Abstract
It has been suggested that immune-pressure-mediated positive selection operates to maintain the antigenic polymorphism on the third variable (V3) loop of the gp120 of human immunodeficiency virus type 1 (HIV-1). Here we present evidence, on the basis of sequencing 147 independently cloned env C2/V3 segments from a single family (father, mother, and their child), that the intensity of positive selection is related to the V3 lineage. Phylogenetic analysis and amino acid comparison of env C2/V3 and gag p17/24 regions indicated that a single HIV-1 subtype E source had infected the family. The analyses of unique env C2/V3 clones revealed that two V3 lineage groups had evolved in the parents. Group 1 was maintained with low variation in all three family members regardless of the clinical state or the length of infection, whereas group 2 was only present in symptomatic individuals and was more positively charged and diverse than group 1. Only virus isolates carrying the group 2 V3 sequences infected and induced syncytia in MT2 cells, a transformed CD4+-T-cell line. A statistically significant excess of nonsynonymous substitutions versus synonymous substitutions was demonstrated only for the group 2 V3 region. The data suggest that HIV-1 variants, possessing the more homogeneous group 1 V3 element and exhibiting the non-syncytium-inducing phenotype, persist in infected individuals independent of clinical status and appear to be more resistant to positive selection pressure.
Nonsynonymous mutations in protein-coding loci are often eliminated from the population because they can cause deleterious effects on protein function or reduce the fitness of the organisms. This functional constraint suppresses nonsynonymous substitution (Ka), and synonymous substitution (Ks) generally exceeds Ka on protein-coding genes (32), including the gag, pol, and env genes of human immunodeficiency virus type 1 (HIV-1) (21, 50). In a host-parasite relationship, however, new nonsynonymous mutations in a particular region can often be maintained in the population, where they confer a selective advantage on the organism (positive selection). The existence of the positive selection force that increases amino acid polymorphism was first noticed on the hypervariable regions of the major histocompatibility complex and immunoglobulin heavy chain (25, 26, 57) and soon afterwards for the surface antigen sites of many pathogens, including HIV-1 (4, 18, 22, 30, 38, 50, 52, 53, 64, 67).
The third variable (V3) loop of the HIV-1 envelope gp120 is highly immunogenic, and neutralizing antibodies from infected individuals can recognize this region (45, 61). Therefore, it is conceivable that new nonsynonymous mutations on the V3 loop-coding locus confer a selective advantage on HIV-1 via avoidance of antibody recognition. Consistently, Ka exceeds Ks in the V3 region (4, 22, 49, 50, 52, 53, 64, 67), and Ka in the V3 region is reported to correlate with the duration of the immunocompetent period (33). These data suggest that immune-pressure-mediated positive selection is operating on the V3 region to maintain antigenic polymorphism.
The V3 loop, on the other hand, is a critical determinant to specify coreceptor usage for HIV-1 entry (8, 9, 48a, 55). This indicates that V3 sequence variation is influenced by a functional constraint as well as by positive selection. In this regard, V3 loop amino acid sequences of non-syncytium-inducing (NSI)/macrophage-tropic variants consist of a less diverse and less positively charged population than that of syncytium-inducing (SI)/T-cell line-tropic variants (7, 17, 36, 37, 51, 65). The NSI variants generally predominate in the asymptomatic period of HIV-1 infection in vivo and use CCR5 as an entry coreceptor of infection (2, 3, 9–11, 14). In contrast, the SI variants often appear in association with disease progression and acquire the ability to infect cells expressing CXCR4 or other chemokine receptors (3, 10, 13, 16, 54).
In this study, we examined whether the intensity of positive selection is related to NSI and SI V3 genotype. To address this issue, we determined the nucleotide sequences of 147 independent V3 clones from uncultured peripheral blood mononuclear cells (PBMCs) and from virus isolates derived from an HIV-1 subtype E-infected Japanese family (48), divided them into two subgroups based on the presence of basic amino acid substitutions and on the extent of variation, and ascertained the NSI and SI phenotypes of virus isolates bearing the two V3 subpopulations in MT2 cell infection assays. Analyses of Ka and Ks values within each group at different sampling points or for different individuals suggest that the V3 subpopulation for the NSI phenotype is maintained with lower variation than that for the SI phenotype in infected individuals and that positive selection operates less extensively on the NSI V3 subpopulation. This is the first systematic comparison of gp120 V3 sequence evolution with the biological changes in HIV-1 isolates, obtained following horizontal and vertical transmission in a single family.
MATERIALS AND METHODS
Epidemiologic and clinical information on the study subjects.
The family consisted of a male index patient (NH1), the female spouse of NH1 (NH2), and their child (NH3). NH1 had no history of blood transfusion, surgical operation, or homosexual activity. He had a history of sexual contacts with female prostitutes in Thailand in 1989 and 1990. He was positive for HIV-1 antibodies in September 1992. He had chronic fatigue and lost 10 kg of weight between 1991 and 1992. He had developed AIDS-related pulmonary complications (Centers for Disease Control and Prevention [CDC] category C3) at the time of blood collection in June 1993 for the present study. The CD4+ lymphocyte count was 40 × 106/liter at the sampling time. NH1 died of AIDS-related pulmonary complications in March 1994.
NH2 had no documented risk factors for HIV-1 infection other than sexual contacts with NH1. She was found to be HIV-1 seropositive in November 1992. The CD4+ lymphocyte counts were 501 and 412 × 106/liter in October 1992 and May 1993, respectively. NH2 was asymptomatic (CDC category A2) at the initial blood collection in June 1993. She developed Pneumocystis carinii pneumonia in February 1996 (CDC category C3) and had been subjected to administration of zidovudine and dideoxyinosine since May 1996. The CD4+ lymphocyte count remained between 26 and 80 × 106/liter in 1996. Blood specimens were collected from NH2 in March 1996 (NH2-II) and January 1997 (NH2-III), and the CD4+ lymphocyte counts were 39 × 106 and 26 × 106/liter in February 1996 and December 1996, respectively.
NH3 was born to NH2 in June 1991. NH3 was positive for HIV-1 antibodies in December 1992. Data for his CD4+ lymphocyte count are limited to December 1992 (3,719 × 106/liter). He had no AIDS-defining illness at the time of blood collection in June 1993.
Nucleotide sequencing of HIV env C2/V3 and p17/p24 gag regions from uncultured PBMCs.
Whole blood of the three family members was collected in June 1993. For NH2, blood specimens were also collected in March 1996 (NH2-II) and January 1997 (NH2-III). PBMCs were isolated by Ficoll-Hypaque (Pharmacia LKB, Uppsala, Sweden) density centrifugation, and an env V3 region and flanking regions (324 bp) were amplified from the PBMCs by nested PCR. The first amplification step was performed for 30 cycles with outer primers MK369 and MK616 (43). Five microliters of the first PCR product were used for the second amplification step with inner primers V3-A and V3-C (23). The PCR products were cloned directly into pCRII cloning vectors (Invitrogen, NV Leek, The Netherlands). Fifteen to 22 of the cloned DNAs for each infected individual at a sampling point were sequenced on both strands with either an ALFII automated DNA sequencer (Pharmacia LKB) or an ABI PRISM 310 automated sequencer (Perkin-Elmer, Norwalk, Conn.).
A region covering a part of the long terminal repeat U5, the whole p17 gag gene, and a part of the p24 gag gene (520 bp) was amplified by nested PCR from uncultured PBMCs in June 1993. The outer primers were JA152 and JA155, and the inner primers were JA153 and JA154 (31). After the PCR, the primers were removed from the PCR products with a Centricon-100 (Amicon). The purified PCR products were sequenced directly on both strands with an ABI PRISM 310 automated sequencer.
HIV isolation, env C2/V3 nucleotide sequencing, and infection of MT2 cells.
Virus isolation was achieved by coculture of freshly isolated PBMCs (5 × 106) from the family members with an equal number of phytohemagglutinin (PHA)-stimulated PBMCs from HIV-1-seronegative individuals. At 2- to 3-day intervals, the cells were fed with RPMI 1640 with 10% fetal bovine serum and 20 U of recombinant interleukin-2/ml. At 1-week intervals, fresh PHA-stimulated PBMCs were added to the culture. Culture supernatants were taken for reverse transcriptase (RT) assay (63) every 2 to 3 days during a 40-day period after coculture.
Total RNA was extracted from 50 μl of the RT-positive culture supernatant (5), and one-fifth of the RNA was subjected to synthesis of complementary DNA with the primer C3E-130B, 5′-AGA AAA ATT CCC CTC TAC AAT TAA-3′, and avian myeloblastosis virus RT (Takara Shuzo Co., Kyoto, Japan) followed by PCR with primers C2E-110A, 5′-TTC AAT GGG ACA GGG CCA TGT-3′, and C3E-130B. The amplified DNAs corresponding to the HIV-1 env C2/V3 region (324 bp) were either sequenced directly or after cloning into pCRII, as described above.
A transformed CD4+-T-cell line, MT2 (2 × 105 cells), was incubated in 0.2 to 0.3 ml of medium containing 2 × 105 to 5 × 105 cpm of 32P-RT activity (63) for 12 h at 37°C, and the cells were grown in 1 to 2 ml of the corresponding medium in 24-well plates. Half of the volume of the culture medium was replaced by fresh medium every 2 to 3 days, and a portion was used for RT assay. The cultures were terminated 25 days after infection, during which time syncytium formation was monitored under a light microscope.
Analyses of molecular evolution of HIV sequences.
For the reference group and the outgroup of the phylogenetic tree, corresponding sequences of HIV-1 subtypes A through H and simian immunodeficiency virus (SIV)CPZGAB (CPZGAB) were collected from the Los Alamos National Laboratory database (39). The nucleotide sequences were aligned with sequences from the NH family with CLUSTAL W version 1.7 (60). The alignment was corrected manually to ensure that gaps did not alter the reading frame. A distance matrix of nucleotide substitutions was estimated from the alignment according to the method of Tajima and Nei (56), and phylogenetic trees were constructed from the matrix by the neighbor-joining method (47) with the NEIGHBOR program of the PHYLIP package version 3.5c (15a). Bootstrap analyses (15) were done for the tree with the DNABOOT and CONSENCE programs of the PHYLIP package, with 100 resamplings. The trees were rooted by CPZGAB or 18 available V3 sequences of HIV-1 subtype E from Thailand, the Central African Republic, and England (39) and were drawn with NJPLOT. The maximum-likelihood and the parsimonious trees were constructed from aligned sequences with the DNAML and DNAPARS programs (J. Felsenstein), respectively. For DNAML, the value for the transition/transversion ratio was set at 1.5 (24). Only the neighbor-joining trees are presented in this article, because trees generated by the other two algorithms gave the same topology.
Nucleotide substitution per site was estimated with PHYLIP for each pairwise sequence comparison on the basis of the Kimura two-parameter method (28). Nonsynonymous and synonymous substitutions (Ka and Ks) were estimated according to the method of Nei and Gojobori (40) with MEGA version 1.0 (29a). Means and variances of Ka and Ks for all pairwise comparisons of a sample population were calculated according to the method of Nei and Jin (41), in which patristic distances were evaluated by unweighted pair group method with arithmetic mean (UPGMA) trees from each value calculated by the NEIGHBOR program. Welch’s t test was used to evaluate the statistical significance of differences between Ka and Ks values (30).
Nucleotide sequence accession numbers.
The DDBJ database accession numbers for the nucleotide sequences reported here are D78024 to D78070 and AB014775 to AB014874 for the env C2/V3 region and AB015938 to AB015942 for the gag p17/p24 region.
RESULTS
Genetic links of HIV-1 within the NH family.
Epidemiological information suggested that HIV-1 infections in the NH family were caused by intrafamilial transmission of viruses of Thai origin (see Materials and Methods). To ascertain whether the HIV-1 quasispecies in the family were evolutionarily closely related, we determined the phylogenetic relationships of the env C2/V3 and p17 gag sequences (1, 24, 42) from the family.
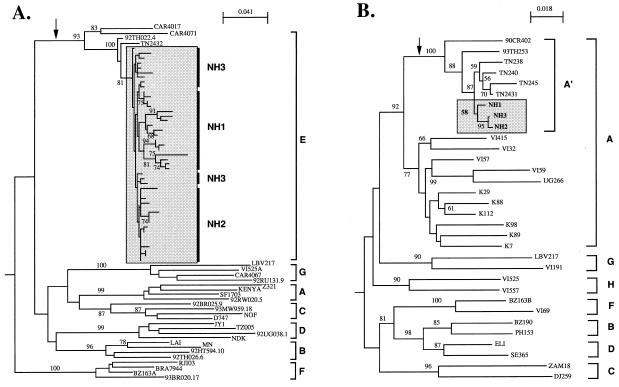
Fifteen to 17 clonal nucleotide sequences were determined for the env C2/V3 region (324 bp) from uncultured PBMCs of each infected individual in June 1993. Figure 1A shows the neighbor-joining tree with the sequences from the family and the reference sequences of HIV-1 group M (subtypes A through G) (39). The tree shows that the env C2/V3 sequences from the family form a monophyletic group (bootstrap value, 81/100). This family cluster was most closely related to HIV-1 subtype E sequences from Thailand (TN2432 and 92TH022.4; bootstrap value, 100/100), whereas it was distant from subtype E from the Central African Republic (CAR4017 and CAR4071) and from the other HIV-1 subtypes. The monophyletic relationship of the family sequences was reproducible when 18 of the other reported HIV-1 subtype E sequences from Thailand, the Central African Republic, and England (39) were included in the tree or when the trees were constructed with other algorithms, including parsimony and maximum likelihood (data not shown).
FIG. 1.
Monophyletic relationship of the env C2/V3 (A) and gag p17/p24 (B) nucleotide sequences from a Japanese family shown as neighbor-joining trees. The trees were constructed with programs of the PHYLIP package and rooted by an CPZGAB as an outgroup. Bootstrap values (15) above 75/100 (A) and 50/100 (B) are indicated at the nodes of the tree. An arrow indicates the branch that leads to the subtype E cluster. The sequences from the family are enclosed by a shaded box. Brackets at the right represent HIV-1 group M reference sequences (39).
Figure 1B shows the neighbor-joining tree with the p17/p24 gag nucleotide sequences (520 bp) from the family and the reported HIV-1 subtype A through H representatives (19, 39). As noted previously (19), the gag region of subtype E from Thailand and the Central African Republic formed a monophyletic group that was related to subtype A (A′; bootstrap value, 100/100). In that A′ cluster, gag sequences from the family had a monophyletic cluster (bootstrap value, 58/100) that was genetically most closely related to the sequences from Thailand (TN238, TN240, TN245, and TN2431) rather than to that of African origin (90CR402). The branch lengths in the NH family cluster were significantly positive, with a P value of less than 0.01, when the gag tree was generated with the maximum-likelihood algorithm with PHYLIP DNAML (data not shown).
The deduced amino acids from the nucleotide sequences were compared to 19 of the subtype E sequences from Thailand (19, 20, 34). Comparison of the env C2/V3 region (105 amino acids) showed that sequences from the family uniquely shared a tryptophan at position 61 and a valine at position 86 in the C2/V3 region (data not shown). In addition, the p17/p24 gag sequences from the family members shared common substitutions when compared with a sequence from Thailand at positions 26, 93, 128, and 143 in the p17/p24 gag region, and a common polymorphic site was identified within the family at position 120 of the p17/p24 gag region by direct sequencing (data not shown).
env V3 genotype of provirus in uncultured PBMCs of the family.
Epidemiological information suggested that NH1 was infected in Thailand during the early epidemic of subtype E infections in the country (62). Subtype E env C2/V3 sequences in this period were highly homogeneous among asymptomatic carriers (12, 27, 34, 43). This enabled us to generate the NSI consensus of the subtype E V3 loop from 21 NSI virus isolates from the early 1990s in Thailand (12, 27). To delineate NSI and SI structural characteristics of the V3 loops from the family members, deduced amino acid sequences were compared to the consensus.
The V3 loop sequences from NH1 with AIDS could be divided into two groups (Fig. 2A, NH1). One population (group 1) was characterized by the presence of a GPGQ motif at the tip of the V3 loop and by the lack of basic amino acid substitutions with respect to the consensus. Group 1 consisted of a minor population (4 of 17) in NH1, whereas it represented the only population detected in the asymptomatic carriers, NH2 (15 of 15) and NH3 (15 of 15) (Fig. 2A, NH2 and NH3). The group 1 sequences were similar to each other or to the NSI consensus. The three family members shared variants carrying an identical V3 loop sequence of group 1 (Fig. 2A).
FIG. 2.
env V3 genotype of provirus in uncultured PBMCs (A) and of virus isolates from the NH family (B). Deduced amino acids of the V3 sequences were aligned with a consensus sequence made from 21 subtype E NSI isolates (12, 27). The open box indicates a potential site for N-linked glycosylation. The shaded box indicates a subgroup of V3 sequences that are characterized by the presence of a GPGR motif at the tip of the V3 loop and basic substitutions with respect to the subtype E NSI consensus. Net charge is the number of positively charged amino acids (R and K) minus the number of negatively charged amino acids (D and E). (A) Sequences from uncultured PBMCs of NH1, NH2, and NH3 in June 1993 and of NH2 in March 1996 (NH2-II) and January 1997 (NH2-III). The arrows indicate a sequence identical among the three family members. A shaded arrowhead indicates a sequence similar to those from NH1 isolates. (B) Sequences from HIV-1 isolates from NH1 (HIV-1NH1), NH2 (HIV-1NH2), and NH3 (HIV-1NH3) in June 1993. The shaded arrowheads indicate sequences identical or similar to those from uncultured PBMCs of the corresponding individuals.
The other group (group 2) was characterized by the presence of a GPGR motif at the tip of the V3 loop and by several substitutions of the subtype E NSI consensus with basic amino acids (Fig. 2A, NH1). The basic substitutions were often seen at positions 8 (T to K or R; 10 of 13), 11 (S to R; 7 of 13), and 18 (Q to R; 17 of 17). Group 2 lost a potential site for N-linked glycosylation in the 5′ half of the V3 loop. Four of 13 group 2 clones had a phenylalanine residue proximal to the lost N glycosylation site in the loop. These characteristics are often found in subtype E SI isolates from individuals with AIDS (66). Group 2 consisted of a major population (13 of 17) in NH1, whereas it was not found in NH2 or NH3. Group 2 was more diversified from the subtype E NSI reference sequence than was group 1.
The two V3 groups were identified in NH2 as well as NH1, after NH2 developed AIDS (Fig. 2A, NH2-II and -III). In March 1996 (NH2-II), group 1 with a GPGQ motif consisted of a major population (16 of 20). After 10 months (NH2-III), group 2 with a GPGR motif became major (18 of 22). Group 2 from NH2-II and NH2-III had basic amino acid substitutions at positions 11 (S to R; 42 of 42) and 18 (Q to R; 42 of 42).
The env C2/V3 nucleotide sequences from NH2-II and NH2-III formed a monophyletic group with those from NH1, NH2, and NH3 in June 1993, outside other subtype E sequences from Thailand on the neighbor-joining tree (data not shown). In addition, all sequences from NH2-II and NH2-III had a tryptophan residue at position 20 in the V3 loop (Fig. 2A, NH2-II and -III) that was uniquely shared in V3 sequences from the family in June 1993 (Fig. 2A, NH1, NH2, and NH3). All of the nucleotide sequences from uncultured PBMCs were subjected to the BLAST 2.0 program (National Center For Biotechnology Information) to search sequence similarity to the previously reported sequences in nucleotide databases. The sequences with the highest scores (ranging from 93 to 97% in the present data sets) were all from HIV-1 env C2/V3 sequences from Southeast Asia.
env V3 genotype and MT2 tropism of HIV-1 isolates from the family.
HIV-1 isolates were obtained from the family, and their V3 nucleotide sequences and abilities to induce syncytia on MT2 cells were determined. A virus isolate from NH1 (HIV-1NH1) in June 1993 had a group 2 V3 genotype with a GPGR motif and basic substitutions at positions 8, 11, and 18 (Fig. 2B, NH1, PBMC). A major population in HIV-1NH1 (Fig. 2B, NH1) was similar to a sequence in uncultured PBMCs of NH1 (Fig. 2A, NH1). No sequences with a GPGQ motif were identified in HIV-1NH1. HIV-1NH1 replicated in PHA-stimulated PBMCs and MT2 cells. RT activity appeared in the MT2 culture supernatant on day 9 after infection and peaked on day 12, and concomitantly syncytia were observed in the culture. V3 sequences in the supernatant of RT-positive MT2 culture also contained a GPGR motif and basic substitutions at positions 8, 11, and 18 (Fig. 2B, NH1, MT2). All of the 58 C2/V3 nucleotide sequences from the virus isolates clustered with the sequences from uncultured PBMCs under the same node in the neighbor-joining tree (data not shown).
Virus isolates from NH2 and NH3 (HIV-1NH2 and HIV-1NH3) in June 1993 had a group 1 V3 genotype with a GPGQ motif and a low net positive charge (Fig. 2B, NH2 and NH3). Major V3 sequence populations in uncultured PBMCs of NH2 and NH3 were still dominant in HIV-1NH2 and HIV-1NH3. The virus isolates grew in PHA-stimulated PBMCs, whereas MT2 was nonpermissive for these isolates: no RT activity in culture supernatant and no syncytium formation were detected during a 25-day cultivation after infection.
Nucleotide diversity in env C2/V3 region and PCR fidelity.
Table 1 summarizes the number of env C2/V3 nucleotide sequences obtained from uncultured PBMCs of the NH family in 1993, 1996, and 1997, along with the nucleotide diversity of the group 1 and group 2 quasispecies. Among a total of 89 C2/V3 clones sequenced, 87 clones had distinct nucleotide sequences over the entire 324-bp region and 86 clones had open reading frames. Among the 86 clones, 51 and 35 sequences encoded group 1 and group 2 V3 loops, respectively. The mean nucleotide substitution values per site (28) in the C2/V3 segments were slightly lower for group 1 than for group 2, i.e., they ranged from 0.006 ± 0.003 to 0.020 ± 0.010 for group 1 and from 0.014 ± 0.011 to 0.041 ± 0.017 for group 2. The total nucleotide diversity in each patient was higher in individuals with AIDS (NH1, NH2-II, and NH2-III) than in asymptomatic carriers (NH2 and NH3).
TABLE 1.
Nucleotide diversity in env C2/V3 region from uncultured PBMCs
| Patient | Clinical status | No. of clones
|
Nucleotide substitution rate in env C2/V3 regionc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequenced | Uniquea | Senseb | Group 1 | Group 2 | Group 1 | Group 2 | Group 1 + 2 | ||
| NH1 | C3 | 17 | 17 | 17 | 4 | 13 | 0.009 ± 0.005 | 0.041 ± 0.017 | 0.038 ± 0.016 |
| NH2 | A2 | 15 | 13 | 12 | 12 | 0 | 0.011 ± 0.006 | 0.011 ± 0.006 | |
| NH2-II | C3 | 20 | 20 | 20 | 16 | 4 | 0.020 ± 0.010 | 0.014 ± 0.011 | 0.029 ± 0.017 |
| NH2-III | C3 | 22 | 22 | 22 | 4 | 18 | 0.006 ± 0.003 | 0.026 ± 0.014 | 0.034 ± 0.018 |
| NH3 | A1 | 15 | 15 | 15 | 15 | 0 | 0.016 ± 0.005 | 0.016 ± 0.005 | |
| Total | 89 | 87 | 86 | 51 | 35 | ||||
Number of distinct nucleotide sequences over the entire env C2/V3 region sequenced (324 bp).
Number of nucleotide sequences with an open reading frame.
Mean values ± standard deviations estimated on the basis of Kimura two-parameter method (28) by using PHYLIP.
We estimated the inherent Taq polymerase error rate under the PCR conditions described here. A plasmid carrying the full genome of HIV-1 (pLAI [44]) was diluted to 102 or 103 copies per reaction mixture and subjected to nested PCR with the same sets of primers under the same thermal cycling conditions described in Materials and Methods. The PCR products were cloned into pCRII, and about 20 clones of the C2/V3 region (324 bp) were sequenced. Within 21 clones from a 102-copy template (6,809 bases sequenced), five substitutions (A→C, A→T, and three A→G) in five different clones were identified, all of which were nonsynonymous substitutions located at different positions. Within 19 clones from a 103-copy template (6,156 bases sequenced), four substitutions (T→C and three A→G) were noted, three of which were nonsynonymous substitutions and one of which was a synonymous substitution, mapped to different positions in the four different clones. The estimated Taq polymerase error rates under our experimental conditions were 6.5 × 10−4 to 7.3 × 10−4 mutation frequency/bp/60 cycles. The values were comparable to previous results obtained for the HIV-1 tat region (3.2 × 10−4 mutation frequency/bp/35 cycles) (35). Thus, the contribution of Taq polymerase to the frequency of base substitutions observed in the present study (Table 1) was less than 12%.
V3 sequence variation and Ka/Ks values.
A neighbor-joining tree was constructed with the V3 nucleotide sequences (105 bp) of the independent C2/V3 clones (Fig. 3). The tree was rooted by 18 of the HIV-1 subtype E V3 sequences from Thailand and the Central African Republic (39). The tree was divided into two major clusters showing distinct branch lengths. The clustering pattern was reproducible regardless of the algorithms used for constructing trees (data not shown). Sequences of one cluster had shorter branches than did those of the other, suggesting the presence of a sibling sequence population that was less diversified from a putative ancestral V3 sequence of the intrafamilial infection. Notably, the cluster with shorter branches consisted of sequences from all clinical stages (NH1, NH2, NH2-II, NH2-III, and NH3), and they all encoded the group 1 V3 loop with a GPGQ motif and a low positive net charge. In contrast, the cluster with longer branches consisted of only those sequences from patients with AIDS (NH1, NH2-II, and NH2-III), and they all encoded the group 2 V3 loop with a GPGR motif and a high positive net charge. When the neighbor-joining tree was constructed on the flanking sequences of the V3 loop, no relation was detected between the branch length and the group 1 or 2 lineages (data not shown).
FIG. 3.
Neighbor-joining tree of V3 nucleotide sequences from uncultured PBMCs of the family members at different sampling points. The tree was constructed with programs of the PHYLIP package and rooted by 18 available V3 sequences of HIV-1 subtype E from Thailand, the Central African Republic, and England (39). Bootstrap values (15) above 50/100 are indicated at the nodes of the tree.
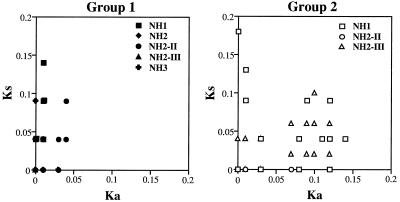
Nonsynonymous and synonymous substitutions (Ka and Ks) (40) were calculated for all pairwise comparisons of the 86 V3 nucleotide sequences from uncultured PBMCs. Then, Ka and Ks values derived from two sequences from the same V3 lineage group of the same sampling point of the same individual were extracted and plotted on a graph with x and y axes for Ka and Ks values, respectively. Thus, pairwise comparisons of group 1 were done for NH1 (n = 6), NH2 (n = 66), and NH3 (n = 105) in June 1993 as well as for NH2-II in March 1996 (n = 120) and NH2-III in January 1997 (n = 6). Comparisons of group 2 were done for NH1 in June 1993 (n = 78), NH2-II in March 1996 (n = 6), and NH2-III in January 1997 (n = 153). In the plots for group 1, the Ks value exceeded the Ka value for most pairs regardless of the sampling time or the individual (Fig. 4A). In contrast, there were many pairs in which the Ka value exceeded the Ks value in the plots for group 2 (Fig. 4B).
FIG. 4.
Ranges of nonsynonymous and synonymous substitutions (Ka and Ks) in the region encoding the V3 loop. A total of 89 cloned V3 segments from the uncultured PBMCs of the NH family were divided into group 1 and group 2, based on the presence of basic amino acid substitutions and on the extent of variation, as shown in Fig. 2A and B and 3. Ka and Ks values (40) for group 1 and group 2 were calculated for all pairwise comparisons of the V3 nucleotide sequences from the same sampling point of the same individual. Sequences encoding stop codons were excluded from the analysis. Each plot shows Ka (abscissa) and Ks (ordinate) values of a pairwise comparison. Overlapping points giving identical values are shown as a single plot.
The statistical significances of differences between means of Ka and Ks with variance (41) within each V3 lineage at each sampling point were evaluated by Welch’s t test (Table 2). In group 1, the means of Ka were lower than those of Ks regardless of the clinical stage of the infected individual (Ka/Ks of 0.091, 0.124, and 0.549 for group 1 from NH1, NH2, and NH3; t > 9.957; df > 22; P < 0.005). Although Ka exceeded Ks for NH2-II group 1, the difference was not statistically significant (t = 1.300; df = 22). The Ka/Ks ratio for NH2-III group 1 could not be calculated because all nucleotide sequences of the V3 region were identical. In contrast to the predominance of Ks for group 1, Ka exceeded Ks for group 2 (Ka/Ks ratios of 1.659 and 2.447 for group 2 from NH1 and NH2-III; P < 0.005). The Ka/Ks ratio of the group 2 sequences from NH2-II could not be calculated because all substitutions of the V3 region were nonsynonymous. Finally, the Ks values of group 1 and group 2 at the same sampling points were not significantly different, whereas the Ka values of group 2 consistently exceeded those of group 1.
TABLE 2.
Means of nonsynonymous (Ka) and synonymous (Ks) substitutions within V3 region at different time pointsa
| V3 genotype | Individual | Clinical stage | No. of sequence comparisons | Ka (± SE) | Ks (± SE) | Ka/Ks | P |
|---|---|---|---|---|---|---|---|
| Group 1 | NH1 | AIDS | 6 | 0.007 ± 0.002 | 0.073 ± 0.006 | 0.091 | <0.005 |
| NH2 | AC | 66 | 0.002 ± 0.000 | 0.013 ± 0.001 | 0.124 | <0.005 | |
| NH2-II | AIDS | 120 | 0.015 ± 0.003 | 0.010 ± 0.003 | 1.496 | NS | |
| NH2-III | AIDS | 6 | 0.000 | 0.000 | |||
| NH3 | AC | 105 | 0.006 ± 0.000 | 0.011 ± 0.000 | 0.549 | <0.005 | |
| Group 2 | NH1 | AIDS | 78 | 0.076 ± 0.002 | 0.046 ± 0.001 | 1.659 | <0.005 |
| NH2-II | AIDS | 6 | 0.040 ± 0.115 | 0.000 ± 0.112 | ∞ | NS | |
| NH2-III | AIDS | 153 | 0.043 ± 0.001 | 0.017 ± 0.001 | 2.447 | <0.005 |
Mean ± standard error was calculated by the method described by Nei and Jin (41). P, statistical significance of differences between the means of Ka and Ks tested by Welch’s t test. NS, not significant.
DISCUSSION
Compared to the mechanisms to yield new mutants via error-prone viral RT, genetic recombination, and rapid turnover of virus in vivo, the selection process that maintains genetic variation is less well understood. The present work has provided for the first time evidence that the intensity of positive selection may be related to the V3 lineage.
To understand V3 evolution in the Japanese family NH, we started to examine the probable infection pathway of virus. Two independent pieces of evidence support the possibility that HIV-1 isolates in the Japanese family had evolved from a common source. First, all of the env and gag nucleotide sequences from the three family members were of HIV-1 subtype E and had a monophyletic relationship (Fig. 1). Second, all of the HIV Env and Gag sequences from this family possessed amino acid residues unique to the family. Taken together with the epidemiological and clinical information, as well as the nucleotide diversity of the env C2/V3 region (Table 1), this evidence caused us to conclude that a single source of HIV-1 subtype E of Thai origin had infected NH1, who had transmitted the viruses to NH2, who had transmitted the viruses to NH3 at some time between 1989 and 1992.
Among the V3 loop amino acid sequences from the family, two major subgroups, group 1 with a GPGQ motif and group 2 with a GPGR motif, were identified (Fig. 2). Structural similarities to the subtype E NSI consensus, predominance in asymptomatic mother and child, and the identity to the sequences of NSI virus isolates from the family all indicate that the group 1 V3 loops are those of NSI variants. Of note is the fact that the group 1 sequences were identified in NH1 with AIDS and were consistently found after NH2 had developed AIDS, suggesting that this quasispecies can persist in infected individuals independent of clinical status. Group 2, on the other hand, was only present in symptomatic individuals and shared many structural features with the reported sequences of the subtype E SI variants. Consistently, virus isolates carrying the group 2 V3 sequences infected and induced syncytia in MT2 cells. Thus, at least two V3 lineage groups had evolved in the two parents during disease progression, one of which was related to the NSI phenotype and the other to the SI phenotype.
Interestingly, all of the group 1 sequences showed relatively low variation irrespective of the clinical status of the infected individuals or the length of infection, whereas all of the group 2 sequences formed a cluster with long branches and were identified only in patients with AIDS (Fig. 3). The low variation for the group 1 sequences were unlikely to be due to template resampling during PCR and cloning processes, because (i) most group 1 sequences were unique over the entire 324-bp C2/V3 region sequenced (Table 1), (ii) Taq polymerase error contributed less than 12% to the observed substitution frequencies in the C2/V3 region, and (iii) the relatively high degree of sequence homogeneity was seen mainly in the V3 loop and not in the flanking regions (48). An alternative and more likely explanation is that the HIV-1 variants possessing group 1 V3 loops existed as a quasispecies with a relatively homogeneous V3 loop element. The conclusion is compatible with the general observation that V3 loop sequences of NSI isolates consist of a less diverse population than those of SI isolates (7, 17, 29, 36, 37, 51, 65).
The difference in variations in the group 1 and group 2 sequence populations could have been caused by two factors. First, mutation rates may be higher in the group 2 V3 region than in the group 1 V3 region. Second, there may be a stronger positive selection in the group 2 V3 sequence population than in that of group 1. Although the former possibility needs to be assessed further, we obtained evidence that the latter mechanism is involved in generating the genotype-dependent difference in V3 variation. Calculation and plotting of Ka and Ks values for each pairwise comparison within the same V3 lineage group revealed that group 1 and group 2 exhibited distinct distribution patterns of the Ks/Ka plots. In group 2, Ka exceeded Ks for most pairs (Fig. 4). Statistical tests of the difference between means of Ka and Ks ruled out the possibility that this observation was due to chance (Table 2). These results strongly argue against the plain model where the group 2 nucleotide diversity would be caused simply by the high error rate of RT or by the rapid turnover of the viruses in patients with little immune response. Although these possibilities alone may increase the nucleotide diversity (Ka + Ks), they should not cause the excess of Ka over Ks because of the functional constraint on the protein-coding locus (32). Thus, the present data indicate that there had been a driving force which increased the probability of fixing nonsynonymous substitutions during the evolution of the group 2 lineage.
In contrast to the group 2 population, Ks exceeded Ka for most pairs in group 1. The Ka/Ks ratios were consistently less than 1.0 in group 1, except at one sampling point, where the difference between Ka and Ks values was not statistically significant. In addition, the mean Ka value for group 1 was consistently lower than that for group 2 at the same sampling point of the same individual. These data indicate that positive selection did not play a major role in the nucleotide diversity of the group 1 V3 lineage. The conclusion is compatible with the observation that the V3 region of NSI gp120 appears to be hidden from the blocking antibodies, being a less effective target for sera of HIV-1-infected individuals (6). In addition, the less rapid and efficient progeny production of NSI than SI variants (58, 59) might have resulted in the smaller extent of antigen presentation or recognition by host immune surveillance. Thus, the differences in intensity of positive selection among the group 1 and 2 V3 lineages may be attained by differences in susceptibility to the host immune responses.
The above-mentioned differences may all be attributed primarily to the differential abilities of viruses to use entry coreceptors. The V3 region has been implicated in the interaction of HIV-1 envelope with chemokine receptors expressed on the target cells (8, 9, 48a, 55). The ability to interact with a particular molecule would become a strong functional constraint which decreases the amino acid variation of the responsible region. However, once the viruses obtain the competence, V3 diversity would be maintained by a balance between the functional constraint and the immune pressure driving positive selection. Experiments assessing the ability of each group 1 or group 2 V3 loop to specify CCR5 and/or CXCR4 usage are currently in progress by constructing a series of infectious V3 recombinants.
Group 2 V3 loop amino acid sequences from NH1, NH2-II, and NH2-III frequently shared unique amino acids, which were not seen in group 1 (Fig. 2A). Either convergent evolution or the direct transmission of SI variants from NH1 to NH2 can explain the observation. In this regard, it is likely that viruses transmitted from NH1 to NH2 were the NSI variants, because neither SI V3 genotypes nor SI viruses were identified when NH2 was asymptomatic (Fig. 2). Therefore, the similarities between groups 2 in the two individuals appear to imply that there is a convergent evolutionary pattern in the V3 loop in the context of subtype E gp120 during disease progression. The possibility is supported by the observation that many of these substitutions also recurred in AIDS patients in Thailand (66), regardless of the predominant role of NSI variants for person-to-person transmission (46, 68).
In conclusion, HIV-1 V3 sequence evolution appears to be bimodal in an infected individual; a V3 sequence lineage for NSI phenotype persists independent of clinical status and is maintained with a relatively low variation due to the dominant role of the functional constraint. In contrast, a V3 sequence lineage for SI phenotype emerges with disease progression and is maintained with a high variation, because of the dominant role of positive selection.
ACKNOWLEDGMENTS
We thank Simeon Aidoo for help with sequencing and Keith Peden for providing pLAI.
This work was supported by grants from the Ministry of Health and Welfare of Japan, the Ministry of Education, Science and Culture of Japan, and the Science and Technology Agency of the Japanese Government.
REFERENCES
- 1.Albert J, Wahlberg J, Leitner T, Escanilla D, Uhlen M. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J Virol. 1994;68:5918–5924. doi: 10.1128/jvi.68.9.5918-5924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C, Feng Y, Kennedy P, Murphy P, Berger E. CC CKR5: a RANTES, MIP-1 α, MIP-1 β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Bjorndal A, Deng H, Jansson M, Fiore J, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D, Fenyo E. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonhoeffer S, Holmes E C, Nowak M A. Causes of HIV diversity. Nature. 1995;376:125. doi: 10.1038/376125a0. [DOI] [PubMed] [Google Scholar]
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou-Habib D, Roderiquez G, Oravecz T, Berman P, Lusso P, Norcross M. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Connor R, Sheridan K, Ceradini D, Choe S, Landau N. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.De Wolf F, Hogervorst E, Goudsmit J, Fenyo E M, Rubsamen-Waigmann H, Holmes H, Galvao-Castro B, Karita E, Wasi C, Sempala S D, Baan E, Zorgdrager F, Lukashov V, Osmanov S, Kuiken C, Cornelissen M the World Health Organization Network for HIV Isolation and Characterization. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res Hum Retroviruses. 1994;10:1387–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 13.Doranz B, Rucker J, Yi Y, Smyth R, Samson M, Peiper S, Parmentier M, Collman R, Doms R. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15a.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 16.Feng Y, Broder C, Kennedy P, Berger E. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganeshan S, Dickover R E, Korber B T, Bryson Y J, Wolinsky S M. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F, Yue L, Craig S, Thornton C L, Robertson D L, McCutchan F E, Bradac J A, Sharp P M, Hahn B H the WHO Network for HIV Isolation and Characterization. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res Hum Retroviruses. 1994;10:1359–1368. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- 21.Gojobori T, Moriyama E, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci USA. 1990;87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gojobori T, Yamaguchi Y, Ikeo K, Mizokami M. Evolution of pathogenic viruses with special reference to the rates of synonymous and nonsynonymous substitutions. Jpn J Genet. 1994;69:481–488. doi: 10.1266/jjg.69.481. [DOI] [PubMed] [Google Scholar]
- 23.Hattori T, Shiozaki K, Eda Y, Tokiyoshi S, Matsushita S, Inaba H, Fujimaki M, Meguro T, Yamada K, Honda M, Nishikawa K, Takatsuki K. Characteristics of the principal neutralizing determinant of HIV-1 prevalent in Japan. AIDS Res Hum Retroviruses. 1991;7:825–830. doi: 10.1089/aid.1991.7.825. [DOI] [PubMed] [Google Scholar]
- 24.Holmes E C, Zhang L Q, Simmonds P, Rogers A S, Brown A J. Molecular investigation of human immunodeficiency virus (HIV) infection in a patient of an HIV-infected surgeon. J Infect Dis. 1993;167:1411–1414. doi: 10.1093/infdis/167.6.1411. [DOI] [PubMed] [Google Scholar]
- 25.Hughes A, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 26.Hughes A, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichimura H, Kliks S, Visrutaratna S, Ou C-Y, Kalish M, Levy J. Biological, serological, and genetic characterization of HIV-1 subtype E isolates from northern Thailand. AIDS Res Hum Retroviruses. 1994;10:263–269. doi: 10.1089/aid.1994.10.263. [DOI] [PubMed] [Google Scholar]
- 28.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 29.Kuiken C L, de Jong J-J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 30.Lamers S, Sleasman J, She J, Barrie K, Pomeroy S, Barrett D, Goodenow M. Independent variation and positive selection in env V1 and V2 domains within maternal-infant strains of human immunodeficiency virus type 1 in vivo. J Virol. 1993;67:3951–3960. doi: 10.1128/jvi.67.7.3951-3960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leitner T, Escanilla D, Marquina S, Wahlberg J, Brostrom C, Hansson H B, Uhlen M, Albert J. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 32.Li W-H, Graur D. Fundamentals of molecular evolution. Sunderland, Mass: Sinauer Associates, Inc.; 1991. [Google Scholar]
- 33.Lukashov V, Kuiken C, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCutchan F E, Hegerich P A, Brennan T P, Phanuphak P, Singharaj P, Jugsudee A, Berman P W, Gray A M, Fowler A K, Burke D S. Genetic variants of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1887–1895. doi: 10.1089/aid.1992.8.1887. [DOI] [PubMed] [Google Scholar]
- 35.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 36.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milich L, Margolin B, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- 38.Mindell D. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proc Natl Acad Sci USA. 1996;93:3284–3288. doi: 10.1073/pnas.93.8.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 40.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 42.Ou C-Y, Ciesielski C A, Myers G, Bandea C I, Luo C-C, Korber B T M, Mullins J I, Schochetman G, Berkelman R L, Economou A N, Witte J J, Furman L J, Satten G A, MacInnes K A, Curran J W, Jaffe H W, Group L I, Group E I. Molecular epidemiology of HIV transmission in a dental practice. Science. 1992;256:1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 43.Ou C-Y, Takebe Y, Weniger B G, Luo C-C, Kalish M L, Auwanit W, Yamazaki S, Gayle H D, Young N L, Schochetman G. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341:1171–1174. doi: 10.1016/0140-6736(93)91001-3. (Erratum, 342:250.) [DOI] [PubMed] [Google Scholar]
- 44.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 45.Profy A, Salinas P, Eckler L, Dunlop N, Nara P, Putney S. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J Immunol. 1990;144:4641–4647. [PubMed] [Google Scholar]
- 46.Roos M T L, Lange J M A, Goede R E Y, Coutinho R A, Schellekens P T A, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Sato H, Taniguchi K, Tomita Y, Shiino T, Miyakuni T, Takebe Y. Evidence for the selective pressure to reduce heterogeneity of HIV-1 subtype E envelope V3-loop sequences in an intrafamilial infection case. AIDS. 1997;11:396–397. [PubMed] [Google Scholar]
- 48a.Sato, H., K. Kato, and Y. Takebe. Functional complementation of the envelope hypervariable V3 loop of human immunodeficiency virus type 1 subtype B by the subtype E V3 loop. Virology, in press. [DOI] [PubMed]
- 49.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H, Malnati M, Plebani A, Siccardi A, Littman D, Fenyo E, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 50.Seibert S, Howell C, Hughes M, Hughes A. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1) Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 51.Shpaer E, Delwart E, Kuiken C, Goudsmit J, Bachmann M, Mullins J. Conserved V3 loop sequences and transmission of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1679–1684. doi: 10.1089/aid.1994.10.1679. [DOI] [PubMed] [Google Scholar]
- 52.Shpaer E G, Mullins J I. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J Mol Evol. 1993;37:57–65. doi: 10.1007/BF00170462. [DOI] [PubMed] [Google Scholar]
- 53.Simmonds P, Balfe P, Ludlam C, Bishop J, Brown A. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons G, Wilkinson D, Reeves J, Dittmar M, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P, Weiss R, Clapham P. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speck R, Wehrly K, Platt E, Atchison R, Charo I, Kabat D, Chesebro B, Goldsmith M. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka T, Nei M. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- 58.Tersmette M, de Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel T, Kurth R, Norley S. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J Immunol. 1994;153:1895–1904. [PubMed] [Google Scholar]
- 62.Weniger B G, Takebe Y, Ou C-Y, Yamazaki S. The molecular epidemiology of HIV in Asia. AIDS. 1994;8(Suppl. 2):S13–S28. [PubMed] [Google Scholar]
- 63.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolinsky S M, Korber B T, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 65.Yamashita A, Yamamoto N, Matsuda J, Koyanagi Y. Cell type-specific heterogeneity of the HIV-1 V3 loop in infected individuals: selection of virus in macrophages and plasma. Virology. 1994;204:170–179. doi: 10.1006/viro.1994.1521. [DOI] [PubMed] [Google Scholar]
- 66.Yu X-F, Wang Z, Beyrer C, Celentano D D, Khamboonruang C, Allen E, Nelson K. Phenotypic and genotypic characteristics of human immunodeficiency virus type 1 from patients with AIDS in northern Thailand. J Virol. 1995;69:4649–4655. doi: 10.1128/jvi.69.8.4649-4655.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Diaz R, Ho D, Mosley J, Busch M, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]