocal microscope, and analyses were performed using NIS Elements 4.6 software. H&E-stained slices were scanned using a slide scanner microscope (Zeiss AxioScan.Z1).
To study the immune memory effect, mice were euthanized 14 days after the first injection, and the spleens were collected. The spleens were triturated using frosted slides and fixed using 4% paraformaldehyde that contained 1% BSA. The spleen suspensions were stained with PE/Cy7-conjugated anti-mouse CD45, Percy/Cy5.5-conjugated anti-mouse CD3, and APC-conjugated anti-mouse CD4 for T cell activation analysis or PE-conjugated anti-mouse CD62L and FITC-conjugated anti-mouse CD44 for immune memory T cell analysis. After staining, the suspensions were analyzed using a flow cytometer.
In addition, when survival rate observation was completed, the spleens of one cured mouse (ARMFUL group) and two healthy mice (the same age as a cured mouse) as donors were collected. T cells were isolated from the spleen via magnetic bead separation (MojoSort Mouse CD8 T Cell Isolation Kit, BioLegend, no. 480031), followed by culturing in the primary T cell culture system (iCell, no. PriMed-iCell-026). Further investigation of the immune memory effect was evaluated by coculturing T cells and B16F10 tumor cells as follows. B16F10 tumor cells were seeded in 48-well plates (2.5 × 104 cells per well) and allowed to adhere for 2 hours. Then, 2.5 × 105 T cells were added and cocultured at 37°C with a primary T cell culture system. The group that was treated with PBS rather than T cells was set as the control. After 24 hours of incubation, suspending T cells were collected, and adhesive cancer cells were left in plates. Then, 500 μl of CCK-8/DMEM [1/9 (v/v)] solution was added. After 3 hours of incubation, the absorbance was measured at the wavelength of 450 nm using a microplate reader, and cell viability was calculated. Meanwhile, the collected T cells were washed twice and fixed using 4% paraformaldehyde that contained 1% BSA. The cells were stained with PE-conjugated anti-mouse TNF-α, FITC-conjugated anti-mouse IFN-γ, or PE-conjugated anti-mouse perforin and FITC-conjugated anti-mouse granzyme B. After staining, the suspensions were analyzed using a flow cytometer.
For safety evaluation, the blood and major organs of mice were collected 14 days after the first injection. The serum of blood was separated by centrifugation at 2500g for 10 min, followed by the detection of alanine aminotransferase, aspartate aminotransferase, albumin, alkaline phosphatase, urea, creatinine, uric acid, and creatine phosphokinase-MB. The organs (heart, liver, spleen, lung, and kidney) were fixed in 4% paraformaldehyde and sliced for H&E staining. The stained slices were scanned using a slide scanner microscope (Zeiss AxioScan.Z1).
Systemic antitumor effect in vivo
To investigate the systemic antitumor effect, the female C57BL/6 mice (8-week-old) were inoculated with 1 × 106 B16F10 cells in 100 μl of PBS by subcutaneous injection into the back of the lower right flanks as the primary tumor. Three days later, mice were inoculated with 1 × 106 B16F10 cells in 100 μl of PBS by subcutaneous injection into the back of the lower left flanks as the distant tumor. Tumors of two sides were measured with calipers, and the tumor volume was calculated according to the formula (1/2 × length × width2). The tumor-bearing mice were randomly allocated to control and treatment groups on day 7 after the first implantation. The primary tumors of mice in the treatment groups received intratumor injected 1 × 106 macrophages in 100 μl of PBS per mouse per injection every 3 days for 12 days. Macrophages were engineered before injection. Briefly, M1 polarized BMDMs were engineered by incubating with PBS and ARMFUL for 4 hours in serum-free DMEM. Then, cells were collected and washed with PBS for further injection. The primary tumors of mice in control were intratumorally injected with 100 μl of PBS. The tumor size and mice weight were monitored. The mice were euthanized when their tumor size reach 2000 mm3 or when rapidly lost weight (15 to 20%). Fifteen days after the first injection, mice were euthanized, and the tumors of two sides and spleens were collected for immune effects analysis as previously.
In vivo antitumor effect combined with aPD-1
Female BALB/c mice (8-week-old) were inoculated with 1 × 106 4T1-Luc cells in 100 μl of PBS by subcutaneous injection into the upper right mammary fat pad. Tumors were measured with calipers, and tumor volume was calculated according to the formula (1/2 × length × width2). The tumor-bearing mice were randomly allocated to the control and treatment groups on day 6 after implantation. For the cell treatment groups, the mice received intravenous injected 1 × 106 engineered macrophages in 100 μl of PBS per mouse per injection every 3 days for 12 days. The day after cell injection, the mice in antibody treatment groups received intraperitoneal injected 50 μg of aPD-1 (InVivoMab anti-mouse PD-1, Bio X Cell) per mouse per injection. Engineering BMDMs were acquired from BALB/c mice as previously described, and M1-polarized macrophages were engineered by incubating with ARMFUL for 4 hours in serum-free DMEM. To monitor tumor growth, the mice received in vivo imaging on days 2, 7, and 12 after the first injection. Briefly, mice were administrated with 200 μl of d-luciferin potassium salt bioluminescence substrate (15 mg/ml; Yeasen) in PBS and 200 μl of pentobarbital sodium (0.6 wt %) in PBS via intraperitoneal injection. Ten minutes after injection, mice were imaged under anesthesia using an in vivo imaging system (AniView600, BLT Photon Technology).
Data analysis
CLSM images were analyzed by Leica Application Suite X (Leica), and immunofluorescence staining images were modulated by NIS-Elements Viewer (Nikon). H&E staining images were present with K-Viewer (K-Tron International). Flow cytometry analysis was performed in CytExpert (Beckman). Gray values and Spearman’s correlation value were analyzed by ImageJ software. Live-cell dynamic images and videos were conducted with zenCELL owl (innoME). In vivo images of mice burdened with tumors were captured by AniView supplied by BLT Photon Technology. Transcriptome analyses were performed in the cloud platform of LC-Bio Technology (www.omicstudio.cn). Columns and line graphs were drawn through GraphPad Prism 9. Optimization of images and typography of figures were performed using Photoshop and Illustrator, and video editing was conducted using Premiere Pro (Adobe).
Statistical analysis
The data were represented as means ± SD using GraphPad Prism 9. Statistical significance is calculated by t test, one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test, two-way ANOVA with Turkey’s multiple comparison tests, and log-rank (Mantel-Cox) test for survival.
Acknowledgments
Funding: This work is supported by the National Key Research and Development Program of China (2019YFA0111300), the National Natural Science Foundation of China (22277155, 51903256, 21907113, 52103197, and 52202359), the Science and Technology Program of Guangzhou (202102010225), the China Postdoctoral Science Foundation (2020M680133 and 2022T150747), the Guangdong Provincial Science and Technology Program (International Scientific Cooperation, 2018A050506035), the China Primary Health Care Foundation (2022-003), the Guangdong Provincial Pearl River Talents Program (2019QN01Y131), the National University of Singapore (NUHSRO/2020/133/Startup/08, NUHSRO/2023/008/NUSMed/TCE/LOA, NUHSRO/2021/034/TRP/09/Nanomedicine, NUHSRO/2021/044/Kickstart/09/LOA), National Medical Research Council (MOH-OFIRG23jan-0005, CG21APR1005), Singapore Ministry of Education (MOE-000387-00), and National Research Foundation (NRF-000352-00).
Author contributions: Chu.Z., Y.T., X.C., and M.L. conceived and designed the project. Chu.Z., Q.Z., K.Y., H.K., and Che.Z. performed the experiments and data analysis. F.C., Y.X., R.S., E.J., and W.S. contributed to the data analysis. Chu.Z., Q.Z., F.C., Y.T., X.C., and M.L. analyzed the results and co-wrote the paper. All the authors have discussed the results and approved the final version.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S24
Legends for movies S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Sadelain M., Rivière I., Riddell S., Therapeutic T cell engineering. Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Cheng H., Peng H., Zhou H., Li P. Y., Langer R., Non-genetic engineering of cells for drug delivery and cell-based therapy. Adv. Drug Del. Rev. 91, 125–140 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Lesch S., Blumenberg V., Stoiber S., Gottschlich A., Ogonek J., Cadilha B. L., Dantes Z., Rataj F., Dorman K., Lutz J., Karches C. H., Heise C., Kurzay M., Larimer B. M., Grassmann S., Rapp M., Nottebrock A., Kruger S., Tokarew N., Metzger P., Hoerth C., Benmebarek M.-R., Dhoqina D., Grünmeier R., Seifert M., Oener A., Umut Ö., Joaquina S., Vimeux L., Tran T., Hank T., Baba T., Huynh D., Megens R. T. A., Janssen K.-P., Jastroch M., Lamp D., Ruehland S., Di Pilato M., Pruessmann J. N., Thomas M., Marr C., Ormanns S., Reischer A., Hristov M., Tartour E., Donnadieu E., Rothenfusser S., Duewell P., König L. M., Schnurr M., Subklewe M., Liss A. S., Halama N., Reichert M., Mempel T. R., Endres S., Kobold S., T cells armed with C-X-C chemokine receptor type 6 enhance adoptive cell therapy for pancreatic tumours. Nat. Biomed. Eng. 5, 1246–1260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalbasi A., Siurala M., Su L. L., Tariveranmoshabad M., Picton L. K., Ravikumar P., Li P., Lin J.-X., Escuin-Ordinas H., Da T., Kremer S. V., Sun A. L., Castelli S., Agarwal S., Scholler J., Song D., Rommel P. C., Radaelli E., Young R. M., Leonard W. J., Ribas A., June C. H., Garcia K. C., Potentiating adoptive cell therapy using synthetic IL-9 receptors. Nature 607, 360–365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Lang S., Tian Y., Zhang J., Yan X., Fang Z., Weng J., Lu N., Wu X., Li T., Cao H., Li Z., Huang X., Glycoengineering of natural killer cells with CD22 ligands for enhanced anticancer immunotherapy. ACS Cent. Sci. 6, 382–389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D., Zheng Y., Lin Z., Liu X., Li J., Yang H., Tan W., Equipping natural killer cells with specific targeting and checkpoint blocking aptamers for enhanced adoptive immunotherapy in solid tumors. Angew. Chem. Int. Ed. 59, 12022–12028 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Hong S., Yu C., Wang P., Shi Y., Cao W., Cheng B., Chapla D. G., Ma Y., Li J., Rodrigues E., Narimatsu Y., Yates Iii J. R., Chen X., Clausen H., Moremen K. W., Macauley M. S., Paulson J. C., Wu P., Glycoengineering of NK cells with glycan ligands of CD22 and selectins for B-cell lymphoma therapy. Angew. Chem. Int. Ed. 60, 3603–3610 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Luo X., Tian Y., Wu T., Weng J., Li Z., Ye F., Huang X., Equipping natural killer cells with Cetuximab through metabolic glycoengineering and bioorthogonal reaction for targeted treatment of KRAS mutant colorectal cancer. ACS Chem. Biol. 16, 724–730 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Zhao Q., Gong Z., Li Z., Wang J., Zhang J., Zhao Z., Zhang P., Zheng S., Miron R. J., Yuan Q., Zhang Y., Target reprogramming lysosomes of CD8+ T cells by a mineralized metal–organic framework for cancer immunotherapy. Adv. Mater. 33, 2100616 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Shi P., Zhao N., Coyne J., Wang Y., DNA-templated synthesis of biomimetic cell wall for nanoencapsulation and protection of mammalian cells. Nat. Commun. 10, 2223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields C. W., Evans M. A., Wang L. L.-W., Baugh N., Iyer S., Wu D., Zhao Z., Pusuluri A., Ukidve A., Pan D. C., Mitragotri S., Cellular backpacks for macrophage immunotherapy. Sci. Adv. 6, eaaz6579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephan M. T., Irvine D. J., Enhancing cell therapies from the outside in: Cell surface engineering using synthetic nanomaterials. Nano Today 6, 309–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q., Wei W., Wang P., Zuo L., Li F., Xu J., Xi X., Gao X., Ma G., Xie H.-Y., Biomimetic magnetosomes as versatile artificial antigen-presenting cells to potentiate T-cell-based anticancer therapy. ACS Nano 11, 10724–10732 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Hao M., Hou S., Li W., Li K., Xue L., Hu Q., Zhu L., Chen Y., Sun H., Ju C., Zhang C., Combination of metabolic intervention and T cell therapy enhances solid tumor immunotherapy. Sci. Transl. Med. 12, eaaz6667 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Nie W., Wei W., Zuo L., Lv C., Zhang F., Lu G. H., Li F., Wu G., Huang L. L., Xi X., Xie H.-Y., Magnetic nanoclusters armed with responsive PD-1 antibody synergistically improved adoptive T-cell therapy for solid tumors. ACS Nano 13, 1469–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ma L., Dichwalkar T., Chang J. Y. H., Cossette B., Garafola D., Zhang A. Q., Fichter M., Wang C., Liang S., Silva M., Kumari S., Mehta N. K., Abraham W., Thai N., Li N., Wittrup K. D., Irvine D. J., Enhanced CAR–T cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 365, 162–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C.-X., Zhang Y., Dong X., Zhang L., Liu M.-D., Li B., Zhang M.-K., Feng J., Zhang X.-Z., Artificially reprogrammed macrophages as tumor-tropic immunosuppression-resistant biologics to realize therapeutics production and immune activation. Adv. Mater. 31, 1807211 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Zheng C., Zhang J., Chan H. F., Hu H., Lv S., Na N., Tao Y., Li M., Engineering nano-therapeutics to boost adoptive cell therapy for cancer treatment. Small Methods 5, 2001191 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Hou X., Zhang X., Zhao W., Zeng C., Deng B., McComb D. W., Du S., Zhang C., Li W., Dong Y., Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 15, 41–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im S., Jang D., Saravanakumar G., Lee J., Kang Y., Lee Y. M., Lee J., Doh J., Yang Z. Y., Jang M. H., Kim W. J., Harnessing the formation of natural killer–tumor cell immunological synapses for enhanced therapeutic effect in solid tumors. Adv. Mater. 32, 2000020 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Tang L., Zheng Y., Melo M. B., Mabardi L., Castano A. P., Xie Y. Q., Li N., Kudchodkar S. B., Wong H. C., Jeng E. K., Maus M. V., Irvine D. J., Enhancing T cell therapy through TCR-signaling-responsive nanoparticle drug delivery. Nat. Biotechnol. 36, 707–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y., Che J., Ji X., Li Y., Xie J., Chen X., Recent advances in biomaterial-boosted adoptive cell therapy. Chem. Soc. Rev. 51, 1766–1794 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Jackson H. J., Rafiq S., Brentjens R. J., Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 13, 370–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou A. J., Chen L. C., Chen Y. Y., Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov. 20, 531–550 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Chan J. D., Lai J., Slaney C. Y., Kallies A., Beavis P. A., Darcy P. K., Cellular networks controlling T cell persistence in adoptive cell therapy. Nat. Rev. Immunol. 21, 769–784 (2021). [DOI] [PubMed] [Google Scholar]
- 26.DePeaux K., Delgoffe G. M., Metabolic barriers to cancer immunotherapy. Nat. Rev. Immunol. 21, 785–797 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah N. N., Fry T. J., Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol. 16, 372–385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S., Kivimäe S., Dolor A., Szoka F. C., Macrophage-based cell therapies: The long and winding road. J. Control. Release 240, 527–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan V., Barber-Rotenberg J. S., Jung I.-Y., Lacey S. F., Rech A. J., Davis M. M., Hwang W.-T., Lal P., Carpenter E. L., Maude S. L., Plesa G., Vapiwala N., Chew A., Moniak M., Sebro R. A., Farwell M. D., Marshall A., Gilmore J., Lledo L., Dengel K., Church S. E., Hether T. D., Xu J., Gohil M., Buckingham T. H., Yee S. S., Gonzalez V. E., Kulikovskaya I., Chen F., Tian L., Tien K., Gladney W., Nobles C. L., Raymond H. E., Frazee D., Truran M., Veloso E., McConville H., Aguedelo J., Hower S., Ngo S., Jadlowsky J., Melenhorst J. J., Roche A., Everett J., Gupta M., Nazimuddin F., Bartoszek C., Koterba N., Reynolds R., Ellington F., Kloss C. C., Lee J., Zhao Y., Scholler J., Riley J. L., Bailey C., White A., Hudson B., Chang P., Maus M. V., Levine B. L., Hexner E. O., Siegel D. L., Bushman F. D., June C. H., Fraietta J. A., Haas N. B., PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: A phase 1 trial. Nat. Med. 28, 724–734 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayan V., Gladney W., Plesa G., Vapiwala N., Carpenter E., Maude S. L., Lal P., Lacey S. F., Melenhorst J. J., Sebro R., Farwell M., Hwang W.-T., Moniak M., Gilmore J., Lledo L., Dengel K., Marshall A., Coughlin C. M., June C. H., Haas N. B., A phase I clinical trial of PSMA-directed/TGFβ-insensitive CAR-T cells in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 37, TPS347 (2019). [Google Scholar]
- 31.Milone M. C., Xu J., Chen S.-J., Collins M. A., Zhou J., Powell D. J., Melenhorst J. J., Engineering-enhanced CAR T cells for improved cancer therapy. Nat. Cancer 2, 780–793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rafiq S., Hackett C. S., Brentjens R. J., Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siriwon N., Kim Y. J., Siegler E., Chen X., Rohrs J. A., Liu Y., Wang P., CAR-T cells surface-engineered with drug-encapsulated nanoparticles can ameliorate intratumoral T-cell hypofunction. Cancer Immunol. Res. 6, 812–824 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Nguyen N. T., Huang K., Zeng H., Jing J., Wang R., Fang S., Chen J., Liu X., Huang Z., You M. J., Rao A., Huang Y., Han G., Zhou Y., Nano-optogenetic engineering of CAR T cells for precision immunotherapy with enhanced safety. Nat. Nanotechnol. 16, 1424–1434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fultang L., Booth S., Yogev O., Martins da Costa B., Tubb V., Panetti S., Stavrou V., Scarpa U., Jankevics A., Lloyd G., Southam A., Lee S. P., Dunn W. B., Chesler L., Mussai F., De Santo C., Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood 136, 1155–1160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickner W., Schekman R., Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsden H. R., Tomatsu I., Kros A., Model systems for membrane fusion. Chem. Soc. Rev. 40, 1572–1585 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Du S., Liew S. S., Li L., Yao S. Q., Bypassing endocytosis: Direct cytosolic delivery of proteins. J. Am. Chem. Soc. 140, 15986–15996 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kong H., Yi K., Zheng C., Lao Y. H., Zhou H., Chan H. F., Wang H., Tao Y., Li M., Membrane-fusogenic biomimetic particles: A new bioengineering tool learned from nature. J. Mater. Chem. B 10, 6841–6858 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Zheng C., Zhong Q., Song W., Yi K., Kong H., Wang H., Tao Y., Li M., Chen X., Membrane-fusion-mediated multiplex engineering of tumor cell surface glycans for enhanced NK cell therapy. Adv. Mater. 35, e2206989 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Kim B., Sun S., Varner J. A., Howell S. B., Ruoslahti E., Sailor M. J., Securing the payload, finding the cell, and avoiding the endosome: Peptide-targeted, fusogenic porous silicon nanoparticles for delivery of siRNA. Adv. Mater. 31, 1902952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim B., Pang H.-B., Kang J., Park J.-H., Ruoslahti E., Sailor M. J., Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus. Nat. Commun. 9, 1969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J., Santos O. A., Park J.-H., Selective photosensitizer delivery into plasma membrane for effective photodynamic therapy. J. Control. Release 191, 98–104 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Lee J., Kim J., Jeong M., Lee H., Goh U., Kim H., Kim B., Park J.-H., Liposome-based engineering of cells to package hydrophobic compounds in membrane vesicles for tumor penetration. Nano Lett. 15, 2938–2944 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Klichinsky M., Ruella M., Shestova O., Lu X. M., Best A., Zeeman M., Schmierer M., Gabrusiewicz K., Anderson N. R., Petty N. E., Cummins K. D., Shen F., Shan X., Veliz K., Blouch K., Yashiro-Ohtani Y., Kenderian S. S., Kim M. Y., O’Connor R. S., Wallace S. R., Kozlowski M. S., Marchione D. M., Shestov M., Garcia B. A., June C. H., Gill S., Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 38, 947–953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L., Li H., Wang J., Zhang J., Liang X.-J., Guo W., Gu Z., Leveraging macrophages for cancer theranostics. Adv. Drug Del. Rev. 183, 114136 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Li J., Jiang X., Li H., Gelinsky M., Gu Z., Tailoring materials for modulation of macrophage fate. Adv. Mater. 33, 2004172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulkarni A., Chandrasekar V., Natarajan S. K., Ramesh A., Pandey P., Nirgud J., Bhatnagar H., Ashok D., Ajay A. K., Sengupta S., A designer self-assembled supramolecule amplifies macrophage immune responses against aggressive cancer. Nat. Biomed. Eng. 2, 589–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramesh A., Brouillard A., Kumar S., Nandi D., Kulkarni A., Dual inhibition of CSF1R and MAPK pathways using supramolecular nanoparticles enhances macrophage immunotherapy. Biomaterials 227, 119559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramesh A., Kumar S., Nandi D., Kulkarni A., CSF1R- and SHP2-inhibitor-loaded nanoparticles enhance cytotoxic activity and phagocytosis in tumor-associated macrophages. Adv. Mater. 31, 1904364 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Chen Q., Wang C., Zhang X., Chen G., Hu Q., Li H., Wang J., Wen D., Zhang Y., Lu Y., Yang G., Jiang C., Wang J., Dotti G., Gu Z., In situ sprayed bioresponsive immunotherapeutic gel for post-surgical cancer treatment. Nat. Nanotechnol. 14, 89–97 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Dammes N., Goldsmith M., Ramishetti S., Dearling J. L. J., Veiga N., Packard A. B., Peer D., Conformation-sensitive targeting of lipid nanoparticles for RNA therapeutics. Nat. Nanotechnol. 16, 1030–1038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng C., Wang Q., Wang Y., Zhao X., Gao K., Liu Q., Zhao Y., Zhang Z., Zheng Y., Cao J., Chen H., Shi L., Kang C., Liu Y., Lu Y., In situ modification of the tumor cell surface with immunomodulating nanoparticles for effective suppression of tumor growth in mice. Adv. Mater. 31, 1902542 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Zheng C., Zhao X., Wang Y., Zhao Y., Zheng Y., Zhang Z., Liu Q., Liu Y., Shi L., Immune modulating nanoparticles depleting tumor-associated macrophages to enhance immune checkpoint blockade therapy. Chem. Eng. J. 435, 134779 (2022). [Google Scholar]
- 55.Zhao Y., Han B., Hao J., Zheng Y., Chai J., Zhang Z., Liu Y., Shi L., Bi-specific macrophage nano-engager for cancer immunotherapy. Nano Today 41, 101313 (2021). [Google Scholar]
- 56.Wang Y.-F., Zhang C., Yang K., Wang Y., Shan S., Yan Y., Dawson K. A., Wang C., Liang X.-J., Transportation of AIE-visualized nanoliposomes is dominated by the protein corona. Natl. Sci. Rev. 8, nwab068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X., Liu C., Zheng Z., Chen S., Pang X., Xiang X., Tang J., Ren E., Chen Y., You M., Wang X., Chen X., Luo W., Liu G., Xia N., Vesicular antibodies: A bioactive multifunctional combination platform for targeted therapeutic delivery and cancer immunotherapy. Adv. Mater. 31, 1808294 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Adolfsen K. J., Callihan I., Monahan C. E., Greisen P. J., Spoonamore J., Momin M., Fitch L. E., Castillo M. J., Weng L., Renaud L., Weile C. J., Konieczka J. H., Mirabella T., Abin-Fuentes A., Lawrence A. G., Isabella V. M., Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan H., Jiang W., von Roemeling C. A., Qie Y., Liu X., Chen Y., Wang Y., Wharen R. E., Yun K., Bu G., Knutson K. L., Kim B. Y. S., Multivalent bi-specific nanobioconjugate engager for targeted cancer immunotherapy. Nat. Nanotechnol. 12, 763–769 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S24
Legends for movies S1 and S2
Movies S1 and S2