Abstract
Background
Prior observation has shown differences in COVID-19 hospitalization risk between SARS-CoV-2 variants, but limited information describes hospitalization outcomes.
Methods
Inpatients with COVID-19 at 5 hospitals in the eastern United States were included if they had hypoxia, tachypnea, tachycardia, or fever, and SARS-CoV-2 variant data, determined from whole-genome sequencing or local surveillance inference. Analyses were stratified by history of SARS-CoV-2 vaccination or infection. The average effect of SARS-CoV-2 variant on 28-day risk of severe disease, defined by advanced respiratory support needs, or death was evaluated using models weighted on propensity scores derived from baseline clinical features.
Results
Severe disease or death within 28 days occurred for 977 (29%) of 3369 unvaccinated patients and 269 (22%) of 1230 patients with history of vaccination or prior SARS-CoV-2 infection. Among unvaccinated patients, the relative risk of severe disease or death for Delta variant compared with ancestral lineages was 1.30 (95% confidence interval [CI]: 1.11–1.49). Compared with Delta, the risk for Omicron patients was .72 (95% CI: .59–.88) and compared with ancestral lineages was .94 (.78–1.1). Among Omicron and Delta infections, patients with history of vaccination or prior SARS-CoV-2 infection had half the risk of severe disease or death (adjusted hazard ratio: .40; 95% CI: .30–.54), but no significant outcome difference by variant.
Conclusions
Although risk of severe disease or death for unvaccinated inpatients with Omicron was lower than with Delta, it was similar to ancestral lineages. Severe outcomes were less common in vaccinated inpatients, with no difference between Delta and Omicron infections.
Keywords: SARS-CoV-2, COVID-19, Delta variant, Omicron variant, inpatient outcomes
Unvaccinated adults hospitalized with COVID-19 with Delta infections had increased risk of requiring advanced respiratory support or dying within 28 days, compared with Omicron and ancestral lineages, which were similar. Vaccination lowered severe disease risk, with no Omicron versus Delta difference.
In December 2020, the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.1.7, now known as the Alpha variant, was identified as the first variant of concern [1]. Emerging SARS-CoV-2 variants continue to cause waves of infections and hospitalizations. Differences in the transmissibility and clinical severity of SARS-CoV-2 variants have the potential to greatly impact public health as large segments of the population simultaneously become infected and require hospitalization. By mid-January 2022, daily coronavirus disease 2019 (COVID-19) hospitalizations in the United States reached a new peak of 150 000 [2].
Omicron and Delta variants are more transmissible than earlier circulating SARS-CoV-2 lineages [3,4]. Compared with prior lineages, Alpha [5,6] and Delta [7–11] variants are associated with greater hospitalization risk and, for Delta, worse inpatient outcomes, but these findings have been limited by sample size and confounders [7,8,12]. Recent studies report decreased risk of hospitalization, severe illness, and death for Omicron compared with Delta variants [11,13–17], but limited data are available to describe the clinical trajectory of patients once hospitalized in context with previously circulating ancestral lineages [14,18].
Changing demographics, comorbidities, vaccination status, and therapeutics confound interpretation of the impact of SARS-CoV-2 lineage on COVID-19 severity. There remains a need to understand the clinical demands and outcomes of patients hospitalized with current variants. Therefore, we used a clinical registry to observe the impact of SARS-CoV-2 variants on clinical trajectory and outcome.
METHODS
Ethical Considerations and Data Availability
Research was conducted under Johns Hopkins Institutional Review Board protocol IRB00300364 with a waiver of consent. SARS-CoV-2 genomes were uploaded to the Global Initiative on Sharing All Influenza Data (GISAID) [19].
Setting and Data Source
The data source used for this study is JH-CROWN: The COVID Precision Medicine Analytics Platform Registry, a registry of electronic medical record (EMR) data from Johns Hopkins Medicine (JHM), which includes 5 acute-care hospitals in Maryland and the District of Columbia [20]. The EMR includes SARS-CoV-2 test results and vaccination records from the Chesapeake Regional Information System for our Patients (CRISP) [21] and other registries. Baseline comorbidities, including those to determine organ transplantation or immunocompromised status and Elixhauser score, were determined from International Classification of Diseases, Tenth Revision (ICD-10), codes [22,23]; conditions recorded in prehospitalization encounters or with start dates less than 48 hours after hospitalization were considered.
Cohort Composition and Outcome Determination
Cohort criteria aimed to include patients requiring inpatient care for COVID-19 and to exclude inpatients with insufficiently severe SARS-CoV-2 infections to independently warrant hospitalization. Inpatients with SARS-CoV-2 infection were identified based on positive JHM laboratory test results, record of positive SARS-CoV-2 test results from outside JHM recorded via CRISP, and identification by local infection control. Hospitalizations more than 14 days after diagnosis, more than 24 hours prior to diagnosis, and subsequent hospitalizations after an index hospitalization were excluded. Symptomatic COVID-19 requiring inpatient care was defined by a pulse oximetry recording of less than 95%, temperature of 38°C or greater, heart rate greater than 110 beats/minute, respiratory rate greater than 24 breaths/minutes, or use of supplemental oxygen within the first 24 hours of admission. Inpatients transferred from outside hospitals or admitted to surgical or psychiatric services were excluded.
Patients who received a dose of Ad26.COV2.S (Johnson & Johnson [Janssen]) more than 4 weeks or a second dose of mRNA-1273 (Moderna) or BNT162b2 (Pfizer-BioNTech) more than 2 weeks prior to hospitalization were classified as vaccinated; additional vaccine doses were not considered due to the small sample size. Prior SARS-CoV-2 infection was defined by a positive SARS-CoV-2 test more than 60 days or ICD-10 code more than 30 days prior to hospitalization indicating a positive SARS-CoV-2 test or COVID-19. Unvaccinated patients with a history of prior SARS-CoV-2 infection were grouped with vaccinated patients for stratified analyses based on reports showing similar protective effects [24].
Severe COVID-19 was defined as respiratory support with high-flow nasal cannula, noninvasive positive-pressure ventilation, or mechanical ventilation [25]. The primary outcome was severe disease or death within 28 days of hospital presentation, chosen because severe COVID-19 onset occurs within this time frame for nearly all patients who develop severe disease [20]. Patients who were discharged alive and without record of death were assumed to survive through day 28 without severe disease.
SARS-CoV-2 Whole-Genome Sequencing
Whole-genome sequencing (WGS) surveillance was performed on remnant positive SARS-CoV-2 clinical samples, chosen irrespective of hospitalization status with priority for samples with cycle threshold values less than 20. RNA was extracted using Chemagic 360 (Perkin Elmer). Whole-genome sequencing was performed on the Oxford Nanopore GridION using the V3 primer ARTIC SARS-CoV-2 sequencing protocol or NEBNext ARTIC SARS-CoV-2 Companion Kit (VarSkip Short SARS-CoV-2 #E7660-L). Each WGS run pooled 94 samples plus a negative and positive control [26]. Alignment and variant calling were performed with the artic-ncov2019 medaka protocol [27]. Questionable mutations were visually inspected. Clades were determined using Nextclade [28] and lineages with Pangolin COVID-19 Lineage Assigner [29]. Sequencing results with insufficient depth and coverage for clade and lineage assignment were excluded.
Variant Inference
The rapid spread of novel variants after local introduction defines periods when a single viral clade dominates community transmission. Using our health system's WGS surveillance, we determined the rolling 2-week distribution of viral lineages classified as follows: Alpha, Delta, Omicron, ancestral lineages (emerging before Alpha), and other [30]. When the rolling 2-week average identified 95% or greater of WGS results as a single variant, patients without WGS results who were diagnosed on that day were inferred to have dominant lineage infections.
Statistical Methods
Propensity score weighting was used to infer the causal effect of variant on outcomes. This approach infers the outcomes that might be observed if the entire cohort were infected with 1 variant (eg, Delta) as compared with another (eg, Omicron), or the average treatment effect of a particular variant. Only pre-infection baseline clinical features were considered as covariates in regression and propensity models to avoid adjustment for intermediate variables in the causal pathway from the variant to outcomes that would bias estimates of the variants' effects. Admission clinical characteristics known to be severe COVID-19 risk factors, including vital signs, ratio of oxygen saturation by pulse oximetry to the fraction of inspired oxygen (SpO2:FiO2), basic laboratory values, and inflammatory markers, were reported by variant [23].
Propensity for a particular variant was assessed using gradient boosted logistic regression using the R twang package [31]. Weighted regression models were fit using the survey package [32]. Propensity models included age, sex, body mass index (BMI), Elixhauser score [33], and history of organ transplantation or immunocompromised status. Age, sex, BMI, and a composite comorbidity score were chosen for model inclusion due to their known association with illness severity [20]. Organ transplantation or immunocompromised status was chosen a priori for model inclusion as it is not included in the Elixhauser score [33] as well as observations of poor outcomes and variable response to vaccination in this population [34,35].
Log binomial regression assessed the average treatment effect on the risk difference and relative risk scales. Models estimated remdesivir use, steroid use, and hypoxia at presentation, reported by SpO2:FiO2. The probability of surviving and remaining free of severe disease was assessed using the Kaplan–Meier estimator. Convergence, positivity, and balance were assessed in propensity score analyses.
Sensitivity analysis restricted to patients with confirmed variants was performed using age, sex, and Elixhauser score to determine propensity scores. The association of vaccination or prior SARS-CoV-2 infection with severe disease was limited to patients with Omicron or Delta infections and assessed using a Cox proportional hazard model adjusted for baseline clinical features, variant, and the interaction of variant and history of vaccination or prior SARS-CoV-2 infection.
RESULTS
Patient Population
From 1 September 2020 to 7 May 2022, 7586 adults were hospitalized within 2 weeks of COVID-19 diagnosis; 271 patients transferred from other hospitals and 236 admitted to surgical or psychiatric services were excluded (Supplementary Figure 1). Among 7079 remaining patients, 6095 (86%) with vital-sign abnormalities of tachycardia (n = 2249, 37%), tachypnea (n = 3028, 50%), fever (n = 2126, 35%), hypoxia (n = 4815, 79%), or supplemental oxygen requirement (n = 3978, 65%) were considered for inclusion.
Whole-Genome Sequencing Results and Variant Inference
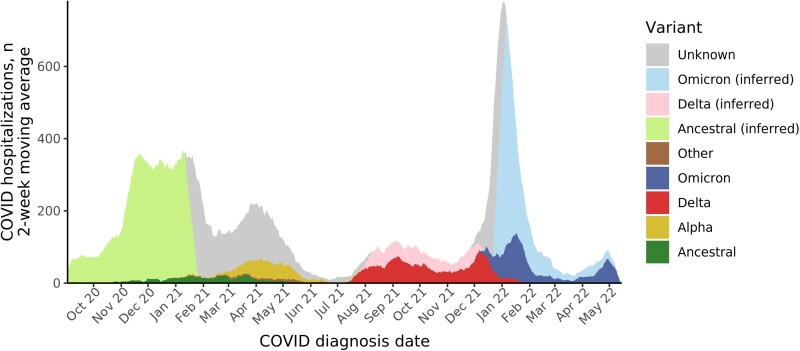
For 386 of 613 (63%) study days, a single lineage accounted for 95% or more of the rolling 2-week average (Figure 1 and Supplementary Figure 2), including ancestral lineages (139 days), Delta (125 days), and Omicron (122 days). Among 6260 patients meeting inclusion criteria, WGS results were available for 1311 (22%) inpatients; variant was inferred for an additional 3288 (54%) inpatients. Confirmed or inferred variant data were available for 3369 of 4616 (73%) patients without a prior history of SARS-CoV-2 vaccination or infection and 1147 of 1369 (84%) vaccinated or previously infected patients. Inferred variant matched WGS data for 789 of 813 (97%) patients with results during a period of single lineage dominance. During Omicron community dominance, 355 of 374 (95%) patients with available WGS results had the Omicron variant, including 84 of 94 (89%) patients with severe disease. Omicron subvariants included BA.1 (n = 293), BA.2 (n = 69), and BA.2.12.1 (n = 27).
Figure 1.
SARS-CoV-2 variants among hospitalized patients: 2-week moving average by date of COVID-19 diagnosis and variant. COVID-19–related hospitalizations with available confirmed SARS-CoV-2 variant results determined by WGS are shaded in darker colors. WGS surveillance of positive SARS-CoV-2 samples within Johns Hopkins Medicine was used to determine a community prevalence of SARS-CoV-2 variants (Supplementary Figure 2). During time periods when the 2-week moving distribution of variants reached 95%, variant was inferred for hospitalized patients who did not have WGS results. During periods when no single variant reached 95%, no inference of variant for patient without WGS was attempted. Abbreviations: COVID-19/COVID, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WGS, whole-genome sequencing.
Clinical and Demographic Features
Among 3369 patients without prior SARS-CoV-2 vaccination or infection, the median age was 62 years (interquartile range [IQR]: 49–75 years) (Table 1) and 1687 (50%) were male. Vaccinated or previously infected patients were older (median age: 69 years; IQR: 57–80 years; P < .001) and had more comorbidities (median Elixhauser score: 6 [IQR: 4–9] compared to 5 [IQR: 3–7]) (Table 2). Among patients without a prior history of SARS-CoV-2 vaccination or infection, those infected with ancestral lineages were older than other groups (median age: 65 years [IQR: 53–78] compared to 58 years [IQR: 43–70]). The proportion of patients who were Black was higher in Alpha (64%), Delta (42%), and Omicron (45%) variant groups than in ancestral lineages (31%).
Table 1.
Demographics and Clinical Characteristics by Variant Among 3369 Unvaccinated Patients Hospitalized With COVID-19 Without a Prior History of SARS-CoV-2 Infection
| Characteristics | All (N = 3369) | Ancestral (n = 2039) | Alpha (n = 201) | Delta (n = 546) | Omicron (n = 549) | Other (n = 34) |
|---|---|---|---|---|---|---|
| Age, years | 62.0 (49.0–75.0) | 65.0 (53.0–78.0) | 59.0 (47.0–69.0) | 55.0 (41.0–66.0) | 61.0 (45.0–73.0) | 61.0 (50.2–66.2) |
| Sex | ||||||
| ȃFemale | 1682 (49.9%) | 1009 (49.5%) | 104 (51.7%) | 272 (49.8%) | 279 (50.8%) | 18 (52.9%) |
| ȃMale | 1687 (50.1%) | 1030 (50.5%) | 97 (48.3%) | 274 (50.2%) | 270 (49.2%) | 16 (47.1%) |
| BMI, kg/m2 | 28.5 (24.3–34.1) | 28.5 (24.5–33.9) | 29.8 (25.7–35.6) | 29.1 (24.6–35.6) | 26.8 (23.0–32.7) | 31.8 (26.4–37.4) |
| Race/ethnicity | ||||||
| ȃAsian | 155 (4.6%) | 123 (6.0%) | 6 (3.0%) | 17 (3.1%) | 8 (1.5%) | 1 (2.9%) |
| ȃBlack | 1253 (37.2%) | 637 (31.2%) | 128 (63.7%) | 229 (41.9%) | 245 (44.6%) | 14 (41.2%) |
| ȃHispanic | 374 (11.1%) | 276 (13.5%) | 7 (3.5%) | 45 (8.2%) | 45 (8.2%) | 1 (2.9%) |
| ȃNon-Hispanic White | 1430 (42.4%) | 908 (44.5%) | 52 (25.9%) | 231 (42.3%) | 224 (40.8%) | 15 (44.1%) |
| ȃOther or unknown | 157 (4.7%) | 95 (4.7%) | 8 (4.0%) | 24 (4.4%) | 27 (4.9%) | 3 (8.8%) |
| Nursing home admission | 217 (6.4%) | 189 (9.3%) | 2 (1.0%) | 7 (1.3%) | 19 (3.5%) | 0 (0.0%) |
| Elixhauser score | 5.0 (3.0–7.0) | 5.0 (3.0–7.0) | 5.0 (3.0–8.0) | 4.0 (2.0–6.0) | 5.0 (3.0–8.0) | 4.0 (3.0–8.5) |
| Immunocompromised/transplant | 287 (8.5%) | 159 (7.8%) | 34 (16.9%) | 46 (8.4%) | 46 (8.4%) | 2 (5.9%) |
| Diabetes | 1348 (40.0%) | 843 (41.3%) | 87 (43.3%) | 182 (33.3%) | 223 (40.6%) | 13 (38.2%) |
| Cardiovascular disease | 1611 (47.8%) | 1015 (49.8%) | 98 (48.8%) | 199 (36.4%) | 286 (52.1%) | 13 (38.2%) |
| Chronic pulmonary disease | 1104 (32.8%) | 678 (33.3%) | 74 (36.8%) | 155 (28.4%) | 185 (33.7%) | 12 (35.3%) |
| Malignancy | 947 (28.2%) | 617 (30.3%) | 63 (31.3%) | 112 (20.5%) | 143 (26.1%) | 12 (35.3%) |
| Pregnant | 25 (0.7%) | 12 (0.6%) | 2 (1.0%) | 7 (1.3%) | 4 (0.7%) | 0 (0.0%) |
| SpO2:FiO2ratio | 435.0 (275.0–459.0) | 430.0 (277.0–454.0) | 435.0 (287.0–459.0) | 430.0 (258.0–454.0) | 444.0 (293.0–464.0) | 442.0 (336.0–459.0) |
| Pulse | 99.0 (86.0–113.0) | 97.0 (84.0–111.0) | 102.0 (90.0–113.0) | 104.0 (88.0–116.0) | 101.0 (86.0–115.0) | 93.5 (85.0–107.2) |
| Respiratory rate, breaths/minute | 20.0 (18.0–27.0) | 21.0 (18.0–27.0) | 20.0 (18.0–26.0) | 21.0 (18.0–27.0) | 20.0 (18.0–26.0) | 21.0 (18.0–27.5) |
| Temperature, °C | 37.2 (36.8–37.9) | 37.2 (36.8–38.0) | 37.3 (36.8–38.2) | 37.1 (36.7–37.9) | 36.9 (36.6–37.5) | 37.5 (37.0–38.2) |
| White blood cell count | 6.7 (5.0–9.2) | 6.6 (5.0–9.1) | 6.3 (4.8–8.3) | 6.2 (4.7–8.6) | 7.8 (5.5–10.7) | 6.0 (3.9–6.8) |
| GFR, mL/minute per 1.73 m2 | 70.0 (45.0–94.0) | 67.0 (44.0–91.0) | 64.0 (37.0–92.0) | 82.0 (54.0–99.0) | 69.0 (46.8–99.0) | 83.0 (63.5–90.0) |
| CRP level, mg/L | 6.2 (2.6–11.6) | 6.2 (2.5–11.4) | 5.8 (3.0–11.5) | 7.4 (3.5–12.6) | 5.4 (1.8–11.4) | 3.5 (1.3–10.1) |
| IL-6 level, pg/mL | 28.5 (11.4–62.8) | 27.0 (11.3–59.0) | 17.8 (10.7–53.8) | 31.1 (11.3–93.0) | 77.2 (30.5–98.4) | 36.5 (26.4–11 026.9) |
Data are presented as median (IQR) or no. (%). Vital signs and laboratory results are the first observation within 36 hours of hospital admission. Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; GFR, glomerular filtration rate; IL-6, interleukin 6; IQR, interquartile range; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SpO2:FiO2, ratio of oxygen saturation by pulse oximetry to the fraction of inspired oxygen.
Table 2.
Demographics and Clinical Characteristics by Variant Among 1230 Patients Hospitalized With COVID-19 With a History of SARS-CoV-2 Infection or Vaccination
| Characteristics | All (N = 1230) | Ancestral (n = 26) | Alpha (n = 15) | Delta (n = 295) | Omicron (n = 891) | Other (n = 3) |
|---|---|---|---|---|---|---|
| Age, years | 69.0 (57.0–79.8) | 57.0 (50.8–69.8) | 59.0 (54.0–70.5) | 68.0 (59.0–80.0) | 69.0 (57.0–80.0) | 74.0 (65.5–82.5) |
| Sex | ||||||
| ȃFemale | 564 (45.9%) | 11 (42.3%) | 8 (53.3%) | 143 (48.5%) | 400 (44.9%) | 2 (66.7%) |
| ȃMale | 666 (54.1%) | 15 (57.7%) | 7 (46.7%) | 152 (51.5%) | 491 (55.1%) | 1 (33.3%) |
| BMI, kg/m2 | 27.1 (23.1–32.6) | 28.4 (25.3–33.5) | 26.5 (21.3–34.6) | 28.4 (24.4–33.8) | 26.7 (22.8–32.0) | 32.0 (29.2–33.5) |
| Race/ethnicity | ||||||
| ȃAsian | 54 (4.4%) | 3 (11.5%) | 1 (6.7%) | 15 (5.1%) | 35 (3.9%) | 0 (0.0%) |
| ȃBlack | 396 (32.2%) | 8 (30.8%) | 9 (60.0%) | 88 (29.8%) | 290 (32.5%) | 1 (33.3%) |
| ȃHispanic | 74 (6.0%) | 6 (23.1%) | 1 (6.7%) | 11 (3.7%) | 56 (6.3%) | 0 (0.0%) |
| ȃNon-Hispanic White | 658 (53.5%) | 8 (30.8%) | 4 (26.7%) | 170 (57.6%) | 474 (53.2%) | 2 (66.7%) |
| ȃOther or unknown | 48 (3.9%) | 1 (3.8%) | 0 (0.0%) | 11 (3.7%) | 36 (4.0%) | 0 (0.0%) |
| Vaccinated | 1181 (96.0%) | 3 (11.5%) | 9 (60.0%) | 291 (98.6%) | 876 (98.3%) | 2 (66.7%) |
| Prior SARS-CoV-2 infection | 83 (6.7%) | 23 (88.5%) | 6 (40.0%) | 10 (3.4%) | 43 (4.8%) | 1 (33.3%) |
| Nursing home admission | 83 (6.7%) | 2 (7.7%) | 0 (0.0%) | 12 (4.1%) | 69 (7.7%) | 0 (0.0%) |
| Elixhauser score | 6.0 (4.0–9.0) | 6.5 (5.0–10.5) | 8.0 (5.0–10.5) | 6.0 (4.0–9.0) | 6.0 (4.0–9.0) | 3.0 (2.0–6.5) |
| Immunocompromised/transplant | 238 (19.3%) | 6 (23.1%) | 5 (33.3%) | 67 (22.7%) | 159 (17.8%) | 1 (33.3%) |
| Diabetes | 564 (45.9%) | 13 (50.0%) | 7 (46.7%) | 147 (49.8%) | 397 (44.6%) | 0 (0.0%) |
| Cardiovascular disease | 793 (64.5%) | 13 (50.0%) | 9 (60.0%) | 187 (63.4%) | 582 (65.3%) | 2 (66.7%) |
| Chronic pulmonary disease | 502 (40.8%) | 16 (61.5%) | 7 (46.7%) | 124 (42.0%) | 354 (39.7%) | 1 (33.3%) |
| Malignancy | 512 (41.7%) | 11 (42.3%) | 10 (66.7%) | 137 (46.6%) | 353 (39.7%) | 1 (33.3%) |
| Pregnant | 4 (0.3%) | 0 (0.0%) | 0 (0.0%) | 1 (0.3%) | 3 (0.3%) | 0 (0.0%) |
| SpO2:FiO2 ratio | 449.0 (328.0–464.0) | 449.0 (354.0–464.0) | 435.0 (240.5–449.0) | 444.0 (339.0–459.0) | 449.0 (326.5–464.0) | 357.0 (353.0–396.0) |
| Pulse | 96.0 (82.0–111.0) | 97.5 (87.5–112.0) | 99.0 (94.0–109.5) | 93.0 (80.5–108.0) | 98.0 (82.0–112.0) | 102.0 (82.5–122.5) |
| Respiratory rate, breaths/minute | 20.0 (18.0–24.0) | 21.0 (18.0–25.8) | 20.0 (18.0–34.0) | 20.0 (18.0–24.0) | 20.0 (18.0–24.0) | 18.0 (17.0–21.0) |
| Temperature, °C | 36.9 (36.6–37.4) | 37.1 (36.6–38.2) | 36.9 (36.6–37.9) | 37.0 (36.6–37.7) | 36.8 (36.5–37.3) | 37.2 (36.5–37.9) |
| White blood cell count | 7.6 (5.5–10.7) | 6.0 (4.6–9.4) | 5.0 (3.3–6.3) | 7.0 (5.4–9.3) | 7.9 (5.7–11.2) | 4.2 (3.9–4.7) |
| GFR, mL/minute per 1.73 m2 | 63.0 (37.0–89.0) | 67.0 (39.5–96.0) | 59.0 (43.0–104.5) | 61.0 (37.0–87.0) | 64.0 (37.0–87.5) | 80.0 (69.5–91.0) |
| CRP level, mg/L | 4.2 (1.3–10.9) | 3.3 (1.7–13.9) | 5.9 (3.5–6.9) | 5.5 (2.0–11.9) | 3.6 (1.0–10.2) | 4.2 (4.0–4.5) |
| IL-6 level, pg/mL | 42.4 (22.5–77.4) | 51.4 (34.7–82.5) | 166.7 (90.6–242.8) | 44.7 (28.6–60.5) | 33.0 (20.0–86.1) | . (.-.) |
Data are presented as median (IQR) or no. (%). Vital signs and laboratory results are the first observation within 36 hours of hospital admission. Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; GFR, glomerular filtration rate; IL-6, interleukin 6; IQR, interquartile range; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SpO2:FiO2, ratio of oxygen saturation by pulse oximetry to the fraction of inspired oxygen.
The presenting SpO2:FiO2 was higher for unvaccinated patients with Omicron infections (median: 444; IQR: 293–464) compared with patients with Delta (median: 430; IQR: 258–454) and ancestral lineage (median: 430; IQR: 277–454) infections. Among 2554 (76%) unvaccinated patients with C-reactive protein (CRP) recorded less than 36 hours after admission, patients with Omicron infections had lower (5.4; IQR: 1.9–11.4) and Delta patients had higher (7.4; IQR: 3.5–12.6) median CRP compared with patients with ancestral lineage infections (6.2; IQR: 2.5–11.4).
Inpatient Outcomes, Patients Without a History of SARS-CoV-2 Vaccination or Prior Infection
Among patients without a history of SARS-CoV-2 vaccination or prior infection, 977 (29%) developed severe disease or death within 28 days of hospitalization (median time to event: 0.45 days; IQR: 0.07–2.35 days), including 583 (29%) ancestral, 66 (33%) Alpha, 179 (33%) Delta, 140 (26%) Omicron, and 9 (27%) other variant group infections (Table 3 and Supplementary Table 1). By 28 days of follow-up, almost all patients were discharged or developed severe disease; only 12 (<1%) patients remained hospitalized without having developed severe disease. There were 541 (16%) unvaccinated patients who required mechanical ventilation or died by day 28 (median time to event: 2.8 days; IQR: 0.5–7.6 days) (Supplementary Figure 3). By day 28, there were 182 (9%) ancestral, 9 (4%) Alpha, 46 (8%) Delta, 41 (7%) Omicron, and 3 (9%) other patients who died (median time to death: 9.9 days; IQR: 5.1–173 days).
Table 3.
Severe Disease or Death by Day 28 of Hospitalization Among Hospitalized Unvaccinated Patients Without a Prior History of SARS-CoV-2 Infection
| All Patients (N = 3369) | No Severe Disease or Death by Day 28 (n = 2392) | Severe Disease or Death by Day 28 (n = 977) | P | |
|---|---|---|---|---|
| Variant, inferred or confirmed | .067 | |||
| ȃAncestral | 2039 (60.5%) | 1456 (71.4%) | 583 (28.6%) | |
| ȃAlpha | 201 (6.0%) | 135 (67.2%) | 66 (32.8%) | |
| ȃDelta | 546 (16.2%) | 367 (67.2%) | 179 (32.8%) | |
| ȃOmicron | 549 (16.3%) | 409 (74.5%) | 140 (25.5%) | |
| ȃOther | 34 (1.0%) | 25 (73.5%) | 9 (26.5%) | |
| Variant, confirmed only | .236 | |||
| ȃAncestral | 141 (16.7%) | 101 (71.6%) | 40 (28.4%) | |
| ȃAlpha | 201 (23.8%) | 135 (67.2%) | 66 (32.8%) | |
| ȃDelta | 334 (39.5%) | 216 (64.7%) | 118 (35.3%) | |
| ȃOmicron | 136 (16.1%) | 101 (74.3%) | 35 (25.7%) | |
| ȃOther | 34 (4.0%) | 25 (73.5%) | 9 (26.5%) |
Data are presented as no. (%). Percentages in the first column are calculated by column; the remaining percentages are calculated by row. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
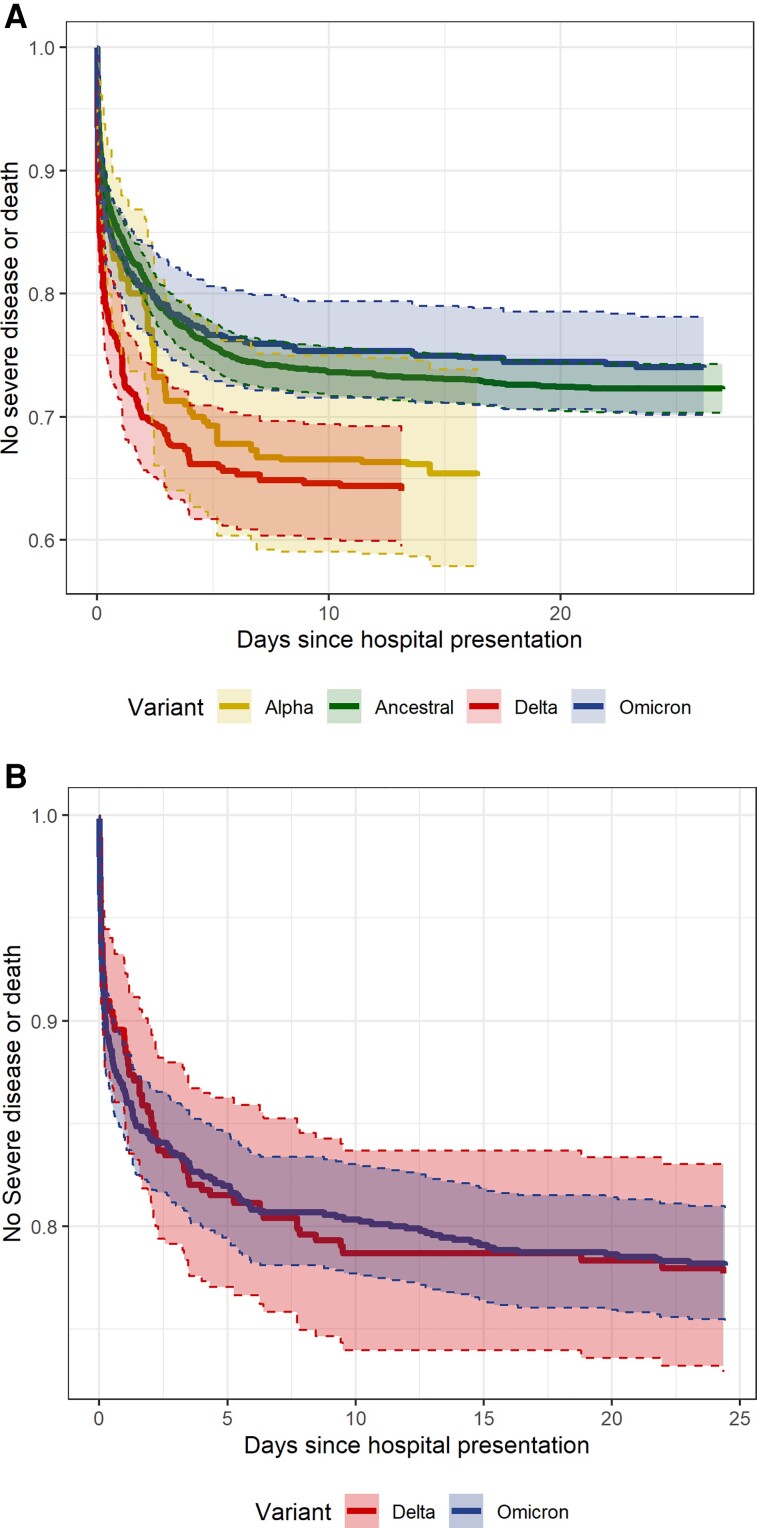
Average treatment effect models estimated a 28-day probability of severe disease or death by variant group of 0.28 for ancestral, 0.35 for Alpha, 0.36 for Delta, and 0.26 for Omicron infections (Table 4; model balance shown in Supplementary Table 2 and Supplementary Figure 4). Figure 2A plots the inverse probability weighted risk of severe disease or death by variant over the course of hospitalization. The relative risk of developing severe disease or death within 28 days for the Delta variant compared with ancestral lineages was 1.3 (95% confidence interval [CI]: 1.12–1.51). Compared with the Delta variant, the 28-day relative risk of severe disease or death for Omicron was .72 (95% CI: .59–.88) and compared with ancestral lineages was .94 (.78–1.11). Compared with ancestral lineages, the weighted mean presenting SpO2:FiO2 for the Delta variant was lower by 21.53 (95% CI: −35.26 to −7.8). Patients with Delta infections were more likely to receive remdesivir (relative risk: 1.12; 95% CI: 1.02–1.22) and patients with Omicron infectious were less likely to receive steroids (relative risk: .88; 95% CI: .82–.95) than patients with ancestral lineage infections. A sensitivity analysis of 2610 unvaccinated patients that excluded inference of Alpha, Delta, and Omicron variants showed similar estimates for the relative risk of severe disease or death, but with wider CIs (Supplementary Table 3 and Supplementary Figure 5).
Table 4.
Clinical Outcomes and Features by Variant in an Average Treatment Effect Model Adjusted for Risk Factors for Severe COVID-19 Among Unvaccinated Patients With No Prior History of SARS-CoV-2 Infection
| Ancestral | Alpha | Delta | Omicron | |
|---|---|---|---|---|
| Mean presenting SpO2:FiO2 | 363.54 (358.31–368.77) | 359.45 (339.73–379.17) | 342.01 (329.32–354.7) | 364.58 (353.48–375.68) |
| ȃMean difference | Ref | −4.09 (−24.49 to 16.31) | −21.53 (−35.26 to −7.8) | 1.04 (−11.23 to 13.31) |
| 28-Day risk of severe disease or death | ||||
| ȃEstimate | 0.28 (0.26–0.3) | 0.35 (0.27–0.43) | 0.36 (0.31–0.41) | 0.26 (0.22–0.3) |
| ȃRelative risk, ancestral as reference | Ref | 1.26 (0.95–1.56) | 1.3 (1.11–1.49) | 0.94 (0.78–1.1) |
| ȃRelative risk, Delta as reference | 0.77 (0.66–0.89) | 0.97 (0.74–1.26) | Ref | 0.72 (0.59–0.88) |
| ȃRisk difference, ancestral as reference | Ref | 0.07 (−0.01 to 0.15) | 0.08 (0.03–0.13) | −0.02 (−0.06 to 0.03) |
| Treatment with remdesivir | ||||
| ȃEstimate | 0.53 (0.51–0.55) | 0.61 (0.53–0.69) | 0.59 (0.54–0.64) | 0.51 (0.46–0.55) |
| ȃRelative risk, ancestral as reference | Ref | 1.16 (1–1.31) | 1.12 (1.02–1.22) | 0.96 (0.87–1.06) |
| Treatment with dexamethasone | ||||
| ȃEstimate | 0.71 (0.69–0.74) | 0.79 (0.73–0.85) | 0.76 (0.72–0.8) | 0.63 (0.58–0.67) |
| ȃRelative risk, ancestral as reference | Ref | 1.1 (1.01–1.2) | 1.06 (1–1.13) | 0.88 (0.81–0.94) |
The average treatment effect model uses inverse probability weighting to estimate clinical features and outcomes if the entire cohort was composed of a single variant. Weighting was performed using risk factors for severe COVID-19 including age, sex, body mass index, Elixhauser comorbidity score, and presence of immunosuppression or history of transplantation. Values in parentheses represent the 95% confidence interval of the estimates. Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Ref, reference; SpO2:FiO2, ratio of oxygen saturation by pulse oximetry to the fraction of inspired oxygen.
Figure 2.
Inverse probability weighted risk of severe disease or death by days since hospital presentation and variant for patients hospitalized with COVID-19 including (A) 3207 unvaccinated patients without a prior history of SARS-CoV-2 infection and (B) 1159 patients with a history of SARS-CoV-2 infection or vaccination. The shaded area surrounding the estimate expresses 95% CIs bound by dashed lines. Among 3369 unvaccinated patients without a prior history of SARS-CoV-2 infection with available inferred variant data, 34 with other variants and 128 with missing BMI were excluded from the model. Among 1230 patients with a prior history of SARS-CoV-2 infection or vaccination with available inferred variant data, 44 with variants other than Omicron or Delta and 27 with missing BMI were excluded from the model. Abbreviations: BMI, body mass index; CI, confidence interval; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Inpatient Outcomes, Patients With a History of Vaccination or Prior SARS-CoV-2 Infection
The 1230 inpatients with a history of vaccination or prior SARS-CoV-2 infection included 295 (35%) Delta, 891 (62%) Omicron, and 44 Alpha, Ancestral, or other lineage infections. There were 66 (23%) vaccinated patients with Delta and 194 (22%) vaccinated patients with Omicron who developed severe disease or who died within 28 days (Table 5 and Supplementary Table 4), with a median time to event of 0.5 (IQR: 0.08–3.24) days. By 28 days of follow-up, there remained 15 (1%) vaccinated persons hospitalized without having developed severe disease.
Table 5.
Severe Disease or Death by Day 28 of Hospitalization Among Hospitalized Patients With a Prior History of SARS-CoV-2 Vaccination or Infection
| All Patients (N = 1230) |
No Severe Disease or Death by Day 28 (n = 961) |
Severe Disease or Death by Day 28 (n = 269) | P | |
|---|---|---|---|---|
| Variant, inferred or confirmed | .686 | |||
| ȃAncestral | 26 (2.1%) | 22 (84.6%) | 4 (15.4%) | |
| ȃAlpha | 15 (1.2%) | 10 (66.7%) | 5 (33.3%) | |
| ȃDelta | 295 (24.0%) | 229 (77.6%) | 66 (22.4%) | |
| ȃOmicron | 891 (72.4%) | 697 (78.2%) | 194 (21.8%) | |
| ȃOther | 3 (0.2%) | 3 (100%) | 0 (0.0%) | |
| Variant, confirmed only | .378 | |||
| ȃAncestral | 6 (1.3%) | 6 (100%) | 0 (0.0%) | |
| ȃAlpha | 15 (3.2%) | 10 (66.7%) | 5 (33.3%) | |
| ȃDelta | 188 (40.4%) | 138 (73.4%) | 50 (26.6%) | |
| ȃOmicron | 253 (54.4%) | 197 (77.9%) | 56 (22.1%) | |
| ȃOther | 3 (0.6%) | 3 (100%) | 0 (0.0%) |
Data are presented as no. (%). Percentages in the first column are calculated by column; the remaining percentages are calculated by row. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Among patients with Omicron and Delta infections, inpatients who were vaccinated or had a history of SARS-CoV-2 infection had less than half the 28-day risk of severe disease or death compared with unvaccinated inpatients (adjusted hazard ratio: 0.4; IQR: 0.3–0.54) (Supplementary Table 5). The adjusted 28-day risk of severe disease or death was similar between patients with Delta and Omicron variant infections (Figure 2B and Supplementary Figure 6). There were 35 (12%) patients with Delta and 113 (13%) patients with Omicron with a history of SARS-CoV-2 vaccination or prior infection who required mechanical ventilation or who died within 28 days of hospitalization (median time to event: 1.92 days; IQR: 0.18–7.59 days).
DISCUSSION
Leveraging on-site SARS-CoV-2 genomic surveillance and real-time EMR linkage, we report clinically relevant differences in severe disease risk among patients hospitalized with symptomatic COVID-19 by SARS-CoV-2 variant. Compared with previously circulating lineages, unvaccinated patients with the Delta variant were more likely to develop severe illness or death when adjusted for known risk factors for severe COVID-19. Although unvaccinated patients hospitalized with the Omicron variant had a lower risk of severe disease compared with those with the Delta variant, the risk of severe disease or death was similar compared with SARS-CoV-2 lineages that circulated earlier in the pandemic. Among vaccinated patients, there was no difference in the risk of developing severe illness between Delta and Omicron variants once patients were hospitalized.
The finding that unvaccinated individuals hospitalized with Omicron infections have a similar risk of severe disease compared with cases prior to the emergence of variants of concern undercuts public perception that Omicron is a mild disease. Outpatients with Omicron infections in prior reports had a 0.2–0.5-fold risk of requiring hospitalization compared with those with Delta infections [11,14–16]. Once hospitalized, patients with Omicron in a South African cohort had a 0.3-fold risk of requiring oxygen therapy compared with patients with Delta [15]. Other comparisons of Omicron to Delta infections have shown a 0.34-fold risk of intensive care unit (ICU) admission and a 0.31-fold risk of death [36,37]. Adjustment for unascertained prior infection and vaccination in some population-based cohorts reduces the magnitude of intrinsic severity estimate differences between Omicron and Delta variants [37,38]. Comparison of disease severity between Omicron and Delta alone overlooks the increased severity of the Delta variant compared with prior lineages that were responsible for millions of deaths. Using stricter criteria for cohort entry to exclude asymptomatic SARS-CoV-2 infection, we found a 0.72-fold risk of severe disease for patients with Omicron compared with Delta, but no significant difference compared with ancestral lineages.
Patients who were vaccinated or who previously had SARS-CoV-2 infection were less than half as likely as unvaccinated patients to develop severe disease in the Delta and Omicron era, and there was no difference in this risk by variant. We found that vaccinated patients requiring hospitalization were older and had more comorbidities. Vaccines remain largely effective in preventing COVID-19–related mortality and population-based surveillance continues to show a more than 5-fold protection against hospitalization [39–41].
Comparison of severe disease risk between variants is critical as the evidence base for understanding disease severity, therapeutic efficacy, and health system burden is primarily based on pre–Delta variant observations. The perception that healthy persons hospitalized for COVID-19 are unlikely to require ICU-level care formed early in the pandemic underestimated their risk after the emergence of the Delta variant. As it is infeasible to re-evaluate every COVID-19 therapeutic each time a new variant emerges, data comparing severe disease risk by variant are necessary for clinicians to regauge the risks and benefits of COVID-19 therapeutics.
Study limitations include partial reliance on community prevalence to infer SARS-CoV-2 lineage and increased likelihood of unrecognized prior SARS-CoV-2 infection over the observation period. However, surveillance was performed locally with 97% concordance of inferred to confirmed variant. Model estimates from sensitivity analyses limited to confirmed variants were similar to primary analyses. Low-level co-circulation of the comparatively severe Delta variant during Omicron-dominant transmission may overrepresent Delta among hospitalized patients, especially the severely ill. Although inferred-confirmed variant concordance decreased to 95% during Omicron-dominant transmission, this misclassification biases against finding a severity difference between Omicron and Delta.
Reliance on EMR data likely undercounted the cohort prevalence of prior SARS-CoV-2 infection. A longitudinal serosurvey suggests that unreported SARS-CoV-2 is common [42], which may convey increasing unrecognized immunity that underestimates the intrinsic severity of recently emerging variants [43]. Unrecognized prior SARS-CoV-2 infection in our cohort may bias towards underestimating Omicron severity, likely greatest for Omicron to ancestral lineage comparisons given the shorter interval between Delta and Omicron waves. Our focus on hospitalized patients does not report differences in intrinsic severity between variants and benefits of vaccination conveyed by reducing hospitalization risk. However, our cohort provides a comparison of variant severity within the frame of reference of clinicians providing inpatient COVID-19 care. Differences in patient characteristics and care patterns may limit the generalizability of our findings. Defining symptomatic COVID-19 by vital sign abnormalities likely includes patients hospitalized for exacerbation of chronic medical conditions temporally related to a positive SARS-CoV-2 test and misses others with subtle symptoms. A limited sample size may have failed to detect a small, severe disease risk difference between the Omicron variant and ancestral lineages.
Study strengths include strict, objective inclusion criteria for symptomatic COVID-19, detailed clinical information from the EMR, extensive WGS results, and robust causal inference methods. Our findings show that, for vaccinated patients who are hospitalized with COVID-19, their risk of severe disease or death is low—reduced by half compared to unvaccinated patients—and equally low for both Omicron and Delta infections. Despite the comparatively lower risk of hospitalization for Omicron infections, for unvaccinated patients who require hospitalization there remains a risk of severe disease nearly equivalent to the pre-Delta waves of COVID-19.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Matthew L Robinson, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
C Paul Morris, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Joshua F Betz, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Yifan Zhang, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Robert Bollinger, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Natalie Wang, Krieger School of Arts & Sciences, Johns Hopkins University, Baltimore, Maryland, USA.
David R Thiemann, Division of Cardiology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Amary Fall, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Raghda E Eldesouki, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Julie M Norton, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
David C Gaston, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Michael Forman, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Chun Huai Luo, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Scott L Zeger, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Amita Gupta, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Brian T Garibaldi, Division of Pulmonary and Critical Care, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Heba H Mostafa, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Notes
Acknowledgments. The authors acknowledge the Johns Hopkins Precision Medicine Analytics Platform (PMAP) and Core for Clinical Research Data Acquisition (CCDA) for preparing the EMR data for analysis. This study was only possible with the efforts of the Johns Hopkins Clinical Microbiology Laboratory and clinicians and staff caring for patients with COVID-19.
Disclaimer. The contents are solely the responsibility of the authors and do not necessarily represent the official view of the sponsors.
Financial support. Support for this work was provided by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), to the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C), and the NIH RADx-UP initiative (R01DA045556-04S1), and with the National Institute on Drug Abuse to the HIV Prevention Trials Network Laboratory Center (UM1AI068613), National Heart, Lung, and Blood Institute, NIH, and National Institute of Biomedical Imaging and Bioengineering, NIH, to the NIH RADx-Tech program (3U54HL143541-02S2 and U54EB007958-12S1), Centers for Disease Control and Prevention (contract number 75D30121C11061), Maryland Department of Health, the Johns Hopkins University President’s Fund Research Response, the John Templeton Foundation, the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, John Hopkins inHealth, and the Johns Hopkins Precision Medicine Initiative. R. B. also reports support for this work from the Food and Drug Administration, paid to their institution. S. L. Z. also reports the following support for this work: U54CA260492-01 (S. Klein; paid to their institution); NIH/National Cancer Institute, Johns Hopkins Excellence in Pathogenesis and Immunity Center for SARS-CoV-2 (JH-EPICS) (30 September 2020–31 August 2022); 90077243 (Rosen) Scleroderma Research Foundation Trajectory Modeling (1 October 2017–30 June 2020).
References
- 1. Public Health England . Investigation of novel SARS-COV-2 variant, variant of concern 202012/01. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf. Accessed 30 January 2022.
- 2. US Department of Health and Human Services . Hospital utilization. Available at: https://protect-public.hhs.gov/pages/hospital-utilization. Accessed 13 January 2022.
- 3. Allen H, Vusirikala A, Flannagan J, et al. Household transmission of COVID-19 cases associated with SARS-CoV-2 Delta variant (B.1.617.2): national case-control study. Lancet Reg Health Eur 2021; 12:100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. UK Health Security Agency . SARS-CoV-2 variants of concern and variants under investigation in England, Technical briefing 32. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042688/RA_Technical_Briefing_32_DRAFT_17_December_2021_2021_12_17.pdf. Accessed 20 December 2021.
- 5. Nyberg T, Twohig KA, Harris RJ, et al. Risk of hospital admission for patients with SARS-CoV-2 variant B.1.1.7: cohort analysis. BMJ 2021; 373:n1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bager P, Wohlfahrt J, Fonager J, et al. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect Dis 2021; 21:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butt AA, Dargham SR, Chemaitelly H, et al. Severity of illness in persons infected with the SARS-CoV-2 Delta variant vs Beta variant in Qatar. JAMA Intern Med 2021; 182:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fisman DN, Tuite AR. Evaluation of the relative virulence of novel SARS-CoV-2 variants: a retrospective cohort study in Ontario, Canada. CMAJ 2021; 193:E1619–E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheikh A, McMenamin J, Taylor B, Robertson C;Public Health Scotland and the EAVE II Collaborators . SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021; 397:2461–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Twohig KA, Nyberg T, Zaidi A, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 Delta (B.1.617.2) compared with Alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis 2022; 22:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheikh A, Kerr S, Woolhouse M, et al. Severity of Omicron variant of concern and effectiveness of vaccine boosters against symptomatic disease in Scotland (EAVE II): a national cohort study with nested test-negative design. Lancet Infect Dis 2022; 22:959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis 2022; 75:e1128–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skarbinski J, Wood MS, Chervo TC, et al. Risk of severe clinical outcomes among persons with SARS-CoV-2 infection with differing levels of vaccination during widespread Omicron (B.1.1.529) and Delta (B.1.617.2) variant circulation in Northern California: a retrospective cohort study. Lancet Reg Health Am 2022; 12:100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes associated with Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat Med 2022; 28:1933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet 2022; 399:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sievers C, Zacher B, Ullrich A, et al. SARS-CoV-2 Omicron variants BA.1 and BA.2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Eurosurveillance 2022; 27:2200396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menni C, Valdes AM, Polidori L, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of Omicron and Delta variant dominance: a prospective observational study from the ZOE COVID study. Lancet 2022; 399:1618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Espenhain L, Funk T, Overvad M, et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Euro Surveill 2021; 26:2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Global Initiative on Sharing All Influenza Data (GISAID) . hCoV-19 tracking of variants. Available at: https://www.gisaid.org/hcov19-variants/. Accessed 1 February 2022.
- 20. Garibaldi BT, Fiksel J, Muschelli J, et al. Patient trajectories among persons hospitalized for COVID-19: a cohort study. Ann Intern Med 2021; 174:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chesapeake Regional Information System for our Patients . Clinical data. Available at: https://www.crisphealth.org/applications/clinical-data/. Accessed 2 February 2022.
- 22. Gasparini A. Comorbidity: an R package for computing comorbidity scores. J Open Source Softw 2018; 3:648. [Google Scholar]
- 23. Wongvibulsin S, Garibaldi BT, Antar AAR, et al. Development of severe COVID-19 adaptive risk predictor (SCARP), a calculator to predict severe disease or death in hospitalized patients with COVID-19. Ann Intern Med 2021; 174:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leon TM, Dorabawila V, Nelson L, et al. COVID-19 cases and hospitalizations by COVID-19 vaccination status and previous COVID-19 diagnosis—California and New York, May-November 2021. MMWR Morb Mortal Wkly Rep 2022; 71:125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marshall JC, Murthy S, Diaz J, et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis 2020; 20:e192–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quick J. nCoV-2019 sequencing protocol v3 (LoCost) V.3. Available at: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye. Accessed 3 January 2022.
- 27. Nick Loman WR, Rambaut A. nCoV-2019 novel coronavirus bioinformatics protocol. Available at: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html. Accessed 1 February 2022.
- 28. Aksamentov I, Roemer C, Hodcroft E, Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
- 29. Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Centers for Disease Control and Prevention . SARS-CoV-2 variant classifications and definitions. Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html. Accessed 14 January 2022.
- 31. Griffin BA, Morral AR, Burgette LF, et al. Toolkit for Weighting and Analysis of Nonequivalent Groups (TWANG). Available at: https://www.rand.org/statistics/twang.html. Accessed 30 January 2022.
- 32. Lumley T. Analysis of complex survey samples. J Stat Softw 2004; 9:1–19. [Google Scholar]
- 33. Elixhauser A, Steiner C, Harris DR, Coffey RM. Comorbidity measures for use with administrative data. Med Care 1998; 36:8–27. [DOI] [PubMed] [Google Scholar]
- 34. Shields AM, Tadros S, Al-Hakim A, et al. Impact of vaccination on hospitalization and mortality from COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. Front Immunol 2022; 13:984376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stock PG, Henrich TJ, Segev DL, Werbel WA. Interpreting and addressing suboptimal immune responses after COVID-19 vaccination in solid organ transplant recipients. J Clin Invest 2021; 131:e151178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med 2022; 28:1933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet 2022; 399:1303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davies MA, Kassanjee R, Rousseau P, et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection in the omicron-driven fourth wave compared with previous waves in the Western Cape Province, South Africa. Trop Med Int Health 2022; 27:564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collie S, Champion J, Moultrie H, Bekker LG, Gray G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med 2022; 386:494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021; 385:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Centers for Disease Control and Prevention . COVID data tracker. Available at: https://covid.cdc.gov/covid-data-tracker/#covidnet-hospitalizations-vaccination. Accessed 15 November 2022.
- 42. Clarke KEN, Jones JM, Deng Y, et al. Seroprevalence of infection-induced SARS-CoV-2 antibodies—United States, September 2021-February 2022. MMWR Morb Mortal Wkly Rep 2022; 71:606–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhattacharyya RP, Hanage WP. Challenges in inferring intrinsic severity of the SARS-CoV-2 Omicron variant. N Engl J Med 2022; 386:e14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.