Abstract
Background
We assessed how laboratories use and handle reporting of results of rapid diagnostics performed on positive blood culture broths, with a focus on antimicrobial resistance (AMR) markers.
Methods
A survey assembled by the Antibacterial Resistance Leadership Group Diagnostics Committee was circulated from December 2020 to May 2021. The survey was sent to local hospitals, shared on the ClinMicroNet and Division C listservs, and included in a College of American Pathologists proficiency testing survey.
Results
Ninety-six laboratories of various sizes across the United States (95%) and outside of the United States (5%) participated. Of the laboratories that had at least 1 rapid diagnostic in place (94%), significant heterogeneity in methods used and reporting practices was found across community (52%) and academic (40%) laboratories serving hospitals of various sizes. Respondents had implemented 1 to 6 different panels/platforms for a total of 31 permutations. Methods of reporting rapid organism identification and AMR results varied from listing all targets as “detected”/“not detected” (16–22%) without interpretive guidance, to interpreting results (23–42%), or providing therapeutic guidance comments to patient-facing healthcare teams (3–17%).
Conclusions
Current approaches to reporting molecular AMR test results from positive blood culture vary significantly across clinical laboratories. Providing interpretative comments with therapeutic guidance alongside results reported may assist clinicians who are not well-versed in genetic mechanisms of AMR. However, this is currently not being done in all clinical laboratories. Standardized strategies for AMR gene result reporting are needed.
Keywords: antimicrobial resistance test result reporting, multiplex molecular panels, blood cultures, AMR marker reporting
Approaches to reporting molecular antibacterial resistance test results from positive blood culture broths vary across clinical laboratories. Some current practices may undermine the value of the tests used. We provide recommendations for reporting identification and resistance from positive blood cultures.
With an estimated burden of >575 000 episodes of bloodstream infections (BSIs) associated with >79 000 North American deaths/year, BSIs are a public health concern [1]. Delays in effective therapy are associated with poor outcomes, including mortality [2]. Thus, rapid detection of pathogens with associated antimicrobial susceptibility testing (AST) is essential for guiding effective treatment, especially given increasing antimicrobial resistance (AMR).
Blood cultures (BCs) are standard tests for BSIs, with rapid pathogen identification and antimicrobial susceptibility methods often used (Table 1). Rapid microbial identification has been achieved using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) performed on growth from solid media after short incubation durations or directly from positive BCs [3–5]. For rapid AST, rapid disk diffusion and novel technologies to assess growth through digital microscopy or microbial weighing have been used [6–10]. Multiplex molecular panels performed directly from positive BCs are commonly used [11, 12].
Table 1.
Rapid Molecular and/or Phenotypic Methods Performed on Positive Blood Culture Broths
| Manufacturer/Platform/Method | Panel | Organism Groups Detecteda | No. of Organism Targets | Antimicrobial Resistance Marker Targets | ||||
|---|---|---|---|---|---|---|---|---|
| Extended-Spectrum β-Lactamase Family | Carbapenemase Family | Colistin Resistance | Methicillin Resistance | Vancomycin Resistance | ||||
| Genotypic methods | ||||||||
| BioFire (bioMérieux) Filmarray | BCID | Gram-positive bacteria, Gram-negative bacteria and yeast | 24 | … | bla KPC | … | mecA | vanA/B |
| … | BCID2 | Gram-positive bacteria, Gram-negative bacteria and yeast | 33 | bla CTX-M | bla KPC bla NDM bla VIM bla IMP bla OXA-48 | mcr-1 |
mecA/C
mecA/C and MREJb |
vanA/B |
| GenMark (Roche) ePlex | BCID-GP | Gram-positive bacteria | 20 | … | … | … | mecA/C | vanA/B |
| … | BCID-GN | Gram-negative bacteria | 21 | bla CTX-M | bla KPC bla NDM bla VIM bla IMP bla OXA-48/blaOXA-23 | … | … | … |
| … | BCID-FP | Yeast and Fusarium species | 15 | … | … | … | … | … |
| Luminex Verigene | BC-GP | Gram-positive bacteria | 12 | … | … | … | mecA | vanA/B |
| … | BC-GN | Gram-negative bacteria | 9 | bla CTX-M | bla KPC bla NDM bla VIM bla IMP bla OXA-48 | … | … | … |
| Great Basin diagnostic system | Staph ID/R PCR | Staphylococcus aureus, Staphylococcus lugdunensis, Staphylococcus species other than aureus and lugdunensis | 3 | … | … | … | mecA | … |
| Becton Dickinson BD MAX | StaphSR | S. aureus | 1 | … | … | … | mecA/C and MREJ | … |
| Cepheid GeneXpert | Xpert MRSA/SA BC | S. aureus | 1 | … | … | … | mecA and MREJ | … |
| Proteomic method | ||||||||
| Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry performed directly from positive blood culture broths (e.g., SepsiTyper) | NA | NAc | NA | … | … | … | … | … |
| Phenotypic methods | ||||||||
| Accelerate— fluorescent in situ hybridization based identification and phenotypic AST | PhenoTest BC | Gram-positive bacteria,Gram-negative bacteria and yeast | 16 | … | … | … | … | … |
| Rapid disk diffusion AST method performed directly from positive blood culture broth | NA | NA | NA | … | … | … | … | … |
Abbreviations: AST, antimicrobial susceptibility testing; BC, blood culture; BCID, blood culture identification; FP, fungal panel; GN, Gram-negative; GP, Gram-positive; ID, identification; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MRSA, methicillin-resistant Staphylococcus aureus; NA, not applicable; PCR, polymerase chain reaction; SA: Staphylococcus aureus.
Individual targets differ based on the panel and platform.
MREJ is a molecular target for the junction of the staphylococcal cassette chromosome mec element (SCCmec; a chromosomal cassette harboring mecA or mecC) and orfX in order to specifically link the mecA/C gene to Staphylococcus aureus in order to identify methicillin-resistant Staphylococcus aureus.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry is capable of detecting an array of Gram-positive bacteria, Gram-negative bacteria, acid-fast bacilli, and fungi. The reliability of identification is dependent on the database applied.
Molecular panels simultaneously detect common BSI pathogens and some consequential AMR genes; implementation in combination with antimicrobial stewardship (AS) interventions has demonstrated benefits to patient care [13–15]. Multiplex panels are increasingly viewed as a diagnostic standard, but there is little guidance as to how results should be reported to best guide patient care. This is increasingly important as the number of AMR gene targets increases and as many healthcare providers are not well-versed in AMR genes. The purpose of the REPORTing of Antimicrobial resistance from Blood Cultures (REPORT-ABC) survey was to assess how laboratories have implemented and handle reporting of rapid diagnostics performed on positive BCs, with a focus on AMR markers from molecular panels.
METHODS
The REPORT-ABC survey was assembled by the Antibacterial Resistance Leadership Group (ARLG) Diagnostics Committee (DC). Following circulation of a pilot survey to a small number of laboratories to provide feedback as to clarity of survey questions, the final online version (Supplementary Materials) was circulated from December 2020 to May 2021 through dispersal by the authors to local hospitals, ClinMicroNet and Division C listservs (managed by the American Society for Microbiology), and the College of American Pathologists (CAP) Gram Positive/Negative Blood, Molecular (GPBC/GNBC) proficiency testing (PT) survey. CAP accredits approximately 40% of laboratories in the United States and administers PT programs for laboratories globally to ensure that laboratories achieve accurate test results. There were 774 participants enrolled in the voluntary GPBC/GNBC CAP PT to which the REPORT-ABC survey was circulated, representing 40% of approximately 2000 laboratories participating in the Bacteriology D survey.
Analysis and Statistics
Survey responses were collated into a summary report; a formal response rate not calculatable due to diversity of dissemination methods. Frequencies of responses were calculated and associations between responses and laboratory characteristics assessed using the Fisher exact test. Each response category was analyzed for questions where multiple responses were allowed. The following laboratory characteristics were analyzed: size (<100, 101–500, 501–900, and >900 beds), type of laboratory (academic medical center, community-based hospital, and other/reference), leadership training (medical microbiology trained director, director who oversaw anatomic and/or clinical pathology, and other), and test panel/platform type (BioFire BCID/BCID2 panels [bioMérieux] vs other). Statistical analyses were performed using Stata software (StataCorp, 2021; Stata Statistical Software Release17).
RESULTS
Participating Laboratories
A total of 96 surveys were completed by laboratory managers/supervisors (36%), laboratory directors (30%), medical technologists (18%), and those with other leadership roles (16%; e.g., technical supervisor/managers, leads), representing approximately 5%–10% of laboratories performing blood cultures in the United States based on estimates of numbers of CAP-accredited laboratories and PT enrollment. Respondents represented community-based hospitals (51%), academic medical centers (40%), and reference (4%) laboratories across the United States (95%; 30% Midwest, 24% South, 23% West, 18% Northeast) and outside the United States (5%). Responding laboratories provided services to hospitals of varying sizes, with 101–500 beds (44%) being most common, followed by >900 beds (26%), 501–900 beds (20%), and ≤100 beds (8%).
Laboratory Directorship and AS Involvement
Most participating laboratories had a medical microbiology trained director (53%) or a director who oversaw anatomic and/or clinical pathology (33%) available for guidance; however, 6% did not have a director with an advanced degree. Nonlaboratory-based AS staff were commonly involved in deciding how results were reported (43%), made aware of availability and report configurations as the laboratory implemented them (27%), or involved throughout the process, from selecting the diagnostic platform to reporting results (20%).
Rapid Diagnostic Platforms
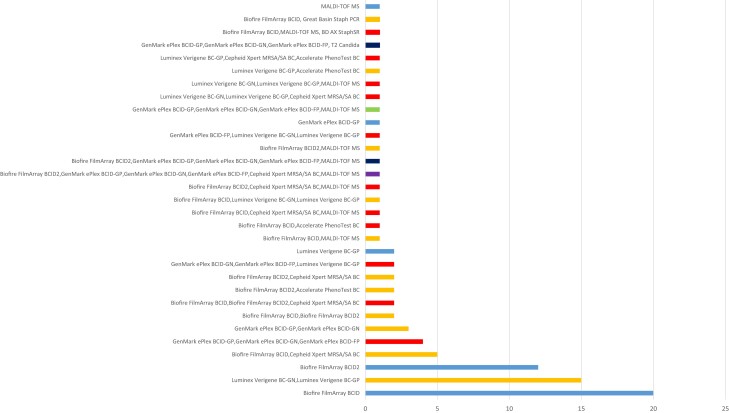
Only 6 (6%) laboratories indicated that they did not use a rapid method. A variety of rapid molecular and/or phenotypic platforms were performed by laboratories using at least 1 rapid diagnostic method (Table 1). Respondents applied 1–6 panels/platforms for up to 31 permutations of tests (Figure 1). Most used 1 panel/platform (40%), followed by 2 (38%), 3 (18%), 4 (2%), and 5 to 6 (1%). Figure 1 shows the most common tests/test combinations used. Combined platform approaches were most common using broad panels (e.g., Blood Culture Identification [BCID]/BCID2) combined with narrow Staphylococcus aureus/mecA specific panels (i.e., Cepheid Xpert methicillin-resistant S. aureus [MRSA]/SA BC, Great Basin Staph ID/R PCR [Great Basin Scientific], BD MAX StaphSR [BD]) and use of molecular panels with rapid MALDI-TOF MS or combination of a Gram-positive multiplex or BCID/BCID2 panel with the Accelerate Pheno system. Xpert, Accelerate Pheno, Great Basin Staph ID/R, BD MAX Staph SR, and T2 Biosystems platforms were only used in combined platform approaches. There were no differences between platforms used based on level of directorship, type of laboratory, or size of hospital.
Figure 1.
Breakdown of different combinations of rapid molecular and/or phenotypic panels/platforms performed for testing positive blood culture broths at participating laboratories. Length of bar reflects the number of laboratories performing the combinations of rapid molecular and/or phenotypic tests. Numbers of panels/platforms used by laboratories: light blue, 1; orange, 2; red, 3; green, 4; dark blue, 5; and purple, 6. MALDI-TOF MS directly from positive blood culture broths (e.g., SepsiTyper). Abbreviations:BC, blood culture; BCID, blood culture identification; FP, fungal panel; GN, Gram-negative; GP, Gram-positive; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MRSA, methicillin-resistant Staphylococcus aureus; PCR, polymerase chain reaction; SA: Staphylococcus aureus.
Rapid Diagnostic Workflows
Rapid diagnostic tests performed on positive BCs were mostly performed 24 hours/7 days a week (82%), with only a few laboratories limiting testing to day and evening shifts (8%), day shifts (7%), or workweek day shifts (2%). Rapid diagnostic tests were set up in most laboratories following Gram stain (GS) (78%), with 19%, all of which were BCID/BCID2 users (P < .001), processing positive BCs with GS. Interestingly, 2 BCID/BCID2 laboratories that set up molecular tests immediately with GS also used the Xpert assay. Rapid BC results were reported to patient-facing healthcare providers by calling GSs initially, with a second call to report rapid results once available (29%) or GS and rapid results were called together once both were available (27%). For the latter, 22 of 24 laboratories use BCID/BCID2 (P = .001), with the remaining 2 using GenMark or Luminex panels. The rest called GSs upfront, with 17% informing the clinical team of the availability of rapid results within a defined time frame after calling (e.g., 4 hours) and 20% not mentioning forthcoming rapid results or providing a second call upon test result availability. A higher proportion of community hospitals (41%) called GS and molecular results together, with 37% of academic centers calling the GS result initially (P < .001).
Reporting mecA and Staphylococcus Species
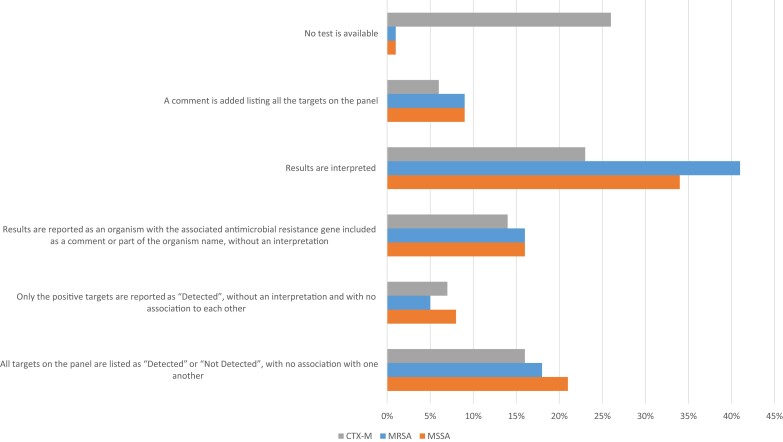
Two scenarios were provided: S. aureus and mecA both detected and S. aureus detected but mecA not (Figure 2; see Supplementary Material for survey questions). Overall, 42% would have interpreted scenario 1 as MRSA, whereas only 34% would have reported scenario 2 as methicillin-susceptible S. aureus (MSSA). Interpretation of the result helps associate the AMR marker to the organism to appropriately guide patient management. Interpretation of scenario 2 was more common among laboratories using diagnostic platforms other than BCID/BCID2 (51% vs 33%; P = .009). Academic laboratories, reference laboratories, and hospitals with larger bed sizes were more likely to interpret scenario 2 as MSSA than were community or smaller hospitals (type of laboratory, P = .011; hospital bed size, P = .002). A small proportion (20%) would have associated an AMR gene with organism identification without an interpretation (eg, S. aureus, mecA detected or S. aureus, mecA not detected). The remainder would have reported results as “detected” or “not detected” whether all targets were listed or only positive targets were listed as “detected,” with no association to one another. Overall, 12% indicated that they would have provided a comment listing all panel targets; 3%–6% indicated that they would provide a therapeutic comment in either scenario. Respondents provided free text comments that indicated recommendation of infectious disease consultation or pointing to treatment guidelines in a booklet, app, or active link in the report or initiation of active interventions by the AS team based on results. Addition of a therapeutic comment or other intervention was more common among academic and reference laboratories and laboratories that serve hospitals with more beds than community-based hospitals or hospitals with smaller bed numbers, respectively (hospital size, P < .002; type of laboratory, P = .001). For Staphylococcus species other than S. aureus, 40% reported mecA, with 18% limiting mecA reporting to select species (eg, Staphylococcus epidermidis, Staphylococcus lugdunensis). The latter mostly applied to Verigene BC-GP and BCID/BCID2 users.
Figure 2.
Reporting strategies used by laboratories (N = 90) applying rapid molecular diagnostics from positive blood culture broths. Three scenarios were provided and responses were tabulated for the following: Staphylococcus aureus and mecA were both detected (interpreted as MRSA; blue bars); S. aureus was detected but mecA was not (interpreted as MSSA; orange bars); and Escherichia coli and blaCTX-M were both detected (CTX-M) interpreted as extended-spectrum β-lactamase-producing E. coli; gray bars). See Supplementary Materials for details on the survey questions. Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus.
Reporting AMR Markers Among Gram-Negative Bacteria
Participants were provided a scenario where Escherichia coli and blaCTX-M were detected by a molecular assay and asked how it would be reported (Figure 2). Only 29% of laboratories would have interpreted the result in the report (e.g., extended-spectrum β-lactamase [ESBL]–producing E. coli, ESBL producer), with 17% associating the AMR gene with organism identification with no interpretation (eg, E. coli, blaCTX-M detected). Similar to MSSA/MRSA, the remaining laboratories would have reported results as detected or not detected whether all targets on the panel were listed (19%) or only positive targets were listed as detected (9%) with no association to one another. Interestingly, a larger proportion (17%) would have provided treatment guidance when blaCTX-M was detected compared with not detected (2%). Therapeutic guidance was more commonly provided by academic/reference laboratories than community laboratories (P = .031). Most therapeutic comments associated with detection of blaCTX-M that were provided indicated carbapenems as drugs of choice. Others indicated they would not report blaCTX-M but would notify an AS or clinical team member (14%).
In scenarios where E. coli is detected but blaCTX-M is not or Pseudomonas aeruginosa is detected but blaKPC is not, less than half (47%) would report the absence of an AMR marker. Furthermore, if multiple organisms were detected with a single AMR marker (e.g., E. coli, Klebsiella pneumoniae, and blaCTX-M), an array of responses were provided. The most common responses included listing results as detected or not detected whether all targets on the panel were listed (25%) or only positive targets listed as detected (16%), with no association to one another.
Discrepant Phenotypic and Genotypic Results
Participants were asked how discordant results between genotypic and phenotypic susceptibility results were addressed. Of 96 participants, 87 responded, with 128 responses received. Of these, 74% indicated that further testing would be performed to try to identify the reason for the discordance. Once the reason was identified, results would be reported and/or amended based on discrepant analysis. If discrepancy persisted, 25% favored leveraging the genotypic result (e.g., AMR marker is detected and predicted phenotype is forced resistant), with 18% favoring the phenotypic result as correct (e.g., AMR marker result is amended based on phenotypic AST result). Some participants elaborated that the approach was handled on a case-by-case basis or that the most conservative approach was taken (e.g., resistance reported whether detected by genotypic or phenotypic methods). A minority indicated that no further testing would be pursued and that both results would be reported with (3%) or without (3%) a comment highlighting the discrepancy. The remaining 5% would not perform any discordant analysis but would report the phenotype as resistant if an AMR marker was detected, despite testing phenotypically susceptible. A statistically significant association between the type of laboratory leadership and performance of discordant analysis was noted (P = .045). Specifically, laboratories with PhD/MD/DO medical microbiology trained leadership were more likely to perform discordant analyses (78%) prior to reporting results.
AS and Infection Prevention and Control Interventions
Laboratories were asked what type of AS interventions were in place for addressing rapid BC diagnostic results; 89 responded. Many (n = 43) chose more than 1 response, for a total of 165 responses. Common stewardship interventions included antibiotic treatment guidelines (45%), active intervention on all results (36%) or on select results (33%), inclusion of templated results and comments (27%), and clinical decision support tools built into the electronic medical record (10%). Various combinations of approaches were identified where the most common included antibiotic treatment guidelines with active intervention on all results (22%) or templated results and comments, antibiotic treatment guidelines, and active intervention on select results (9%). Additional comments indicated involvement of pharmacy and infectious diseases fellows/faculty in communicating results. Few laboratories (13%) indicated that AMR markers from rapid molecular diagnostic tests were incorporated into annual blood isolate–specific antibiograms. When asked about how results were communicated by the laboratory when AMR markers that would require patient isolation were detected, electronic notification to infection prevention and control (IPAC) (53%), phone call to IPAC (34%), and automatic communication of the need for isolation added to the electronic medical record (30%) were most common. In addition, 42% provided a response of “other,” with the most common response indicating a call to the primary care giver was made. Use of fax (2%) or no communication (8%) was uncommon.
DISCUSSION
As rapid diagnostics from positive BCs are increasingly commonly applied for clinical care and panels continue to grow in complexity, understanding best approaches to reporting is required. Based on REPORT-ABC survey results, combined with available literature, the ARLG-DC assembled best practice recommendations for implementing, testing, and reporting results of rapid molecular panels that detect AMR markers in positive BCs (Table 2). It is imperative that patient-facing clinicians can easily interpret the results to beneficially impact patient care. If reported results are unclear, not concise, or contain terminology unfamiliar to patient-facing healthcare providers, misinterpretation or lack of understanding of results will obviate benefits of rapid pathogen and AMR detection.
Table 2.
Antibacterial Resistance Leadership Group Diagnostics Committee Best Practice Recommendations for Reporting Identification and Antibacterial Resistance Markers From Positive Blood Culture Broths
| Topic | Best Practice Recommendation |
|---|---|
| Use of testing | Laboratories should adopt rapid diagnostic methods for microorganism identification and antibacterial resistance detection from positive blood culture broths |
| Timing of testing | Rapid methods for microorganism identification and antibacterial resistance detection from positive blood culture broths should be performed following Gram stain results 24 hours a day, 7 days a week |
| Reporting Gram stain results | Laboratories should report positive Gram stain results from positive blood culture broths as rapidly as possible (and not longer than 60 minutes after the blood culture signals positive) and not delay reporting Gram stain results if a rapid molecular test cannot be performed and resulted within 60 minutes of the blood culture signaling positive |
| Reporting antibacterial resistance marker results from rapid diagnostics | Laboratories should not report results from rapid molecular panels as “detected” or “not detected” for all targets on the panel, with no association to each other and no further interpretation provided Laboratories should work with their antimicrobial stewardship colleagues to develop reports that interpret findings and provide therapeutic guidance in a way that results are easily understood by patient-facing clinicians from diverse medical and surgical specialties and with varying levels of experience and that facilitates appropriate real-time changes in patient management |
| Handling of discordant phenotypic and genotypic susceptibility results | Laboratories should have a process in place to resolve discordant phenotypic and genotypic susceptibility results |
| Antimicrobial stewardship team membership | Microbiology leadership, preferably a medical microbiology trained laboratory director, should be included as a member of the institutional antimicrobial stewardship team to serve as the liaison from the laboratory and to aid in selection and implementation of novel diagnostics, such as rapid diagnostic methods from positive blood culture broths |
| Antimicrobial stewardship involvement | The institutional antimicrobial stewardship team should participate in selecting a rapid diagnostic method (or methods) for microorganism identification and antibacterial resistance detection from positive blood culture broths, developing associated reporting strategies, and establishing antimicrobial stewardship interventions specific to the diagnostic(s) |
Consistent with rapid diagnostics from positive BCs becoming standard of care, very few participating laboratories (6%) indicated that they did not use them. However, this may reflect selection bias, as those with rapid molecular panels may have been more likely to respond to the survey based on the method of dissemination, which likely enriched for laboratories that had implemented rapid diagnostics. Among laboratories with at least 1 rapid diagnostic in place (94%), heterogeneity in methods and reporting practices was apparent. The platforms applied ranged from 3% using Accelerate Pheno to 35% using BCID/BCID2. These differences may reflect, in part, timing of the arrival to the market and cost.
Rapid diagnostic tests performed on positive BC broths were mostly performed 24/7 (82%), with only a few laboratories limiting testing to certain shifts or days. When performing testing, most laboratories began testing following BC broths signaling positive after GS results were read, consistent with instructions for use (78%). Certain BioFire consumers processed testing with the GS. Although logical, this is technically off-label. Because false-positive results may be encountered due to DNA from nonviable organisms in BC broth media, it is important to correlate rapid identification with GS [16]. Furthermore, these panels may be unreliable in detecting polymicrobial infections and cannot detect off-target organisms, which may be observed on GS. Rapid BC diagnostic results were most commonly reported by calling GSs initially with a second call to report rapid results once available (29%) or GS and rapid results were called together once both results were available (26%). Initial positive BC results should be communicated as critical values to clinical teams as soon as possible or within 60 minutes of confirmation of an abnormal result (e.g., GS results indicating the presence of a microorganism) [17]. Laboratories that wait for molecular panel results prior to calling GSs may be delaying appropriate therapy, in turn, affecting patient safety. Laboratories should report the GS as soon as possible and not delay reporting GSs if the molecular test cannot be performed within a similar time frame.
Discordant results between genotypic and phenotypic susceptibility results may occur; these are more likely for Gram-negative AMR markers [18]. For Gram-positive organisms, accuracies of prediction are often higher (98%–100%) given that singular mechanisms of resistance account for most clinically significant resistance (e.g., mecA/mecC for MRSA, vanA/B for vancomycin resistant enterococci). Alternatively, Gram-negative bacterial resistance mechanisms are heterogeneous, resulting in lower susceptibility prediction accuracies [19–22]. The absence of blaCTX-M or blaKPC was not reported by 53% of laboratories, likely to prevent assumptions that expanded-spectrum cephalosporins or carbapenems would test susceptible due to their absence, respectively. Thus, some laboratories only reported AMR markers among Gram-negative organisms when present. When participating laboratories encountered discordant phenotype to genotype results, only 49% pursued additional testing to resolve discordances. Aside from good laboratory practice, it is now a CAP requirement (CAP checklist item MIC.21855) [23] to link AMR determinants and phenotypic susceptibility results to a specific organism in the final patient report. Additional discordant analysis should be pursued when genotype and phenotype do not align to rule out the possibility of analytical error. Interestingly, laboratories with medical microbiology trained leadership were more likely to perform discordant analyses than laboratories with other levels of directorship.
Methods for reporting of rapid AMR and organism results to clinical teams varied from listing all targets as detected/not detected to interpreting results and providing therapeutic comments to the patient-facing teams. Depending on the AMR gene/organism scenario provided in the survey, 16%–22% reported results of AMR markers and organisms as detected or not detected with no association to each other. A recent study evaluating physician BCID result interpretation and prescribing was performed via an electronic survey at a single institution. The laboratory reported BCID results as detected or not detected for all panel targets with no association to each other or further interpretation on the patient report. BCID results were misinterpreted 50% of the time, and only 60% of physicians stated that they adjusted therapy based on BCID results [24]. Thus, the approach of listing all targets as detected/not detected should be discouraged as it does not relay meaningful information that is impactful to patient management.
Ideally, laboratories should work with AS, infectious diseases, and IPAC teams to devise reporting structures that aid in interpretation of results that target patient-facing clinicians of various specialties and backgrounds. For example, 42% and 34% of respondents interpreted the presence and absence of mecA with S. aureus as MRSA and MSSA, respectively. Despite fewer laboratories interpreting the association of blaCTX-M and E. coli as an ESBL-producing E. coli, a larger proportion provided a therapeutic comment when blaCTX-M was detected compared with when MSSA or MRSA was detected. Therapeutic comments associated with detection of blaCTX-M can guide clinicians to carbapenems as the preferred treatment, consistent with recent Infectious Diseases Society of America treatment guidance for multidrug-resistant Gram-negative infections [25].
Active AS interventions, coupled with rapid molecular diagnostics from positive BCs, improve antibiotic use and clinical outcomes [14, 15]. Additionally, templated comments provided with BCID results improved antibiotic use but were not as effective as direct AS intervention in terms of antibiotic deescalation [13]. AS interventions can be resource-intensive and may not be available in all hospitals. Therefore, at a minimum, molecular AMR results should be interpreted for clinicians and additional tools, such as templated comments, treatment guidelines, and/or decision support resources, put into place to help guide therapeutic management. These general principles will likely apply to rapid testing directly from other specimen types (e.g., testing respiratory secretions and synovial and/or spinal fluid). Looking forward, future studies to compare different reporting practices and the impacts of these practices on patient care should be pursued.
Limitations of this study include the small number of laboratory participants. However, the survey results provide valuable data on reporting practices across various laboratory and hospital sizes. As this was a voluntary survey, there is the potential for selection bias due to the absence of a stratified approach that would have included a representative sample of different hospitals/locations to respond to the survey.
As rapid diagnostic panels from positive BC broths grow in complexity of targets, methods of reporting results become more critical. The REPORT-ABC survey demonstrated that there is significant heterogeneity in reporting structures. The ARLG-DC recommends that AMR and microorganism results be interpreted to a level at which patient-facing clinicians can apply results to guide care. AS and IPAC teams should work with microbiology laboratory leadership to ensure that there is appropriate implementation of rapid diagnostics, to develop interpretive comments for reports, and to ensure appropriate interventions are made.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Patricia J Simner, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Department of Medicine, Division of Infectious Diseases, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Jennifer Dien Bard, Department of Pathology and Laboratory Medicine, Children's Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Christopher Doern, Department of Pathology, Virginia Commonwealth University Health System, Richmond, Virginia, USA.
J Kristie Johnson, Department of Pathology, University of Maryland School of Medicine, Baltimore, Maryland, USA.
Lars Westblade, Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York, USA; Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine, New York, New York, USA.
Gayane Yenokyan, Johns Hopkins Biostatistics Center, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Robin Patel, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA; Division of Public Health, Infectious Diseases and Occupational Medicine, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA.
Kimberly E Hanson, Department of Medicine, Infectious Diseases Division, University of Utah School of Medicine, Salt Lake City, Utah, USA; Department of Pathology, Clinical Microbiology Division, University of Utah Associated Regional and University Pathologists (ARUP) Laboratories, Salt Lake City, Utah, USA.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). P. J. S. and K. E. H. report support for this work from the Antibacterial Resistance Leadership Group.
Financial support. The research presented here was supported in part by the National Institute of Allergy and Infectious Diseases of the NIH under award UM1AI104681. Statistical support was provided by the Johns Hopkins Institute for Clinical and Translational Research, which is funded in part by grant UL1 TR003098 from the National Center for Advancing Translational Sciences (NCATS), a component of the NIH, and the NIH Roadmap for Medical Research.
References
- 1. Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 2013; 19:501–9. [DOI] [PubMed] [Google Scholar]
- 2. Seymour CW, Gesten F, Prescott HC, et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med 2017; 376:2235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arroyo MA, Denys GA. Parallel evaluation of the MALDI Sepsityper and Verigene BC-GN assays for rapid identification of gram-negative bacilli from positive blood cultures. J Clin Microbiol 2017; 55:2708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faron ML, Buchan BW, Ledeboer NA. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for use with positive blood cultures: methodology, performance, and optimization. J Clin Microbiol 2017; 55:3328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kayin M, Mert B, Aydemir S, Ozenci V. Comparison of rapid BACpro® II, Sepsityper® kit and in-house preparation methods for direct identification of bacteria from blood cultures by MALDI-TOF MS with and without Sepsityper® module analysis. Eur J Clin Microbiol Infect Dis 2019; 38:2133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akerlund A, Jonasson E, Matuschek E, et al. EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: validation in 55 European laboratories. J Antimicrob Chemother 2020; 75:3230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandrasekaran S, Abbott A, Campeau S, et al. Direct-from-blood-culture disk diffusion to determine antimicrobial susceptibility of gram-negative bacteria: preliminary report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol 2018; 56:e01678–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datar R, Orenga S, Pogorelcnik R, Rochas O, Simner PJ, van Belkum A. Recent advances in rapid antimicrobial susceptibility testing. Clin Chem 2021; 68:91–8. [DOI] [PubMed] [Google Scholar]
- 9. Jonasson E, Matuschek E, Kahlmeter G. The EUCAST rapid disc diffusion method for antimicrobial susceptibility testing directly from positive blood culture bottles. J Antimicrob Chemother 2020; 75:968–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pancholi P, Carroll KC, Buchan BW, et al. Multicenter evaluation of the Accelerate PhenoTest BC Kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J Clin Microbiol 2018; 56:e01329–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peker N, Couto N, Sinha B, Rossen JW. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect 2018; 24:944–55. [DOI] [PubMed] [Google Scholar]
- 12. Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R. Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev 2018; 31:e00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Messacar K, Parker SK, Todd JK, Dominguez SR. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol 2017; 55:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. [DOI] [PubMed] [Google Scholar]
- 16. Butler-Wu S, Davis R.. 2020. Genotypic false detections from blood culture bottles: are we only seeing the tip of the iceberg? Washington, DC: American Society for Microbiology. Available at:https://asm.org/ASM/media/Policy-and-Advocacy/BlCx-contaminating-DNA-FINAL.pdf?ext=.pdf. Accessed January 2023.
- 17. Clinical and Laboratory Standards Institute . Principals and procedures for blood cultures; M47, 2nd ed. Approved guideline. Wayne, PA: CLSI, 2022. [Google Scholar]
- 18. Yee R, Dien Bard J, Simner PJ. The genotype-to-phenotype dilemma: how should laboratories approach discordant susceptibility results? J Clin Microbiol 2021; 59:e00138-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buchan BW, Ginocchio CC, Manii R, et al. 2013. Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med 10:e1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mestas J, Polanco CM, Felsenstein S, Dien Bard J. Performance of the Verigene gram-positive blood culture assay for direct detection of gram-positive organisms and resistance markers in a pediatric hospital. J Clin Microbiol 2014; 52:283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siu GK, Chen JH, Ng TK, et al. Performance evaluation of the Verigene gram-positive and gram-negative blood culture test for direct identification of bacteria and their resistance determinants from positive blood cultures in Hong Kong. PLoS One 2015; 10:e0139728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamma PD, Sharara SL, Pana ZD, et al. Molecular epidemiology of ceftriaxone non-susceptible Enterobacterales isolates in an academic medical center in the United States. Open Forum Infect Dis 2019; 6:ofz353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. College of American Pathologists. Microbiology checklist. Northfield, IL: CAP, 2021. [Google Scholar]
- 24. Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol 2017; 55:1496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America guidance on the treatment of extended-spectrum beta-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72:1109–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.