Abstract
Summary
In any population under selective pressure, a central challenge is to distinguish the genes that drive adaptation from others which, subject to population variation, harbor many neutral mutations de novo. We recently showed that such genes could be identified by supplementing information on mutational frequency with an evolutionary analysis of the likely functional impact of coding variants. This approach improved the discovery of driver genes in both lab-evolved and environmental Escherichia coli strains. To facilitate general adoption, we now developed ShinyBioHEAT, an R Shiny web-based application that enables identification of phenotype driving gene in two commonly used model bacteria, E.coli and Bacillus subtilis, with no specific computational skill requirements. ShinyBioHEAT not only supports transparent and interactive analysis of lab evolution data in E.coli and B.subtilis, but it also creates dynamic visualizations of mutational impact on protein structures, which add orthogonal checks on predicted drivers.
Availability and implementation
Code for ShinyBioHEAT is available at https://github.com/LichtargeLab/ShinyBioHEAT. The Shiny application is additionally hosted at http://bioheat.lichtargelab.org/.
1 Introduction
Escherichia coli and Bacillus subtilis are ideal model organisms for genotype–phenotype studies due to their unique advantages. They grow fast and benefit from a vast array of genetic editing techniques (Swings et al. 2018, Choudhury et al. 2020, Csörgő et al. 2020, Zhang et al. 2020) and bioinformatics databases (Keseler et al. 2021, Szklarczyk et al. 2019, Tierrafría et al. 2022). Increasingly, studies that seek to pinpoint the driver genes of phenotypes of interest combine adaptive laboratory experiments (ALEs) with next-generation sequencing (Tenaillon et al. 2016, Zeigler and Nicholson 2017, van den Bergh et al. 2018, Bruckbauer et al. 2019, Karve and Wagner 2022). Typically, these studies rank genes based on their relative mutational frequency in parallel streams of replications. This sole use of mutational frequency ignores additional information on the functional impact of coding variants, however, and reduces the power to detect secondary diver genes.
To improve the identification of driver genes, we recently developed a new EA integration approach (Marciano et al. 2022), which exploits the Evolutionary Action (EA) score (Katsonis and Lichtarge 2014) for the likely impact of any missense mutation in any given protein from past evolutionary history. EA scores tend to correlate well with experimental mutagenesis studies in objective, blinded challenges evaluated by third parties (Katsonis and Lichtarge 2019) and to predict the harmful effect of mutations in diverse applications (Katsonis et al. 2022). In a direct test of its potential for elucidating ALE-induced phenotypes in E.coli, EA integration improved phenotype driver gene discovery compared with frequency-based method, especially so in the clinical/environmental datasets (Marciano et al. 2022).
To broaden access to our method, we developed a user-friendly R Shiny (Chang et al. 2022) package, ShinyBioHEAT (Biodetection of High Evolutionary Action Targets), using golem framework (Fay et al. 2022) which allows easy installation across platforms and running locally. The main feature for ShinyBioHEAT is to identify phenotype driving genes in E.coli and B.subtilis from sequencing data by combining EA scores with frequency statistics (Marciano et al. 2022). Additional modules are developed to allow sequential analysis through STRING for the top predicted genes and visualization of mutational profiles on protein structures.
2 Features
2.1 Driver gene analysis module
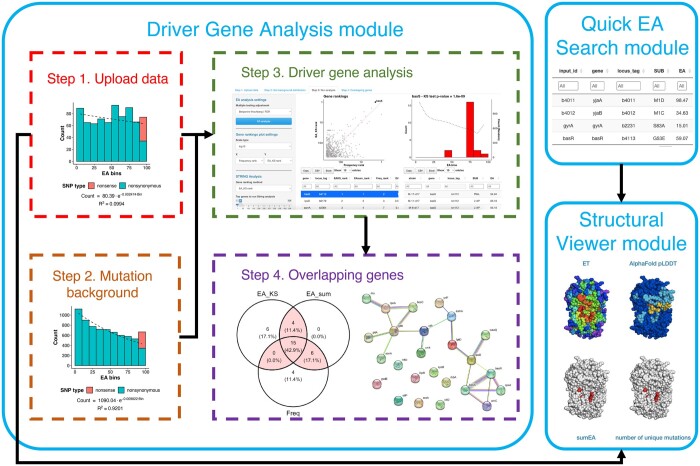
This is the main module of ShinyBioHEAT application (Fig. 1), which allows the identification of driver genes from E.coli and B.subtilis sequencing data using EA integration and a frequency-based method. It currently supports three reference genomes: E.coli MG1655 (RefSeq: NC_000913.3), E.coli REL606 (RefSeq: NC_012967.1), and B.subtilis 168 (RefSeq: NC_000964.3). Sequencing data can be uploaded as variant call format (VCF), amino acid substitutions, or breseq GenomeDiff (GD) format (Deatherage and Barrick 2014). The amino acid substitutions will be determined if VCF format is used. EA scores are then assigned to each missense mutation, which will be compared with a mutation background to identify the driver genes. Mutation background can be generated through randomly simulated mutations in the selected reference genome or a custom set of mutations.
Figure 1.
Graphical overview of the functional modules in ShinyBioHEAT. Data flow is indicated with black arrows.
To account for functional impact of mutations in driver gene prediction, EA integration was implemented with two different approaches: EA_KS and EA_sum. They compare the EA distribution of mutations for each gene in the evolve strains against the mutation background, and then prioritize genes that accumulate more impactful mutations during the adaptation. As an orthogonal control, a frequency-based method is also implemented, which ranks the genes based on mutation count and gene length.
To further evaluate the top-ranked genes and narrow down the genes for experimental validation, an interactive Venn diagram is implemented to allow identification of overlapping predictions by the three approaches. Genes that are highly ranked by different methods are more likely driver genes. In addition, driver genes tend to cluster well in protein–protein interaction networks. We utilize the STRING API (Szklarczyk et al. 2019) to allow quick STRING PPI enrichment test on the top or overlapping predictions.
2.2 Quick EA search
The Quick EA search module allows user to identify the EA scores for missense mutations in the selected reference genome on-the-fly. EA consistently predicts well the protein mutational impact in objective challenges (Katsonis and Lichtarge 2019), which makes it a useful resource.
2.3 Structure viewer
Visualizing mutations on the protein structure provides valuable insights on the molecular mechanism of protein function and can guide mutagenesis studies. The recent advances in protein structure predictions give access to high-quality protein structures for nearly all E.coli and B.subtilis proteins (Jumper et al. 2021). The structure viewer displays the AlphaFold protein structures using r3dmol library (Rego and Koes 2015, Su and Johnston 2022) with four different coloring schemes: Evolutionary Trace (ET), pLDDT, sumEA, and number of unique mutations. ET estimates the importance of a residue position in a protein by examining its evolutionary history (Lichtarge et al. 1996). Clustering of important ET residues is a hallmark for protein functional site (Wilkins et al. 2013; Wang et al. 2021). PLDDT is the structure prediction accuracy score from AlphaFold. SumEA and number of unique mutations project the evolutionary burden in the evolved strains. A Pymol session file with the same coloring scheme is also generated to allow closer examination on Pymol (Schrödinger LLC 2015).
An example study case using ShinyBioHEAT is provided in the Supplementary Data.
3 Conclusion
ShinyBioHEAT is a user-friendly Shiny interface to identify phenotype driver genes in adapted E.coli with minimal coding experience. It also provides downstream analyses through STRING database and color mapping to AlphaFold protein structures. It is freely distributed as an R package under the MIT license at https://github.com/LichtargeLab/ShinyBioHEAT.
Supplementary Material
Acknowledgements
The authors would like to thank David Marciano, Amanda Williams and Saeid Parvandeh for feedbacks and testing.
Contributor Information
Chen Wang, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Harikumar Govindarajan, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Panagiotis Katsonis, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Olivier Lichtarge, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States; Quantitative and Computational Biosciences Graduate Program, Baylor College of Medicine, Houston, TX 77030, United States; Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, United States; Cancer and Cell Biology Graduate Program, Baylor College of Medicine, Houston, TX 77030, United States; Computational and Integrative Biomedical Research Center, Baylor College of Medicine, Houston, TX 77030, United States.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work has been supported by the National Science Foundation [DBI-2032904 to O.L.] and the National Institutes of Health [GM066099, AG074009, AG061105, and AG068214 to O.L.]. This work has also been supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA) under BAA-17-01 [contract #2019-19071900001 to O.L.].
Data availability
The data underlying this article are available in its online supplementary material and its GitHub repository.
References
- Bruckbauer ST, Trimarco JD, Martin J et al. Experimental evolution of extreme resistance to ionizing radiation in Escherichia coli after 50 cycles of selection. J Bacteriol 2019;201:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire J et al. shiny: Web Application Framework for R 2022. https://shiny.posit.co.
- Choudhury A, Fenster JA, Fankhauser RG et al. CRISPR/Cas9 recombineering-mediated deep mutational scanning of essential genes in Escherichia coli. Mol Syst Biol 2020;16:e9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csörgö B, Nyerges A, Pál C. Targeted mutagenesis of multiple chromosomal regions in microbes. Curr Opin Microbiol 2020;57:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. In: Sun, L., Shou, W. (eds) Engineering and Analyzing Multicellular Systems. Methods in Molecular Biology, New York, NY: Humana Press 2014, 165–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay C, Guyader V, Rochette S et al. golem: A framework for robust shiny applications. 2022. https://github.com/ThinkR-open/golem.
- Jumper J, Evans R, Pritzel A et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve S, Wagner A. Environmental complexity is more important than mutation in driving the evolution of latent novel traits in E. coli. Nat Commun 2022;13:5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, Wilhelm K, Williams A et al. Genome interpretation using in silico predictors of variant impact. Hum Genet 2022;141:1549–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the evolutionary action of protein-coding variations on fitness. Genome Res 2014;24:2050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, Lichtarge O. CAGI5: Objective performance assessments of predictions based on the evolutionary action equation. Hum Mutat 2019;40:1436–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Gama-Castro S, Mackie A et al. The EcoCyc database in 2021. Front Microbiol 2021;12:711077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE et al. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 1996;257:342–58. [DOI] [PubMed] [Google Scholar]
- Marciano DC, Wang C, Hsu T-K et al. Evolutionary action of mutations reveals antimicrobial resistance genes in Escherichia coli. Nat Commun 2022;13:3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego N, Koes D. 3Dmol.js: Molecular visualization with WebGL. Bioinformatics 2015;31:1322–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 1.8, 2015.
- Su W, Johnston B. r3dmol: An R package for visualizing molecular data in 3D, 2022.
- Swings T, Marciano DC, Atri B et al. CRISPR-FRT targets shared sites in a knock-out collection for off-the-shelf genome editing. Nat Commun 2018;9:2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Barrick JE, Ribeck N et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 2016;536:165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierrafría VH, Rioualen C, Salgado H et al. RegulonDB 11.0: Comprehensive high-throughput datasets on transcriptional regulation in Escherichia coli K-12. Microb Genom 2022;8(5):mgen000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bergh B, Swings T, Fauvart M et al. Experimental design, population dynamics, and diversity in microbial experimental evolution. Microbiol Mol Biol Rev 2018;82:e00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Konecki DM, Marciano DC et al. Identification of evolutionarily stable functional and immunogenic sites across the SARS-CoV-2 proteome and greater coronavirus family. Bioinformatics 2021;37:4033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AD, Venner E, Marciano DC et al. Accounting for epistatic interactions improves the functional analysis of protein structures. Bioinformatics 2013;29:2714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Nicholson WL. Experimental evolution of Bacillus subtilis. Environ Microbiol 2017;19:3415–22. [DOI] [PubMed] [Google Scholar]
- Zhang K, Su L, Wu J et al. Recent advances in recombinant protein production by Bacillus subtilis. Annu Rev Food Sci Technol 2020;11:295–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in its online supplementary material and its GitHub repository.