Abstract
Aims
The nuclear factor-κB (NF-κB) signalling pathway plays a critical role in the pathogenesis of multiple vascular diseases. However, in endothelial cells (ECs), the molecular mechanisms responsible for the negative regulation of the NF-κB pathway are poorly understood. In this study, we investigated a novel role for protein tyrosine phosphatase type IVA1 (PTP4A1) in NF-κB signalling in ECs.
Methods and results
In human tissues, human umbilical artery ECs, and mouse models for loss of function and gain of function of PTP4A1, we conducted histological analysis, immunostaining, laser-captured microdissection assay, lentiviral infection, small interfering RNA transfection, quantitative real-time PCR and reverse transcription-PCR, as well as luciferase reporter gene and chromatin immunoprecipitation assays. Short hairpin RNA-mediated knockdown of PTP4A1 and overexpression of PTP4A1 in ECs indicated that PTP4A1 is critical for inhibiting the expression of cell adhesion molecules (CAMs). PTP4A1 increased the transcriptional activity of upstream stimulatory factor 1 (USF1) by dephosphorylating its S309 residue and subsequently inducing the transcription of tumour necrosis factor-alpha-induced protein 3 (TNFAIP3/A20) and the inhibition of NF-κB activity. Studies on Ptp4a1 knockout or transgenic mice demonstrated that PTP4A1 potently regulates the interleukin 1β-induced expression of CAMs in vivo. In addition, we verified that PTP4A1 deficiency in apolipoprotein E knockout mice exacerbated high-fat high-cholesterol diet-induced atherogenesis with upregulated expression of CAMs.
Conclusion
Our data indicate that PTP4A1 is a novel negative regulator of vascular inflammation by inducing USF1/A20 axis-mediated NF-κB inactivation. Therefore, the expression and/or activation of PTP4A1 in ECs might be useful for the treatment of vascular inflammatory diseases.
Keywords: Protein phosphatase, Endothelial cells, Cell adhesion molecules, Vascular inflammation, Atherosclerosis
Graphical Abstract
Graphical Abstract.
1. Introduction
Vascular inflammation plays a central role in the pathophysiology of a number of diseases, including atherosclerosis and septic shock.1 Complementary adhesion molecules on the surface of endothelial cells (ECs) and leucocytes are regulated by inflammatory conditions and govern cell–cell adhesion.2 ECs are activated by pro-inflammatory molecules and express cell adhesion molecules (CAMs), including selectins and members of the immunoglobulin superfamily (IgSF), initiating leucocyte recruitment to sites of inflammation.3 IgSF CAMs, such as vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1), have been implicated in vascular inflammation.4–6
Nuclear factor-κB (NF-κB) is a major transcription factor that induces pro-inflammatory gene expression that plays a pivotal role in the pathophysiology of vascular inflammation.7 The NF-κB signalling cascade is prominently regulated by the IκB kinase (IKK) complex and tumour necrosis factor-alpha-induced protein 3 (TNFAIP3, also known as A20) via posttranslational modifications.8,9 Destroying the IKK complex or overexpressing A20 attenuates atherosclerosis progression in hyperlipidemic mouse models.10,11A20 is a primary response gene to inflammatory stimuli in human umbilical vein ECs and plays a negative role in NF-κB-mediated inflammation.12–14 Genome-wide association studies have demonstrated that single nucleotide polymorphisms (SNPs) at the A20 gene locus are risk factors for various inflammatory diseases.15–19 The A20 expression in ECs was shown to be protective for tumour necrosis factor-α (TNF-α)-mediated apoptosis and transplant arteriosclerosis and exhibited anti-atherogenic effects in both mice and humans.11,12,20
The promoter of the human A20 gene contains binding sites for NF-κB and upstream stimulatory factor 1 (USF1), and a transcriptional repressor, downstream regulatory element antagonist modulator.21,22 USF1 contains a basic helix–loop–helix leucine zipper (bZIP) domain and interacts with E-box consensus sites (CANNTG) on the promoters of genes involved in stress and immune responses through recruitment of chromatin remodelling enzymes and co-activators.23,24 Previous studies have demonstrated that USF1 is phosphorylated at several residues and is activated by various kinases.23 However, it is unknown whether phosphatase is involved in the regulation of the transcriptional activity of USF1 by dephosphorylation, especially in terms of its effects on A20.
The protein tyrosine phosphatase (PTP) 4A (PTP4A, also known as PRL) family are dual-specificity PTPs consisting of three members, namely, PTP4A1, PTP4A2, and PTP4A3. PTP4As possess a PTP active site (CX5R), a polybasic region and a consensus C-terminal CAAX sequence for prenylation.25–27Ptp4a1 is an immediate-early gene contributing to liver regeneration and cell growth and differentiation that regulates the bZIP transcription factor ATF-5 (also known as ATF-7).27,28 PTP4A1 plays a positive role in cell signalling, unlike other protein phosphatases that antagonize protein kinases.25,29 A recent study revealed the protective association of the local ancestry region harboring PTP4A1 gene with fatal coronary heart disease (CHD) among African Americans from the Atherosclerosis Risk in Communities Study using a genome-wide admixture mapping.30 In a study evaluating mouse strain-specific differentially regulated genes, Ptp4a1 has been identified as a gene with potential athero-protective function by comparison with genes expression in apolipoprotein E-deficient (ApoE−/−) mice fed a normal or high-fat diet. Although PTP4A1 has been suggested as a potential athero-protective gene in humans and mice; however, it remains unknown whether PTP4A1 directly participates in atherosclerosis.
Here, we identified PTP4A1 as a novel negative regulator of NF-κB-mediated vascular inflammation through activation of the USF1/A20 axis. Using in vitro and in vivo genetic approaches, we demonstrated that PTP4A1 attenuates the expression of CAMs in ECs by increasing the USF1 transcriptional activity on the A20 promoter and verified that PTP4A1 deficiency in ApoE−/− mice exacerbates the development of atherosclerosis. Therefore, controlling the expression or activity of PTP4A1 in ECs is a promising strategy for mitigating the progression of vascular inflammatory diseases.
2. Methods
2.1. Human tissue studies
This study was conformed to the principles outlined in the Declaration of Helsinki and was approved by the Institutional Review Board of Severance Hospital, Seoul, Korea (IRB No.4–2013-0688). All patients provided written informed consent. Marking information was eliminated from all samples before analysis for the protection of privacy. Aortic tissue samples were obtained from patients with a thoracic aortic aneurysm during the surgery of aortic graft replacement. Isolated aortic tissue samples were classified with normal or atheroma according to the absence or presence of atherosclerotic plaques by the pathologist, respectively.
2.2. Experimental animals
All researches were following the Guidelines on the Care Use of Laboratory Animals (National Institutes of Health Publication no. 85-23, revised 1996), and animal study protocols were approved by the Institutional Animal Care and Use Committee of the Korea Research Institute of Bioscience and Biotechnology (KRIBB-AEC-18203). Ptp4a1−/− mice were generated using the CRISPR/Cas9 system. The ApoE−/− mice on a C57BL/6 background were obtained from The Jackson Laboratory (Bar Harbor, Maine, USA). To generate double knockout ApoE−/−, Ptp4a1−/− mice, Ptp4a1−/− mice were crossed with ApoE−/− mice. To generate Ptp4a1 transgenic (Tg) mice, Tie2 promoter/enhancer was used in a C57BL/6 background produced by Macrogen (Daejeon, Korea). All mice were genotyped by PCR. Primers were shown in Supplementary material online, Table S1. All experiments were performed with generations F4–F6 using littermate. Six-week-old male mice were fed either a normal chow (NC) diet contained 13.2% fat, 24.6% protein, or high-fat high cholesterol (HFHC) diet contained 1.5% cholesterol, and 0.5% cholic acid (atherogenic cholate-containing diet; all w/w; D12336; Research Diets Inc., New Brunswick, NJ, USA). For some experiments, C57BL/6 background male mice at 8 weeks of age were intraperitoneally injected with phosphate-buffered saline (PBS), mouse interleukin-1β (IL-1β) (2 μg/20 g; 401ML; R&D systems, MN, U.S.A) for 8 h. After the study, all animals were anaesthetized by isoflurane inhalation (3%) plus 1 L/min O2 and euthanized by exsanguination.
2.3. Histological analysis
After hemodynamic measurements, mice were euthanized and hearts, aortas, and pulmonary vasculature were perfused with PBS through the left ventricle. Serum cholesterol levels were measured using an automatic blood chemical analyser (Hitachi, Tokyo, Japan). For morphological analysis, hearts were fixed with 10% (v/v) phosphate-buffered formalin overnight and embedded in OCT (3801480; Leica, Wetzlar, Germany) and frozen on dry ice. To analyse aortic sinus plaque lesions, cross-sections (10 μm) were prepared using Cryostat (CM 1860; Leica, Wetzlar, Germany). Each section was histochemically stained with Oil-Red-O (O0625; Sigma-Aldrich, St Louis, USA) and hematoxylin and eosin (H&E). After staining, photographs were captured under a light microscope (BX53F2; Olympus Corp, Tokyo, Japan). The whole aortas were dissected from the proximal ascending aorta to the bifurcation of the iliac artery, and adventitial fat was removed. For en face analysis, aortas were split longitudinally, pinned onto flat, black silicone plates, and fixed in 10% (v/v) phosphate-buffered formalin overnight. Fixed aortas were stained with Oil-Red-O for 4 h. After washing with PBS briefly, digitally photographed at a fixed magnification. Total aortic areas and lesion areas were calculated by AxioVision (Carl Zeiss, Jena, Germany).
2.4. Immunostaining
For en face confocal microscope imaging, isolated thoracic aortas fixed in 10% (v/v) phosphate-buffered formalin for overnight. After permeabilization with 0.25% (v/v) Triton-X-100 in PBS, thoracic aortas were washed, blocked for 1.5 h in 10% (v/v) chicken serum, and incubated with anti-VCAM-1, ICAM-1 (R&D Systems), and anti-PTP4A1 (Everest) Ab in 10% (v/v) chicken serum (1:100 dilution) for overnight at 4°C. After additional washing, aortas were incubated with Alexa Fluor-594 anti-goat IgG (Invitrogen) in PBS (1:100 dilution) for 2 h at room temperature, and with DAPI (Sigma-Aldrich) in PBS for 20 min. For the en face confocal microscopic ICAM-1 and VCAM-1 images with an endothelial marker, the piece of aortas was stained with Mouse VE-cadherin/CD144 Antibody (AF1002, R&D Systems) in 10% (v/v) chicken serum overnight at 4°C. After washing, the aortas were incubated with Alexa Fluor-488 anti-goat IgG (Invitrogen) in PBS for 2 h at room temperature. After washing, the aortas were incubated with Mouse ICAM-1/CD54 Biotinylated Antibody (BAF796, R&D Systems), Mouse VCAM-1/CD106 Biotinylated Antibody (BAF643, R&D Systems) or Normal Goat IgG Biotinylated Control (BAF108, R&D Systems) preincubated with Streptavidin, Alexa Fluor-594 conjugate (ab272189, Abcam), respectively. Fluorescence was imaged using a confocal microscope (ZEISS LSM 800; Leica, Wetzlar, Germany). For aortic sinus staining, sections were incubated with primary antibody overnight at 4°C. The primary antibodies used were goat anti-VCAM-1, ICAM-1 Ab (R&D Systems), and rat anti-MOMA-2 (BIO-RAD). As a negative control, tissues were stained with rat and goat mouse IgG isotype control antibodies (Vector Laboratories). Nuclei were stained with DAPI. Slides were viewed with a microscope (BX53F2; Olympus Corp, Tokyo, Japan). Stained analysed quantification for relative fluorescence intensity (RFI) using ZEN blues. Primary antibodies were shown in Supplementary material online, Table S2.
2.5. Cell culture
Human umbilical artery endothelial cells (HUAECs) (C-12202; Promo Cell, Heidelberg, Germany) were grown in M199 medium (SH30253.01; HyClone, Logan, UT) supplemented with 20% fetal bovine serum (FBS) (SH30084.03; HyClone, Australian), 1% antibiotic-antimycotic (15240-062; Gibco, Billings, MT), 3 ng/mL bFGF (01-106; Millipore, Billerica, MA), and 5 units/mL heparin (H3149; Sigma, St. Louis, USA). HUAECs were starved with culture media containing 1% FBS for 6 h before stimulation with human IL-1β (10 ng/mL; 201-LB; R&D systems) or human TNF-α (10 ng/mL; 210-TA; R&D systems). In some experiments, cells were starved with culture media containing 1% FBS for 6 h before pretreated with dimethyl sulfoxide (DMSO) or 10 μM pyrrolidine dithiocarbamate (PDTC), NF-κB inhibitor antioxidant (ab141406; Abcam, Cambridge, UK). DMSO or PDTC was incubated for 30 min before adding IL-1β (10 ng/mL, 3 h). Two hundred and ninety-three T cells (CRL-3216; ATCC, VA, USA) were maintained in DMEM (SH30243.01; HyClone, Logan, UT) and 10% FBS containing 1% antibiotic-antimycotic. Two hundred and ninety-three T cells were stimulated with IL-1β (10 ng/mL) after starvation with serum-free media for 6 h. Cells were cultured at 37°C under 5% CO2 and 95% relative humidity.
2.6. Immunoblot analysis and enzyme-linked immunosorbent assay
Cells were washed with PBS and lysed with RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 1% Triton-X-100) or NP-40 buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, pH 7.4) containing protease and phosphatase inhibitor cocktail (GenDEPOT, Huston, TX, USA). Cell lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then protein was electrotransferred onto an Immobilon-P Membrane, PVDF. Blocked membranes were then incubated with the indicated primary antibodies (1:1000 dilution) for 2 h at room temperature. The membranes were subjected to the horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000 dilution; Invitrogen, Carlsbad, CA, USA) reaction for 1 h at room temperature. The HRP was visualized using a chemiluminescent substrate. Primary antibodies were shown in Supplementary material online, Table S2. For sandwich enzyme-linked immunosorbent assay, Duoset Ab pairs detecting mouse monocyte chemoattractant protein-1 (MCP-1; DY479) and mouse TNF-α (DY410) were purchased from R&D Systems.
2.7. Quantitative real-time PCR (qRT-PCR) and reverse transcription-PCR (RT-PCR)
The tissues or cultured cells were homogenized in TRIzol reagent (15596018; Life Technologies, MD, USA) and cDNA was synthesized with the M-MLV Reverse Transcriptase kit (N1705, C1101, N2515, U1518; Promega, Madison, USA). qRT-PCR was performed using quantiMix SYBR kit (QS105; PKT Co., Seoul, Korea). The absolute number of gene copies was normalized using human GAPDH and mouse 18s standardized using a sample standard curve. RT-PCR was performed using 2X GoTaq Master Mix (M7123; Promega, Madison, USA) and primers. The level of human GAPDH and mouse 18s served as a normalization. Primers were shown in Supplementary material online, Table S3.
2.8. Cell adhesion assay
PTP4A1 alteration cell lines were assessed through a gelatin-coated six-well plate. HUAECs were seeded at 5 × 105 in culture media and cultured for about 48 h. When the monolayer of HUAECs confluency formed, monocyte THP-1 cells stained with Cell-Tracker Green CMFDA (Invitrogen, Carlsbad, CA, USA) were seeded on the HUVECs at the final density of 1 × 106 cells in culture media. The cells were incubated at 37°C for 24 h. Non-adhesive cells were washed three times in PBS. Attached THP-1 cells to HUAECs were visualized by fluorescence microscopy using an Olympus IX81-ZDC inverted fluorescence microscope, and the green fluorescence cells were measured and counted.
2.9. Luciferase reporter gene assay
Two hundred and ninety-three T cells or HUAECs were seeded in six-well plates (1.5–3 × 105 cells/well) using DMEM and 10% FBS or 20% FBS with supplements. The next day, the media changed by 1% FBS each media. To determine the effect of treatment PBS or pro-inflammatory cytokines, cells were transfected with 2 μg pGL3_ICAM-1 or pGL3_VCAM-1 or pGL4_A20 (−1080 to +520, −520 to +289, E-box mutant; wild-type A20 E-box sequence: GTC ACG TGA CTT T, mutant A20 E-box sequence: GTG TGA CGA GGA C) respectively, and 0.2 μg pRL-TK (Renilla TK) using 5 μL of the transfection reagent Lipofectamine 2000. After incubation for 16 h, cytokine was treated for indicated times, and cells were treated with 1× lysis buffer (100 μL/well) supplied in the Dual-Luciferase Reporter Assay kit (E1960; Promega). Each sample was moved into 96-well plates in a 30 μL solution. Firefly and Renilla luciferase activities were measured separately on a fluorescence spectrophotometer (GloMax 96; Promega) in triplicate according to the manufacturer’s instructions. The relative transcriptional activity was normalized by the corresponding control sample.
2.10. Chromatin immunoprecipitation (ChIP) assay
All cells (1–0.3 × 107) were processed using the EZ-Chip Kit (17-371; Millipore, Darmstadt, Germany) according to the manufacturer’s instructions. Briefly, genomic DNA was crosslinked with 1% formaldehyde and fragmented into 500 ± 100 bp fragments by sonicating for 10 s with 5 μm/wave 10 times. Soluble chromatin was incubated overnight with each antibody (human RNA polymerase 2 antibody, mouse IgG antibody, p-p65 antibody, HA antibody, Flag antibody). Finally, immunoprecipitated DNA fragments were amplified and quantified by RT-PCR and qRT-PCR using the following PCR primers specific to the A20 gene promoter (USF1 binding activity on E-box of A20) and ICAM-1 or VCAM-1 gene promoter. Primers were shown in Supplementary material online, Table S3.
2.11. Statistics
All data are expressed as the mean ± standard error of the mean (S.E.M.). Statistical tests included the two-tailed Student’s t-test, two-way analysis of variance (ANOVA), and the Mann–Whitney U statistical test were used where appropriate. Statistical tests are described in the figure legends for each experiment. P values less than 0.05 denoted statistical significance.
3. Results
3.1. PTP4A1 regulates the expression of ICAM-1 and VCAM-1 in response to pro-inflammatory cytokines in ECs
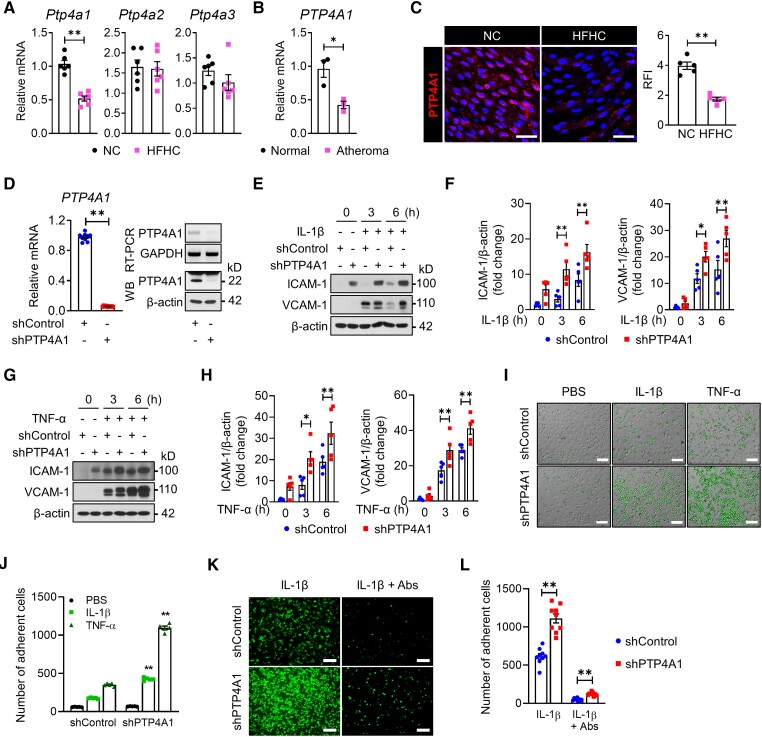
To explore whether the PTP4A family is associated with vascular inflammatory disease, we examined the mRNA levels of the Ptp4a1–3 genes in mice aorta with or without atheroma. The mRNA level of Ptp4a1, but not Ptp4a2 and Ptp4a3, was decreased by 50% in the aorta of ApoE−/− mice fed an HFHC diet compared with those fed an NC diet (Figure 1A). In addition, we identified that PTP4A1 expression was downregulated by 50% in human aortas with atheroma compared with those of normal controls (Figure 1B). To avoid the effects of immune cell recruitment in the atherosclerotic plaques when evaluating the Ptp4a1 expression levels, we conducted en face immunostaining to assess the PTP4A1 expression levels in the aortic endothelium. The PTP4A1 expression in ECs was downregulated in the artery wall in C57BL/6 mice fed an HFHC diet compared with mice fed an NC diet (Figure 1C).
Figure 1.
Downregulation of PTP4A1 increases the expression levels of ICAM-1 and VCAM-1 on endothelial cells. (A) The mRNA level of Ptp4a1–3 was measured via qRT-PCR analysis in the aorta of 8-week-old ApoE−/− mice fed an NC or HFHC diet for 2 weeks (n = 6). (B) The mRNA levels of PTP4A1 were assessed via qRT-PCR in the human artery wall with or without atheroma (n = 3). (C) Representative en face immunostaining confocal images for the PTP4A1 protein (red) on the endothelium of the descending aorta from 8-week-old C57BL/6 mice (n = 5 mice per group) fed an NC or HFHC diet for 2 weeks. The nuclei were stained with DAPI. Scale bars, 50 μm. RFI was compared between the groups (right panel). (D) Downregulation of PTP4A1 was evaluated via qRT-PCR, RT-PCR, and immunoblot analysis (n = 9). PTP4A1 protein levels in lysates of shControl- or shPTP4A1-treated HUAECs were determined via western blot analysis after immunoprecipitation by PTP4A1 antibody. β-actin in whole-cell lysates served as a loading control. (E–H) shControl- and shPTP4A1-treated HUAECs were stimulated with IL-1β (E and F) or TNF-α (G and H). The ICAM-1 and VCAM-1 expression levels in the cell lysates were determined via immunoblot analysis. Bar diagrams represent normalized protein levels (n = 5). (I and J) Representative images (I) and a quantification graph (J) of the monocyte adhesion assay in shControl- or shPTP4A1-treated HUAECs after treatment with PBS, IL-1β, or TNF-α for 3 h (n = 5). Scale bars, 25 μm. (K and L) Representative images (K) and a quantification graph (L) of the monocyte adhesion assay with or without the neutralization of ICAM-1 and VCAM-1 (n = 9). Scale bars, 25 μm. All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (Mann–Whitney U test for A–C; two-tailed Student’s t-test for D; two-way ANOVA for F, H, J, and L).
Next, we established stable HUAECs with knockdown of PTP4A1 by PTP4A1-specific short hairpin RNA (shRNA) to investigate the role of PTP4A1 in vascular inflammation (Figure 1D). After IL-1β or TNF-α treatment, the expression levels of ICAM-1 and VCAM-1 were significantly enhanced in PTP4A1-downregulated HUAECs compared with control HUAECs (Figure 1E–H). With the increase in the ICAM-1 and VCAM-1 expression levels, the number of adherent monocytes dramatically increased on stimulated HUAECs with PTP4A1 knockdown compared with control cells; this effect was blocked by the neutralization of ICAM-1 and VCAM-1 (Figure 1I–L). To further determine the protective role of PTP4A1 in vascular inflammation, we generated HUAECs stably overexpressing PTP4A1 and confirmed an increase in the mRNA and protein levels of PTP4A1 (see Supplementary material online, Figure S1A). In contrast to PTP4A1 knockdown in HUAECs, the PTP4A1 overexpression significantly decreased the ICAM-1 and VCAM-1 expression levels via treatment of HUAECs with IL-1β or TNF-α for 3 or 6 h (see Supplementary material online, Figure S1B–E) and reduced monocyte adhesion on HUAECs treated with IL-1β or TNF-α (see Supplementary material online, Figure S1F and G). These results indicate that PTP4A1 negatively regulates the expression of ICAM-1 and VCAM-1 in response to pro-inflammatory cytokines in ECs.
3.2. Deficiency of PTP4A1 exacerbates the expression of ICAM-1 and VCAM-1 in the vasculature of mice via Il-1β administration
To determine the pathophysiological role of PTP4A1 in vascular inflammation, we generated Ptp4a1−/− mice using CRISPR/Cas9 and verified the deletion of the Ptp4a1 gene via sequencing and PCR of genomic DNA (see Supplementary material online, Figure S2A). As PTP4A1 is ubiquitously expressed in tissues and cells, including immune cells, we verified whether PTP4A1 deficiency in mice has effects on hematopoietic and myeloid cells. A complete blood count of 8–10-week-old Ptp4a1+/+ and Ptp4a1−/− male mice revealed the counts for white blood cells and the percentages of neutrophils, lymphocytes, and monocytes were comparable between Ptp4a1+/+ and Ptp4a1−/− mice (see Supplementary material online, Figure S2B). The major indices for erythrocytes, including red blood cell count, hemoglobin content and hematocrit, and platelet numbers, were not changed by PTP4A1 deficiency in mice (see Supplementary material online, Figure S2C).
Next, we tested the role of PTP4A1 in IL-1β-stimulated vascular inflammation and found that the ICAM-1 and VCAM-1 expression levels were upregulated in both Ptp4a1+/+ and Ptp4a1−/− mice 8 h after IP injection of IL-1β. Notably, loss of PTP4A1 significantly enhanced the IL-1β-induced expression of these CAMs in the endothelial layer of the aorta, which were identified by en face confocal microscopic imaging (Figure 2A and B, and see Supplementary material online, Figure S2D) and by qRT-PCR (Figure 2C). Consistently, IL-1β stimulation induced the ICAM-1 and VCAM-1 expression levels in the pulmonary microvessels in both Ptp4a1+/+ and Ptp4a1−/− mice compared to PBS controls, and lack of PTP4A1 further increased the levels of ICAM-1 and VCAM-1 in the lungs of IL-1β injected mice (Figure 2D–F).
Figure 2.
PTP4A1 deficiency enhances IL-1β-mediated expression of ICAM-1 and VCAM-1 in the vasculature of mice. (A–F) Ptp4a1+/+ and Ptp4a1−/− mice (8 weeks old) were intraperitoneally injected with PBS or IL-1β (2 μg/20 g for 8 h). (A and B) Representative en face immunostaining confocal images (left) and quantification of the RFI (right) of ICAM-1 (A) and VCAM-1 (B) in the endothelium of the descending aorta from Ptp4a1+/+ and Ptp4a1−/− male mice (n = 5 mice per group). CD144 was stained as an endothelial cells marker. Scale bars, 50 μm. (C) qRT-PCR analysis of ICAM-1 (left) and VCAM-1 (right) mRNA levels in the aortas of Ptp4a1+/+ and Ptp4a1−/− male mice (n = 6 mice per group). (D and E) Representative immunostaining (D) and quantification of RFI (E) of ICAM-1 and VCAM-1 on the pulmonary vasculature (n = 5). Scale bars, 150 μm. (F) qRT-PCR analysis of ICAM-1 (left) and VCAM-1 (right) mRNA levels in the lungs of Ptp4a1+/+ and Ptp4a1−/− male mice (n = 5 mice per group). All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (two-way ANOVA for A–C, E, and F).
To assess the anti-inflammatory effect of PTP4A1 in vivo, we generated Ptp4a1 Tg mice expressing PTP4A1 under the control of the Tie2 promoter/enhancer and selected a transgenic line with a high PTP4A1 expression in the artery wall (see Supplementary material online, Figure S3A and B). PTP4A1 upregulation in the artery wall significantly inhibited ICAM-1 and VCAM-1 expression levels after IL-1β administration, as assessed by en face immunostaining (see Supplementary material online, Figure S3C and D). An IL-1β challenge upregulated ICAM-1 and VCAM-1 mRNA and the protein in the lungs of C57BL/6 mice; however, their upregulation was comparatively reduced in the lungs of Ptp4a1 Tg mice (see Supplementary material online, Figure S3E and F). These results indicate that PTP4A1 suppresses the expression levels of ICAM-1 and VCAM-1 in the vasculature of mice with acute inflammation.
3.3. PTP4A1 deficiency exacerbates the development of atherosclerosis in ApoE−/− mice
To investigate the role of PTP4A1 in the pathogenesis of atherosclerosis, ApoE−/− Ptp4a1−/− mice were produced and given an HFHC diet for 8 weeks. Compared with ApoE−/− mice, ApoE−/− Ptp4a1−/− mice had larger atherosclerotic plaques on their aortic arches and demonstrated a 25% increase in plaque area on the aorta after being fed an HFHC diet for 8 weeks (Figure 3A and B). Consistently, the plaque areas on the aortic sinuses were higher in ApoE−/− Ptp4a1−/− mice than in control mice (81 vs. 111, × 103 µm2) (Figure 3C). Immunohistochemical analysis revealed increased ICAM-1 and VCAM-1 expression levels and an increased number of infiltrated macrophages in the aortic sinuses of ApoE−/−Ptp4a1−/− mice compared with ApoE−/− mice after being fed an HFHC diet for 8 weeks (Figure 3D and see Supplementary material online, Figure S4A). After laser captured microdissection (LCM) of the plaque areas on the aortic sinuses of ApoE−/− and ApoE−/− Ptp4a1−/− mice fed an HFHC diet, we analysed the mRNA levels of Icam-1, Vcam-1, and F4/80 and confirmed the increased expression of these genes in ApoE−/− Ptp4a1−/− mice (Figure 3E and F). However, the levels of total cholesterol (T-chol), low-density lipoprotein-cholesterol (LDL-C), high-density lipoprotein-cholesterol (HDL-C), MCP-1, and TNF-α in plasma were similar between the groups before and after being fed an HFHC diet (Figure 3G and H). These results indicate that PTP4A1 plays a partial negative role in atherosclerosis development in mice via the regulation of ICAM-1 and VCAM-1 expression levels.
Figure 3.
Deletion of PTP4A1 accelerates the development of atherosclerosis in ApoE−/− mice. (A–H) ApoE−/− and ApoE−/− Ptp4a1−/− littermate mice were fed an HFHC diet for 8 weeks. (A) Representative images of atherosclerotic plaques on the aortic arch (n = 9–11). Scale bars, 0.2 cm. (B) Representative en face images of Oil-Red-O-stained aorta (left) and quantification of plaque area (%) on the whole aorta (right) (n = 9–11). (C) Representative Oil-Red-O-stained plaque images of aortic sinuses (left). Scale bars, 500 μm. Quantification of lesion areas of atherosclerotic plaques on the aortic sinus (right) (n = 8 mice per group). (D) Immunostaining of adhesion molecules (ICAM-1 and VCAM-1) and macrophages (MOMA-2) on the aortic sinus (n = 7–8 mice per group). Nuclei were stained with DAPI. Scale bars, 200 μm. Quantification of ICAM-1 (RFI), VCAM-1 (RFI), and macrophages (percentage of positive area) in atherosclerotic lesions. (E and F) Expression analysis of plaque-specific genes via LCM and qRT-PCR. Representative images (E) of H&E-stained aortic sinuses. Scale bars, 250 μm. Red-dotted lines indicate the plaques captured using LCM. The mRNA levels (F) of Icam-1, Vcam-1, F4/80, and Ptp4a1 on laser-captured plaques of aortic sinuses (n = 5 mice per group). (G) Plasma levels of T-chol, LDL-C, and HDL-C in ApoE−/− and ApoE−/− Ptp4a1−/− mice fed an HFHC diet for 0 and 8 weeks (n = 12 mice per group). (H) The levels of MCP-1 and TNF-α in plasma were quantified via ELISA in ApoE−/− and ApoE−/− Ptp4a1−/− mice fed an HFHC diet for 0 and 8 weeks (n = 7–12 per group). All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (Mann–Whitney U test for B, C, D, and F; two-way ANOVA for G and H).
As PTP4A1 is expressed in immune cells, we verified whether PTP4A1 deficiency affects macrophage-derived foam cell formation. Isolated peritoneal macrophages from Ptp4a1+/+ and Ptp4a1−/− mice injected with 3% thioglycollate medium were incubated with or without Dil-oxLDL for 24 h. The uptake of Dil-oxLDL on peritoneal macrophages was assessed by flow cytometry and fluorescence microscopy analysis and showed no difference between the two groups (see Supplementary material online, Figure S4B and C). In addition, the transcriptional levels of the scavenger receptor and inflammatory genes including CD36, Il-1β, and Tnf-α were not changed by PTP4A1 deficiency in macrophages; however, treatment of oxLDL increased the levels of mRNA encoding from CD36, Il-1β, and Tnf-α compared to media alone controls (see Supplementary material online, Figure S4D). Next, we verified whether PTP4A1 deficiency affects the polarization of macrophages. Bone marrow cells were collected from femurs and tibiae of Ptp4a1+/+ and Ptp4a1−/− mice, and cultured with the differentiation and polarization medium. Macrophages were characterized by the expression of inducible nitric oxide synthase (iNOS) and IL-1β for the M1 population, while the M2 population was characterized by the expression of arginase 1 (ArgI) (see Supplementary material online, Figure S4E and F). The levels of iNOS and IL-1β in M1 population and the levels of ArgI in M2 population were markedly increased than others; however, deficiency of PTP4A1 did not affect the polarization of macrophages.
3.4. PTP4A1 inhibits NF-κB activation through its phosphatase activity in ECs
To examine the mechanism by which PTP4A1 negatively regulates CAM expression in ECs under atherosclerotic conditions, the ICAM-1 and VCAM-1 expression levels were measured using a luciferase reporter assay. The ICAM-1 and VCAM-1 promoter activities in IL-1β-stimulated HUAECs were significantly enhanced upon PTP4A1 knockdown but reduced upon PTP4A1 overexpression (Figure 4A and see Supplementary material online, Figure S5A). Since NF-κB is a major transcription factor for ICAM-1 and VCAM-1 expression levels, we measured the activity of p65, a key component of the NF-κB complex. p65 phosphorylation in HUAECs was enhanced upon PTP4A1 knockdown under basal and IL-1β-stimulated conditions, whereas it was decreased upon PTP4A1 overexpression (Figure 4B and C and see Supplementary material online, Figure S5B and C). Consistently, the binding activity of p65 on the promoters of the ICAM-1 and VCAM-1 genes was increased in PTP4A1-knockdown HUAECs compared with the control (Figure 4D). Inhibition of NF-κB signalling by PDTC reversed the increase in ICAM-1 and VCAM-1 expression levels as well as p65 phosphorylation in PTP4A1-knockdown HUAECs (Figure 4E). These results indicate that PTP4A1 downregulates the expression levels of ICAM-1 and VCAM-1 by inhibiting the NF-κB signalling pathway under basal and inflammatory conditions in ECs.
Figure 4.
Phosphatase activity of PTP4A1 is required to inhibit the NF-κB-mediated expression of cell adhesion molecules. (A–H) HUAECs were stably transfected with shControl, shPTP4A1, mock or PTP4A1 using Lentivirus. (A) HUAECs were transiently transfected with a luciferase reporter plasmid of ICAM-1 or VCAM-1 and a Renilla luciferase internal control plasmid. The cells were stimulated with PBS or IL-1β, and luciferase activity was detected using the dual-luciferase assay system (n = 6). (B and C) HUAECs were stimulated with PBS or IL-1β for 2 h. Immunoblot analysis in cell lysates of shControl- or shPTP4A1-treated HUAECs (B) and bar diagrams representing normalized protein levels (n = 5) (C). (D) ChIP assays were performed to measure the binding activity of p-p65 to the promoters of ICAM-1 and VCAM-1 in shControl- or shPTP4A1-treated HUAECs stimulated with IL-1β for 2 h (n = 5). GAPDH was used as an internal control after immunoprecipitation by antibodies for RNA polymerase II (RNAP II, positive control) or IgG (negative control). (E) HUAECs were stimulated with PBS or IL-1β for 3 h after DMSO or PDTC treatment. Immunoblot analysis in cell lysates from shControl- or shPTP4A1-treated HUAECs. Bar diagrams representing normalized protein levels (n = 5). (F) Scheme for WT PTP4A1, DN-PTP4A1 (defective phosphatase activity; D74A plus C104S point mutations), and ΔCAAX (a truncated PTP4A1 at the prenylation motif). (G and H) HUAECs were stimulated with PBS or IL-1β. (G) Immunoblot analysis on cell lysates from mock-, PTP4A1-, or DN-PTP4A1-treated HUAECs. Bar diagrams representing normalized protein levels (n = 5). (H) Immunoblot analysis in cell lysates from mock-, PTP4A1-, or ΔCAAX-treated HUAECs. Bar diagrams representing normalized protein levels (n = 5). All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (two-way ANOVA for A, C, E, G, and H; two-tailed Student’s t-test for D).
Phosphatase activity and the prenylation motif are required for PTP4A1 subcellular localization and biological activity.27 Thus, we constructed two mutated forms of PTP4A1, a dominant-negative (DN)-PTP4A1 (D72A/C104S, with defective phosphatase activity) and a truncated form of PTP4A1 without the prenylation motif (ΔCAAX) (Figure 4F). Compared with wild-type (WT) PTP4A1, overexpression of DN-PTP4A1 did not inhibit IL-1β-mediated upregulation of CAMs and p65 phosphorylation (Figure 4G). Contrarily, the overexpression of ΔCAAX still exerted inhibitory effects on IL-1β-mediated CAM expression and p65 phosphorylation (Figure 4H), suggesting that phosphatase activity but not prenylation of PTP4A1 is essential for its anti-inflammatory function in the endothelium.
3.5. Endothelial PTP4A1 suppresses NF-κB signalling through A20 upregulation
We next investigated how PTP4A1 regulates NF-κB activity. Based on the NF-κB signalling cascade, we examined the activation of IKKα/β and observed that PTP4A1 negatively regulates IKKα/β phosphorylation in IL-1β-stimulated HUAECs (Figure 5A and see Supplementary material online, Figure S6A). A20 has emerged as a negative feedback regulator of NF-κB signalling by editing ubiquitin chains on upstream molecules of IKK,14 and we confirmed that the A20 expression is inversely correlated with the activation of IKKα/β and p65 in IL-1β- and TNF-α-treated HUAECs (see Supplementary material online, Figure S6B and C). Moreover, the small interfering RNA (siRNA)-induced knockdown of A20 in HUAECs activated the NF-κB signalling pathway, as determined by the increased phosphorylation of IKKα/β and expression of CAMs under both basal and IL-1β-stimulated conditions (see Supplementary material online, Figure S6D), confirming that A20 is a critical negative regulator of NF-κB signalling. Notably, the A20 expression was significantly downregulated upon PTP4A1 knockdown in HUAECs under basal and IL-1β-stimulated conditions, whereas it was upregulated upon PTP4A1 overexpression (Figure 5B–E). Knockdown of A20 in PTP4A1-overexpressing HUAECs reversed the inhibitory effects of PTP4A1 on the expression of CAMs and the phosphorylation of p65 (Figure 5F). As evaluated via RT-PCR and luciferase reporter analyses, we determined that PTP4A1-mediated A20 expression was regulated at the transcriptional level in IL-1β-stimulated ECs (Figure 5G–I and see Supplementary material online, Figure S6E–G). Compared with WT PTP4A1, DN-PTP4A1 failed to induce the expression of the A20 luciferase reporter gene (Figure 5J), demonstrating that the phosphatase activity of PTP4A1 is required for the transcriptional regulation of A20.
Figure 5.
PTP4A1 negatively regulates NF-κB transcriptional activity by controlling the A20 expression. (A–J) HUAECs were stably transfected with shControl, shPTP4A1, mock, PTP4A1, or DN-PTP4A1 using Lentivirus. (A) Immunoblot analysis in cell lysates of shControl- or shPTP4A1-treated HUAECs stimulated by IL-1β for the indicated times. Bar diagrams representing normalized protein levels (n = 5). (B–E) HUAECs were stimulated with PBS or IL-1β for 2 h. Immunoblot analysis in cell lysates from shControl- or shPTP4A1-treated HUAECs (B) and bar diagrams representing normalized protein levels (n = 5) (C). Immunoblot analysis in cell lysates from mock or PTP4A1-overexpressing HUAECs (D) and bar diagrams representing normalized protein levels (n = 5) (E). (F) Mock or PTP4A1 HUAECs were transfected with control siRNA or A20 siRNA (siA20) before PBS or IL-1β treatment for 3 h. Immunoblot analysis in cell lysates from mock or PTP4A1 HUAECs. Bar diagrams representing normalized protein levels (n = 5). (G and H) Stable HUAECs were stimulated with PBS or IL-1β for 2 h. The mRNA levels of A20 were detected via RT-PCR (G) and qRT-PCR (H) analysis in shControl- or shPTP4A1-treated HUAECs (n = 5). GAPDH was used as a loading control. (I and J) A20 luciferase reporter assays in shControl- or shPTP4A1-treated HUAECs (n = 5) (I) and A20 luciferase reporter assays in mock, PTP4A1, or DN-PTP4A1 HUAECs (n = 6) (J). All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (two-way ANOVA for A, C, E, F, H, I, and J).
3.6. PTP4A1 enhances the activity of USF1 for A20 transcription
The transcription factor USF1 is the dominant E-box-binding protein that initiates A20 gene transcription.22 We validated the USF1-mediated transcriptional upregulation of the A20 gene using a promoter assay (see Supplementary material online, Figure S7A) and found that the E-box sequence of the A20 gene is important for inducing A20 gene expression by USF1 (see Supplementary material online, Figure S7B). The binding activity of USF1 to the E-box region of the A20 promoter peaked at 60 min after IL-1β treatment (see Supplementary material online, Figure S7C). PTP4A1, but not DN-PTP4A1, exerted a synergistic effect with USF1 on the promoter activity of A20 (Figure 6A). The siRNA-mediated knockdown of USF1 abolished the PTP4A1-increased promoter activity of A20 after IL-1β stimulation in PTP4A1-overexpressing HUAECs (Figure 6B). Moreover, USF1 knockdown in PTP4A1-overexpressing HUAECs significantly impaired PTP4A1-mediated anti-inflammatory effects by regulating ICAM-1 and downregulating A20, implying that PTP4A1-mediated A20 gene induction requires USF1 (Figure 6C and D). The overexpression of PTP4A1 and HA-USF1 increased the binding activity of USF1 to the E-box region of the A20 promoter under the basal condition, and binding significantly increased after stimulation with IL-1β (Figure 6E). In line with previous results, only WT PTP4A1, but not DN-PTP4A1, enhanced the binding activity of USF1 to the E-box region of the A20 promoter (Figure 6F).
Figure 6.
PTP4A1 increases the activity of USF1 for A20 transcription. (A) A20 luciferase reporter assays in USF1 with PTP4A1- or DN-PTP4A1-overexpressing HUAECs stimulated with PBS or IL-1β for 4 h (n = 6). (B) A20 luciferase reporter assays in mock- or PTP4A1-overexpressing HUAECs treated with control siRNA or USF1 siRNA (siUSF1). Cells were stimulated with PBS or IL-1β for 4 h (n = 6). (C and D) HUAECs were stimulated with PBS or IL-1β for 3 h. Immunoblot analysis in cell lysates from mock- or PTP4A1-overexpressing HUAECs treated with siControl or siUSF1 (C). Bar diagrams representing normalized protein levels (n = 5) (D). (E) ChIP assay for PTP4A1-mediated USF1-binding activity on the E-box of the A20 promoter in PBS- or IL-1β-stimulated 293 T cells for 1 h (n = 5). qRT-PCR data (top) and RT-PCR (bottom) are presented. (F) ChIP assay for USF1-binding activity on the E-box of the A20 promoter in the presence of PTP4A1 or DN-PTP4A1. Two hundred and ninety-three T cells were stimulated with PBS or IL-1β for 1 h (n = 5). qRT-PCR data (top) and RT-PCR (bottom) are presented. All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (two-way ANOVA for A, B, D, E, and F).
3.7. PTP4A1 enhances A20 transcription by dephosphorylating the S309 residue of USF1
We then examined whether PTP4A1 physically interacts with USF1. The co-immunoprecipitation (co-IP) and ChIP assay results indicated that PTP4A1 was physically bound to USF1 (see Supplementary material online, Figure S8), and the interaction between PTP4A1 and USF1 was maintained at the E-box region of the A20 promoter under IL-1β stimulation (Figure 7A). To identify the binding region of USF1 for PTP4A1, we constructed two truncated forms of USF1: HA-USF1 (Δ256–310, loss of the ZIP domain) and HA-USF1 (Δ201–310, loss of the bHLH and ZIP domains). By co-IP analysis, we found that the bHLH domain (amino acids 201–255) of USF1 is important for the interaction with PTP4A1 (Figure 7B). Finally, to address whether phosphorylation and/or dephosphorylation of USF1 is involved in the regulation of A20 promoter activity, we constructed mutants with a point mutation on putative phosphorylation sites of USF1 at residue 153 by replacing Thr with Ala, at residue 185 by replacing Tyr with Phe, and at residues 186, 230, 233, 262, or 309 by replacing Ser with Ala. As presented in Figure 7C, T153A, Y185F, S233A, and S262A had nearly the same A20 promoter activity as WT USF1, whereas S186A and S230A were about 50% less active. Interestingly, the S309A mutant significantly enhanced the promoter activity of A20 compared with WT USF1, suggesting that dephosphorylation of the Ser residue at 309 in USF1 may be important for enhancing the promoter activity of A20 at least partially. To confirm the importance of the phosphorylation of S309, we constructed the mimetic constitutive phosphorylation form (S309D) of USF1 and confirmed that USF1 (S309D) completely blocked the PTP4A1-mediated synergistic effects of the A20 promoter assay (Figure 7D). The results of the ChIP assay indicated that the changes in the transcriptional activity of USF1 mutant forms at S309 resulted from the alteration of binding activity on the promoter of the A20 gene (Figure 7E). Taken together, dephosphorylation of the S309 residue of USF1 by PTP4A1 is critical for the regulation of the binding activity of USF1 at the E-box promoter region of the A20 gene, followed by an increase in A20 expression and the mitigation of the inflammatory response in ECs.
Figure 7.
PTP4A1 increases A20 transcription by dephosphorylating the Ser residue at 309 on USF1. (A) ChIP assay for USF1 or PTP4A1-binding activity on the A20 promoter region in 293 T cells (n = 5). qRT-PCR data (top) and RT-PCR (bottom) are presented. (B) Co-IP of flag-PTP4A1 with WT HA-USF1 or truncated forms in 293 T cells. Data were obtained from five independent experiments. (C) A20 luciferase reporter assays in HUAECs transfected with A20 luciferase reporter plasmids and Renilla luciferase internal control plasmids, as well as PTP4A1 with the indicated USF1 constructs. Cells were stimulated with PBS or IL-1β for 4 h (n = 5). (D) A20 luciferase reporter assays with USF1 constructs (WT, S309A, S309D) in parental or PTP4A1-overexpressing HUAECs. The cells were stimulated with PBS or IL-1β for 4 h (n = 5). (E) ChIP assay for USF1 (WT, S309A, S309D)-binding activity on the A20 promoter region in PBS- or IL-1β-stimulated 293 T cells for 1 h (n = 5). qRT-PCR data (top) and RT-PCR (bottom) are presented. All data are expressed as mean ± S.E.M. *P < 0.05 and **P < 0.01 (two-way ANOVA for A, C, D, and E).
4. Discussion
Kinase- and phosphatase-mediated posttranslational modification of signalling molecules is critical for the impact of intracellular signalling pathways on cellular functions.31 Kinase-mediated phosphorylation of members of the NF-κB signalling pathway enhances its transactivation potential, but phosphatases that participate in controlling signalling are relatively not studied. Here, we found that the phosphatase PTP4A1 is a vital factor that regulates the expression of ICAM-1 and VCAM-1 in HUAECs. In vivo studies using Ptp4a1 knockout or transgenic mice demonstrated the potent regulation of PTP4A1 on IL-1β-induced expression of ICAM-1 and VCAM-1. In addition, we verified that ApoE−/− Ptp4a1−/− mice fed an HFHC diet revealed the increment of atherosclerotic plaques with the augmentation of the CAM expression and macrophage recruitment. Mechanistically, PTP4A1, but not the defective phosphatase form of PTP4A1, inhibited the expression of CAMs and the activation of NF-κB in ECs via an increase in the transcriptional activity of USF1, inducing A20 gene transcription. USF1 or A20 knockdown mitigated the PTP4A1 inhibitory effects on CAM expression and NF-κB activation in ECs. Finally, we demonstrated that PTP4A1 physically interacts with USF1 and dephosphorylates the S309 residue of USF1, leading the increased binding activity to the E-box of the A20 gene promoter region. Although a few studies have demonstrated the roles of phosphatases in the regulation of the NF-κB signalling pathway,32,33 the impact of PTP4A1 on the NF-κB signalling pathway and vascular diseases has not yet been reported. Therefore, our findings suggest a protective role of PTP4A1 in vascular inflammation and that PTP4A1 is a novel negative regulator of the NF-κB signalling pathway in ECs.
In humans, PTP4A1, PTP4A2, and PTP4A3 are located on chromosomes 6q12, 1p35, and 8q24, respectively. PTP4A1 and PTP4A2 are ubiquitously expressed in various tissues, but PTP4A3 is mainly expressed in the heart and skeletal muscle.34,35PTP4A1 is highly expressed in the liver, adrenal gland, colon, and bone marrow, and the PTP4A1 expression is upregulated by early growth response factor 1 and p5336,37 and is downregulated by several microRNAs (miRNAs).38–40 Here, we observed PTP4A1 downregulation in ECs after stimulation with pro-inflammatory cytokines or oxidized LDL (data not shown), which may be associated with an increase in miRNA expression. Indeed, miR-29 enhancement has been implicated in cardiovascular disease pathogenesis,41,42 and miR-339-5p is upregulated in ischaemic heart disease,43 suggesting the possibility of miRNA-mediated PTP4A1 downregulation in ECs by atherogenic stimuli. Notably, Tabibiazar et al. reported that Ptp4a1 was involved in a potentially atheroprotective cluster that was highly expressed in atherosclerosis-resistant C3H/HeJ mice compared with atherosclerosis-susceptible C57BL/6 mice.44 They also demonstrated decreased Ptp4a1 expression in high-fat diet-challenged ApoE−/− mice,44 which was also demonstrated in the present study.
Contrary to PTP4A1 downregulation, PTP4A2 knockdown in HUAECs revealed no difference in the expression levels of ICAM-1 and VCAM-1 compared with controls (data not shown), suggesting a specific role of PTP4A1 in the regulation of inflammatory responses in ECs. Notably, PTP4A1 knockdown in ECs induced high ICAM-1 expression and mild VCAM-1 expression without an inflammatory mediator. ICAM-1 is constitutively expressed on various cell surfaces, including ECs,45 but VCAM-1 is expressed only after ECs are stimulated by several inflammatory mediators.46 Thus, we speculated that PTP4A1 downregulation triggers intracellular signalling, resulting in ICAM-1 and VCAM-1 expression levels. Based on this hypothesis, PTP4A1-downregulated HUAECs also experienced downregulation of A20 levels and increased p65 phosphorylation even under the basal condition, suggesting that PTP4A1 per se is a pivotal negative regulator of the NF-κB signalling pathway.
Here, we highlight endothelial PTP4A1 as an effective therapeutic approach for increasing A20 expression and function. A20 was originally identified in human ECs and is acutely induced in response to TNF-α, IL-1β, and LPS, suggesting its importance in the role of the vessel wall in vascular inflammation.14 The increased expression of NF-κB target genes, including Vcam-1 and Icam-1, in ApoE−/− mice induced by A20 haploinsufficiency accelerated atherosclerotic lesion development.11 Human genetic studies have demonstrated direct implications of abnormal A20 function or expression due to polymorphisms and mutations in the pathogenesis of inflammatory and autoimmune diseases.15,16,18,19 Two tag SNPs (rs5029930 and rs610604) in human A20 on chromosome 6q23 are independently associated with CHD,19 and the difference in a single amino acid residue (E627A; C57BL/6 vs. FVB) in A20 increases the susceptibility of C57BL/6 mice to atherosclerosis.47 Indeed, A20 overexpression in the vessel wall or endothelial progenitor cells prevents xenograft-induced atherosclerosis, and genetic approaches modifying A20 levels demonstrated its protective effect against atherosclerosis in ApoE−/− mice,11,48 suggesting its potential as a therapeutic target.
As in a previous study,22 using siUSF1 treatment, a ChIP assay and an A20 luciferase reporter assay, we demonstrated that USF1 is a pivotal transcriptional factor for A20 transcription. The DNA binding of USF1 is modulated by posttranslational modification via multiple signal transduction pathways.23 In this study, we first identified that the bZIP domain (amino acids 201–292), especially the basic helix–loop–helix region (amino acids 201–255), of USF1 is important for physical interaction with PTP4A1, validating previous reports that the bZIP-containing transcriptional factor, ATF5/7, physically interacts with PTP4A1.28 The A20 luciferase reporter assay with WT and mutant USF1 forms revealed that phosphorylation of the S186 residue and dephosphorylation of the S309 residue of USF1 are important for the transcriptional regulation of A20. PTP4A1-mediated dephosphorylation of the S309 residue of USF1 is critical for increasing A20 transcription. However, whether PTP4A1-mediated dephosphorylation of the USF1 S309 residue affects the transcriptional activity of other USF1 target genes needs to be elucidated.
Although the present study focused on the PTP4A1-mediated ECs function in inflammatory conditions, we used the Ptp4a1 global knockout mice and the Tie2 promoter/enhancer-mediated Ptp4a1 Tg mice. As PTP4A1 is ubiquitously expressed in various tissues and cells, including immune cells, the limitation of the present study would be confirmed by endothelial-specific knockout or transgenic mice in a future study.
Overall, our study demonstrates the novel function and molecular mechanism of the regulation of vascular inflammation by endothelial PTP4A1, enhancing the impact of USF1 on the transcriptional activity of A20. Our results indicate that the anti-inflammatory effects of PTP4A1 are required for vascular inflammatory diseases, including atherosclerosis. As current therapeutic approaches, such as the use of antibodies or small molecules, have more potential for antagonizing the activities of target molecules rather than agonizing them, this study could increase A20 expression and function by PTP4A1 as a therapeutic approach.
Supplementary material
Supplementary Material is available at Cardiovascular Research online.
Supplementary Material
Contributor Information
Min Ji Cho, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Dong Gwang Lee, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Jeong Woong Lee, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Byungtae Hwang, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Sung-Jin Yoon, Environmental Disease Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Seon-Jin Lee, Environmental Disease Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Young-Jun Park, Environmental Disease Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Seung-Ho Park, Environmental Disease Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Hee Gu Lee, Immunotherapy Convergence Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Yong-Hoon Kim, Laboratory Animal Resource Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Chul-Ho Lee, Laboratory Animal Resource Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Jangwook Lee, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Nam-Kyung Lee, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Tae-Su Han, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Hyun-Soo Cho, Stem Cell Convergence Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Jeong Hee Moon, Disease Target Structure Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Ga Seul Lee, Disease Target Structure Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Kwang-Hee Bae, Metabolic Regulation Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Geum-Sook Hwang, Integrated Metabolomics Research Group, Western Seoul Centre, Korea Basic Science Institute, 150 Bugahyeon-ro, Seodaemun-gu, Seoul 03759, Republic of Korea.
Sang-Hak Lee, Division of Cardiology, Department of Internal Medicine, Severance Hospital, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Republic of Korea.
Sang J Chung, School of Pharmacy, Sungkyunkwan University, 2066 Seobu-ro, Jangan-gu, Suwon 16419, Republic of Korea.
Sungbo Shim, Department of Biochemistry, Chungbuk National University, 1 Chungdae-ro, Seowon-gu, Cheongju 28644, Republic of Korea.
Jaehyung Cho, Division of Haematology, Department of Medicine, Washington University School of Medicine, St. Louis, MO 63110, USA; Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Goo Taeg Oh, Heart-Immune-Brain Network Research Centre, Ewha Womans University, 52 Ewhayeodae-gil, Seodaemun-gu, Seoul 03760, Republic of Korea.
Young-Guen Kwon, Department of Biochemistry, College of Life Science and Biotechnology, Yonsei University, 50 Yonsei-ro, Seodaemun-gu, Seoul 03722, Republic of Korea.
Jong-Gil Park, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bioscience, KRIBB School of Bioscience, Korea University of Science and Technology (UST), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Jeong-Ki Min, Biotherapeutics Translational Research Centre, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bioscience, KRIBB School of Bioscience, Korea University of Science and Technology (UST), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Author contributions
M.J.C. and D.G.L. designed the study, performed experiments, analysed the data, prepared the figures, and wrote the manuscript. J.W.L. generated gene-targeted mice. B.H., S.-J.Y., S.-J.L., Y.-J.P., S.-H.P., H.G.L., Y.-H.K., C.-H.L., J.L., N.-K.L., T.-S.H., H.-S.C., J.H.M., G.S.L., K.-H.B., G.-S.H., S.-H.L., S.J.C., and S.S. helped to conduct experiments. J.C., G.T.O., and Y.-G.K. helped to write the manuscript and interpreted data. J.-G.P. and J.-K.M. provided general supervision, designed the study, supported the experiments, and assisted with the critical discussion, preparation, and submission of the manuscript.
Funding
This study was supported by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Programme (KGM5272151), by the National Research Council of Science and Technology (CAP-12-02-KBSI), by the National Research Foundation of Korea grants (NRF-2018R1C1B6005004), and by the Bio-Synergy Research Project (NRF- 2015M3A9C4070489) through the National Research Foundation of Korea, which was funded by the Ministry of Science, ICT and Future Planning.
Data availability
The data related to this article are available in the article itself or in the Supplementary Material Online.
Translational perspective.
The inhibitory effect of PTP4A1 in the dysfunction of vascular endothelial cells could apply to develop a drug for vascular inflammatory diseases including atherosclerosis. This study highlights an alternative as well as an effective therapeutic approach to increase A20 expression, which is an endogenous negative regulator of NF-κB signalling in vascular inflammatory disorders.
References
- 1. Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol 2012;32:2045–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol 2007;7:678–689. [DOI] [PubMed] [Google Scholar]
- 3. Cotran RS, Mayadas-Norton T. Endothelial adhesion molecules in health and disease. Pathol Biol 1998;46:164–170. [PubMed] [Google Scholar]
- 4. Park JG, Ryu SY, Jung IH, Lee YH, Kang KJ, Lee MR, Lee MN, Sonn SK, Lee JH, Lee H, Oh GT, Moon K, Shim H. Evaluation of VCAM-1 antibodies as therapeutic agent for atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis 2013;226:356–363. [DOI] [PubMed] [Google Scholar]
- 5. Kitagawa K, Matsumoto M, Sasaki T, Hashimoto H, Kuwabara K, Ohtsuki T, Hori M. Involvement of ICAM-1 in the progression of atherosclerosis in APOE-knockout mice. Atherosclerosis 2002;160:305–310. [DOI] [PubMed] [Google Scholar]
- 6. Nakashima Y, Raines EW, Plump AS, Breslow JL, Ross R. Upregulation of VCAM-1 and ICAM-1 at atherosclerosis-prone sites on the endothelium in the ApoE-deficient mouse. Arterioscler Thromb Vasc Biol 1998;18:842–851. [DOI] [PubMed] [Google Scholar]
- 7. de Winther MP, Kanters E, Kraal G, Hofker MH. Nuclear factor kappaB signaling in atherogenesis. Arterioscler Thromb Vasc Biol 2005;25:904–914. [DOI] [PubMed] [Google Scholar]
- 8. Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol 2010;2:a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol 2012;12:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gareus R, Kotsaki E, Xanthoulea S, van der Made I, Gijbels MJ, Kardakaris R, Polykratis A, Kollias G, de Winther MP, Pasparakis M. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab 2008;8:372–383. [DOI] [PubMed] [Google Scholar]
- 11. Wolfrum S, Teupser D, Tan M, Chen KY, Breslow JL. The protective effect of A20 on atherosclerosis in apolipoprotein E-deficient mice is associated with reduced expression of NF-kappaB target genes. Proc Natl Acad Sci U S A 2007;104:18601–18606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferran C, Stroka DM, Badrichani AZ, Cooper JT, Wrighton CJ, Soares M, Grey ST, Bach FH. A20 inhibits NF-kappaB activation in endothelial cells without sensitizing to tumor necrosis factor-mediated apoptosis. Blood 1998;91:2249–2258. [PubMed] [Google Scholar]
- 13. Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004;430:694–699. [DOI] [PubMed] [Google Scholar]
- 14. Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol 2014;35:22–31. [DOI] [PubMed] [Google Scholar]
- 15. Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol 2009;30:383–391. [DOI] [PubMed] [Google Scholar]
- 16. Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet 2008;40:1059–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, Bruinenberg M, Heap GA, Platteel M, Ryan AW, de Kovel C, Holmes GK, Howdle PD, Walters JR, Sanders DS, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, Kelleher D, Barisani D, Bardella MT, McManus R, van Heel DA, Wijmenga C. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut 2009;58:1078–1083. [DOI] [PubMed] [Google Scholar]
- 18. Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, Reizis B, DeFranco A, Criswell LA, Nakamura MC, Ma A. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol 2011;12:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boonyasrisawat W, Eberle D, Bacci S, Zhang YY, Nolan D, Gervino EV, Johnstone MT, Trischitta V, Shoelson SE, Doria A. Tag polymorphisms at the A20 (TNFAIP3) locus are associated with lower gene expression and increased risk of coronary artery disease in type 2 diabetes. Diabetes 2007;56:499–505. [DOI] [PubMed] [Google Scholar]
- 20. Bach FH, Ferran C, Hechenleitner P, Mark W, Koyamada N, Miyatake T, Winkler H, Badrichani A, Candinas D, Hancock WW. Accommodation of vascularized xenografts: expression of “protective genes” by donor endothelial cells in a host Th2 cytokine environment. Nat Med 1997;3:196–204. [DOI] [PubMed] [Google Scholar]
- 21. Amir-Zilberstein L, Dikstein R. Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF). J Biol Chem 2008;283:1317–1323. [DOI] [PubMed] [Google Scholar]
- 22. Tiruppathi C, Soni D, Wang DM, Xue J, Singh V, Thippegowda PB, Cheppudira BP, Mishra RK, Debroy A, Qian Z, Bachmaier K, Zhao YY, Christman JW, Vogel SM, Ma A, Malik AB. The transcription factor DREAM represses the deubiquitinase A20 and mediates inflammation. Nat Immunol 2014;15:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res 2005;18:337–348. [DOI] [PubMed] [Google Scholar]
- 24. Baxevanis AD, Vinson CR. Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Genet Dev 1993;3:278–285. [DOI] [PubMed] [Google Scholar]
- 25. Bessette DC, Qiu D, Pallen CJ. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev 2008;27:231–252. [DOI] [PubMed] [Google Scholar]
- 26. Sun JP, Wang WQ, Yang H, Liu S, Liang F, Fedorov AA, Almo SC, Zhang ZY. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry 2005;44:12009–12021. [DOI] [PubMed] [Google Scholar]
- 27. Sun JP, Luo Y, Yu X, Wang WQ, Zhou B, Liang F, Zhang ZY. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. J Biol Chem 2007;282:29043–29051. [DOI] [PubMed] [Google Scholar]
- 28. Peters CS, Liang X, Li S, Kannan S, Peng Y, Taub R, Diamond RH. ATF-7, a novel bZIP protein, interacts with the PRL-1 protein-tyrosine phosphatase. J Biol Chem 2001;276:13718–13726. [DOI] [PubMed] [Google Scholar]
- 29. Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL Phosphatases as potential molecular targets in cancer. Mol Cancer Ther 2005;4:1653–1661. [DOI] [PubMed] [Google Scholar]
- 30. Shendre A, Irvin MR, Wiener H, Zhi D, Limdi NA, Overton ET, Shrestha S. Local ancestry and clinical cardiovascular events among African Americans from the atherosclerosis risk in communities study. J Am Heart Assoc 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther 1999;82:111–121. [DOI] [PubMed] [Google Scholar]
- 32. Koch E, Pircher J, Czermak T, Gaitzsch E, Alig S, Mannell H, Niemeyer M, Krotz F, Wornle M. The endothelial tyrosine phosphatase SHP-1 plays an important role for vascular haemostasis in TNFalpha-induced inflammation in vivo. Mediators Inflamm 2013;2013:279781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chew J, Biswas S, Shreeram S, Humaidi M, Wong ET, Dhillion MK, Teo H, Hazra A, Fang CC, Lopez-Collazo E, Bulavin DV, Tergaonkar V. WIP1 Phosphatase is a negative regulator of NF-kappaB signalling. Nat Cell Biol 2009;11:659–666. [DOI] [PubMed] [Google Scholar]
- 34. Hardy S, Kostantin E, Hatzihristidis T, Zolotarov Y, Uetani N, Tremblay ML. Physiological and oncogenic roles of the PRL phosphatases. FEBS J 2018;285:3886–3908. [DOI] [PubMed] [Google Scholar]
- 35. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng Y, Du K, Ramirez S, Diamond RH, Taub R. Mitogenic up-regulation of the PRL-1 protein-tyrosine phosphatase gene by Egr-1 Egr-1 activation is an early event in liver regeneration. J Biol Chem 1999;274:4513–4520. [DOI] [PubMed] [Google Scholar]
- 37. Min SH, Kim DM, Heo YS, Kim YI, Kim HM, Kim J, Han YM, Kim IC, Yoo OJ. New p53 target, phosphatase of regenerating liver 1 (PRL-1) downregulates p53. Oncogene 2009;28:545–554. [DOI] [PubMed] [Google Scholar]
- 38. Hu JY, Yi W, Wei X, Zhang MY, Xu R, Zeng LS, Huang ZJ, Chen JS. miR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed Pharmacother 2016;79:247–253. [DOI] [PubMed] [Google Scholar]
- 39. Zhou C, Liu G, Wang L, Lu Y, Yuan L, Zheng L, Chen F, Peng F, Li X. MiR-339-5p regulates the growth, colony formation and metastasis of colorectal cancer cells by targeting PRL-1. PLoS One 2013;8:e63142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, Kung HF, Zeng YX, Xie D. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of beta-catenin signaling. Ann Oncol 2014;25:2196–2204. [DOI] [PubMed] [Google Scholar]
- 41. Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, van Heijningen P, Essers J, Brandes RP, Zeiher AM, Dimmeler S. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res 2011;109:1115–1119. [DOI] [PubMed] [Google Scholar]
- 42. Sassi Y, Avramopoulos P, Ramanujam D, Gruter L, Werfel S, Giosele S, Brunner AD, Esfandyari D, Papadopoulou AS, De Strooper B, Hubner N, Kumarswamy R, Thum T, Yin X, Mayr M, Laggerbauer B, Engelhardt S. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating Wnt signaling. Nat Commun 2017;8:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saddic LA, Chang TW, Sigurdsson MI, Heydarpour M, Raby BA, Shernan SK, Aranki SF, Body SC, Muehlschlegel JD. Integrated microRNA and mRNA responses to acute human left ventricular ischemia. Physiol Genomics 2015;47:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tabibiazar R, Wagner RA, Spin JM, Ashley EA, Narasimhan B, Rubin EM, Efron B, Tsao PS, Tibshirani R, Quertermous T. Mouse strain-specific differences in vascular wall gene expression and their relationship to vascular disease. Arterioscler Thromb Vasc Biol 2005;25:302–308. [DOI] [PubMed] [Google Scholar]
- 45. van de Stolpe A, van der Saag PT. Intercellular adhesion molecule-1. J Mol Med (Berl )1996; 74:13–33. [DOI] [PubMed] [Google Scholar]
- 46. Cook-Mills JM, Marchese ME, Abdala-Valencia H. Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxid Redox Signal 2011;15:1607–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Idel S, Dansky HM, Breslow JL. A20, a regulator of NFkappaB, maps to an atherosclerosis locus and differs between parental sensitive C57BL/6J and resistant FVB/N strains. Proc Natl Acad Sci U S A 2003;100:14235–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu C, Ying D, Mi J, Li L, Zeng W, Hou C, Sun J, Yuan W, Wen C, Zhang W. Development of anti-atherosclerotic tissue-engineered blood vessel by A20-regulated endothelial progenitor cells seeding decellularized vascular matrix. Biomaterials 2008;29:2628–2636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data related to this article are available in the article itself or in the Supplementary Material Online.