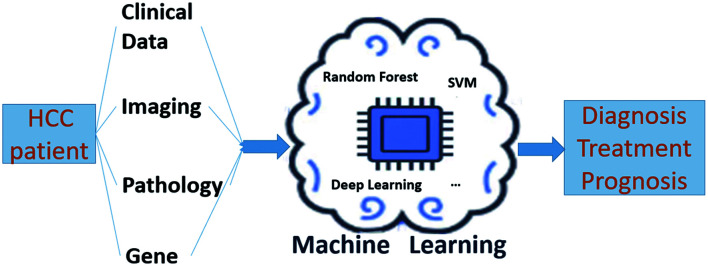
Abstract
Hepatocellular carcinoma (HCC) is a common tumor. Although the diagnosis and treatment of HCC have made great progress, the overall prognosis remains poor. As the core component of artificial intelligence, machine learning (ML) has developed rapidly in the past decade. In particular, ML has become widely used in the medical field, and it has helped in the diagnosis and treatment of cancer. Different algorithms of ML have different roles in diagnosis, treatment, and prognosis. This article reviews recent research, explains the application of different ML models in HCC, and provides suggestions for follow-up research.
Keywords: Machine learning, Hepatocellular carcinoma, Artificial intelligence, Prognosis
Graphical abstract
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy and one of the four most common causes of cancer-related death worldwide.1,2 HCC is the fastest-growing cause of cancer-related deaths in the USA, and it is possible that HCC will become the third largest cause in 2030.3 In recent years, hepatitis B vaccine and antiviral treatment have been widely used,4,5 and the treatments of HCC are various, including surgery, ablation, transcatheter arterial chemoembolization (TACE), chemotherapy targeted immunotherapy and others.3,6 However, the symptoms of HCC are not easy to detect, and the overall prognosis is poor.7 HCC needs early detection, accurate prediction, individualized treatment and follow-up.
Artificial intelligence (AI) is a new subject that studies and develops theories, methods, technologies, and application systems for simulating, extending, and expanding human intelligence. Machine learning (ML) is the core of AI. ML can make the computer simulate or realize human learning behavior to acquire new knowledge or skills and reorganize existing knowledge to improve its own performance.8 In the past decade, ML has been gradually applied to medical research, and has made progress in many aspects.9 In particular, cancer-related research, including lung cancer, breast cancer and so on, and HCC-related research is increasing. ML studies in HCC involve not only diagnosis, treatment, prognosis, and other aspects, but also a variety of algorithm models including decision trees, support vector machines (SVMs), random forest and deep learning.10 The application of ML in HCC can reveal the relationship between AI and HCC and also be instrumental in the prevention and treatment of HCC. This review focuses on the application of ML in the aspects of diagnosis, treatment, and prognosis of HCC.
ML for the Diagnosis of HCC
The diagnosis of HCC depends on pathology, for patients with chronic hepatitis B and liver cirrhosis, radiology can also help with diagnosis.3 However, radiologic diagnosis requires typical imaging features,11 but more than 10% of tumors lack imaging hallmarks of HCC. If the imaging is not typical, a biopsy or second contrast-enhanced study should be performed.12 Biopsy is an invasive procedure with a sensitivity of about 70%, and lower for tumors with a diameter <2 cm. Sometimes it is difficult to distinguish well-differentiated HCC from dysplastic nodules. The diagnosis model of HCC can be established through ML, which can help to diagnose and treat the disease in clinic early and easily. For the diagnosis, it is convenient to obtain clinical data, like albumin, platelet (PLT), total bilirubin, alpha-fetoprotein (AFP), alkaline phosphatase (ALP), γ-glutamyl transferase (GGT), aspartate transaminase (AST), portal vein thrombosis, and others. Phan et al.13 established a convolutional neural network (CNN) model to predict the occurrence of HCC in HBV infected patients by selecting clinical data from Taiwan Health database. The AUC of the model was 0.886 and the accuracy was 0.980. Nam et al.14 constructed a deep neural network to predict the incidence rate of HCC in patients with HBV-related cirrhosis who received entecavir antiviral treatment. The c-index of the model was 0.782, which was significantly better than the traditional six scores (PAGE-B, CU-HCC, HCC-RESCUE, ADRESS-HCC, mPAGE-B, and THRI).
In addition, the accuracy of ML models varies significantly. Sato et al.15 collected relevant clinical data from patients diagnosed with HCC at the first visit and HBV infected patients who developed HCC during the follow-up period. They used logistic regression model for linear classification, SVM, gradient boosting, random forest, neural network, deep learning, and other algorithms for nonlinear classification, and established HCC diagnosis prediction model based on clinical data. Then, all the models were verified in the test set. They found that the gradient boosting model had the highest accuracy. Similarly, Angelis et al.16 used six algorithms including decision tree, random forest, SVM, k-nearest neighbor (KNN) classification, AdaBoost, and gradient boosting to make models based on the collected clinical data. They also found that the gradient boosting had the highest accuracy of 84% and a sensitivity of 92%. Kim et al.17 used gradient boosting machine (referred to as GBM), which is one of the boosting algorithms, to establish a model for the follow-up results of patients with HBV hepatitis treated with entecavir or tenofovir. The model predicted high or low risk of HCC in patients with HBV hepatitis, and it has been externally verified in Western cohorts. However, the study of Wong et al.18 on HCC prediction models of HBV and HCV patients in Hong Kong reported that among logistic regression model, ridge regression model, AdaBoost algorithm model, decision tree model, and random forest model, the accuracy of ridge regression [area under the receiver operating characteristic (AUROC) 0.844] and random forest model (AUROC: 0.837) were stable, and better than other traditional scores (CU-HCC, GAG-HCC, REACH-B, PAGE-B, and REAL-B) (Table 1).19–27
Table 1. Details of traditional scores mentioned in this article.
| Scores | Author and year | Function | Based indicators | Research center | Results |
|---|---|---|---|---|---|
| PAGE-B | Papatheodoridis et al. 201619 | Score for prediction of the 5-year HCC risk in Caucasian CHB patients under entecavir/tenofovir | Age, sex, and platelets | Multicenter | c-index :0.82 |
| CU-HCC | Wong et al. 201020 | Clinical score in predicting the risk of HCC among HBV carriers | Age, albumin, bilirubin, HBV DNA, and cirrhosis | Multicenter | Negative predictive value: 97.8% and 97.3% in the training and validation cohorts |
| HCC-RESCUE | Sohn et al. 201721 | Prediction model for the development of HCC in treatment-naïve patients receiving oral antiviral treatment for CHB | Age, sex, and cirrhosis. | Multicenter | AUROCs :1 year, 3 years, and 5 years were 0.798, 0.788, and 0.817 in the testing cohort and 0.817, 0.810 and 0.809 in the validation cohort |
| ADRESS-HCC | Flemming et al. 201422 | Risk prediction model to estimate the 1-year probability of HCC | Age, diabetes, race, etiology of cirrhosis, sex, and severity of liver dysfunction | Multicenter | |
| mPAGE-B | Kim et al. 201823 | Modified PAGE-B scores to improve the predictive performance | Age, sex, platelet counts, and serum albumin levels | Multicenter | c-index 0.704 and 0.691 in the testing cohort and the validation cohort |
| THRI | Sharma et al. 201724 | Scoring system to predict HCC risk for patients with cirrhosis | Age, sex, etiology, and platelets | Multicenter | AUROC: 0.82 and 0.72 in the testing cohort and the validation cohort |
| GAG-HCC | Yuen et al. 200925 | Score to identify high-risk CHB patients for treatment and screening of HCC | Age, sex, HBV DNA levels, core promoter mutations, and cirrhosis | Multicenter | c-index: 0.77 |
| REACH-B | Yang et al. 201126 | Score to estimate the risk of developing HCC at 3, 5, and 10 years in patients with chronic hepatitis B | Sex, age, serum alanine aminotransferase concentration, HBeAg status, and serum HBV DNA level | Multicenter | AUROC: 0.902, 0.783 and 0.806 |
| REAL-B | Yang et al. 202027 | HCC risk score using routine clinical variables among a treated Asian cohort | Sex, age, alcohol use, diabetes, baseline cirrhosis, platelet count, and alpha-fetoprotein | Multicenter | AUROC: >0.80 |
AUROC, area under the receiver operating characteristic; HCC, hepatocellular carcinoma.
For high-risk patients with chronic hepatitis B and cirrhosis, the diagnosis can be established by imaging. However, it is difficult to identify when the image characteristics are not typical. ML is good at processing images, so it has advantages in imaging identification. Bharti et al.28 obtained 754 regions of interest (ROI) through the echotexture and roughness of the liver surface in the ultrasound imaging, and constructed a CNN model to distinguish normal liver, chronic hepatitis, cirrhosis, and HCC. The classification accuracy of the model was 96.6%. Similarly, other studies have suggested that the model established by ultrasound imaging features has good accuracy in distinguishing benign and malignant liver nodules.29,30 Moreover, Brehar et al.31 compared HCC detection models based on ultrasound imaging. They compared the CNN model with the traditional multilayer perceptron, SVM, random forest, and AdaBoost algorithm, and found that the accuracy, sensitivity and specificity of CNN were good, and it was significantly better than the traditional ML algorithm. Recently, Jin et al.32 established a deep learning model through two-dimensional shear wave elastography and corresponding ultrasound images, which can predict the possibility of hepatitis B patients developing into HCC within 5 years. This provides an important reference for the treatment and follow-up of patients with chronic hepatitis B. HCC and intrahepatic cholangiocarcinoma (ICC) both occur in the liver, but their biological behavior, treatment methods, and prognosis are very different. The overall prognosis of ICC is poor. Most patients present with advanced tumors, and only 15% of patients with ICC underwent resection.33 Even for patients who are indicated for surgical resection, the study suggests that the probability of cure is about 10%.34 The resection mode, chemotherapy and targeted treatment of ICC are very different from those of HCC.35,36 Therefore, it has significant to be able to distinguish HCC and ICC in a noninvasive manner. Ren et al.37 established a SVM model by selecting the ROI of lesion on the ultrasound imaging to identify HCC and ICC. The results showed that the accuracy, specificity, and sensitivity of the model were above 0.800, and it had good generalization ability.
Enhanced CT is of great significance in the diagnosis of HCC. When it is controversial to discriminate the nature of nodules with CT images, a good ML model can improve the reliability of diagnosis. The CNN model established by Yasaka et al.38 effectively identified the types of liver masses through enhanced CT, and masses can be divided into five categories using this model. They are category A, classic HCCs; category B, malignant liver tumors other than classic and early HCCs; category C, indeterminate masses or mass-like lesions including early HCCs and dysplastic nodules and rare benign liver masses other than hemangiomas and cysts; category D, hemangiomas; and category E, cysts. Mokrane et al.39 extracted quantitative imaging features from CT images to establish candidate models for diagnosing uncertain liver nodules in patients with liver cirrhosis using three ML algorithms (KNN, SVM, and random forest). They selected the best model using the AUC and Youden index. The model helped to judge uncertain liver nodules in a noninvasive manner. MRI has a similar role. Hamm et al.40 established a CNN model using MRI images, and it divided liver lesions into six categories (simple cyst, cavernous hemangioma, focal nodular hyperplasia (FNH), HCC, ICC, and colorectal cancer metastasis). The discrimination result of the model was better than that of the radiologist, and the specificity and sensitivity were greater than 90%. Liu et al.41 made a SVM model to distinguish combined hepatocellular cholangiocarcinoma, ICC, and HCC using the radiological characteristics of MRI and CT. At the same time, they found that enhanced phase MRI and nonenhanced phase and portal vein phase CT were more helpful for differentiation. Because of the development of ML, it is also possible to predict the pathological grade of HCC by noninvasive evaluation by imaging. Mao et al.42 manually extracted radiomics features and synthesized features using recursive feature elimination, and then established a prediction model of HCC pathological grade with AUC of 0.8014 using the XGBoost model. Nebbia et al.43 established a ML model using multiparameter MRI images to achieve preoperative prediction of microvascular infiltration (MVI) status. The researchers also compared the effects of extracting only from the tumor region, extracting only from the peritumor edge region and combining them. The result showed that preoperative MRI is feasible to predict MVI, and multiparameter MRI sequences are complementary in recognition.
The results of pathological examination depend to a certain extent on the selection of specimens and pathologist judgment. Using ML can not only reduce the error of results but also shorten the time to diagnosis. Lin et al.44 established a CNN model using multiphoton microscopic imaging of unstained specimens to judge the degree of HCC differentiation. Chen et al.45 established a CNN model using HE stained pathological images. The accuracy of the model in distinguishing benign and malignant HCC was 96.0%, and it predicted HCC mutated genes (including CTNNB1, FMN2, TP53 and ZFX4) from the images. Kiani et al.46 made a CNN model based on hematoxylin and eosin stained specimen images that effectively helped in the pathological differentiation of HCC and ICC. In addition, on the molecular level, ML training through mutations in related genes can also assist in HCC diagnosis. The research of Zhang et al.47 established a SVM model based on 11 selected genes that distinguished HCC, adjacent noncancerous tissues, and hepatitis cirrhosis.
Chen et al.48 explored the significance of HBV reverse transcriptase (RT) gene for HCC patients. They used four ML methods to establish HBV RT sequences to predict HCC. The results show that the random forest model based on 10 combined features had the best predictive performance, and the individual HCC risk score obtained by the random forest model distinguished HCC and HBV patients. Circulating tumor gene (ctDNA) detection makes it possible to detect tumors early and noninvasively and helps to match suitable targeted drugs. The study of Tao et al.49 established a random forest model that distinguished HCC and HBV patients by somatic copy number aberrations of ctDNA through low-depth whole-genome sequencing of plasma samples from HBV-related HCC patients and cancer-free HBV patients. The diagnosis of HCC is not limited to “diagnosis.” Diagnosis, including etiology and disease severity, greatly affects the treatment plan and prognosis. Especially in clinical practice, diseases are often atypical. The application of ML of HCC has a huge impact on the diagnosis and differential diagnosis of HCC (Table 2).13–15,17,18,29–32,37–49
Table 2. Details of machine learning for the diagnosis of hepatocellular carcinoma.
| Author and year | Data type | Sample number | Machine learning model/algorithm | Results |
|---|---|---|---|---|
| Phan et al. 202013 | Clinical data | N: 6,052 (training set: 70%; test set: 30%) | Convolutional neural network | AUC: 0.886 |
| Nam et al. 202014 | Clinical data | Training set: 424; validation set (independent external cohort): 316 | Deep neural network | c-index: 0.782 |
| Sato et al. 201915 | Clinical data | N: 1,580 (training set: 80%; development set and test set: 20%) | SVM, gradient boosting, random forest, neural network, deep learning, and other algorithms | Gradient boosting model had the highest accuracy (87.34%) AUC: 0.94 |
| Kim et al. 202117 | Clinical data | Training set: 6,051; validation set (external validation cohorts): (5,817 patients from Korean centers and 1,640 from Western centers) | GBM | c-index: 0.79 |
| Wong et al. 202218 | Clinical data | N: 124,006 (training set: 70%; test set: 30%) | AdaBoost, decision tree and random forest | Accuracy of random forest (AUROC: 0.837) was stable |
| Schmauch et al. 201929 | Imaging | Training set: 367; validation set: 177 | Deep learning | Weighted mean ROC-AUC scores of 0.891 |
| Li et al. 202130 | Imaging | N: 226 (training set: 80%; test set: 20%) | SVM | AUC: 0.86 |
| Brehar et al. 202031 | Imaging | N: 268 (training set: 66%; test set: 20%; validation set: 14%) | CNN, SVM, random forest, and AdaBoost | CNN was the best (accuracy of 91% with AUC of 95%) |
| Jin et al. 202132 | Imaging | Training set: 262; validation set: 86; testing set: 86 | Deep learning | AUCs: 0.981, 0.942 and 0.900 in training, validation, and testing cohorts |
| Ren et al. 202137 | Imaging | Training set: 149; test set: 38; validation set: 39 | SVM | AUC: 0.936 |
| Yasaka et al. 201838 | Imaging | Training set: 460; test set: 100 | CNN | AUC: 0.92 |
| Mokrane et al. 202039 | Imaging | Discovery set: 142; validation set: 36 | KNN, SVM, and random forest | AUC: 0.70 and 0.66 in discovery and validation cohorts |
| Hamm et al. 201940 | Imaging | Training set: 434; test set: 60 | CNN | AUC: 0.992 |
| Liu et al. 202141 | Imaging | N: 86 | SVM | AUC: 0.77 |
| Mao et al. 202042 | Imaging | Training set: 237; test set: 60 | XGBoost | AUC: 0.8014 |
| Nebbia et al. 202043 | Imaging | N: 99 | SVM | Highest AUC: 0.8669 (multiparametric MRI combination yield) |
| Lin et al. 201944 | Pathology | N: 113 | CNN | Accuracy>90% |
| Chen et al. 202045 | Pathology | Training set: 261; test set: 50; internal validation set: 155; external validation set: 101 | CNN | Accuracy: 96.0% |
| Kiani et al. 202046 | Pathology | Training set: 70; test set: 80; validation set: 26 | CNN | Accuracy: 0.885 |
| Zhang et al. 202047 | Gene | Training set: 1,333; test set: 336 | SVM | Sensitivity: 91.93%, specificity: 100%, and AUC: 0.9597 |
| Chen et al. 202148 | Genes | Training set: 361; validation set: 183 | Random forest, SVM, KNN | Best predictive performances: random forest (AUC: 0.96; accuracy, 0.90) |
| Tao et al. 202049 | Genes | Training set: 209; validation sets: 76/99 | Random forest | AUC>0.800 |
AUROC, area under the receiver operating characteristic; CNN, convolutional neural network; GBM, gradient boosting machine; SVM, support vector machine.
ML for the treatment of HCC
The preferred treatment for HCC is surgical resection, and R0 resection should be performed in patients who can undergo surgery. TACE or radiofrequency ablation (RFA) is recommended for nonresectable HCC patients, and targeted or immunotherapy and other systemic treatment schemes can be used for patients who cannot undergo the above treatment.3 In clinical practice, doctors may encounter some patients whose treatment methods are difficult to decide. For individual patients, there is only one choice, which therefore needs to be made carefully. Properly used, ML can help patients to choose treatment methods.
Choi et al.50 established a clinical decision support system based on 20 clinical indicators selected using a random forest model. The system recommended the initial treatment plan for HCC patients and predicted the overall survival of the corresponding treatment methods. Liu et al.51 established a radiomics model using ultrasound images of HCC patients to predict the efficacy of TACE. The model AUC was 0.93. It predicted progression-free survival of patients and optimized their treatment. On the basis of predicting the first TACE treatment response of HCC patients, Dong et al.52 used six ML models and compared them to select the most appropriate model. The results showed that the random forest model performed best and accurately predicted the early response to the first TACE treatment. With the development of targeted therapy and immunotherapy, the application of ML for the treatment may tend to the selection of targeted Immunologic drugs for HCC patients. ML will provide reference for patients to select suitable targeted drugs in the future (Table 3).50–52
Table 3. Details of machine learning for the treatment of hepatocellular carcinoma.
| Author and year | Data type | Sample number | Machine learning model/algorithm | Results |
|---|---|---|---|---|
| Choi et al. 202050 | Clinical data | Training set: 813; validation set: 208 | Random forest | c-index: 0.725 (RFA/PEIT), 0.695 (resection), 0.803 (TACE), 0.676 (TACE + EBRT), 0.684 (sorafenib), 0.710 (supportive care), 0.959 (transplantation), 0.850 (other therapies) |
| Liu et al. 202051 | Imaging | N: 419 (training and validation cohorts by a ratio of 2:1) | CNN | AUC: 0.93 |
| Dong et al. 202152 | Clinical data& Imaging | N: 110 (training set: 80%; validation set: 20%) | XGBoost, decision tree, SVM, random forest, KNN, fully convolutional networks | Best performance: random forest (AUC: 0.802 accuracy: 0.784, sensitivity: 0.904, and specificity: 0.480) |
AUROC, area under the receiver operating characteristic; CNN, convolutional neural network; TACE, transcatheter arterial chemoembolization.
ML for the prognosis of HCC
Since the 21st century, HCC has been the fastest-growing cause of cancer-related death in the USA, and it is expected that HCC will become the third largest cause by 2030.53 The long-term prognosis of liver transplantation is better than that of hepatectomy, with a recurrence rate of 70% and a 10-year survival rate of 7–15%.54 Liver transplantation is an ideal surgical method for HCC patients, but it is still limited by a small number of donors and high medical costs. How to choose these two treatment methods for people with appropriate indications? Schoenberg et al.55 established a random forest model based on clinical data. The predictive value of the model for early disease-free survival was 0.788. The model divides the patients into high-risk and low-risk groups. The low-risk patients are suitable for liver resection, and the high-risk patients are considered suitable for liver transplantation, so as to guide the selection of treatment.
Ji et al.56 established a prediction model for the prognosis of patients with tumors ≤ 5 cm and no evidence of extrahepatic disease or large vessel invasion after resection. The model determined a critical value using eight clinical characteristics including age, race, AFP, tumor size, tumor number, vascular invasion, histological grade and fibrosis score, and divided the prognosis into low risk, medium risk, and high risk. The results showed that there was no significant difference in the prognosis of low-risk patients undergoing tumor resection or liver transplantation. The model provided a reference for patients as to whether they should undergo neoadjuvant therapy. Huang et al.57 also used the clinical data of patients after hepatectomy to establish a model, but their study compared multiple models (DeepSurv, XGBoost and Random Survival Forest) and found that XGBoost was the best one. They used a heat map to individualize the recurrence risk. The study also divided the prognostic variables of patients in more detail, according to time. Within 1 year after surgery, the importance of cancer thrombus was the highest. At 1 to 2 years after surgery, the number of tumors was the most important variable related to the prognosis of patients, followed by the type of resection, tumor thrombus, and tumor diameter. In the two periods of 2 to 3 years and 3 to 5 years, in addition to the number of tumors, HBV infection was a relatively important variable. Smoking was also associated with late recurrence. A model established by Jiang et al.58 using CT radiomics features not only predicted the MVI status of patients before surgery, but also judged the difference in recurrence-free survival of patients by grouping. Regarding RFA, an SVM model established by Liang et al.59 can effectively identify HCC patients with relatively high recurrence risk after ablation therapy, which is helpful for postoperative follow-up and management of patients.
There are also many studies that used clinical data, pathological information, radiomics characteristics and other data to establish ML models.60–65 They effectively predicted the prognosis of patients and provided great help in the selection of treatment methods, the requirements of postoperative review, and the avoidance of high-risk factors. For patients with Barcelona Clinic Liver Cancer stage B, the international guidelines recommend TACE. However, there is great heterogeneity in patients at that stage, and the efficacy of TACE is different. Lin et al.66 selected the clinical data of patients with BCLC stage B, and extracted five indicators including tumor size, tumor number, BCLC-B substage, AFP, and ALB to establish a random forest model. The model can predict the prognosis of patients after TACE treatment, and distinguish the middle-term HCC patients who are suitable for TACE. The CNN model established by Peng et al.67 also effectively predicted the efficacy of TACE. A model established by Jin et al.68 by extracting the features of enhanced CT effectively predicted the possibility of extrahepatic diffusion or vascular invasion of the patients after the initial TACE treatment (EVIT).
In terms of genes, many studies have explored the model of predicting the prognosis of HCC patients. Chaudhary et al.69 used deep learning for the first time to explore the difference in survival time of HCC patients. They established a model using RNA sequencing (RNA Seq), microRNA sequencing (miRNA Seq), and methylation data that reliably predicted the survival times of six different cohorts. Liu et al.70 selected immune genes with differences between normal and HCC. The model established with those genes predicted the 5-year survival HCC patients. Bedon et al.71 classified HCC patients with progression-free survival with methylation maps, and constructed a model. High-risk and low-risk patients with early cancer progression were classified. Prognosis is a common concern of patients and doctors. The extensive application of ML makes the prognosis more specific, and provides great help for follow-up guidance of patients (Table 4).55–59,66,67,69–71
Table 4. Details of machine learning for the prognosis of hepatocellular carcinoma.
| Author and year | Data type | Sample number | Machine learning model/algorithm | Results |
|---|---|---|---|---|
| Schoenberg et al. 202055 | Clinical data | Training set: 127; test set: 53 | Random forest | AUC: 0.788 |
| Ji et al. 202156 | Clinical data | Training/validation set: 1,899; test set: 879 | GBM | c-index: >0.72 |
| Huang et al. 202157 | Clinical data | Training set: 5,928; internal validation set: 1,483; external validation set: 508 | DeepSurv, XGBoost, random survival forest | Best performance: XGBoost (c-index: 0.713) |
| Jiang et al. 202158 | Clinical data & imaging | Training set: 324; validation set: 81 | XGBoost, 3D-CNN | AUROCs: training set 0.952 and 0.980; validation: 0.887 and 0.906 |
| Liang et al. 201459 | Clinical data | N: 83 | SVM | AUC: 0.69 |
| Lin et al. 202166 | Clinical data | Training set: 602; internal validation set: 301; external validation set: 343 | Random forest | c-index: 0.69, AUROC>0.71 |
| Peng et al. 202067 | Imaging | Training set: 562; validation sets: 89/138 | CNN | AUC:>0.95 |
| Chaudhary et al. 201869 | Gene | Training set: 360; validation sets (5 external datasets): 230/221/166/40/27 | Deep learning | c-index: 0.68 |
| Liu et al. 202170 | Gene | N (3 databases): TCGA 365; ICGC 232; GSE14520 209 | Random forest | AUC:>0.7 |
| Bedon et al. 202171 | Gene | Training set: 300; test set: 74 | Random forest | Accuracy: 0.80 |
AUROC, area under the receiver operating characteristic; CNN, convolutional neural network; GBM, gradient boosting machine; ICGC, International Cancer Genome Consortium; TCGA, The Cancer Genome Atlas.
Conclusion
The study of ML in HCC involves a variety of data such as patient clinical information, imaging information, pathological information, and gene loci. ML can provide guidance and help in the diagnosis of HCC, the selection of patient treatment methods, and prognosis prediction. Especially for noninvasive diagnosis. Its advantages include accuracy in processing images. ML can avoid the contraindications and complications of biopsy, as well as the possibility of tumor rupture and disseminated metastasis. Because of the different treatment methods of HCC and ICC, preoperative differentiation of HCC and ICC by ML can help preoperative assessment of whether surgery can be performed as well as the surgical procedure.
ML has brought great guiding significance to the diagnosis and treatment of HCC from many aspects. In particular, it is not based on subjective assessment and experience to determine the diagnosis and treatment method, but is based on actual data and accuracy to provide evidence. Currently, as described above, there are many types of data available for the application of ML of HCC, including basic clinical information (sex, hepatitis history, blood biochemical examination, and others), imaging data including ultrasound, CT, and MRI, pathology data, and gene data. Moreover, there are many models and algorithms that can be used in the application of ML for HCC. For example, random forest, SVM, deep learning, and so on. It is uncertain which model is suitable for the research problem, but ML models can be selected according to the type of research data. SVM, random forest, artificial neural network, boosting, and bagging algorithms are common models in ML, which are more suitable for the traditional “learning mode,” and so are more suitable for processing numerical data. While the essence of deep learning, including CNN, and others, is complex, along with the complexity of learning and training models, the algorithm are closer to human brain models. Deep learning may be more suitable for processing complex data types. However, there are shortcomings of ML. The learning process is still a black box. It is hard to understand its essence, which may have potential harm. The interpretability of AI is still a problem that needs to be solved. In addition, models are always based on a part of the population. Then the extensive application of the models is facing a huge test and needs to be constantly improved.
There are many types of ML algorithms, different data types, and research methods. However, researchers have been exploring suitable algorithms and models, and have achieved much. It is believed that with the continuous development of AI and ML, HCC-related research models of ML will also be improved and bring good news to HCC patients.
Abbreviations
- AFP
alpha-fetoprotein
- AI
artificial intelligence
- AUROC
area under the receiver operating characteristic
- CNN
convolutional neural network
- GBM
gradient boosting machine
- HCC
hepatocellular carcinoma
- ICC
intrahepatic cholangiocarcinoma
- KNN
k-nearest neighbor
- ML
machine learning
- MVI
microvascular infiltration
- RFA
radiofrequency ablation
- ROI
regions of interest
- SVM
support vector machine
- TACE
transcatheter arterial chemoembolization
References
- 1.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1–61. doi: 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 4.Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Liang W, Jing W, Liu M. Countdown to 2030: eliminating hepatitis B disease, China. Bull World Health Organ. 2019;97(3):230–238. doi: 10.2471/BLT.18.219469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371:m3544. doi: 10.1136/bmj.m3544. [DOI] [PubMed] [Google Scholar]
- 7.Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188314. doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael H, Andreas K. A Brief History of Artificial Intelligence: On the Past, Present, and Future of Artificial Intelligence. Calif Manage Rev. 2019;61(4):5–14. doi: 10.1177/0008125619864925. [DOI] [Google Scholar]
- 9.Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021;11(4):900–915. doi: 10.1158/2159-8290.CD-21-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bansal M, Goyal A, Choudhary A. A comparative analysis of K-Nearest Neighbor, Genetic, Support Vector Machine, Decision Tree, and Long Short Term Memory algorithms in machine learning. Decision Analytics Journal. 2022;3:100071. doi: 10.1016/j.dajour.2022.100071. [DOI] [Google Scholar]
- 11.van der Pol CB, Lim CS, Sirlin CB, McGrath TA, Salameh JP, Bashir MR, et al. Accuracy of the Liver Imaging Reporting and Data System in Computed Tomography and Magnetic Resonance Image Analysis of Hepatocellular Carcinoma or Overall Malignancy-A Systematic Review. Gastroenterology. 2019;156(4):976–986. doi: 10.1053/j.gastro.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 13.Phan DV, Chan CL, Li AA, Chien TY, Nguyen VC. Liver cancer prediction in a viral hepatitis cohort: A deep learning approach. Int J Cancer. 2020;147(10):2871–2878. doi: 10.1002/ijc.33245. [DOI] [PubMed] [Google Scholar]
- 14.Nam JY, Sinn DH, Bae J, Jang ES, Kim JW, Jeong SH. Deep learning model for prediction of hepatocellular carcinoma in patients with HBV-related cirrhosis on antiviral therapy. JHEP Rep. 2020;2(6):100175. doi: 10.1016/j.jhepr.2020.100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato M, Morimoto K, Kajihara S, Tateishi R, Shiina S, Koike K, et al. Machine-learning Approach for the Development of a Novel Predictive Model for the Diagnosis of Hepatocellular Carcinoma. Sci Rep. 2019;9(1):7704. doi: 10.1038/s41598-019-44022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelis I, Exarchos T. Hepatocellular Carcinoma Detection Using Machine Learning Techniques. Adv Exp Med Biol. 2021;1338:21–29. doi: 10.1007/978-3-030-78775-2_4. [DOI] [PubMed] [Google Scholar]
- 17.Kim HY, Lampertico P, Nam JY, Lee HC, Kim SU, Sinn DH, et al. An artificial intelligence model to predict hepatocellular carcinoma risk in Korean and Caucasian patients with chronic hepatitis B. J Hepatol. 2022;76(2):311–318. doi: 10.1016/j.jhep.2021.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Wong GL, Hui VW, Tan Q, Xu J, Lee HW, Yip TC, et al. Novel machine learning models outperform risk scores in predicting hepatocellular carcinoma in patients with chronic viral hepatitis. JHEP Rep. 2022;4(3):100441. doi: 10.1016/j.jhepr.2022.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papatheodoridis G, Dalekos G, Sypsa V, Yurdaydin C, Buti M, Goulis J, et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J Hepatol. 2016;64(4):800–806. doi: 10.1016/j.jhep.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 20.Wong VW, Chan SL, Mo F, Chan T, Loong HH, Wong GL, et al. Clinical scoring system to predict hepatocellular carcinoma in chronic hepatitis B carriers. J Clin Oncol. 2010;28(10):1660–1665. doi: 10.1200/JCO.2009.26.2675. [DOI] [PubMed] [Google Scholar]
- 21.Sohn W, Cho JY, Kim JH, Lee JI, Kim HJ, Woo M, et al. Risk score model for the development of hepatocellular carcinoma in treatment-naïve patients receiving oral antiviral treatment for chronic hepatitis B. Clin Mol Hepatol. 2017;23(2):170–178. doi: 10.3350/cmh.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemming JA, Yang JD, Vittinghoff E, Kim WR, Terrault NA. Risk prediction of hepatocellular carcinoma in patients with cirrhosis: the ADRESS-HCC risk model. Cancer. 2014;120(22):3485–3493. doi: 10.1002/cncr.28832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Kim YD, Lee M, Jun BG, Kim TS, Suk KT, et al. Modified PAGE-B score predicts the risk of hepatocellular carcinoma in Asians with chronic hepatitis B on antiviral therapy. J Hepatol. 2018;69(5):1066–1073. doi: 10.1016/j.jhep.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Sharma SA, Kowgier M, Hansen BE, Brouwer WP, Maan R, Wong D, et al. Toronto HCC risk index: A validated scoring system to predict 10-year risk of HCC in patients with cirrhosis. J Hepatol. 2018;68(1):92–99. doi: 10.1016/j.jhep.2017.07.033. [DOI] [PubMed] [Google Scholar]
- 25.Yuen MF, Tanaka Y, Fong DY, Fung J, Wong DK, Yuen JC, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol. 2009;50(1):80–88. doi: 10.1016/j.jhep.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Yang HI, Yuen MF, Chan HL, Han KH, Chen PJ, Kim DY, et al. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol. 2011;12(6):568–574. doi: 10.1016/S1470-2045(11)70077-8. [DOI] [PubMed] [Google Scholar]
- 27.Yang HI, Yeh ML, Wong GL, Peng CY, Chen CH, Trinh HN, et al. Real-World Effectiveness From the Asia Pacific Rim Liver Consortium for HBV Risk Score for the Prediction of Hepatocellular Carcinoma in Chronic Hepatitis B Patients Treated With Oral Antiviral Therapy. J Infect Dis. 2020;221(3):389–399. doi: 10.1093/infdis/jiz477. [DOI] [PubMed] [Google Scholar]
- 28.Bharti P, Mittal D, Ananthasivan R. Preliminary Study of Chronic Liver Classification on Ultrasound Images Using an Ensemble Model. Ultrason Imaging. 2018;40(6):357–379. doi: 10.1177/0161734618787447. [DOI] [PubMed] [Google Scholar]
- 29.Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, et al. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100(4):227–233. doi: 10.1016/j.diii.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Lv XZ, Zheng X, Ruan SM, Hu HT, Chen LD, et al. Machine Learning-Based Ultrasomics Improves the Diagnostic Performance in Differentiating Focal Nodular Hyperplasia and Atypical Hepatocellular Carcinoma. Front Oncol. 2021;11:544979. doi: 10.3389/fonc.2021.544979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brehar R, Mitrea DA, Vancea F, Marita T, Nedevschi S, Lupsor-Platon M, et al. Comparison of Deep-Learning and Conventional Machine-Learning Methods for the Automatic Recognition of the Hepatocellular Carcinoma Areas from Ultrasound Images. Sensors (Basel) 2020;20(11):3085. doi: 10.3390/s20113085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin J, Yao Z, Zhang T, Zeng J, Wu L, Wu M, et al. Deep learning radiomics model accurately predicts hepatocellular carcinoma occurrence in chronic hepatitis B patients: a five-year follow-up. Am J Cancer Res. 2021;11(2):576–589. [PMC free article] [PubMed] [Google Scholar]
- 33.Amini N, Ejaz A, Spolverato G, Kim Y, Herman JM, Pawlik TM. Temporal trends in liver-directed therapy of patients with intrahepatic cholangiocarcinoma in the United States: a population-based analysis. J Surg Oncol. 2014;110(2):163–170. doi: 10.1002/jso.23605. [DOI] [PubMed] [Google Scholar]
- 34.Spolverato G, Vitale A, Cucchetti A, Popescu I, Marques HP, Aldrighetti L, et al. Can hepatic resection provide a long-term cure for patients with intrahepatic cholangiocarcinoma? Cancer. 2015;121(22):3998–4006. doi: 10.1002/cncr.29619. [DOI] [PubMed] [Google Scholar]
- 35.Mazzaferro V, Gorgen A, Roayaie S, Droz Dit Busset M, Sapisochin G. Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72(2):364–377. doi: 10.1016/j.jhep.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Kelley RK, Bridgewater J, Gores GJ, Zhu AX. Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72(2):353–363. doi: 10.1016/j.jhep.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Ren S, Li Q, Liu S, Qi Q, Duan S, Mao B, et al. Clinical Value of Machine Learning-Based Ultrasomics in Preoperative Differentiation Between Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma: A Multicenter Study. Front Oncol. 2021;11:749137. doi: 10.3389/fonc.2021.749137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286(3):887–896. doi: 10.1148/radiol.2017170706. [DOI] [PubMed] [Google Scholar]
- 39.Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30(1):558–570. doi: 10.1007/s00330-019-06347-w. [DOI] [PubMed] [Google Scholar]
- 40.Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29(7):3338–3347. doi: 10.1007/s00330-019-06205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Khalvati F, Namdar K, Fischer S, Lewis S, Taouli B, et al. Can machine learning radiomics provide pre-operative differentiation of combined hepatocellular cholangiocarcinoma from hepatocellular carcinoma and cholangiocarcinoma to inform optimal treatment planning? Eur Radiol. 2021;31(1):244–255. doi: 10.1007/s00330-020-07119-7. [DOI] [PubMed] [Google Scholar]
- 42.Mao B, Zhang L, Ning P, Ding F, Wu F, Lu G, et al. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning-based radiomics. Eur Radiol. 2020;30(12):6924–6932. doi: 10.1007/s00330-020-07056-5. [DOI] [PubMed] [Google Scholar]
- 43.Nebbia G, Zhang Q, Arefan D, Zhao X, Wu S. Pre-operative Microvascular Invasion Prediction Using Multi-parametric Liver MRI Radiomics. J Digit Imaging. 2020;33(6):1376–1386. doi: 10.1007/s10278-020-00353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H, Wei C, Wang G, Chen H, Lin L, Ni M, et al. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophotonics. 2019;12(7):e201800435. doi: 10.1002/jbio.201800435. [DOI] [PubMed] [Google Scholar]
- 45.Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. doi: 10.1038/s41698-020-0120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. doi: 10.1038/s41746-020-0232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang ZM, Tan JX, Wang F, Dao FY, Zhang ZY, Lin H. Early Diagnosis of Hepatocellular Carcinoma Using Machine Learning Method. Front Bioeng Biotechnol. 2020;8:254. doi: 10.3389/fbioe.2020.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, Zhang Z, Wang Y, Fang M, Zhou J, Li Y, et al. Using Quasispecies Patterns of Hepatitis B Virus to Predict Hepatocellular Carcinoma With Deep Sequencing and Machine Learning. J Infect Dis. 2021;223(11):1887–1896. doi: 10.1093/infdis/jiaa647. [DOI] [PubMed] [Google Scholar]
- 49.Tao K, Bian Z, Zhang Q, Guo X, Yin C, Wang Y, et al. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma. EBioMedicine. 2020;56:102811. doi: 10.1016/j.ebiom.2020.102811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi GH, Yun J, Choi J, Lee D, Shim JH, Lee HC, et al. Development of machine learning-based clinical decision support system for hepatocellular carcinoma. Sci Rep. 2020;10(1):14855. doi: 10.1038/s41598-020-71796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, et al. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9(4):397–413. doi: 10.1159/000505694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong Z, Lin Y, Lin F, Luo X, Lin Z, Zhang Y, et al. Prediction of Early Treatment Response to Initial Conventional Transarterial Chemoembolization Therapy for Hepatocellular Carcinoma by Machine-Learning Model Based on Computed Tomography. J Hepatocell Carcinoma. 2021;8:1473–1484. doi: 10.2147/JHC.S334674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 54.Franssen B, Jibara G, Tabrizian P, Schwartz ME, Roayaie S. Actual 10-year survival following hepatectomy for hepatocellular carcinoma. HPB (Oxford) 2014;16(9):830–835. doi: 10.1111/hpb.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoenberg MB, Bucher JN, Koch D, Börner N, Hesse S, De Toni EN, et al. A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma. Ann Transl Med. 2020;8(7):434. doi: 10.21037/atm.2020.04.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji GW, Fan Y, Sun DW, Wu MY, Wang K, Li XC, et al. Machine Learning to Improve Prognosis Prediction of Early Hepatocellular Carcinoma After Surgical Resection. J Hepatocell Carcinoma. 2021;8:913–923. doi: 10.2147/JHC.S320172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y, Chen H, Zeng Y, Liu Z, Ma H, Liu J. Development and Validation of a Machine Learning Prognostic Model for Hepatocellular Carcinoma Recurrence After Surgical Resection. Front Oncol. 2020;10:593741. doi: 10.3389/fonc.2020.593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang YQ, Cao SE, Cao S, Chen JN, Wang GY, Shi WQ, et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J Cancer Res Clin Oncol. 2021;147(3):821–833. doi: 10.1007/s00432-020-03366-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang JD, Ping XO, Tseng YJ, Huang GT, Lai F, Yang PM. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput Methods Programs Biomed. 2014;117(3):425–434. doi: 10.1016/j.cmpb.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine. 2019;50:156–165. doi: 10.1016/j.ebiom.2019.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shan QY, Hu HT, Feng ST, Peng ZP, Chen SL, Zhou Q, et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19(1):11. doi: 10.1186/s40644-019-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, et al. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72(6):2000–2013. doi: 10.1002/hep.31207. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita R, Long J, Saleem A, Rubin DL, Shen J. Deep learning predicts postsurgical recurrence of hepatocellular carcinoma from digital histopathologic images. Sci Rep. 2021;11(1):2047. doi: 10.1038/s41598-021-81506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao H, Xiong T, Peng J, Xu L, Liao M, Zhang Z, et al. Classification and Prognosis Prediction from Histopathological Images of Hepatocellular Carcinoma by a Fully Automated Pipeline Based on Machine Learning. Ann Surg Oncol. 2020;27(7):2359–2369. doi: 10.1245/s10434-019-08190-1. [DOI] [PubMed] [Google Scholar]
- 65.Saito A, Toyoda H, Kobayashi M, Koiwa Y, Fujii H, Fujita K, et al. Prediction of early recurrence of hepatocellular carcinoma after resection using digital pathology images assessed by machine learning. Mod Pathol. 2021;34(2):417–425. doi: 10.1038/s41379-020-00671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin H, Zeng L, Yang J, Hu W, Zhu Y. A Machine Learning-Based Model to Predict Survival After Transarterial Chemoembolization for BCLC Stage B Hepatocellular Carcinoma. Front Oncol. 2021;11:608260. doi: 10.3389/fonc.2021.608260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng J, Kang S, Ning Z, Deng H, Shen J, Xu Y, et al. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur Radiol. 2020;30(1):413–424. doi: 10.1007/s00330-019-06318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin Z, Chen L, Zhong B, Zhou H, Zhu H, Zhou H, et al. Machine-learning analysis of contrast-enhanced computed tomography radiomics predicts patients with hepatocellular carcinoma who are unsuitable for initial transarterial chemoembolization monotherapy: A multicenter study. Transl Oncol. 2021;14(4):101034. doi: 10.1016/j.tranon.2021.101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu J, Chen Z, Li W. Machine Learning for Building Immune Genetic Model in Hepatocellular Carcinoma Patients. J Oncol. 2021;2021:6676537. doi: 10.1155/2021/6676537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bedon L, Dal Bo M, Mossenta M, Busato D, Toffoli G, Polano M. A Novel Epigenetic Machine Learning Model to Define Risk of Progression for Hepatocellular Carcinoma Patients. Int J Mol Sci. 2021;22(3):1075. doi: 10.3390/ijms22031075. [DOI] [PMC free article] [PubMed] [Google Scholar]