Summary
Background
Genomic alterations in DNA damage response (DDR) genes are common in metastatic castration-resistant prostate cancer (mCRPC). Understanding how these genomic events impact prognosis and/or treatment response is vital for optimising clinical outcomes.
Methods
Targeted sequencing was performed on 407 plasma samples from 375 men with mCRPC. Using the CLIA-certified PredicineCARE™ cell-free DNA (cfDNA) assay, pathogenic alterations in 152 key genes (including 27 DDR-related genes) were assessed, as was the presence and mechanisms of biallelic loss in BRCA2.
Findings
At least one DDR alteration was present in 34.5% (129/375) of patients (including monoallelic alterations). The most frequently altered DDR genes were BRCA2 (19%), ATM (13%), FANCA (5%), CHEK2 (5%) and BRCA1 (3%). Patients with BRCA alterations, especially BRCA2, had significantly worse progression-free survival (PFS) (Hazard ratio (HR) 3.3 [95% CI 1.9–6.0]; Cox regression p < 0.001), overall survival (HR 2.2 [95% CI 1.1–4.5]; Cox regression p = 0.02) and PSA response rates to androgen receptor (AR) pathway inhibitors (32% vs 60%, chi-square p = 0.02). BRCA-deficient tumours were also enriched for alterations within multiple genes including in the AR and PI3K pathways. Zygosity of BRCA2 alterations had no discernible impact on clinical outcomes, with similarly poor PFS for monoallelic vs biallelic loss (median 3.9 months vs 3.4 months vs copy neutral 9.8 months).
Interpretation
These data emphasise that the BRCA genes, in particular BRCA2, are key prognostic biomarkers in mCRPC. The clinical utility of BRCA2 as a marker of poor outcomes may, at least in cfDNA assays, be independent of the zygosity state detected. Enrichment of actionable genomic alterations in cfDNA from BRCA-deficient mCRPC may support rational co-targeting strategies in future clinical trials.
Funding
Several funding sources have supported this study. A full list is provided in the Acknowledgments. No funding was received from Predicine, Inc. during the conduct of the study.
Keywords: Biomarker, Cell-free DNA, Prostate, Prostate cancer, ctDNA, BRCA
Research in context.
Evidence before this study
Literature published prior to this study includes analysis of DDR genes using both solid tumour samples and liquid biopsies. The frequency and therapeutic sensitivity of DDR defects in advanced prostate cancer has been extensively reported, providing the rationale behind recent trials demonstrating efficacy of inhibitors of poly-(ADP ribose) polymerase (PARP) in metastatic castration-resistant prostate cancer (mCRPC). Unfortunately, durable responses to PARP inhibitors in mCRPC remain elusive and primary resistance is an additional major challenge. In addition, smaller prior studies have produced divergent data on whether DDR alterations confer better or worse clinical outcomes on non-PARP inhibitor based systemic therapies in mCRPC.
Added value of this study
To the best of our knowledge, this is the only study that has performed an in-depth genomic analysis of DDR defects using highly sensitive targeted liquid biopsy techniques in a large cohort of advanced prostate cancer patients receiving contemporary systemic therapies. This study provides an atlas of pathogenic somatic and germline DDR defects observed in a large cohort of mCRPC patients. In particular, we investigated the prevalence and mechanisms of biallelic loss of BRCA2 alterations to determine their relative importance on clinical outcomes in mCRPC, as well as the effect of any type of deleterious BRCA alteration. In a cohort of patients with durable follow-up and extensive clinical annotation, we were able to successfully identify markers associated with poor prognosis as well as shorter responses to contemporary therapies. In addition to this, we also show that patients with deleterious BRCA alterations tend to display a distinct genomic phenotype, with tumours enriched for several genes that can be targeted therapeutically.
Implications of all the available evidence
Our results provide insights into the landscape of DDR defects in advanced prostate cancer in the context of contemporary systemic therapies. We propose that the identification of a pathogenic BRCA2 alteration on at least one allele within plasma cfDNA is sufficient to identify patients with inferior clinical outcomes, and that a distinct genomic phenotype is present in patients with BRCA1/2 alterations, with a high prevalence of potentially actionable co-targets i.e. in the AR and PIK3CA pathways. Clinical trials that leverage rational co-targeting strategies may provide an opportunity to enhance the efficacy of PARPi in mCRPC.
Introduction
Genomic alterations in DNA damage response (DDR) genes are present in 20–30% of metastatic castration-resistant prostate cancer (mCRPC) patients,1 and are of particular therapeutic relevance as they can confer sensitivity to PARP (poly-(ADP ribose) polymerase) inhibitors (PARPi).2, 3, 4, 5 Many of these DDR alterations, including in the homologous recombination repair (HRR) gene BRCA2, are early truncal events in prostate tumorigenesis and are readily detected in untreated primary prostate tissue.6 Thus, mCRPC patients will often receive standard-of-care treatments in the context of having DDR alterations. The relationship between DDR status and treatment outcomes with these agents has been contentious, with both positive and negative outcomes reported for the same therapy, including in androgen receptor pathway inhibitor (ARPI)-treated patients.7, 8, 9, 10, 11, 12 Understanding how these DDR alterations potentially impact their prognosis and/or response to non-PARPi treatments is vital to optimising the clinical outcomes of these patients. Similarly, although PARPi are now approved for mCRPC with HRR alterations, durable responses remain elusive and primary resistance is an additional major challenge.13 As a result, dissecting therapeutic vulnerabilities in DDR-altered mCRPC is crucial and may reveal novel PARPi-based combinations for evaluation in future clinical trials.
In the past decade, plasma circulating tumour DNA (ctDNA) analysis has emerged as a minimally-invasive approach for the genomic assessment of patient tumours that is readily amenable to temporal evaluation.14 Detectable in at least 60–85% of mCRPC patients,15 ctDNA is capable of recapitulating the complex intra- and inter-tumour heterogeneity typically seen in advanced prostate cancer and providing a contemporaneous molecular profile that is not necessarily captured in primary prostate biopsies. In this multi-institutional study, we used the CLIA-certified PredicineCARE™ cell-free DNA (cfDNA) assay that assesses 152 key genes, including 27 DDR-related genes, to profile 375 patients from Australia and the United States with mCRPC. Our objectives were to: i) investigate the prognostic significance of DDR alterations; ii) correlate DDR alterations with clinical outcomes from ARPIs and taxane chemotherapy; and iii) define co-occurring genomic alterations that may identify candidate therapeutic vulnerabilities in BRCA-deficient mCRPC.
Methods
Study cohorts and sample processing
Targeted sequencing data from 407 pre-treatment plasma and matched leukocyte DNA samples was collected from 375 men with mCRPC who were enrolled across two different liquid biopsy programs (note 28 and 2 men received second and third lines of therapy, respectively) for this retrospective study. Of these, 162 samples were collected at two Australian institutions (Monash Health and Chris O'Brien Lifehouse), with 245 samples collected at the Mayo Clinic Hospital (Rochester, Minnesota, USA). Of the Australian subgroup 145 samples were from patients commencing ARPI therapy or taxane chemotherapy (n = 90 ARPI, n = 55 taxanes) (Supplementary Figure S1a). Progression-free survival (PFS) and OS data were available for 145 samples and 371 patients, respectively. Details of sample collection and processing have been published previously.16,17 Baseline clinical characteristics and prior treatment exposure are presented in Supplementary Tables S1 and S2. The median follow-up time for non-deceased patients was 26.3 months.
A separate cohort of metastatic hormone-sensitive prostate cancer (mHSPC) patients was also recruited (n = 18) at Monash Health, Melbourne, Australia. Samples were collected and processed using the same approach for the mCRPC cohort, although a smaller targeted panel was used (DDR genes included ATM, ATR, BRCA2, BRCA1, CDK12, CHEK2, FANCA, FANCD2, and FANCI only). Baseline clinical characteristics and targeted panel information for this cohort are provided in Supplementary Tables S3 and S4
Ethics statement
For the Mayo Clinic Hospital cohort, patients were prospectively enrolled between September 2009 and March 2014 to an advanced prostate cancer biomarker registry prior to blood collection at the time of androgen deprivation therapy failure. For the Australian samples, patients with mCRPC or mHSPC were prospectively enrolled between September 2016 and August 2018. All patients provided written informed consent with ethics approval obtained from each institution's human research ethics committee (HREC/14/MH/342 and IRB# 09-007355).
Library preparation, sequencing and bioinformatic analysis
Plasma cfDNA and matching germline DNA (gmDNA) from leukocytes was extracted, underwent library construction and hybrid capture-based targeted sequencing according to previously validated methodology.16,18 The CLIA-certified PredicineCARE™ panel assesses 152 key genes (Supplementary Table S5), including 27 DDR-related genes (ATM, ATR, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, FANCA, FANCC, FANCD2, FANCG, FANCI, FANCL, FANCM, MLH1, MRE11A, MSH2, MSH6, NBN, PALB2, PMS2, RAD50, RAD51B, RAD51C, RAD51D, and RAD54L). Identification of pathogenic somatic variants, rare germline variants, and estimation of plasma tumour content (i.e. circulating tumour DNA, ctDNA fraction) was performed using published methods.16,18 Characterisation of copy number alterations (heterozygous loss vs homozygous loss) was performed as previously described.18 Other mechanisms of mutant allele-specific imbalance (loss of heterozygosity [LOH] or non-deletion LOH) were assumed if a patient had >1 type of alteration in the same gene. Throughout the manuscript, instances where only one allele is affected are referred to as monoallelic loss, whilst the loss/alteration of both alleles (via mutation and/or copy loss) is termed biallelic loss (Supplementary Figure S1b). Somatic mutations were considered clonal if the variant allele frequency (VAF) was ≥25% of the estimated ctDNA fraction. Briefly, pathogenicity was defined as any copy loss event (heterozygous or homozygous) in a DDR gene, and exonic single-nucleotide alterations/indels <10bp that were either truncating or had a ‘pathogenic/likely pathogenic’ assignation on Clinvar. Additionally, cell-free RNA (cfRNA) data from 279 patients using the PredicineRNA™ targeted sequencing assay16 was integrated into this study to test for co-occurrence of measures (specifically the frequency of AR splicing variants in DDR-altered patients; data is provided in Supplementary Table S6).
Validation of copy number analysis using low-pass whole genome sequencing
Low pass whole genome sequencing (LP-WGS) with an average coverage of 2.5x was performed on a subset of patient samples with sufficient additional plasma cfDNA following targeted sequencing (n = 46), using a previously described methodology.18 Briefly, reads were normalised for GC content and mappability and processed via the ichorCNA tool19 to estimate plasma copy number alterations using a hidden Markov model with the following parameter settings: genome partitioning at 1 Mb, using expectation-maximization initialization and a minimum threshold of >5% and >10% change to call a copy number change in autosomal and sex chromosomes, respectively. In total, using targeted sequencing, 34 DDR CNVs in 26 patients were identified across the 46 samples that underwent concurrent LP-WGS. Of these 34 CNVs, 31 were validated with LP-WGS, including 100% concordance for all BRCA2 copy number losses (Supplementary Figure S1c). The three discordant CNVs were not included in outcome analyses.
Clinical outcomes analysis and statistical methodology
Kaplan–Meier survival estimates (log-rank test) and Cox regression models (multivariable regression covariates: ctDNA fraction, presence of cancer-related pain at enrolment, presence of visceral metastases, ECOG performance status, prior taxane chemotherapy, and prior ARPI therapy) were used to assess the association between DDR alterations and clinical outcomes. All somatic and germline pathogenic DDR alterations were included in the analyses. Clinical outcomes assessed were OS (time from treatment commencement until death from any cause), PSA response (PSA decline from baseline of ≥50%, confirmed ≥3 weeks later), and PFS (time from treatment commencement to first confirmed PSA progression, clinical or radiographic progression, treatment reallocation, or death from prostate cancer). Overall survival analyses included unique patients only whilst PSA response and PFS analyses included all available samples. Statistical significance was defined as p < 0.05, except for analyses involving the BRCA2 gene, which were adjusted using the Bonferroni correction method to counteract simultaneous multiple testing.
Role of funders
No specific funding was provided for the study, and no funders had a role in study design, data collection, data analyses, interpretation, or writing of the report.
Results
Landscape of cfDNA DDR alterations in mCRPC
Across the 407 samples, patient cfDNA and matched gmDNA were sequenced to a median unique read depth of 5733X and 323X, respectively. ctDNA was detectable in 314 of 407 (77%) samples, with a median ctDNA fraction in ctDNA positive samples of 16.3% (data in Supplementary Table S7). Consistent with prior findings,15,20 patients with higher plasma ctDNA (≥2%) had significantly shorter OS (median 19.1 vs 40.4 months, p < 0.001; Mann–Whitney U, Supplementary Figure S2). Men with DDR-altered tumours also had higher plasma ctDNA fraction compared to those without any DDR alterations (median 22% vs 3%, p < 0.001; 95% CI 17.7–26.3% and 0.13–6.13% respectively, Mann–Whitney U) however this was not observed when comparing patients with/without DDR mutations only (and not copy number loss) (median 3.5% vs 6.5%, p = 0.061; 95% CI 0.93–6.1 and 2.9–10.1 respectively, Mann–Whitney U).
Of the 375 unique patients in this study, 146 (39%) had a deleterious germline or somatic alteration (copy number variation or mutation) on at least one allele in ≥1 of 27 DDR genes (Fig. 1a and Supplementary Table S8). A subset (4.5%, n = 17) of this cohort harboured a mutation in mismatch repair (MMR) genes (MLH1, MSH2 and MSH6) and were subsequently excluded from downstream analysis due to the propensity of these tumours to accumulate multiple passenger mutations.21 This observation was reflected in our own cohort, where all somatic non-MMR DDR point mutations were classed as subclonal in MMR-altered patients (vs 38% in MMR-unaltered patients). Following the removal of these patients, 34.5% (n = 129) of the total cohort exhibited DDR alterations (including monoallelic alterations), with 6.7% (32 patients) harbouring a deleterious germline mutation.
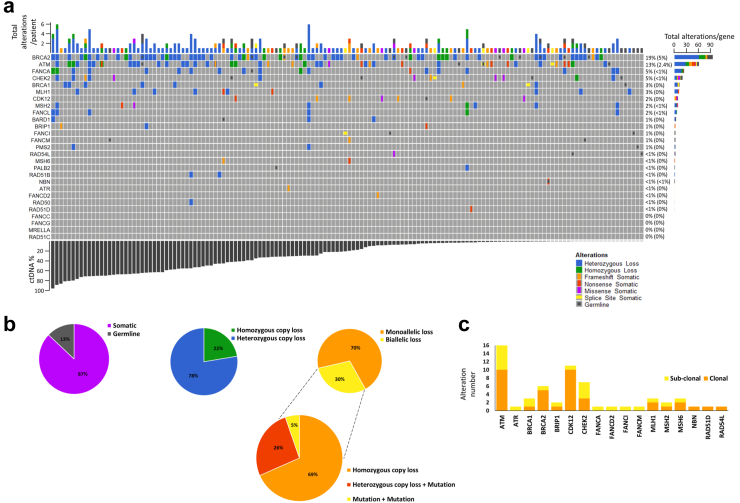
Fig. 1.
The prevalence and characteristics of pathogenic DDR alterations identified in a cohort of mCRPC patients. a Oncoprint of all deleterious DDR alterations and their frequency in the overall cohort (n = 375), ordered by patient ctDNA fraction. Right hand side percentages show frequency of monoallelic alterations in the cohort, with frequency of biallelic alterations shown within the brackets. b Further analysis of BRCA2 alteration types identified in the cohort (n = 65 with BRCA2 alterations), including mechanisms of two-copy loss (biallelic loss), which were classed as either homozygous copy loss, heterozygous loss combined with a pathogenic mutation, or two pathogenic mutations (assumed to be on either allele). c Clonality estimates of all pathogenic somatic mutations (SNVs and small indels) identified in DDR genes (n = 375).
After excluding patients with MMR alterations, the most frequently altered DDR genes were BRCA2 (17%, n = 65), ATM (13%, n = 46), FANCA (5%, n = 18), CHEK2 (5%, n = 18) and BRCA1 (3%, n = 10). Regarding BRCA2-altered patients (n = 65), most defects were heterozygous (78% vs 22% homozygous loss, monoallelic (70% vs 30% biallelic) and somatic (87% vs 13% germline) (Fig. 1b). Somatic BRCA2 alterations were more frequent with prior enzalutamide or abiraterone (35% vs 24%, p = 0.1, Fisher's exact test). All somatic point mutations in Fanconi Anemia (FA) complex genes and ATR were subclonal, but most in CDK12 and BRCA2 were clonal (Fig. 1c). Germline DDR alterations were most prevalent in ATM and BRCA2 (in 3% and 1.3% of patients, respectively; Fig. 1a.
DDR alterations and clinical outcomes in mCRPC
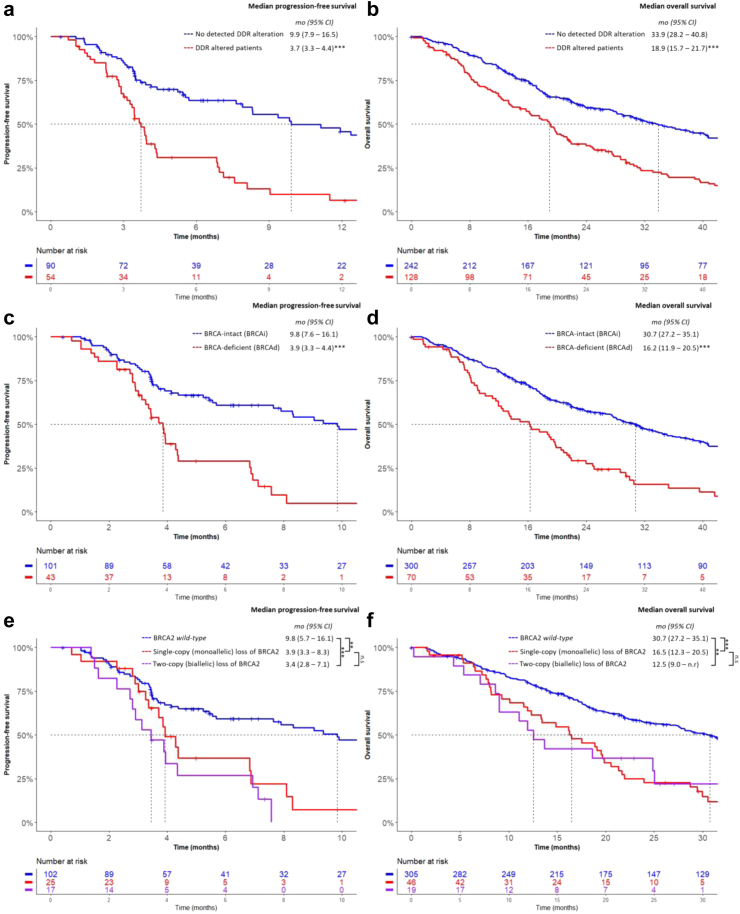
Median OS and PFS for the cohort were 23.7 and 6.9 months, respectively, although PFS data was only available for 146 patient samples. Patients with any detectable pathogenic DDR alteration (≥1) had significantly shorter median PFS and OS compared to DDR-intact cases (3.7 vs 9.9 months, p < 0.001; 18.9 vs 33.9 months, p < 0.001, respectively, log-rank test, Fig. 2a and b). These findings remained significant on multivariable analyses after adjustment for ctDNA fraction and other known poor prognostic factors (Table 1). Any DDR defect was also associated with a lower PSA response rate in the overall cohort (37% vs 66, chi-square p < 0.001; Supplementary Table S9). When DDR alteration types were categorised into BRCA or non-BRCA defects, however, the presence of a non-BRCA DDR alteration was not associated with OS, PFS or PSA response rate (Table 1, Supplementary Table S9). The same observation was made when ATM and CDK12 were individually assessed, although it should be noted that we may be underpowered to assess the significance of CDK12 alterations due to low prevalence (8 patients) in this cohort.
Fig. 2.
Kaplan–Meier analysis of progression-free survival and overall survival in months according to a, b the presence of any pathogenic DDR alteration, c, dBRCA alteration status within the cohort and e, fBRCA2 zygosity. Patients with co-occurring MMR alterations were removed prior to analysis. Note that not all patients had associated progression-free survival data, resulting in a smaller cohort size compared to overall survival analyses. n.s = p > 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, log regression analysis.
Table 1.
Univariable and multivariable Cox proportional hazards analysis of clinical endpoints based on known poor clinical prognosticators and commonly altered DNA damage response genes.
| Variable | Progression-free survival (n = 145) |
Overall survival (n = 285b) |
||||||
|---|---|---|---|---|---|---|---|---|
| na | HR | 95% CI | p | na | HR | 95% CI | p | |
| UNIVARIABLE ANALYSIS | ||||||||
| Any DNA damage response gene | 54 | 3.0 | 1.9–4.7 | <0.001 | 128 | 1.8 | 1.4–2.2 | <0.001 |
| non-BRCAd | 15 | 2.1 | 0.93–4.6 | 0.1 | 63 | 1.4 | 0.93–1.9 | 0.7 |
| BRCA1/2 (BRCAd) | 43 | 3.1 | 1.9–5.0 | <0.001 | 70 | 2.1 | 1.6–2.8 | <0.001 |
| ATM | 19 | 1.3 | 0.66–2.5 | 0.5 | 46 | 1.6 | 1.1–2.3 | 0.01 |
| BRCA1 | 7 | 4.8 | 2.2–11.0 | <0.001 | 10 | 1.9 | 1.0–3.7 | 0.039 |
| BRCA2 | 41 | 2.9 | 1.8–4.6 | <0.001 | 65 | 2.1 | 1.6–2.9 | <0.001 |
| CDK12 | 2 | 5.1 | 0.64–41 | 0.12 | 8 | 1.2 | 0.57–2.6 | 0.6 |
| Low ctDNA (<2%) | 40 | 0.44 | 0.26–0.74 | 0.002 | 142 | 0.6 | 0.45–0.74 | <0.001 |
| Prior chemotherapy | 64 | 1.5 | 0.98–2.3 | 0.06 | 76/204 | 1.5 | 1.1–2.1 | 0.04 |
| Prior ARPI therapy | 39 | 1.9 | 1.2–3.0 | 0.009 | 27/125 | 1.4 | 0.8–2.4 | 0.2 |
| ECOG performance status | 145 | 1.6 | 1.1–2.3 | 0.01 | 124 | 1.8 | 1.2–2.8 | 0.007 |
| Visceral metastasis at baseline | 21 | 1.3 | 0.7–2.3 | 0.4 | 15/124 | 1.7 | 1.0–3.1 | 0.051 |
| Cancer-related pain at baseline | 59 | 2.0 | 1.3–3.1 | <0.001 | 42/124 | 1.6 | 1.01–2.6 | 0.04 |
| MULTIVARIABLE ANALYSISc | ||||||||
| Any DNA damage response gene | 54 | 2.9 | 1.7–4.7 | <0.001 | 128 | 2.7 | 1.6–4.6 | <0.001 |
| BRCA1/2 (BRCAd) | 43 | 2.9 | 1.7–4.8 | <0.001 | 70 | 2.2 | 1.3–3.7 | 0.004 |
| ATM | 19 | – | – | - | 46 | 1.6 | 0.72–3.7 | 0.2 |
| BRCA1 | 7 | 3.3 | 1.4–7.5 | 0.005 | 10 | 2.1 | 0.60–7.6 | 0.2 |
| BRCA2 | 41 | 2.6 | 1.5–4.3 | <0.001 | 65 | 1.9 | 1.1–3.3 | 0.02 |
Clinical variables included in the multivariable analysis: ctDNA fraction (continuous), prior chemotherapy, prior ARPI therapy, presence of visceral metastasis, presence of pain at baseline and ECOG PS > 2. Bonferroni adjustment for multiple testing of BRCA2 variables allows for p-values of <0.005 to be accepted as significant and are highlighted in bold. ARPI = androgen receptor pathway inhibitor, ctDNA = circulating tumour DNA, ECOG PS = Eastern Cooperative Oncology Group performance status.
Whilst overall survival data was available for the whole cohort, progression-free survival data was only available for Australian patient samples.
When <285 of the patients had data available for a variable, the denominator is shown in the ‘n’ column of the table.
Only genes/groups with p < 0.05 in univariable analysis were included in multivariable analysis.
The ‘non-BRCA’ variable includes all patients without BRCA1/2 alterations but do have other DDR alterations present.
Patients with BRCA1/2 alterations had higher mean ctDNA fractions compared to patients with non-BRCA alterations (29% vs 19%, chi-square p = 0.002; Mann–Whitney U). BRCA1/2 alterations were associated with significantly lower median PFS and OS (3.9 vs 9.8 months, p < 0.001; and 16.2 vs. 30.7 months, p < 0.001, respectively; log-rank, Fig. 2c and d), retaining significance on multivariable analyses after adjustment for ctDNA fraction (Table 1). Additionally, these BRCA-deficient (BRCAd) patients had lower PSA response rates compared to the BRCA-intact (BRCAi) cohort (37% vs 63%, chi-square p = 0.005; Supplementary Table S9). We also evaluated outcomes for BRCA1 and BRCA2 patients separately. BRCA1 was independently associated with shorter PFS but not OS (Table 1). BRCA2-deficient (BRCA2d) patients had shorter PFS and OS (3.9 vs 9.4 months, p < 0.0001; and 16.1 vs 30.7 months, p < 0.0001, respectively; log-rank, Supplementary Figures S3a and b), which maintained significance on multivariable analyses (Table 1). Altogether, these data establish BRCA alterations as key biomarkers linked to inferior outcomes in mCRPC.
BRCA2 zygosity as a biomarker of patient outcomes in mCRPC
In prostate and other cancers, monoallelic losses in tumour suppressor genes, including genes involved in DDR, are not considered to exert pathogenic effects except in cases of haploinsufficiency.22,23 Nevertheless, prior data indicates that monoallelic BRCA1/2 loss is associated with attenuated responses to PARPi in mCRPC.3,24 Whether this applies to other treatments is unknown and consequently, we investigated BRCA zygosity and its association with outcomes on non-PARPi therapies. Due to the low frequency of BRCA1 alterations with associated clinical data (n = 10), only BRCA2 underwent this more in-depth zygosity analysis.
Of the 375 unique patients in this study, 65 had evidence of a pathogenic BRCA2 alteration (Fig. 1a). Pathogenic BRCA2 SNVs were most often clonal in nature (83% of all BRCA2 altered patients, Fig. 1c) and 4% of all plasma samples in this study exhibited homozygous BRCA2 loss, similar to the frequency reported in tissue sequencing studies.10 Almost one third of BRCA2d patients (n = 19) had evidence of biallelic BRCA2 alterations, either by a second SNV (n = 1), LOH (a heterozygous loss combined with an SNV, n = 5), or homozygous gene loss (n = 13) (Fig. 1b). Notably, there was no significant difference in plasma tumour content in patients with single vs two-copy loss of BRCA2 (median ctDNA fraction for heterozygous loss 34.5 vs 30.9% for homozygous loss, p = 0.83, and median ctDNA for monoallelic loss 21.8% vs 31% for biallelic loss, p = 0.71; Mann–Whitney testing).
Importantly, the presence of any pathogenic BRCA2 alteration (both monoallelic and biallelic) was independently associated with inferior PFS and OS (Table 1). Additionally, both heterozygous and homozygous BRCA2 losses were associated with shorter PFS (HR 2.8, [95% CI 1.6–4.8]; p < 0.001 and HR 3.8, [95% CI 1.8–8.0]; p < 0.001, respectively, Cox regression, Table 2). Interestingly, BRCA2 zygosity (monoallelic vs biallelic alterations) had similar effects on outcomes, with comparably shorter median PFS and OS vs BRCA2-intact patients (median PFS for monoallelic 3.9 months vs biallelic 3.4 months vs intact 9.8 months, and median OS 16.5 vs 12.5 vs 30.7 months; Fig. 2e and f). Collectively, these data indicate that identification of specific BRCA2 zygosity status in cfDNA may not be necessary for patient stratification in non-PARPi treated mCRPC. Rather, the detection of any anomality in BRCA2 is indicative of poor outcomes.
Table 2.
Cox proportional hazards analysis of clinical endpoints based on genetic variance of deleterious BRCA2 alterations.
| Variable | Progression-free survival |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|---|
| n | HR | 95% CI | p | n | HR | 95% CI | p | |
| BRCA2 point mutation | 13 | 2.2 | 1.1–4.5 | 0.024 | 14 | 1.2 | 0.62–2.2 | 0.6 |
| BRCA2 deletion (heterozygous) | 22 | 2.8 | 1.6–4.8 | <0.001 | 43 | 2.3 | 1.6–3.3 | <0.001 |
| BRCA2 deletion (homozygous) | 11 | 3.8 | 1.8–8.0 | <0.001 | 13 | 3.4 | 1.8–6.3 | <0.001 |
| BRCA2 loss (monoallelic) | 24 | 2.5 | 1.4–4.4 | 0.002 | 46 | 2.1 | 1.5–3.0 | <0.001 |
| BRCA2 loss (biallelic) | 17 | 3.3 | 1.8–6.0 | <0.001 | 19 | 2.2 | 1.3–3.8 | 0.003 |
| BRCA2 (germline) | 10 | 2.3 | 1.0–5.3 | 0.043 | 7 | 1.4 | 0.53–3.9 | 0.5 |
| BRCA2 (somatic) | 37 | 3.0 | 1.9–4.8 | <0.001 | 60 | 2.3 | 1.7–3.2 | <0.001 |
Bonferroni adjustment for multiple testing of BRCA2 variables allows for p-values of <0.005 to be accepted as significant and are highlighted in bold.
BRCA alterations are linked to clinical utility of AR pathway inhibitors
We next evaluated whether any DDR defects were linked to outcomes according to the type of systemic therapy received. Whilst the presence of any detectable pathogenic DDR defect (≥1) was associated with inferior PFS on taxane chemotherapy, no specific DDR alteration was significantly associated with outcomes in chemotherapy-treated individuals, nor were any sub-types of BRCA2 alteration (Supplementary Tables S10 and S11). However, a key limitation of these analyses is the small numbers in many subgroups.
The DDR landscape for ARPI-treated patients (n = 90) stratified by length of PFS is shown in Supplementary Figure S4. For these patients, any deleterious DDR defect (≥1) was linked to significantly shorter PFS (HR 3.0, [95% CI 1.7–5.1]; p < 0.001) and OS (HR 3.3, [95% 1.7–6.3 CI]; p < 0.001, Cox regression, Table 3), and lower PSA response rates (32% vs 64%, chi-square p = 0.004, Supplementary Table S12). Likewise, decreased PFS (HR 3.3, [95% CI 1.9–6.0]; Cox regression p < 0.001), OS (HR 2.2, [95% CI 1.1–4.5]; Cox regression p = 0.02) and PSA response rates (32% vs 60%, chi-square p = 0.02) were observed in BRCAd patients. As separate gene defects, BRCA1 and BRCA2 were significantly associated with shorter PFS (Table 3) and lower PSA response rates (0% vs 56% p = 0.04, and 33% vs 59% p = 0.04, respectively, chi-square, Supplementary Table S12). Additionally, PFS was significantly shorter for ARPI-treated patients with homozygous BRCA2 deletion, monoallelic loss and biallelic loss compared to BRCA2-intact patients (Table 3).
Table 3.
Cox proportional hazards analysis of clinical endpoints based on commonly altered DNA damage response genes and zygosity of BRCA2 in patients who received an androgen receptor pathway inhibitor (ARPI).
| Variable | Progression-free survival |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|---|
| n | HR | 95% CI | p | n | HR | 95% CI | p | |
| PATIENTS WHO RECEIVED AN ARPI | ||||||||
| Any DNA damage response gene | 31 | 3.0 | 1.7–5.1 | <0.001 | 24 | 3.3 | 1.7–6.3 | <0.001 |
| non-BRCAa | 9 | 1.4 | 0.58–3.2 | 0.5 | 8 | 4.4 | 1.9–10 | <0.001 |
| BRCA1/2 (BRCAd) | 22 | 3.3 | 1.9–6.0 | <0.001 | 17 | 2.2 | 1.1–4.5 | 0.02 |
| ATM | 8 | 1.1 | 0.45–2.8 | 0.8 | 5 | 2.8 | 1.0–7.8 | 0.06 |
| BRCA1 | 4 | 11 | 3.6–34 | <0.001 | 1 | – | – | – |
| BRCA2 | 21 | 3.1 | 1.7–5.5 | <0.001 | 16 | 2.0 | 1.0–4.0 | 0.052 |
| BRCA2 point mutation | 7 | 2.6 | 1.1–6.1 | 0.02 | 5 | 0.86 | 0.20–3.7 | 0.8 |
| BRCA2 deletion (heterozygous) | 12 | 2.6 | 1.3–5.3 | 0.01 | 9 | 2.2 | 0.93–5.1 | 0.07 |
| BRCA2 deletion (homozygous) | 4 | 7.0 | 2.2–22 | <0.001 | 3 | 3.9 | 1.2–13 | 0.029 |
| BRCA2 loss (monoallelic) | 14 | 2.7 | 1.4–5.4 | 0.004 | 11 | 2.2 | 1.0–4.7 | 0.047 |
| BRCA2 loss (biallelic) | 7 | 3.8 | 1.6–8.9 | 0.002 | 5 | 1.7 | 0.52–5.9 | 0.4 |
| BRCA2 (somatic) | 20 | 3.0 | 1.6–5.4 | <0.001 | 16 | 2.0 | 1.0–4.0 | 0.052 |
Bonferroni adjustment for multiple testing of BRCA2 variables allows for p-values of <0.005 to be accepted as significant and are highlighted in bold.
The ‘non-BRCA’ variable includes all patients without BRCA1/2 alterations but do have other DDR alterations present.
Pathogenic androgen receptor alterations co-occur with BRCA defects in mCRPC
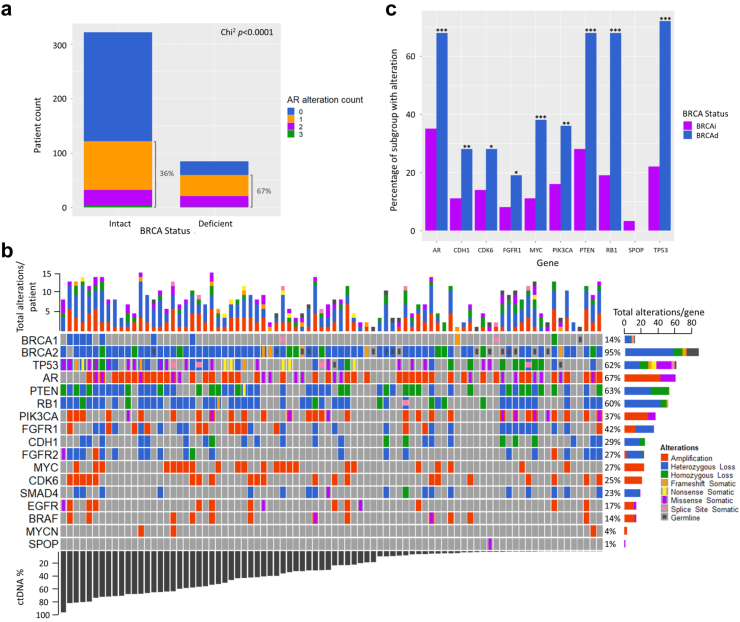
Although PARP inhibitors show impressive clinical activity in mCRPC harbouring BRCA defects, treatment outcomes are highly variable and durability of efficacy can be short-lived.13 Therefore, it is essential to identify potential drivers of primary and acquired resistance to PARP inhibitors. To test whether pathogenic BRCA defects in mCRPC associate with other classes of driver alterations, we compared the frequency of other panel-assessed genes in BRCAd vs BRCAi. Given critical cross-talk between the AR and HRR pathways,25 we initially analysed AR gene alterations in BRCAd vs BRCAi patients, both in cfDNA (AR amplification and AR mutations) and cfRNA (AR splice variants AR-V3, AR-V7 and AR-V9, which are biomarkers of therapeutic resistance in mCRPC).26 We found that 67% (n = 47/70) of BRCAd patients had ≥1 AR alteration compared to 36% (n = 109/305) of BRCAi patients (p < 0.001, chi-square; Fig. 3a), indicating significant activation of AR pathway signalling in the context of deficient homologous recombination repair. These data are supported by prior reports that demonstrated unfavourable outcomes in mCRPC patients harbouring compound AR alterations.16,27
Fig. 3.
Genomic landscape of common driver genes in BRCA-deficient patients. a The prevalence of AR alterations in BRCA-intact (BRCAi) and BRCA-deficient (BRCAd) patients. AR alteration types included point mutations in the ligand-binding domain, gene amplification (as detected in cfDNA), and/or expression of a constitutively active splicing variant as detected in cell-free RNA (AR-V3, V7, V9) and b Oncoprint of deleterious alterations in driver genes identified in the sub-cohort of mCRPC patients that are BRCAd, ordered by ctDNA fraction. The percentages on the right indicate the frequency of alterations for each specific gene across the entire cohort. c Analysis of mutation prevalence based on BRCA status in patients with ctDNA fraction >20%. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, Pearson's Chi2 test.
Enrichment of specific genomic alterations in BRCA-deficient mCRPC
We next investigated differences in prevalence of other commonly-altered genes in mCRPC between BRCAd and BRCAi patients using chi-square analysis (Supplementary Table S13). The genomic landscape of these two subgroups are shown in Fig. 3b and Supplementary Figure S5. Since BRCAd patients had higher ctDNA fractions (median 29% vs 19%), only patients with ctDNA fractions greater than 20% were included in the statistical analysis. Notably, alterations in genes associated with aggressive prostate cancer and neuroendocrine differentiation (TP53, RB1 and MYC)28 were enriched in BRCAd vs BRCAi patients (72% (34/47), 68% (32/47) and 38% (18/47) vs 22% (43/192), 18% (35/192) and 11% (21/192) in BRCAi patients; all chi-square p < 0.001, Fig. 3b and c, Supplementary Figure S5 and Supplementary Tables S13 and S14). The phosphoinositide 3-kinase (PI3K) pathway is frequently dysregulated in prostate cancer, most commonly via PTEN loss/mutations or PIK3CA amplification/mutations.18 The PI3K pathway was preferentially activated in BRCAd patients, with PIK3CA and PTEN alterations present in 36% (17/47) of BRCAd vs 16% (30/192) of BRCAi, and 68% (32/47) vs 28% (53/192) respectively (chi-square p-values 0.02 and <0.001, respectively). Fibroblast Growth Factor Receptor 1 (FGFR1), a driver of prostate cancer metastatic progression,29 was altered (via copy number gain) at twice the frequency in BRCAd patients (19% [9/47] vs 8% [16/192], chi-square p = 0.03). Amplification or mutation of the cyclin-dependent kinase 6 (CDK6) gene, a G1 cell cycle phase regulator that potentiates AR transcriptional activity,30 was also significantly enriched in BRCAd vs BRCAi patients (28% [13/47] vs 14% [26/192], chi-square p = 0.02).
Interestingly, no SPOP mutations were identified in BRCAd patients with >20% ctDNA fraction, lower than prior reports,31 but was at expected frequency (3.2% [6/192]) in BRCAi patients. Given SPOP mutations sensitise to ARPI therapy,32 these data may be a contributing factor in the worse outcomes from ARPI in BRCAd patients in our cohort. Similarly, CDH1 loss (via deletion or inactivating mutation), which is associated with primary resistance to chemotherapy,33 was more prevalent in BRCAd vs BRCAi patients (28% [13/47] vs 11% [22/192], chi-square p = 0.005). This may partially account for the worse PFS and OS, albeit non-significant, observed in BRCAd patients treated with taxanes (Supplementary Table S10).
BRCA2 alterations arise before castrate-resistance and are identifiable in plasma DNA in the hormone-sensitive setting
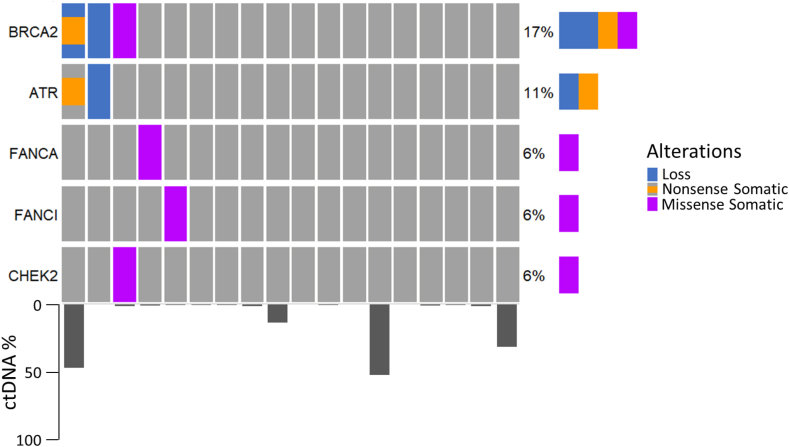
As earlier stages of disease are often associated with a lower tumour burden,34 it may not be feasible to utilise plasma cfDNA assays to identify deleterious somatic alterations due to the high technical sensitivity required, especially if the alterations are subclonal. To investigate the viability of ctDNA assays in earlier disease settings of prostate cancer, we performed targeted plasma sequencing on 18 patients with mHSPC prior to commencement of androgen deprivation therapy. Of these patients, only two had undetectable ctDNA, although unsurprisingly the median ctDNA fraction for the group was lower than in the mCRPC cohort (1.22%, range 0–51.9%). As in the mCRPC cohort, BRCA2 was the most commonly altered DDR gene (Fig. 4), although no germline alterations were identified. We found that 17% (3/18) of patients with hormone-sensitive disease were BRCA2d (no BRCA1 alterations were identified), comparable to our findings in mCRPC (BRCA2d 19%). This confirms that BRCA2 alterations are an early event in prostate cancer, supporting both genomic profiling of hormone-sensitive disease as well as upfront systemic therapy trials for mHSPC incorporating PARPi including Amplitude (NCT04497844) and Talapro-3 (NCT04821622).
Fig. 4.
Oncoprint of deleterious DDR alterations and their frequency in the metastatic hormone-sensitive prostate cancer setting, with accompanying ctDNA fraction data (n = 18). Other DDR genes that were assessed but had no identifiable mutations were ATM, BRCA1, CDK12, and FANCD2.
Discussion
Results from multiple pivotal clinical trials1, 2, 3, 4, 5 have heralded a new era of personalised medicine for mCRPC, with PARPi emerging as a new standard of care for patients with an HRR defect. These trials highlight the critical role that profiling the DDR pathway has in the management of mCRPC patients.
Across a cohort of 375 patients with mCRPC, the CLIA-certified PredicineCARE™ cfDNA assay detected a wide repertoire of alterations in 27 individual DDR-related genes. Interestingly, the frequency of a DDR alteration (excluding MMR altered patients) of 34.5% in our cohort was somewhat higher than prior studies using tissue-based assays that reported DDR alterations in 23–28%.35,36 Potential reasons for this difference include higher detection rates of somatic alterations with contemporaneous cfDNA samples compared to primary archival tissue, use of a larger DDR panel than prior studies (e.g. PROfound, which used 15 genes1), and the inclusion of monoallelic events as DDR-altered. Irrespective, we and others have shown high concordance between cfDNA and tumour tissue in mCRPC,10,37,38 reinforcing the reliability of using cfDNA for molecular profiling in this cohort.
We observed that having any deleterious DDR defect was linked to significantly worse PFS, OS and PSA response rates. Exploring this further, the effect appeared to be largely driven by BRCA2 rather than non-BRCA genes. These data emphasise that the BRCA genes, in particular BRCA2, are key prognostic biomarkers in mCRPC.10,15 The poorer prognosis conferred by BRCA2 is consistent with an aggressive disease phenotype driven, at least in part, by more genomically unstable tumours harbouring higher somatic mutation rates and widespread aneuploidy.39, 40, 41
We next investigated for any link between DDR genes and outcomes on specific therapy for mCRPC, namely taxanes and ARPI. With the exception of lower PFS for patients with any DDR alteration, no other endpoints for individual genes reached statistical significance in patients treated with taxanes. It is worth noting that our data should not be considered conclusive due to the small number of patients that received chemotherapy, with the heterogeneity of the cohort being a limitation of this study. Furthermore, prior studies have provided conflicting data regarding docetaxel efficacy and DDR alterations42,43 and this issue warrants further evaluation.
Conversely, in ARPI-treated patients we found that presence of any DDR defect or BRCAd conferred significantly worse PFS, OS and PSA response rates. The relationship between DDR status and treatment outcomes with ARPI has been contentious with some studies reporting benefit from ARPI in DDR-positive tumours7, 8, 9 but others demonstrating deleterious outcomes.10,11 More recently, data from the phase III PROpel trial44 showed inferior median radiographic PFS in the control arm (Abiraterone only) for patients with DDR alterations vs. wild-type (13.8 months vs 19.1 months). Altogether, these data point to attenuated benefit from ARPI in DDR altered disease.
Our data indicate that the prognostic and predictive utility of BRCA2 may be independent of zygosity state. Similar PFS and OS data was observed for heterozygous and homozygous BRCA2 deletions, and for monoallelic and biallelic loss. In contrast, the phase III TRITON2 trial reported PSA response rates to Rucaparib of 75% and 11% in biallelic and monoallelic BRCA2d patients respectively.3 Thus it appears that zygosity state of BRCA2 influences outcomes from PARPi but not from ARPI (or taxanes). However, in patients with low ctDNA fraction, we acknowledge a second alteration may have been missed in cases with heterozygous deletion or monoallelic loss, and the majority of monoallelic cases likely have a ‘second hit’, as is seen in biopsy specimens.23 A major challenge of ctDNA assays, including our own, remains the sensitive detection of low frequency somatic variants whilst minimising interference. Nevertheless, we propose that the identification of a pathogenic BRCA2 alteration on at least one allele within pre-treatment ctDNA is sufficient to identify patients with inferior clinical outcomes.
We found significant enrichment of potentially actionable targets in patients with BRCAd cancers. BRCAd patients were almost twice as likely to have an AR alteration compared to BRCAi patients. Although trials have shown benefit of combining ARPI with PARPi,44 AR amplification confers limited benefit from ARPI,16,45 and testing other strategies for targeting the AR pathway in BRCAd mCRPC may be warranted. These include PROTACs,46 AR N-terminal domain inhibitors47 and AR DNA-binding domain inhibitors.48 Alterations in genes associated with neuroendocrine cancer (RB1 and TP53),28 were also more common in BRCAd cancers. These data suggest a potential benefit from co-targeting PARP and either DLL3, which is a key therapeutic target in small cell lung cancer,49 or bromodomain and extra-terminal (BET) proteins, which are a promising target in AR-null mCRPC.50 PIK3CA and PTEN alterations were also more frequent in BRCAd patients, pointing to possible combination strategies with AKT inhibitors, which have demonstrated benefit in PTEN-null mCRPC.51 Similarly, activating mutations in FGFR and CDK6, enriched in BRCAd patients, are targetable with FGFR inhibitors and CDK4/6 inhibitors respectively, both of which have progressed into clinical practice in non-prostate cancers.
Although a strength of our analyses was to include data from both Australia and the US, we acknowledge the possibility of confounders that may have impacted on outcomes for patients in their respective countries. Due to there being very little overlap in clinicopathological factors between the two cohorts, we were not able to formally address this issue. We also acknowledge that US data was collected as far back as 2009, and therefore may have less relevance in the context of contemporaneous available treatments in 2023.
In summary, in a large real-world international cohort, we show that BRCA alterations are associated with worse prognosis and reduced benefit from ARPI in mCRPC and are genomic events occurring prior to the development of castrate-resistant disease. A distinct genomic phenotype was observed in BRCAd disease, with alterations in multiple key genes (AR, TSGs, PIK3CA/PTEN, FGFR, and CDK6). Clinical trials that leverage rational co-targeting strategies could provide an approach to enhance the efficacy of PARPi in mCRPC.
Contributors
A.A.A, L.G.H and S.J conceived and designed the Study. D.C, T.Z and P.D designed and performed the experiments. E.M.K, P.B, N.N, M.D, L.K, S.B, L.K.G, K.M, L.G.H and M.K performed data collection. H.F, D.C, E.M.K, S.F and A.A.A analysed and interpreted data. H.F, E.M.K and A.A.A prepared the manuscript and have verified the underlying data. All authors read and approved the final manuscript.
Data sharing statement
De-identified data is available in the supplementary materials accompanying this manuscript. Raw data can be made available upon request.
Declaration of interests
M.K received travel/accommodation from Celgene; D.C, T.Z, P.D and S.J are stockholders in Predicine, Inc.; L.G.H received research funding from Astellas Pharma, travel/accommodation from Astellas Pharma and Pfizer, honoraria from Janssen and Astellas and is on the scientific advisory board from Imagion; Kate Mahon received travel/accommodation from Astellas Pharma; E.M.K received honoraria from Janssen, research funding from Astellas Pharma and AstraZeneca, and travel/accommodations from Astellas Pharma, Pfizer, and Ipsen; A.A.A is a consultant for Astellas Pharma, Janssen, Novartis and Aculeus Therapeutics, is on the speakers bureau for Astellas Pharma, Janssen, Novartis, Amgen, Ipsen, Bristol Myers Squibb, Merck Serono and Bayer, received honoraria from Astellas Pharma, Janssen, Novartis, Tolmar, Amgen, Pfizer, Telix, Sanofi, Astra Zeneca, Merck Serono, Bristol Myers Squibb, Ipsen, Bayer, Pfizer, Noxopharm, Merck Sharpe Dohme, and Aculeus Therapeutics, is on the Scientific Advisory Board for Astellas Pharma, Novartis, Sanofi, AstraZeneca, Tolmar, Pfizer, Telix, Merck Serono, Janssen, Bristol Myers Squibb, Ipsen, Bayer, Merck Sharpe Dome, Amgen, and Noxopharm, travel/accommodations from Astellas, Merck Serono, Amgen, Novartis, Janssen, Tolmar, Pfizer and Bayer, and received research funding from Astellas (investigator), Merck Serono (investigator), Astra Zeneca (investigator), Bristol Myers Squibb (institutional), Astra Zeneca (institutional), Aptevo Therapeutics (institutional), Glaxo Smith Kline (institutional), Pfizer (institutional), MedImmune (institutional), Astellas (institutional), SYNthorx (institutional), Bionomics (institutional), Sanofi Aventis (institutional), Novartis (institutional), Ipsen (institutional), Exelixis (institutional), Merck Sharpe Dome (institutional), Janssen (institutional), Eli Lilly (institutional), Gilead Sciences (institutional), Merck Serono (institutional), Hinova (institutional).
Acknowledgements
No specific funding was provided for the study, however H.F is supported by a Cancer Council Victoria Postdoctoral Fellowship, a Victorian Cancer Agency Early Career Research Fellowship and a Cancer Council Victoria grant-in-aid. E.M.K is supported by a Prostate Cancer Foundation Young Investigator Award and Killam Postdoctoral Fellowship. M.K is supported by a National Institute of Health Award (RO1-CA212097). A.A.A is supported by an Astellas Investigator-initiated study grant, a National Health and Medical Research council Investigator grant, a Cancer Council Victoria grant-in-aid, and an Astra-Zeneca Investigator-initiated study grant.
We acknowledge BioRender for figure generation.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104738.
Appendix A. Supplementary data
References
- 1.de Bono J., Mateo J., Fizazi K., et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 2.Hussain M., Mateo J., Fizazi K., et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med. 2020;383:2345–2357. doi: 10.1056/NEJMoa2022485. [DOI] [PubMed] [Google Scholar]
- 3.Abida W., Patnaik A., Campbell D., et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol. 2020;38:3763–3772. doi: 10.1200/JCO.20.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bono J.S., Mehra N., Scagliotti G.V., et al. Talazoparib monotherapy in metastatic castration-resistant prostate cancer with DNA repair alterations (TALAPRO-1): an open-label, phase 2 trial. Lancet Oncol. 2021;22:1250–1264. doi: 10.1016/S1470-2045(21)00376-4. [DOI] [PubMed] [Google Scholar]
- 5.Smith M.R., Scher H.I., Sandhu S., et al. Niraparib in patients with metastatic castration-resistant prostate cancer and DNA repair gene defects (GALAHAD): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2022;23:362–373. doi: 10.1016/S1470-2045(21)00757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain M.H.A., Mateo J., Sandhu S.K., et al. Next-generation sequencing (NGS) of tumor tissue from >4000 men with metastatic castration-resistant prostate cancer (mCRPC): the PROfound phase III study experience. J Clin Oncol. 2020;38:195. [Google Scholar]
- 7.Hussain M., Daignault-Newton S., Twardowski P.W., et al. Targeting androgen receptor and DNA repair in metastatic castration-resistant prostate cancer: results from NCI 9012. J Clin Oncol. 2018;36:991–999. doi: 10.1200/JCO.2017.75.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonarakis E.S., Lu C., Luber B., et al. Germline DNA-repair gene mutations and outcomes in men with metastatic castration-resistant prostate cancer receiving first-line abiraterone and enzalutamide. Eur Urol. 2018;74:218–225. doi: 10.1016/j.eururo.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro E., Romero-Laorden N., Del Pozo A., et al. PROREPAIR-B: a prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37:490–503. doi: 10.1200/JCO.18.00358. [DOI] [PubMed] [Google Scholar]
- 10.Warner E., Herberts C., Fu S., et al. BRCA2, ATM, and CDK12 defects differentially shape prostate tumor driver genomics and clinical aggression. Clin Cancer Res. 2021;27:1650–1662. doi: 10.1158/1078-0432.CCR-20-3708. [DOI] [PubMed] [Google Scholar]
- 11.Annala M., Struss W.J., Warner E.W., et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol. 2017;72:34–42. doi: 10.1016/j.eururo.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Annala M., Vandekerkhove G., Khalaf D., et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 13.Prados-Carvajal R., Irving E., Lukashchuk N., Forment J.V. Preventing and overcoming resistance to PARP inhibitors: a focus on the clinical landscape. Cancers (Basel) 2021;14:44. doi: 10.3390/cancers14010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fettke H., Kwan E.M., Azad A.A. Cell-free DNA in cancer: current insights. Cell Oncol (Dordr) 2019;42:13–28. doi: 10.1007/s13402-018-0413-5. [DOI] [PubMed] [Google Scholar]
- 15.Kohli M., Tan W., Zheng T., et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. eBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fettke H., Kwan E.M., Docanto M.M., et al. Combined cell-free DNA and RNA profiling of the androgen receptor: clinical utility of a novel multianalyte liquid biopsy assay for metastatic prostate cancer. Eur Urol. 2020;78:173–180. doi: 10.1016/j.eururo.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fettke H., Kwan E.M., Bukczynska P., et al. Independent prognostic impact of plasma NCOA2 alterations in metastatic castration-resistant prostate cancer. Prostate. 2021;81:992–1001. doi: 10.1002/pros.24194. [DOI] [PubMed] [Google Scholar]
- 18.Kwan E.M., Dai C., Fettke H., et al. Plasma cell–free DNA profiling of PTEN-PI3K-AKT pathway aberrations in metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2021;5:622–637. doi: 10.1200/PO.20.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adalsteinsson V.A., Ha G., Freeman S.S., et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fettke H., Kwan E.M., Bukczynska P., et al. Prognostic impact of total plasma cell-free DNA concentration in androgen receptor pathway inhibitor-treated metastatic castration-resistant prostate cancer. Eur Urol Focus. 2021;7:1287–1291. doi: 10.1016/j.euf.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Ritch E., Fu S.Y.F., Herberts C., et al. Identification of hypermutation and defective mismatch repair in ctDNA from metastatic prostate cancer. Clin Cancer Res. 2020;26:1114–1125. doi: 10.1158/1078-0432.CCR-19-1623. [DOI] [PubMed] [Google Scholar]
- 22.Inoue K., Fry E.A. Haploinsufficient tumor suppressor genes. Adv Med Biol. 2017;118:83–122. [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson P., Bandlamudi C., Cheng M.L., et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokol E.S., Jin D.X., Fine A., et al. PARP inhibitor insensitivity to BRCA1/2 monoallelic mutations in microsatellite instability-high cancers. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.21.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asim M., Tarish F., Zecchini H.I., et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat Commun. 2017;8:374. doi: 10.1038/s41467-017-00393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallio H.M.L., Hieta R., Latonen L., et al. Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases. Br J Cancer. 2018;119:347–356. doi: 10.1038/s41416-018-0172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Laere B., Rajan P., Gronberg H., et al. Androgen receptor burden and poor response to abiraterone or enzalutamide in TP53 wild-type metastatic castration-resistant prostate cancer. JAMA Oncol. 2019;5:1060–1062. doi: 10.1001/jamaoncol.2019.0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinott Mizrahi G., Williams I., Azad A., Lawrentschuk N. Genetics of neuroendocrine prostate cancer: recent progress in genetic understanding is translating into therapeutic opportunities. Curr Opin Urol. 2022;32:462–465. doi: 10.1097/MOU.0000000000001015. [DOI] [PubMed] [Google Scholar]
- 29.Labanca E., Yang J., Shepherd P.D.A., et al. Fibroblast growth factor receptor 1 drives the metastatic progression of prostate cancer. Eur Urol Oncol. 2022;5:164–175. doi: 10.1016/j.euo.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim J.T., Mansukhani M., Weinstein I.B. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc Natl Acad Sci U S A. 2005;102:5156–5161. doi: 10.1073/pnas.0501203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri C.E., Baca S.C., Lawrence M.S., et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boysen G., Rodrigues D.N., Rescigno P., et al. SPOP-Mutated/CHD1-Deleted lethal prostate cancer and abiraterone sensitivity. Clin Cancer Res. 2018;24:5585–5593. doi: 10.1158/1078-0432.CCR-18-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan W., Zheng T., Wang A., et al. Dynamic changes in gene alterations during chemotherapy in metastatic castrate resistant prostate cancer. Sci Rep. 2022;12:4672. doi: 10.1038/s41598-022-08520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandekerkhove G., Struss W.J., Annala M., et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol. 2019;75:667–675. doi: 10.1016/j.eururo.2018.12.042. [DOI] [PubMed] [Google Scholar]
- 35.Robinson D., Van Allen E.M., Wu Y.M., et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo J., Porta N., Bianchini D., et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tukachinsky H., Madison R.W., Chung J.H., et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res. 2021;27:3094–3105. doi: 10.1158/1078-0432.CCR-20-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi K.N., Barnicle A., Sibilla C., et al. Detection of BRCA1, BRCA2, and ATM alterations in matched tumor tissue and circulating tumor DNA in patients with prostate cancer screened in PROfound. Clin Cancer Res. 2022;29(1):81–91. doi: 10.1158/1078-0432.CCR-22-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kensler K.H., Baichoo S., Pathania S., Rebbeck T.R. The tumor mutational landscape of BRCA2-deficient primary and metastatic prostate cancer. NPJ Precis Oncol. 2022;6:39. doi: 10.1038/s41698-022-00284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty G., Armenia J., Mazzu Y.Z., et al. Significance of BRCA2 and RB1 Co-loss in aggressive prostate cancer progression. Clin Cancer Res. 2020;26:2047–2064. doi: 10.1158/1078-0432.CCR-19-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quigley D.A., Dang H.X., Zhao S.G., et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;175:889. doi: 10.1016/j.cell.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Neviere Z., Coquan E., Brachet P.E., et al. Outcomes of patients with metastatic castration-resistant prostate cancer according to somatic damage DNA repair gene alterations. Curr Oncol. 2022;29:2776–2791. doi: 10.3390/curroncol29040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nientiedt C., Heller M., Endris V., et al. Mutations in BRCA2 and taxane resistance in prostate cancer. Sci Rep. 2017;7:4574. doi: 10.1038/s41598-017-04897-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke N.W., Armstrong A.J., Thiery-Vuillemin A., et al. Abiraterone and olaparib for metastatic castration-resistant prostate cancer. NEJM Evidence. 2022;1 doi: 10.1056/EVIDoa2200043. [DOI] [PubMed] [Google Scholar]
- 45.Azad A.A., Volik S.V., Wyatt A.W., et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015;21:2315–2324. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- 46.Gao X., III H.A.B., Vuky J., et al. Phase 1/2 study of ARV-110, an androgen receptor (AR) PROTAC degrader, in metastatic castration-resistant prostate cancer (mCRPC) J Clin Oncol. 2022;40:17. [Google Scholar]
- 47.Myung J.-K., Banuelos C.A., Fernandez J.G., et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–2960. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radaeva M., Ban F., Zhang F., et al. Development of novel inhibitors targeting the D-Box of the DNA binding domain of androgen receptor. Int J Mol Sci. 2021;22:2493. doi: 10.3390/ijms22052493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson M.L., Dy G.K., Mamdani H., et al. Interim results of an ongoing phase 1/2a study of HPN328, a tri-specific, half-life extended, DLL3-targeting, T-cell engager, in patients with small cell lung cancer and other neuroendocrine cancers. J Clin Oncol. 2022;40:8566. [Google Scholar]
- 50.Kim D.H., Sun D., Storck W.K., et al. BET bromodomain inhibition Blocks an AR-Repressed, E2F1-activated treatment-Emergent neuroendocrine prostate cancer lineage plasticity program. Clin Cancer Res. 2021;27:4923–4936. doi: 10.1158/1078-0432.CCR-20-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sweeney C., Bracarda S., Sternberg C.N., et al. Ipatasertib plus abiraterone and prednisolone in metastatic castration-resistant prostate cancer (IPATential150): a multicentre, randomised, double-blind, phase 3 trial. Lancet. 2021;398:131–142. doi: 10.1016/S0140-6736(21)00580-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.