Abstract
Non-DNA-binding Stabilin-2/HARE receptors expressed on liver sinusoidal endothelial cells specifically bind to and internalize several classes of phosphorothioate antisense oligonucleotides (PS-ASOs). After Stabilin-mediated uptake, PS-ASOs are trafficked within endosomes (>97%–99%), ultimately resulting in destruction in the lysosome. The ASO entrapment in endosomes lowers therapeutic efficacy, thereby increasing the overall dose for patients. Here, we use confocal microscopy to characterize the intracellular route transverse by PS-ASOs after Stabilin receptor-mediated uptake in stable recombinant Stabilin-1 and -2 cell lines. We found that PS-ASOs as well as the Stabilin-2 receptor transverse the classic path: clathrin-coated vesicle-early endosome-late endosome-lysosome. Chloroquine exposure facilitated endosomal escape of PS-ASOs leading to target knockdown by more than 50% as compared to untreated cells, resulting in increased PS-ASO efficacy. We also characterize cytosolic galectins as novel contributor for PS-ASO escape. Galectins knockdown enhances ASO efficacy by more than 60% by modulating EEA1, Rab5C, and Rab7A mRNA expression, leading to a delay in the endosomal vesicle maturation process. Collectively, our results provide additional insight for increasing PS-ASO efficacy by enhancing endosomal escape, which can further be utilized for other nucleic acid-based modalities.
Keywords: MT: Delivery Strategies, Stabilin, endosome escape, endocytosis, antisense oligonucleotide, galectin, HARE
Graphical abstract
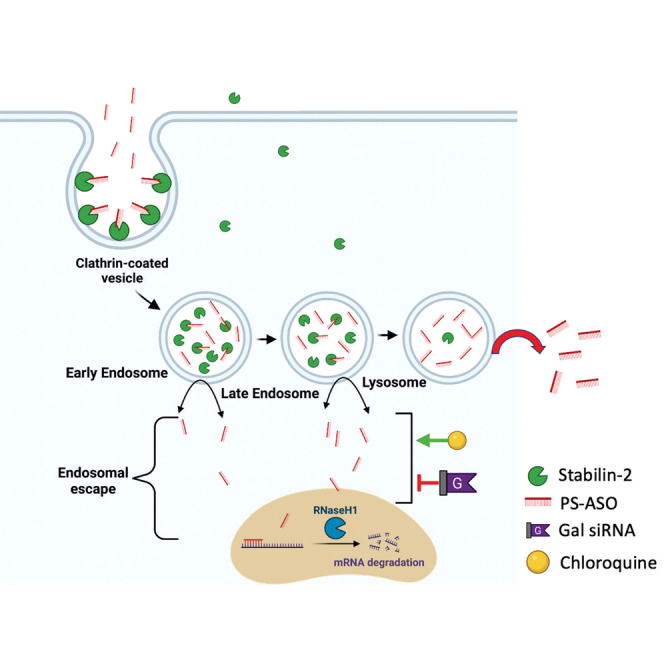
This study was undertaken to investigate how the Stabilin receptors route their cargo, in this case antisense oligonucleotides, into the endolysosomal pathway and to determine what factors that route cargo go directly to the lysosome, by default, or escape into the cytoplasm via suppression of Galectin-1.
Introduction
Antisense oligonucleotides (ASOs) are chemically modified, small (∼18–30 nucleotides), single-stranded deoxyribonucleotides that can be specifically designed to modulate mRNA expression of disease-related genes.1,2,3 ASOs exert their effect via different mechanisms such as RNAse H-mediated degradation of an mRNA target, steric block of translation, and modulation of splicing.4,5 The ASO used in this study utilizes RNAse H-mediated degradation of an mRNA target, malat-1. Over the years, ASOs have undergone several chemical modifications to increase their bioavailability, target binding affinity, and biostability and to reduce their off-target toxicity.1 This has resulted in the FDA approval of ten ASO drugs for clinical use and more than 50 ASOs in clinical trials.6,7,8 Their full potential will be realized as we more fully understand their interactive properties and trafficking within cells.
ASO entrapment in endosomal vesicles impedes their activity; therefore, elucidating the mechanisms for how ASOs are internalized, trafficked, and arrive in the cytoplasm or nucleus of the cell is of critical importance.9,10,11 The gap in knowledge is the molecular mechanisms of how a small minority of ASOs escape the maturing endosome once it is formed after receptor-mediated internalization. Several receptors are theoretically characterized with properties required for efficient oligonucleotide uptake, but so far, only the class H Scavenger receptors (Stabilin-1 and Stabilin-2) have proven to efficiently internalize gymnotic phosphorothioate (PS)-ASOs without conjugation of any targeting moiety or lipid carriers.12,13
Stabilin receptors are type 1 transmembrane receptors involved in the clearance of extracellular matrix components from circulation.14 Their structure is composed of four epidermal growth factor (EGF) clusters, four Fasciclin-1 (Fas-1) domains, one X-link domain, and a single transmembrane domain. Human Stabilin-1 (Stab1) and Stabilin-2 (Stab2) extracellular ectodomains share 55% sequence homology; however, their short intracellular domains are very diverse, leading to differences in their intracellular trafficking and downstream signaling.15 Stabilin-2 is normally expressed as two isoforms: a large 315-kDa receptor (Stab2/315-HARE) and a small 190-kDa receptor (190-HARE). Both receptors offer the same binding and trafficking kinetics for all known ligands,16,17,18 and the cell line used in this study stably expresses the small isoform, 190-HARE.
Stabilin receptors dramatically enhance the bulk uptake of PS-ASOs inside the cell, but not much is known about the intracellular endosomal trafficking of PS-ASOs mediated by these receptors. In vivo studies have shown that only <0.3%–2% of endocytosed oligonucleotides escape from endosomes, leaving more than 98% of internalized cargo for degradation in the lysosomes.19,20 Endosomal maturation is controlled by a plethora of protein/protein complexes and non-protein components, which could influence the trafficking of cargo after receptor-mediated uptake.21 One such family is the Rab GTPases, which serve as specific endosomal compartment markers affecting different aspects of trafficking and endosomal maturation.22 Our previous study has shown the importance of endosomal proteins (EEA1, Rab5C, Rab7A) in facilitating trafficking and escape of ASOs from endosomes.23 However, these are not the only molecules responsible for trafficking of PS-ASOs within the cell. These findings address this shortcoming by investigating the route taken by PS-ASOs after Stabilin receptor-mediated endocytosis.
Several studies have evaluated different small-molecule compounds for enhancing small interfering RNA (siRNA) or ASO escape from endosomes and have observed enhanced target knockdown with small-molecule treatment.21,24,25,26 Galectins have been used as a reporter for endomembrane damage during release of oligonucleotides,24,25 but none of these studies have investigated the endosomal escape-enhancing effects of these small proteins. Galectins belong to a lectin protein family characterized by one or two carbohydrate recognition domains. Galectins are divided into three subfamilies that encompass 15 galectins in mammals, out of which 11 are present in humans.27 Galectins are involved in various cellular processes such as cell growth, cell cycle, apoptosis, and morphogenesis, as well as development and progression of many human diseases.28,29,30 As of this publication, no study has been published investigating the role of galectins related to PS-ASO efficacy by enhancing/diminishing endosomal escape.
In this study, we first investigated the endosomal route taken by PS-ASOs after Stabilin receptor-mediated endocytosis. Next, we evaluated chloroquine (CQ), a membrane-destabilizing agent enhancing PS-ASO escape, as well as its role in rescuing PS-ASO efficacy upon protein knockdown. Lastly, we established the novel role of a cytosolic protein Galectin-1 in ASO escape machinery.
Results
Endosomal trafficking of Stabilin receptor and PS-ASO
Cells internalize cargo via clathrin-dependent and -independent endocytic pathways.31,32 Different oligonucleotide forms are internalized into the cell via either of the pathways. Once internalized, cargos are delivered into the early endosome vesicle, which determines the next subcellular destination for internalized cargo. From the early endosome, cargo can be transported to late endosomes or recycling endosomes or to the trans-Golgi network.33,34 Two different routes are possible from the late endosome: either the lysosome or the trans-Golgi network. This is the simplest suggested pathway for intracellular trafficking of cargo after receptor-mediated internalization. Several different intracellular trafficking pathways are possible, which depends on different receptors involved in oligonucleotide uptake.35
Our previous study has shown that PS-ASO is internalized by the Stabilin receptor using clathrin-mediated endocytosis and is finally degraded into lysosomes.13 Before degradation, PS-ASO must escape, hybridize to targeted mRNA, and be accessible to RNAse H machinery to exert its effect. It has been found that EEA1 and Rab5C in the early endosome, and Rab7A in the late endosome, are key trafficking proteins regulating PS-ASO efficacy.23 This study suggests that Stabilin receptor-mediated PS-ASO uptake is trafficked inside the cell through the early endosome-late endosome-lysosome pathway. Once we know the intracellular route taken by PS-ASO, we would be able to genetically or pharmacologically manipulate the proteins or protein complexes residing in these vesicles to enhance PS-ASO efficacy. To elucidate the endosomal pathway, we carried out a variable time point experiment where simultaneous trafficking of PS-ASO and Stabilins is monitored.
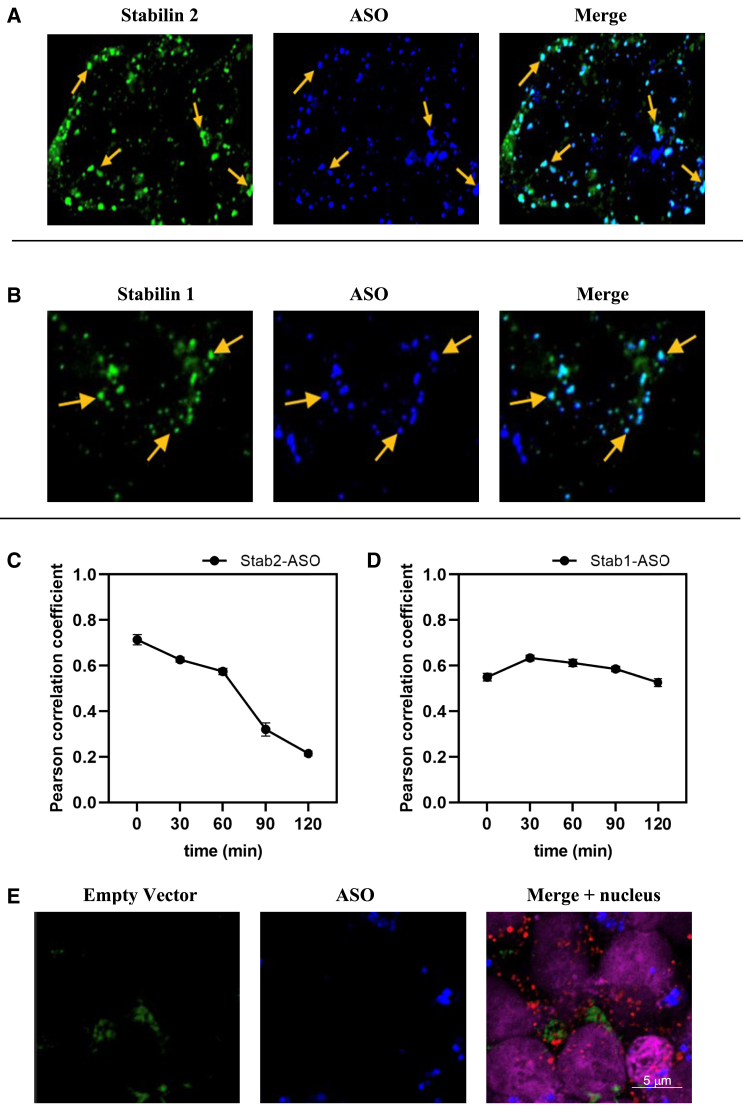
Empty vector (EV), Stabilin-1, and Stabilin-2 (190-HARE) cell lines were used for pulse-chase experiments to assess a cohort of vesicles as they matured. First, we investigated the colocalization pattern between Stabilin receptor-1/-2 and PS-ASO. Confocal microscopy revealed maximum colocalization between Stabilin receptor-1/-2 and PS-ASO at 30- and 0-min chase, respectively (Figures 1A and 1B). Steep descent in Pearson correlation coefficient trend lines was observed between Stabilin-2 and PS-ASO as time progressed (Figure 1C). This suggests that the Stabilin receptor is releasing bound ASO soon after internalization and that it gets recycled back to the plasma membrane. However, a similar decreasing trend was not observed between Stabilin-1 and PS-ASO, suggesting less PS-ASO release (Figure 1D). More colocalization was observed between 190-HARE and PS-ASO as compared with Stabilin-1 and PS-ASO. This is in accordance with our previous study where higher endocytic activity was observed in the Stab2 cells as compared with Stab1 cells taking into account receptor expression levels. We also carried out pulse-chase experiments in the EV cell line and investigated the uptake of PS-ASO. We observed no PS-ASO uptake in this cell line at variable chase time points (Figure 1E), similar to previous data,23 suggesting the importance of Stabilin receptors for uptake and trafficking of PS-ASOs.
Figure 1.
PS-ASO colocalizes with Stabilin receptor after Stabilin-mediated endocytosis
(A) 190-HARE (Stabilin-2), (B) Stabilin-1, and (E) EV cell lines, respectively, were incubated with 0.1 μM PS-ASO for 30 min (pulse), followed by incubation with no PS-ASO media for variable times (0, 30, 60, 90, and 120 min) (chase). Immunocytochemistry was performed and mounted slides were imaged by confocal microscopy. Images were processed with EzColocalization, an ImageJ plugin. Correlation quantification (Pearson correlation coefficient) between (C) Stabilin-2 and PS-ASO and (D) Stabilin-1 and PS-ASO (n = 6 images). PS-ASOs are not internalized in EV cell lines. Some material was adhered on the cell surface, giving blue florescence.
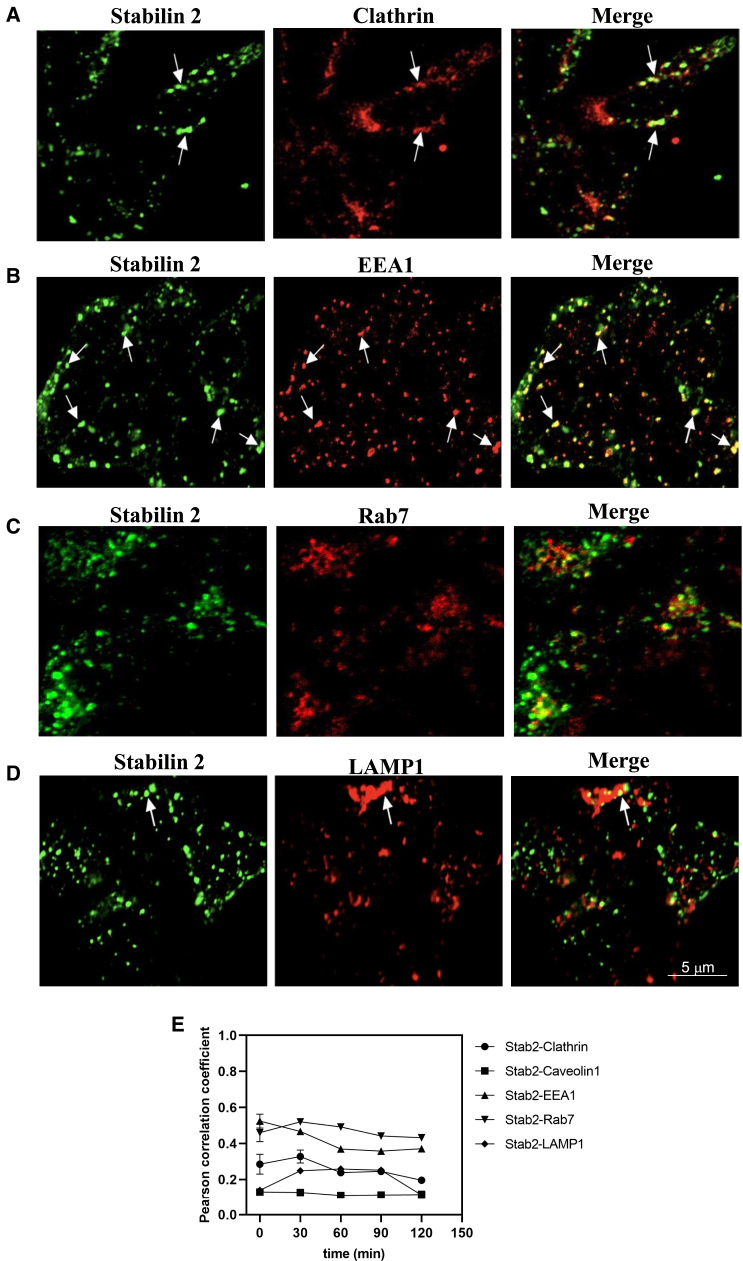
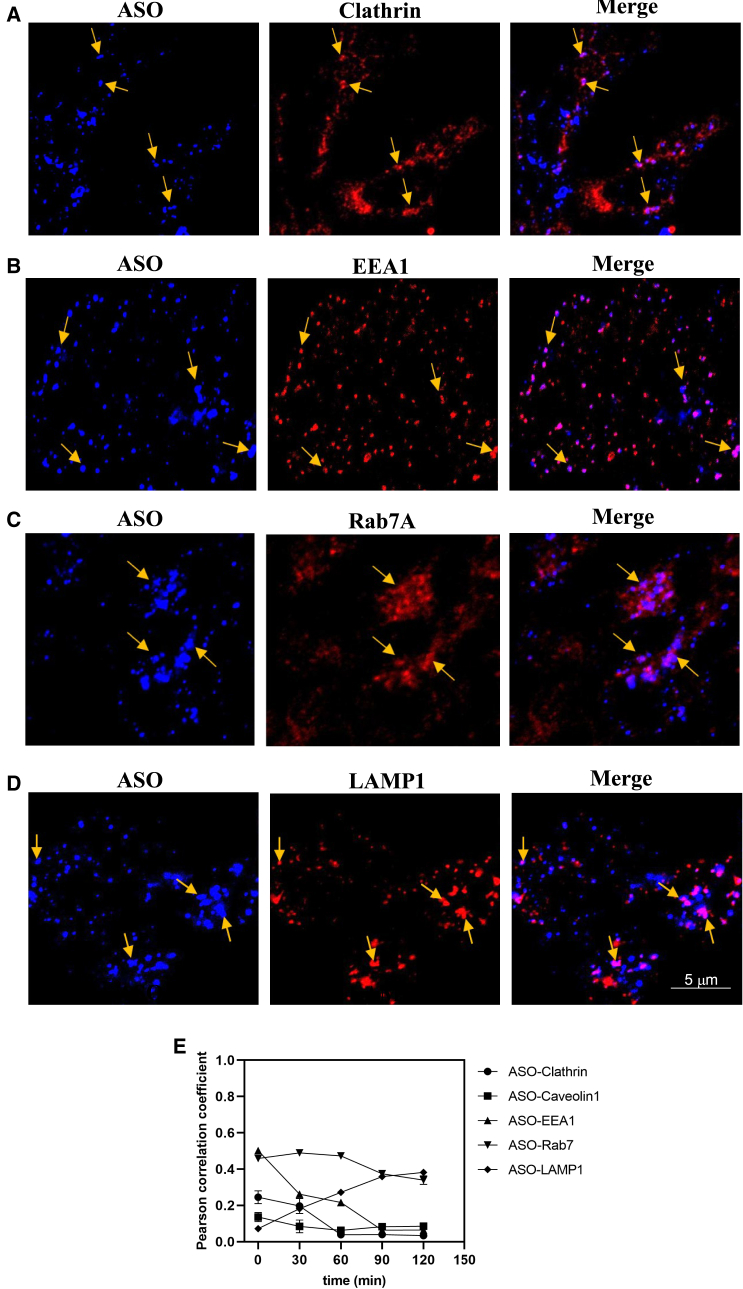
Next, we investigated the trafficking of the Stabilin-2 receptor and PS-ASO. From previous studies, our proposed trafficking pathway for Stabilin-1/-2 receptors and PS-ASO was clathrin-coated vesicle-early endosome-late endosome-lysosome. As per the proposed pathway, we first determined if Stabilin-2 is utilizing a clathrin-dependent or -independent pathway for PS-ASO internalization inside the cell. Our image analysis showed that Stabilin-2 is internalizing PS-ASO through clathrin-coated vesicles (Figure 2A). We observed the best correlation between Stab2 and clathrin-coated vesicle at the 30-min pulse, 0-chase time point, and then correlation trendline decreased (Figure 2E), suggesting that Stab2 has moved forward in the pathway. A similar trend was observed for PS-ASO (Figures 3AF and 3E). However, we did not see any correlation between Stab2 or PS-ASO with Caveolin (Figures S1 and S2E), suggesting that Stab2 is not utilizing caveolin-mediated endocytosis for internalizing PS-ASO. Next, we assessed the colocalization of Stab2 or PS-ASO with an early endosome vesicle (EEA1). Our results showed high correlation between the Stabilin-2 receptor or PS-ASO with EEA1 at 0-min chase (Figures 2B and 3B), suggesting that Stab2 and PS-ASO are being trafficked forward through the early endosomal vesicle. The correlation trend line between Stabilin-2 and EEA1 decreased up to 60-min chase and then became constant (Figure 2E), whereas the correlation trend line reached almost 0 for colocalization between PS-ASO and EEA1 (Figure 3E), suggesting that most of the PS-ASOs have left the early endosome by the end of the 90-min chase. Moving forward, image analysis revealed colocalization between Rab7, a late endosomal marker, and Stab2 (Figure 2C) or PS-ASO (Figure 3C). We observed an increasing trend line for the correlation coefficient between Stabilin-2 and a late endosome vesicle (Rab7) from 0- to 30-min chase (max), followed by a small decreasing trend line from 30- to 60-min chase leading to a constant trend line until it reached 120-min chase (Figure 2E). A similar trend was observed for PS-ASO and Rab7 colocalization where the maximum correlation was seen at 60-min chase (Figure 3E). Last, we investigated the colocalization of Stab2 or PS-ASO in lysosomes, where they will get degraded. Our correlation colocalization analysis results showed that small amounts of Stabilin receptor were trafficked to lysosomes (LAMP1) (Figures 2D and 2E). Since Stab2 is a recycling receptor, most of them are expected to get recycled back to the plasma membrane. Moreover, we observed an increasing trend line for the correlation between PS-ASO and LAMP-1 moving from 0- to 120-min chase, and maximum colocalization was observed at 120-min chase (Figures 3D and 3E), suggesting that most of the PS-ASOs have reached lysosomes for degradation. Since intracellular trafficking is a dynamic process, our results suggests that most Stabilin-2 receptors and PS-ASOs occupied the early endosome 30 min after initial exposure and then trafficked to late endosomes and started accumulating in late endosomes at 30- and 60-min chase. Finally, PS-ASO gets degraded in the lysosomes as previously shown by Miller et al.13
Figure 2.
Trafficking of Stabilin-2 receptor in 190-HARE cell after Stabilin-2-mediated endocytosis
Experiment was performed as explained in Figure 1. Stabilin-2 receptor was found to colocalize in (A) a clathrin vesicle (clathrin), (B) an early endosome (EEA1), (C) a late endosome (Rab7), and a small amount in (D) a lysosome (LAMP1). (E) shows correlation quantification (Pearson correlation coefficient) between the Stabilin-2 vesicle system at different time intervals (0, 30, 60, 90, and 120 min) (n = 6 images).
Figure 3.
Trafficking of PS-ASO in 190-HARE cell after Stabilin-2-mediated endocytosis
Experiment were performed as explained in Figure 1. PS-ASO was trafficked via the same pathway and was found to colocalize in (A) a clathrin vesicle (clathrin), (B) an early endosome (EEA1), and (C) a late endosome (Rab7) and was finally degraded in (D) a lysosome (LAMP1). (E) This is a correlation quantification (Pearson correlation coefficient) between the PS-ASO vesicle system at different time intervals (0, 30, 60, 90, and 120 min) (n = 6 images).
We next investigated trafficking of the Stab1 and ASO after Stab1-mediated PS-ASO internalization. Pulse-chase experiments were carried out with Stabilin-1 cell lines, and confocal microscopy images were analyzed. The results showed that ASOs internalized by Stab1 differ in endolysosomal trafficking path when compared with Stab2 (Figure S2). Like Stab2, Stab1 cells utilized clathrin-mediated endocytosis for ASO uptake instead of caveolin-mediated endocytosis. Stab1 was shown to be localized in the early endosome as well as the late endosome; however, not much colocalization was observed between early endosomal marker EEA1 or late endosomal marker Rab7 and ASOs. Earlier studies have suggested the involvement of Stab1 in the trafficking of endocytic endosomes with the trans-Golgi network (TGN). This could explain less or no localization of ASO in the early or late endosomal vesicle.36,37 Moreover, ASOs started accumulating in lysosomes as shown by LAMP-1/ASO colocalization and reached maximum by the end of 120-min chase, though we did not observe Stabilin-1 colocalization with LAMP-1.
CQ enhances ASO escape from endosomes
From our microscopy imaging results, it is evident that ASOs are being trapped in the endosomal vesicle and later degraded in lysosomes. Previous studies have shown that lysosomotropic compounds promote escape of large molecules from the endosomal vesicle by the “proton sponge effect.”38 These compounds acquire a hydrogen group and become protonated once inside the endosomal vesicle. This leads to an influx of water molecules, which cause swelling and leakage in the endosomal vesicle, aiding in the release of large molecules. CQ, a membrane-destabilizing agent, imparts the same effect by increasing endosomal pH and has been shown to affect receptor-mediated endocytosis.39 Recently, a study showed the effect of various membrane-destabilizing agents on endosomal escape of 3'-cholesterol-modified siRNA (chol-siRNA) and found 47-fold enhanced knockdown with CQ treatment as compared with the control group.24 Based on these studies, we decided to investigate the effect of CQ on endosomal escape of PS-ASOs in our cell lines.
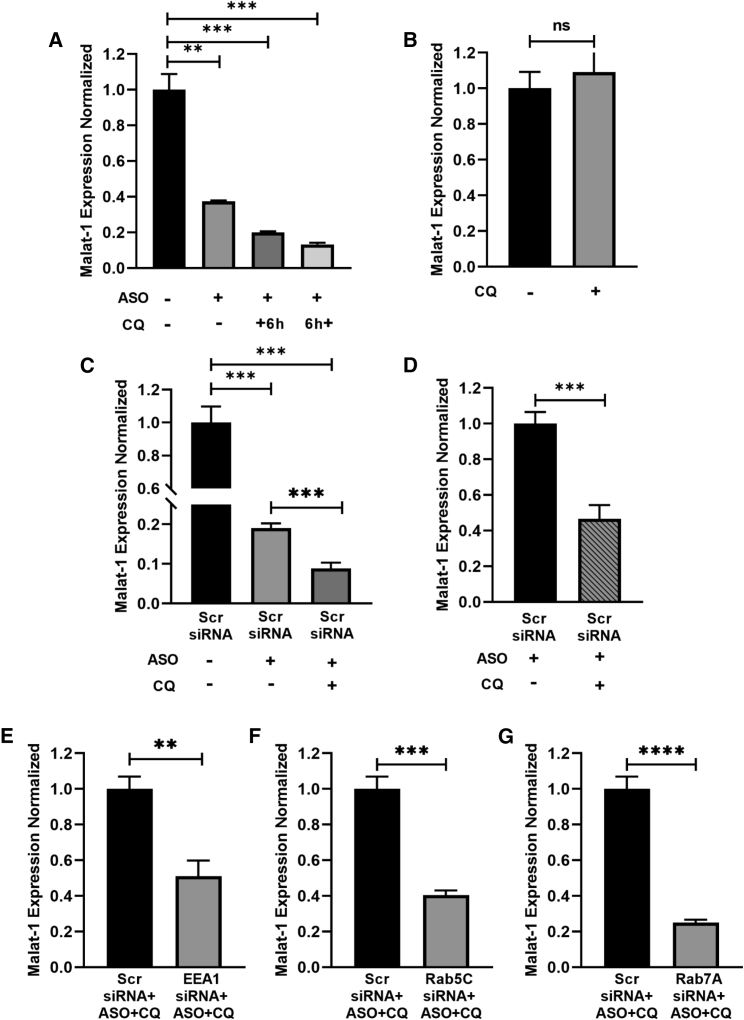
First, we established our controls. Citing previously performed experiments by our group with the assistance of Ionis Pharmaceuticals that were published in 2016,13 we established an effective dose curve of the malat-1 ASO in EV and 190-HARE cells used in all the experiments demonstrated in this paper. The ASO dose we are using in these experiments, and which we used in previous experiments, is 160 nM for gymnotic endocytosis (Figure 4A). Cellular controls determining the effects of three treatments (mock-treated, scrambled siRNA treated, and scrambled siRNA+ASO treated) determined that the effect of Lipofectamine RNAiMax and of the scrambled siRNA did not significantly affect malat-1 knockdown; however, the internalization of ASO contributed to approximately 70%–80% decreased expression of malat-1 on a consistent basis (Figure 4B). Going forward, we next treated cells using one parameter difference to test if specific treatments further modulated malat-1 expression. Cells were treated with 60 μM CQ, 6 h before (+6 h) or after (6 h+) 160 nM PS-ASO treatment. After 24 h of ASO treatment, malat-1 mRNA expression was assessed by qPCR. From our results, cells treated with CQ along with PS-ASO showed 50%–65% further reduction in malat-1 mRNA expression as compared with cells treated with PS-ASO alone (Figure 5). We also evaluated the effect of CQ on malat-1 mRNA expression without PS-ASO treatment to validate that CQ does not affect malat-1 expression on its own (Figure 5B). Next, we decided on a CQ treatment 6 h after PS-ASO treatment because cells looked healthier as compared with cells treated with CQ 6 h before PS-ASO treatment. Previous studies have suggested that CQ treatment damages Rab5+, Rab7+, and LAMP1+ endosomal vesicles.24 EEA1, Rab5C, and Rab7A have also been found to be important endosomal proteins in mediating PS-ASO effectiveness.23 Our study has also observed a similar effect in which endocytic vesicles were enlarged and prone to leakiness (Figure S3A). Next, we investigated if CQ requires these endosomal proteins for mediating its effect. CQ alone affected endosomal escape of ASO (Figure 5C), and the ASO+CQ parameter (bar with the diagonal lines, Figure 5D) will be used as our control to test the effects of CQ in influencing PS-ASO activity after siRNA-mediated knockdown of EEA1/Rab5C/Rab7A. After verifying the knockdown efficiency of siRNAs (Figure S4A–S4C), cells were treated with EEA1, Rab5C, Rab7A, or scrambled siRNA for 48 h, followed by treatment with PS-ASO for 24 h. CQ was added 6 h after PS-ASO treatment for total of 18 h, and then malat-1 mRNA expression was assessed. Cells treated with EEA1 or Rab5C siRNA showed 50%–60% knockdown in malat-1 mRNA expression as compared with scrambled siRNA-treated cells (Figures 5E and 5F). This malat-1 knockdown is comparable to our control treatment group (scrambled [Scr] siRNA+ASO vs. Scr siRNA+ASO+CQ). Similarly, Rab7A siRNA-treated cells showed 70% reduction in mRNA expression as compared with Scr siRNA-treated cells (Figure 5G). From our results, it can be concluded that previously indicated important endosomal proteins are not required by CQ to mediate its effect, nor does CQ have any effect on the functional aspects of these proteins (Figures S3B–S3D).
Figure 4.
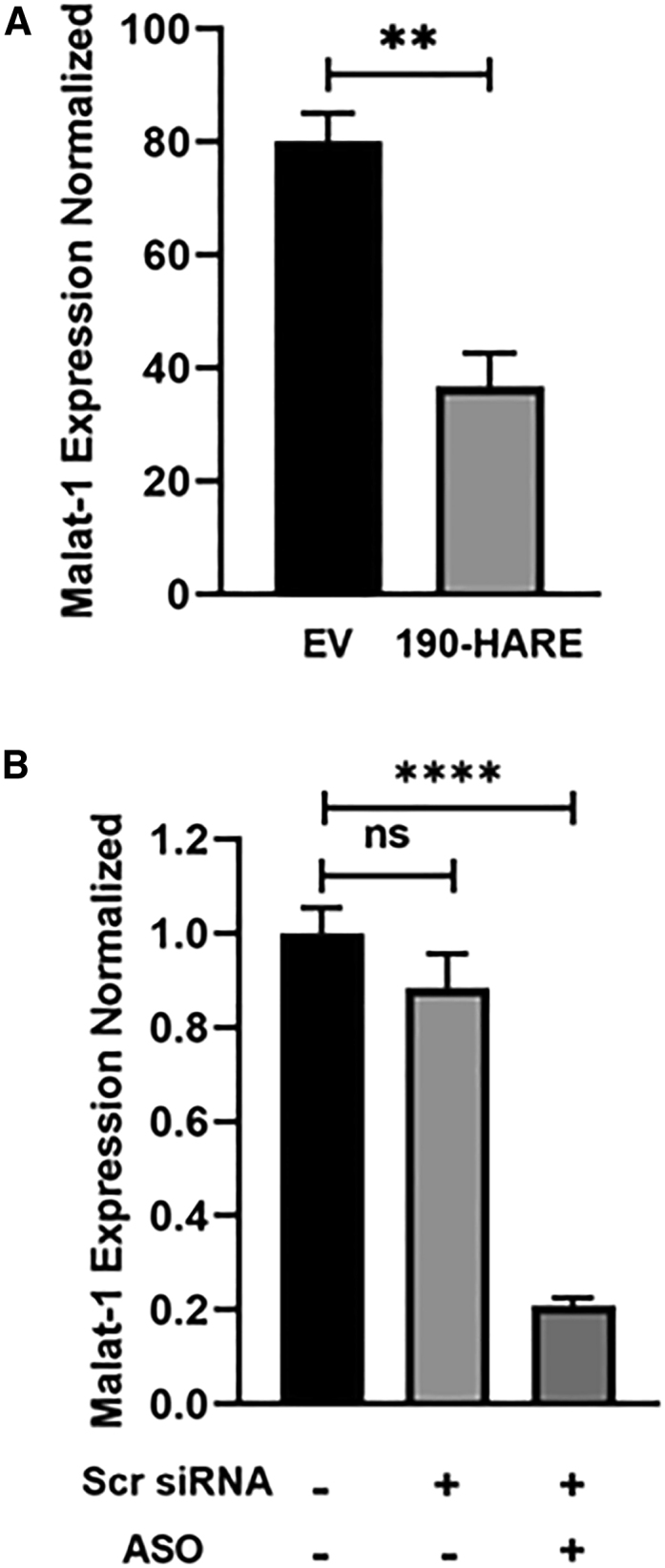
PS-ASO activity in EV and 190-HARE cells
(A) EV or 190-HARE cells were treated with 0.16 μM PS-ASO (24 h). Cells were assessed for malat-1 expression by qPCR. Analysis of PS-ASO activity under different controls. (B) 190-HARE cells were treated with Lipofectamine or Scr siRNA for 48 h, followed by treatment with or without PS-ASO for 24 h as indicated. Cells were assessed for malat-1 expression by qRT-PCR. Statistical analysis was performed using Student’s t test. Data presented as mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. n ≥ 3 in triplicate.
Figure 5.
Chloroquine addition enhances PS-ASO potency
(A) 190-HARE cells were treated with or without 0.16 μM PS-ASO (24 h) followed by 60 μM chloroquine treatment 6 h before (+6 h) or 6 h after (6 h+) PS-ASO treatment. 24 h after PS-ASO treatment, cells were assessed for malat-1 expression by qRT-PCR. Chloroquine addition alone does not affect malat-1 expression. (B) 190-HARE cells were treated with or without 60 μM chloroquine, and malat-1 expression was assessed after 24 h by qRT-PCR. Analysis of PS-ASO activity under different controls. (C) 190-HARE cells were treated with negative control Scr siRNA for 48 h, followed by with or without PS-ASO for 24 h as indicated. 6 h after PS-ASO treatment, cells received 60 μM chloroquine as indicated. 24 h after PS-ASO treatment, cells were assessed for malat-1 expression by qRT-PCR. (D) is an elaborative representation of (C) showing the effect of chloroquine in enhancing PS-ASO activity. Chloroquine acts independently of EEA1/Rab5C/Rab7A. 190-HARE cells were treated with a negative control Scr siRNA and (E) siRNA for EEA1, (F) siRNA for Rab5C, and (G) siRNA for Rab7A respectively for 48 h. PS-ASO and chloroquine treatment was done as explained for Figure 5C. Malat1 mRNA expression was quantified with qRT-PCR. Statistical analysis was performed using Student’s t test. Data presented as mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. n ≥ 3 in triplicate.
In the study done by Colton et al., knocking down EEA1/Rab5C/Rab7A completely abrogated PS-ASO activity, and there was no comparable difference in the malat-1 reduction between PS-ASO treated and non-treated groups.23 Since our results showed that CQ does not require EEA1/Rab5C/Rab7A to mediate its effect, we investigated if the CQ treatment is able to rescue the effect of PS-ASO, which was earlier shown to be abrogated upon EEA1, Rab5C, or Rab7A knockdown. To investigate this, cells were treated with EEA1, Rab5C, or Rab7A siRNA for 48 h, followed by treatment with PS-ASO for 24 h. 6 h after PS-ASO incubation, cells were treated with and without CQ for 18 h, and malat-1 mRNA was assessed. From our results, cells treated with CQ showed 70%–80% enhanced reduction in malat-1 mRNA expression as compared with cells not treated with CQ upon keeping EEA1, Rab5C, or Rab7A knockdown constant (Figures 6A–6C). The reduction in malat-1 mRNA expression in cells treated with CQ was comparable with the reduction in malat-1 mRNA expression in the Scr siRNA+ASO-treated group (Figure 4B). This shows that CQ can restore the PS-ASO activity that was lost after EEA1/Rab5C/Rab7A knockdown, as evidenced by its approximately same knockdown proportion as Scr siRNA+ASO treatment. Our results also suggest that CQ has dominant role in enhancing PS-ASO effectiveness as compared to EEA1/Rab5C/Rab7A endosomal proteins.
Figure 6.
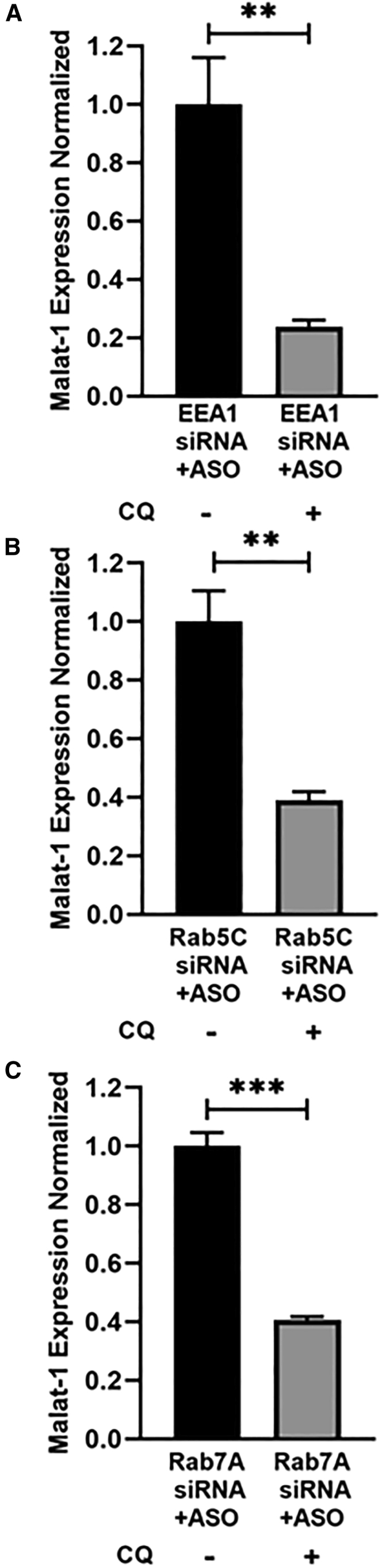
Chloroquine rescues the reduced efficacy of PS-ASO caused by EEA1/Rab5C/Rab7A knockdown (KD)
190-HARE cells were treated with (A) EEA1 siRNA, (B) Rab5C siRNA, and (C) Rab7A siRNA, respectively, for 48 h. PS-ASO and chloroquine treatment was done as explained for Figure 5C. Malat1 mRNA expression was quantified with qRT-PCR. Statistical analysis was performed using Student’s t test. Data presented as mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. n ≥ 3 in triplicate.
Galectin knockdown enhances PS-ASO efficacy
Oligonucleotide release from the endolysosomal system is characterized by endolysosomal membrane damage.40 Membrane damage is detected by the small carbohydrate-binding protein family galectins, which are recruited and clustered at the membrane damage site. Galectins (Gals) recognize the exposed glycosylated membrane components present in the cytosolic compartment. Gals are increasingly utilized as reporters of vesicle damage and endosomal escape of ligand-conjugated siRNAs with or without treatment with membrane-destabilizing compounds.24,25 Gal-8, Gal-9, Gal-3, and, to some extent, Gal-1 and Gal-4 have been previously shown to be recruited around damaged endosomal vesicles.41 Here, we asked the question “do Gals have any functional role in endosomal trafficking machinery, or they are just a sensor for endosomal membrane damage remains unclear?”
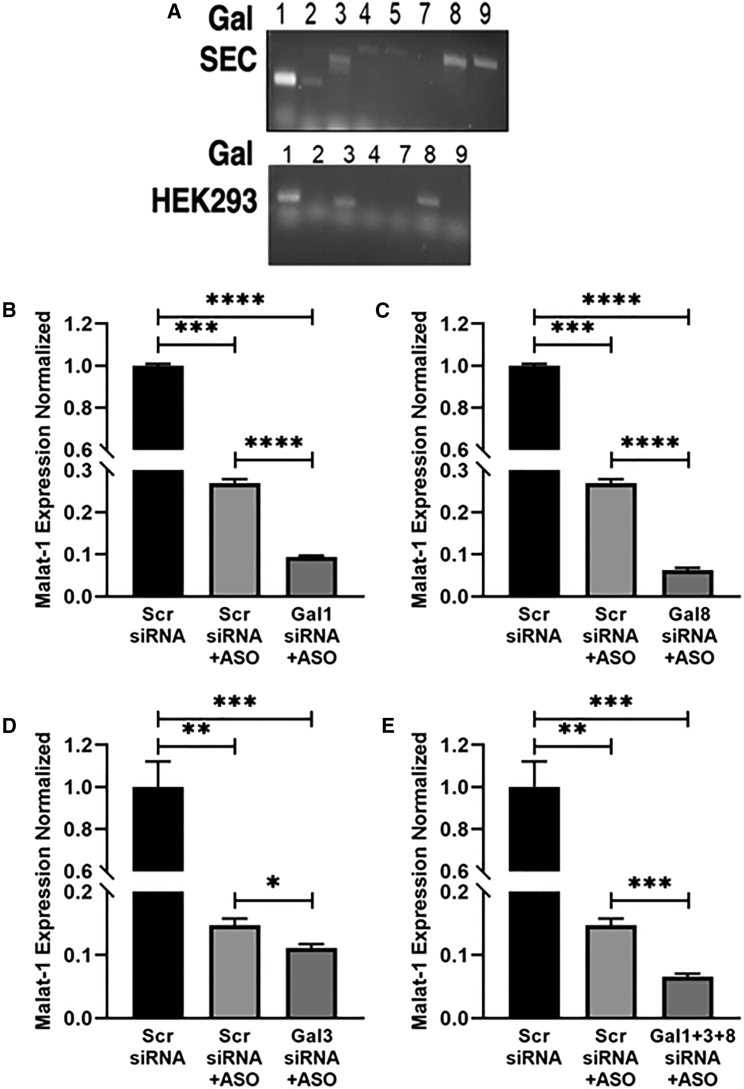
To take one step closer toward understanding the functional role of Gals in ASO escape, we first determined the expression of Gals in sinusoidal endothelial cells (SECs) isolated from murine liver. Results show that SECs express Gal-1 abundantly, have significantly less expression of Gal-8, Gal-9, and Gal-3, and have barely any expression of Gal-2, Gal-4, and Gal-5 (Figure 7A). Next, we verified the expression of Gals in the HEK293 cell line. Results indicate that HEK293 cells expressed Gal-1, Gal-3, and Gal-8 (Figure 7A). Next, we carried out siRNA-mediated knockdown of Gal-1, Gal-8, and Gal-3 in HEK293 cells stably expressing the Stabilin-2 (190 isoform) receptor. Cells were transfected with Gal-1, Gal-8, Gal-3, or Gal-1, Gal-3, and Gal-8 together for 48 h, followed by PS-ASO treatment for 24 h. This was done to investigate if Gal knockdown affects malat-1 expression as a measure of PS-ASO escape. From our results, cells transfected with Gal-1, Gal-8, and Gal-1+Gal-3+Gal-8 siRNA showed 50%–70% enhanced reduction in malat-1 expression as compared with Scr siRNA (control) (Figures 7B, 7C, and 7E). However, cells transfected with Gal-3 siRNA showed only 30% reduction in malat-1 mRNA expression as compared with Scr siRNA control (Figure 7D). An earlier study has shown weak recruitment of Gal-3 upon endosomal damage.41 This suggests less involvement of Gal-3 in endosomal escape, which concurs with our results. To confirm that our qPCR results reflected biological activity, we assessed a pharmacological inhibitor of Gal-1, OTX008, on Gal-1 expression and malat-1 knockdown with PS-ASO. OTX008, which is a Calix[4]arene-derived compound, is an allosteric inhibitor of glycan binding and attenuates Gal-1 binding to membrane-bound glycans.42 Currently, OTX008 is being evaluated as a possible therapeutic against a variety of malignancies.43,44 For our purposes, dosing our 190-HARE cells with 60 μM OTX008 significantly decreased Gal-1 expression while maintaining healthy cellular morphology (Figure 8A). Pretreatment of cells with 60 μM OTX008 for 3 days followed by 24 h treatment of ASO resulted in a further 50% reduction of malat-1 expression, suggesting that decreased Gal-1 expression results in more ASO escape into the cytoplasm (Figure 8B). Altogether, these results suggest that Gals are involved in maintaining endosomal membrane integrity and that their deficiency results in mediating endosomal escape of ASO and, thereby, enhancing ASO efficacy.
Figure 7.
Gal KD enhances PS-ASO efficacy
Gal expression profile in (A) liver sinusoidal endothelial cells (SECs) and HEK293 cells. 190-HARE cells were treated with a negative control Scr siRNA and (B) siRNA for Gal-1 siRNA, (C) siRNA for Gal-8, (D) siRNA for Gal-3, and (E) siRNA for Gal-1+Gal-8+Gal-3, respectively, for 48 h, followed by treatment with or without 0.1 μM PS-ASO for 24 h. Cells were assessed for malat-1 expression by qRT-PCR after 24 h.
Figure 8.
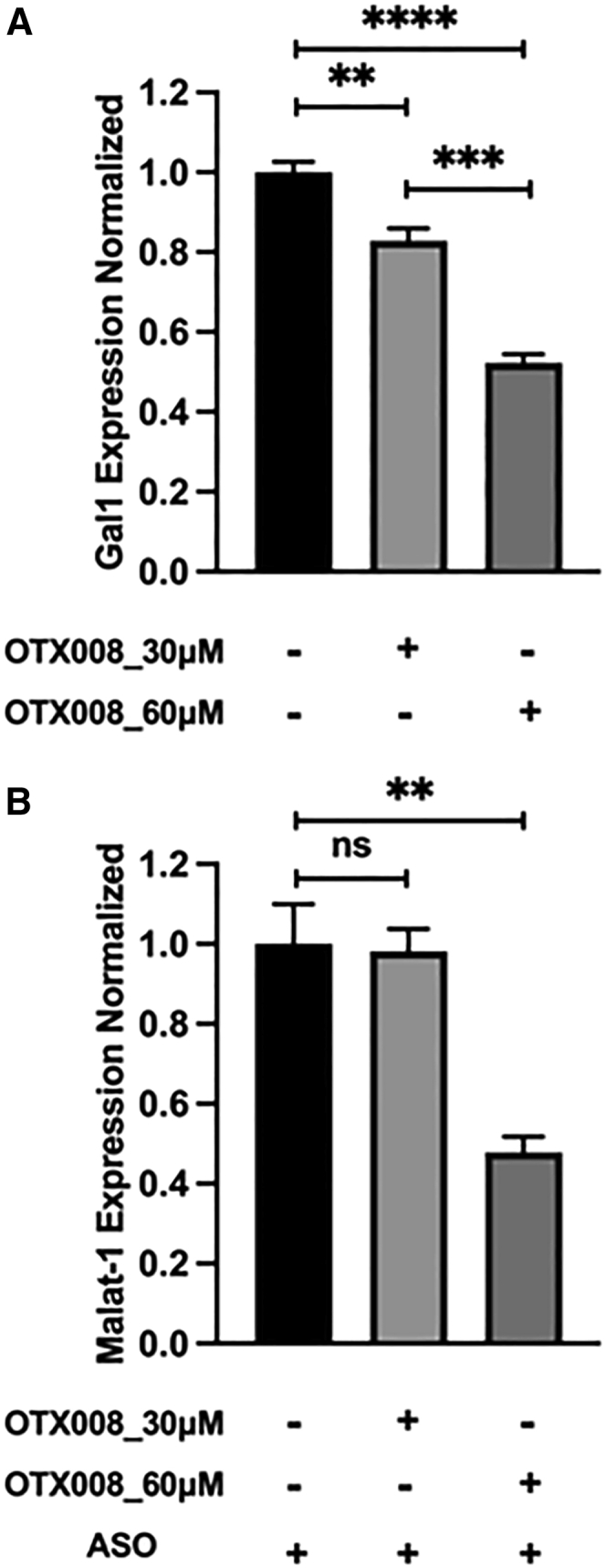
OTX008 inhibitor reduces Gal-1 expression
(A) 190-HARE cells were treated with or without OTX008 inhibitor (30 or 60 μM) for 72 h. After 72 h, cells were assessed for Gal-1 expression by qRT-PCR (n = 3, mean ± SD). (B) 190-HARE cells were treated with OTX008 inhibitor as described in Figure 7A, followed by treatment with or without 0.1 μM ASO for 24 h. Cells were assessed for malat-1 expression by qRT-PCR (n = 3, mean ± SD).
Gals promote endosomal maturation
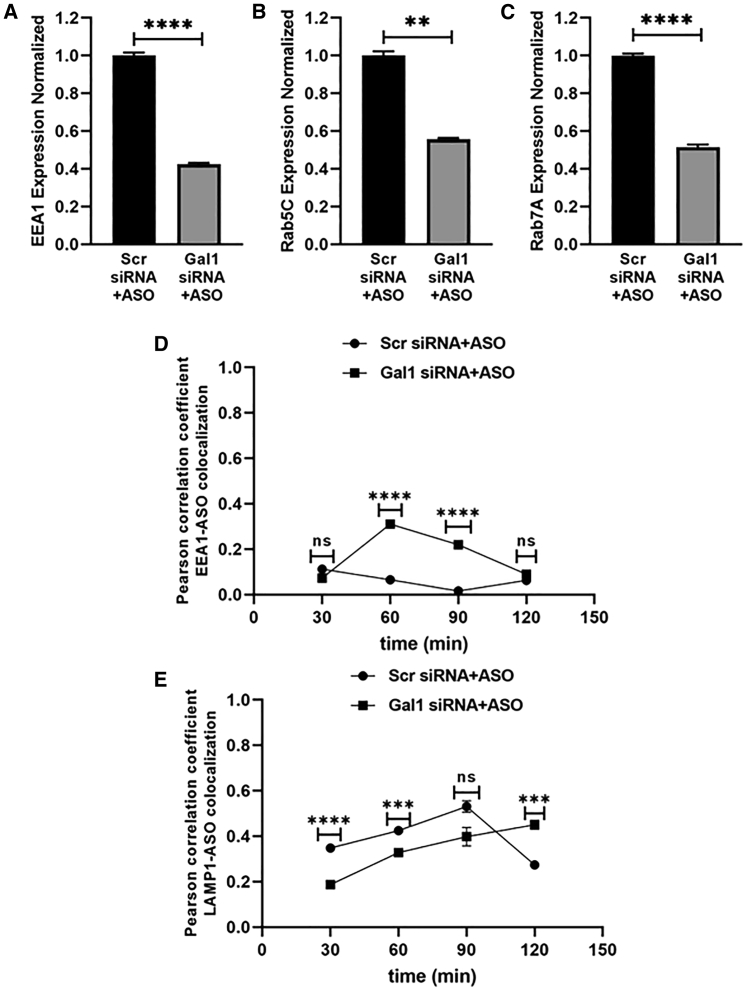
From our confocal microscopy experiments, we delineated the pathway through which ASO is trafficked after Stabilin receptor-mediated endocytosis. Endosomal vesicles contain a plethora of proteins and protein complexes that regulate many aspects of intracellular trafficking.21 One such important family is the Rab protein family and its interacting partners. We have previously shown that siRNA-mediated knockdown of endosomal proteins EEA1, Rab5C, or Rab7A abrogates ASO effectiveness.23 After verifying the functional role of Gal in ASO escape, we wanted to investigate the possible mechanism of action with which Gals enhance ASO effectiveness. Earlier studies have suggested Gal’s role in inhibiting endocytic uptake by lattice formation at the plasma membrane,45 so we performed endocytosis experiments in the recombinant 190-HARE cell line and measured uptake of PS-ASO in the presence and absence of Gals and found no difference in endocytic uptake of PS-ASOs (Figure S5). Next, we investigated the effect of Gal-1 knockdown on the mRNA expression of previously indicated important endosomal proteins EEA1, Rab5C, and Rab7A.23 After transfecting the cells with Gal-1 siRNA or Scr siRNA control for 48 h, followed by treatment with PS-ASO for 24 h, mRNA expression of EEA1, Rab5C, and Rab7A was assessed. Surprisingly, the results showed decreased mRNA expression of EEA1, Rab5C, and Rab7A in Gal-1 siRNA-transfected cells as compared with the Scr siRNA-treated group (Figures 9A–9C). This result suggests the role of Gal-1 in endosomal trafficking regulation. Data presented by Colton et al. demonstrated that knockdown of EEA1, Rab5C, and Rab7A negated PS-ASO activity.23 However, in Gal-1 knockdown-treated cells, we observed reduced expression of EEA1, Rab5C, and Rab7A, which presents a different aspect of PS-ASO regulation by EEA1, Rab5C, and Rab7A mediated by Gal-1, so we investigated whether Gal-1 modulates endosomal vesicle maturation or trafficking. First, we carried out siRNA-mediated knockdown of Gal-1 in 190-HARE cells (Figure S4D) and then performed pulse-chase experiments as described previously with and without Gal-1 siRNA-transfection treatment. Pearson correlation coefficient values between EEA1 and PS-ASO show an increased percentage of PS-ASO-containing early endosomal vesicles at 60- and 90-min chase in the Gal-1 siRNA-treated group as compared with the Scr siRNA-treated group (Figure 9D). Similarly, we found late buildup of PS-ASOs in the lysosomal vesicle in the Gal-1 siRNA-treated group as compared with the Scr siRNA-treated group, evidenced by LAMP1 and PS-ASO colocalization quantification (Figure 9E). We have incorporated representative images for the same (Figures S6 and S7). This suggests that the lack of Gal-1 delays endosomal vesicle maturation, which imparts PS-ASOs with more time to escape; thus, more PS-ASOs reach the target, thereby enhancing their efficacy.
Figure 9.
Gal1 KD reduces EEA1/Rab5c/Rab7A mRNA expression
190-HARE cells were treated with a negative control Scr siRNA and siRNA for Gal-1 for 48 h, followed by treatment with 0.1 μM PS-ASO for 24 h. After 24 h, cells were assessed for (A) EEA1 mRNA expression, (B) Rab5C mRNA expression, and (C) Rab7A mRNA expression by qPCR. Statistical analysis was performed using Student’s t test. Data presented as mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. n ≥ 3 in triplicate. Gal1 KD delays endosomal maturation: 190-HARE cells were treated with a negative control Scr siRNA and siRNA for Gal1 for 48 h, followed by pulse-chase experiment as described in Figure 2 for variable time points (30, 60, 90, and 120 min). Colocalization between (D) early endosome (EEA1) and ASO (E) lysosome (LAMP1) and ASO under different treatment conditions for variable time points was quantified with Pearson correlation coefficient as described in Figure 2 (n = 6 images).
Discussion
Previous studies have established that Stabilin receptors are by far the most efficient receptor for bulk PS-ASO uptake.13 Depending on the cell surface receptors involved in PS-ASO uptake, various internalization pathways could be used resulting in productive (PS-ASO-enhancing effect) or non-productive (PS-ASO-decreasing effect) pathways.12,46,47,48 With the use of other ligands, Stab2 is known to have a rapid recycling rate, though not much is known about the fate of PS-ASOs once they are internalized by the Stabilin receptor or about their destruction in lysosomes. Although several studies have laid out possible mechanisms responsible for escape of PS-ASOs from the endosomal vesicle,49,50,51 a detailed investigation is needed to provide a directional trafficking route followed by the Stabilin receptor and PS-ASO before finally getting degraded in lysosomes. Once we know the endosomal trafficking route, we would be able to exploit the trafficking machinery to enhance PS-ASO escape, thereby increasing PS-ASO efficacy. Our study fills in this gap by providing a detailed trafficking route using confocal microscopy.
Due to the immensity of endosomal escape problems, various approaches have been explored to enhance endosomal escape and thereby advance oligonucleotide therapeutic efficacy. Some of these studies outline ways to manipulate the endosomal machinery by influencing the proteins involved in trafficking processes,23,52,53,54 whereas others utilize small molecules, endolytic peptides, dynamic polyconjugates, and ionizable lipid nanoparticles as well as enveloped viruses to influence oligonucleotide trafficking.55 One small-molecule compound that caught our attention and has been widely studied with siRNAs is CQ, an antimalarial drug.24,56,57 To date, there are no publications detailing its effect on enhancing PS-ASO effect by influencing endosomal escape. Our study addresses this idea by demonstrating the effects of CQ on PS-ASOs in Stab2-expressing cells. We also established its dominant role in enhancing PS-ASO endosomal escape as compared with endosomal proteins involved in regulating trafficking machinery. However, some of the studies are concerned with the cytotoxic effects of high-dose CQ in clinical settings. To overcome this dilemma, Cheng et al. has performed a detailed investigation into the structure-function relationships of CQ and its analogs.58 They came up with a few analogs that are 10 times more efficacious at a lower concentration as compared to CQ. This study could be useful in removing the hindrance posed in the utilization of CQ due to high-dose cytotoxicity and will further help in successful utilization of CQ derivatives in oligonucleotide therapeutics. Theoretically, CQ may be conjugated to PS-ASOs to facilitate escape of the PS-ASOs and to limit disruption of non-PS-ASO-containing endosomal vesicles.
Previous studies have shown the importance of recruitment of vesicles and the proteins that aid the recruitment in facilitating endosomal escape of PS-ASOs from late endosomes. To name a few, COP-II vesicles mediated by Suntaxin-5 (STX5) and M6PR vesicles mediated by GRIP and coiled-coil domain-containing 2 (GCC2) proteins facilitate PS-ASO release by recruiting to late endosomes upon PS-ASO uptake and internalization.53,54 Annexin A2 (ANXA2) and Golgi-58K are another set of proteins that are involved in facilitating PS-ASO release by relocating to late endosomes.59,60 These studies suggested the importance of proteins involved in the recruitment process in facilitating PS-ASO escape, which prompted us to study the role of Gals in enhancing PS-ASO release, as these are also recruited upon endosomal release of PS-ASOs.24,25,41 ANXA2 mediates PS-ASO endosomal release by affecting maturation of early endosomes to late endosomes, which led us to study the mechanism by which Gals decrease PS-ASO efficacy. Surprisingly, Gals also influence endosomal vesicle maturation by stabilizing the mRNA expression of EEA1/Rab5C/Rab7A, important endosomal entities involved in endosomal trafficking and maturation process.
Taken altogether, in this study, we reported the detailed intracellular trafficking route followed by PS-ASO and Stabilin receptors after Stabilin receptor-mediated endocytosis. Furthermore, we evaluated the effect of CQ, an endosomal membrane-destabilizing agent, upon PS-ASO escape and its efficacy. We also investigated the role of CQ in rescuing the lost PS-ASO effect upon important endosomal marker EEA1/Rab5C/Rab7A knockdown and showed that CQ plays a dominant role in enhancing PS-ASO efficacy. Lastly, we showed the novel role of Gals as an important key player in the inhibition of PS-ASO escape by directly promoting endosomal vesicle maturation. Delay in endosomal vesicle maturation imparts more time for PS-ASO escape and provides more PS-ASO accessibility for RNAse H machinery.
Materials and methods
Cell lines
The Stabilin-1, Stabilin-2, and EV stably-expressing cell lines used in this study were generated as previously described.17,61 In brief, a pcDNA5/FRT/V5-6xHIS-TOPO (Life Tech, Carlsbad, CA, USA) mammalian vector was used for integrating the cDNA of interest into the genome of Flp-In HEK293 cells by stable transfection (Life Tech). Cells were cultured in DMEM media containing 8% FBS for optimal growth at 37°C and 5% CO2. Hygromycin B at a working concentration of 50 μg/mL was also added to media to maintain the stable cell lines. The Stab2 cell line used in this study is named 190-HARE in previous papers,18,62 though both the 190-HARE and full-length Stabilin-2 have similar binding and endocytic kinetics.13
Antibodies
The primary antibodies used in this study were either made in house or were purchased from Cell Signaling Technologies. Stabilin-2 primary antibody was made by the Dr. Paul Weigel lab and was labeled as mAb30,63 and Stabilin-1 primary antibody (clone 911) was a kind gift from Dr. Marko Salmi (University of Turku, Turku, Finland). Anti-mouse immunoglobulin G (IgG) Fab2 Alexa Flour 488 (Cell Signaling Technology, catalog #4408S) was used for detecting these primary antibodies. Primary antibodies for clathrin (catalog #2410S), caveolin 1 (catalog #3238S), early endosomal antigen (EEA1) (catalog #2411S), Rab7 (catalog #9367S), and LAMP1 (catalog #9091S) were purchased from Cell Signaling Technology. Goat anti-rabbit IgG Alexa Fluor Plus 647 (Invitrogen, #A32733) and goat anti-mouse IgG Alexa Fluor 488 (Cell Signaling Technologies, #4408S) secondary antibody was used for detecting these primary antibodies.
Oligonucleotide synthesis and delivery
PS-ASOs were obtained from Ionis Pharmaceuticals (Carlsbad, CA, USA). The PS-ASO used in this study was designed against malat-1, a long non-coding RNA (Ionis #395251).13 This PS-ASO (GCTTCAGTCATGACTTCCTT) is a 5-10-5 gapmer that is modified at the ribose 2′ position with a methoxyethyl group (MOE) and a PS backbone on the flanking ends. The delivery of PS-ASO to cultured cells was carried out under gymnotic condition. PS-ASOs were mixed with media containing standard DMEM and 8% FBS to reach a final concentration of 160 nM/well and then were added to cultured cells. The PS-ASO used in microscopy experiments had the same chemistry and was conjugated with Cy3 on the 5′ end and used at a concentration of 100 nM (Ionis #730436).
siRNA delivery
Stab2 cells (190-HARE) were seeded in 24-well plates for 2 days to reach 60% confluency at the time of transfection. The cells were then transfected with siRNA (5 pmol/well) mixed with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, Catalog #13778075) in duplicate and incubated for 48 h. siRNAs against each protein target were ordered from Silencer Select (Ambion). The sense sequences are as follows: EEA1-5′-GCUAAGUUGCAUUCCGAAtt-3′, Rab7A-5′-GAGCUGACUUUCUGACCAAtt-3′, Rab5C-5′-GGACAGGAGCGGUAUCACA-3′, LGALS1-5′-GAUGGAUACGAAUUCAAGUtt-3′, LGALS8-5′-CGAUGUUCCUAGUGACGCAtt-3′, and LGALS3-5′-GACAGUCGGUUUUCCCAUUtt-3′. Negative/Scr control was prestocked (catalog #4390843).
CQ and OTX008 inhibitor treatment
CQ diphosphate salt was purchased from Acros Organics (catalog #455240250). Cells were grown to 80% confluency. We tested different concentrations of CQ (30, 60, 90, and 120 μM), and 60 μM concentration was chosen as working concentration after assessing cell morphology. We followed a stepwise procedure for treatment. First, cells were treated with 160 nM ASO (24 h), followed by CQ treatment after 6 or 18 h. Second, cells were first treated with CQ, and after 6 h or 18 h, ASO treatment followed. After 24 h, cells were processed for RNA isolation and subsequent qPCR.
OTX008 was purchased from MedChemExpress #HY-19756. We tested different concentrations of OTX008 (0.01–60 μM) as previously reported64 and chose 30 and 60 μM as the working concentrations in ethanol after assessing cell morphology and reduction in Gal1 expression. We followed a stepwise procedure for treatment. First, cells were treated with OTX008 or an equal volume of ethanol for 72 h, followed by RNA isolation and subsequent qPCR to assess Gal-1 expression. To assess OTX008’s effect on PS-ASO, cells were first treated with or without OTX008 for 72 h, followed by treatment with or without PS-ASO for 24 h. After 24 h, cells were processed for RNA isolation and subsequent qPCR to assess malat-1 expression.
Purification of RNA and qPCR analysis
48 h after incubation with siRNA, cells were treated with 100 nM PS-ASO against malat-1 for 24 h and washed twice with PBS. Total RNA isolation was carried out using Trizol reagent (Invitrogen, catalog #15596026), according to the manufacturer’s instructions. The RNA was quantified by Nanodrop 2000 spectrophotometer, and the quality was assessed by agarose gel electrophoresis. cDNA was synthesized from an RNA template with the EasyScript cDNA synthesis kit (Lambda Biotech., catalog #G234). Gene expression was analyzed by qRT-PCR using 50 ng cDNA per 20 μL reaction volume. PowerUp SYBR Green Master Mix (Applied Biosystems, catalog #A25742) was used to perform qRT-PCR using the recommended cycling protocol: 2 min at 50°C, 2 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C on a Bio-Rad CFX Connect qPCR machine. Samples were run in at least biological triplicates and technical duplicates. Melt curve was analyzed to evaluate primer specificity. Cq values were used to determine malat-1 mRNA expression relative to internal TBP control; afterward, the relative quantities were normalized to their respective controls for each treatment group to generate reported gene expression levels for each RNA target. The following primers were used for targets: malat-1 (forward: 5′-CACCGAAGGCTTAAAGTAGGAC-3′, reverse: 5′-GCTGACACTTCTCTTGACCTTAG-3′), TBP control (forward: 5′-GATAAGAGAGCCACGAACCAC-3′, reverse: 5′-CAAGAACTTAGCTGGAAAACCC-3′) EEA1.
Pulse-chase experiments
8-well chamber slides were pretreated with poly-L-lysine (Sigma, catalog #P9404) for 5 min, followed by rinsing with water twice. Chamber slides were then dried under the cell culture hood and irradiated with UV light to maintain sterility. Poly-L-lysine facilitates strong attachment between the cell’s plasma membrane and the slide surface. Chamber slides were seeded with cells to reach 60% confluency, and cells in each well were first treated with media containing 100 nM Cy3-labeled ASO for 30 min (pulse), followed by rinsing with PBS twice. Cells were then incubated with media without ASO for 0, 30, 60, 90, and 120 min (chase). After each chase time point, cells were washed once with PBS and fixed with 4× paraformaldehyde (EEA1/LAMP1/caveolin)/formaldehyde (Rab7/clathrin) for 15 min, and immunocytochemistry was performed.
Immunocytochemistry
Fixed cells were permeabilized with 0.1% Saponin (Stabilin-2, Stabilin-1, EEA1, LAMP1) or 0.1% Triton X-100 (clathrin/caveolin/Rab7) for 15 min at room temperature. Next, cells were blocked for 1 h with blocking buffer (1× PBS+5% BSA+0.3% Triton X-100) at room temperature. Multicolor immunostaining was performed using either sequential or simultaneous incubation.
Simultaneous incubation method
For EEA1/LAMP1 antibody with Stabilin-1 or -2 detection, a simultaneous incubation method was used. For simultaneous multicolor staining of EEA1 or LAMP1 with Stabilin-1 or -2, first, cells were incubated with both the primary antibodies in 1× PBS+1% (0.1 g) BSA+0.01% Saponin in a humidified chamber for 1 h at room temperature, followed by washing with PBS thrice (5 min/wash). Next, cells were incubated with both the secondary antibodies in 1× PBS+1% BSA+0.01% Saponin in a humidified chamber for 1 h at room temperature. Next, cells were washed thrice with PBS and then mounted on a coverslip using antifade mounting media (Southern Biotech, catalog #0100-0), and slides were imaged using confocal laser scanning microscopy (CLSM) on a Nikon A1R Ti2 inverted fluorescent microscope using Nikon Elements. A Plan Apo VC 60×/1.40 oil immersion lens was used with a 2× digital zoom for a total magnification of 1,200×. Laser lines used on CLSM were as follows: 405 (nucleus), 488 laser (Stabilin-1/2), 560 laser line (PS-ASO), and 640 laser line (vesicle). Z steps were acquired between 0.7 and 1 μm using channel series/sequence mode. Image analysis was carried out using the EzColocalization plugin in ImageJ.
Sequential incubation method
A sequential incubation method was used for clathrin/caveolin/Rab7 antibody with Stabilin-1/-2 detection. For sequential multicolor staining, cells were first incubated with Stabilin-1 or -2 primary antibodies in 1× PBS+1% (0.1 g) BSA+0.01% Saponin in a humidified chamber for 1 h at room temperature, followed by three washes with PBS (5 min/wash). Next, cells were incubated with secondary antibody in 1× PBS+1% BSA+0.01% Saponin in a humidified chamber for 1 h at room temperature, followed by three washes with PBS (5 min/wash). Cells were blocked a second time with blocking buffer (1× PBS+3% BSA+0.3% Triton X-100) for 1 h to be used with second primary antibody detection. Next, cells were incubated with the second primary antibodies (clathrin/caveolin/Rab7) in 1× PBS+1% BSA+0.3% Triton X-100 in a humidified chamber for 1 h at room temperature, followed by washing with PBS thrice (5 min/wash). Afterward, cells were incubated with the secondary antibodies in 1× PBS+1% BSA+0.01% Saponin in a humidified chamber for 1 h at room temperature. After 1 h, cells were washed thrice with PBS and then mounted on a coverslip using antifade mounting media (Southern Biotech, catalog #0100-0). Slides were imaged using confocal microscopy, and image analysis was carried out using the EzColocalization plugin in ImageJ.
Purification of hepatocytes and liver sinusoidal endothelial cells
Purified hepatocytes and sinusoidal endothelial cells were obtained using our previously published protocol65 which was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Nebraska. Briefly, C57BL6/J mice were anesthetised with 30% isoflurane dissolved in polyethylene glycol 200 with the abdominal cavity exposed. A 25 gauge catheter was inserted into the portal vein and the liver flushed with saline, perfused with collagenase (Collagenase Type I, Worthington Biochemical), and the digested organ was forced through 100 um and 25 um mesh filters. Hepatocytes were purified by low-speed centrifugation. LSECs were purified away from other nonparenchymal cells by Percoll (Cytiva) followed by selective adhesion on polystyrene. Purified cells were then subject to RNA purification (Trizol, Life Technologies), cDNA synthesis (RevertAid First Strand cDNA Synthesis Kit, Thermofisher #1622), and PCR.
Screening galectin expression by PCR
Primer sequences for all PCR reactions used for screening may be found in supplementary Table S1.
Statistics
Statistical analysis was performed using Student’s t test for pairwise comparison with GraphPad Prism 9.4.1. The number of samples used was at least three. For qPCR experiments, first, PS-ASO activity was verified by malat-1 mRNA knockdown assessment in the untreated vs. the ASO-treated group and the Lipofectamine or Scr siRNA- vs. Scr siRNA+ASO-treated group. Afterward, normalization was carried out to deduce any modulation in malat-1 mRNA expression because of added treatment or knockdown when compared with non-treated or Scr siRNA- or Scr siRNA+ASO-treated groups.
Pearson’s correlation coefficient was used for quantitative analysis of confocal microscopy images. At least 6 images were analyzed per dataset.
Acknowledgments
We also commend Dr. You “Joe” Zhou and Terri Fangman from the Morrison Microscopy Center at the University of Nebraska for their assistance with confocal imaging. We are grateful for all PS-ASOs generously supplied by Ionis Pharmaceuticals free of charge. This project was funded by NIH grants HL130864 and GM147913 to E.N.H. and the Nebraska Center for Integrated Biomolecular Communication (NIH National Institute of General Medical Sciences P20 GM113126).
Author contributions
E.P. designed and executed all experiments and wrote the manuscript. E.N.H. assisted in the experimental design and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.07.019.
Supplemental information
Data and code availability
All raw data are available by contacting Prof. Edward Harris at eharris5@unl.edu.
References
- 1.Dhuri K., Bechtold C., Quijano E., Pham H., Gupta A., Vikram A., Bahal R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020;9:2004. doi: 10.3390/jcm9062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 3.Di Fusco D., Dinallo V., Marafini I., Figliuzzi M.M., Romano B., Monteleone G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front. Pharmacol. 2019;10:305. doi: 10.3389/fphar.2019.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossor A.M., Reilly M.M., Sleigh J.N. Antisense oligonucleotides and other genetic therapies made simple. Practical Neurol. 2018;18:126–131. doi: 10.1136/practneurol-2017-001764. [DOI] [PubMed] [Google Scholar]
- 5.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 8.Watts J.K., Corey D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Contr. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Juliano R.L., Ming X., Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjugate Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juliano R.L., Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:35–45. doi: 10.1016/j.addr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller C.M., Donner A.J., Blank E.E., Egger A.W., Kellar B.M., Østergaard M.E., Seth P.P., Harris E.N. Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res. 2016;44:2782–2794. doi: 10.1093/nar/gkw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy J.E., Tedbury P.R., Homer-Vanniasinkam S., Walker J.H., Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 2005;182:1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 15.Harris E.N., Cabral F. Ligand Binding and Signaling of HARE/Stabilin-2. Biomolecules. 2019;9 doi: 10.3390/biom9070273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris E.N., Weigel J.A., Weigel P.H. Endocytic function, glycosaminoglycan specificity, and antibody sensitivity of the recombinant human 190-kDa hyaluronan receptor for endocytosis (HARE) J. Biol. Chem. 2004;279:36201–36209. doi: 10.1074/jbc.M405322200. [DOI] [PubMed] [Google Scholar]
- 17.Harris E.N., Kyosseva S.V., Weigel J.A., Weigel P.H. Expression, processing, and glycosaminoglycan binding activity of the recombinant human 315-kDa hyaluronic acid receptor for endocytosis (HARE) J. Biol. Chem. 2007;282:2785–2797. doi: 10.1074/jbc.M607787200. [DOI] [PubMed] [Google Scholar]
- 18.Harris E.N., Weigel P.H. The ligand-binding profile of HARE: hyaluronan and chondroitin sulfates A, C, and D bind to overlapping sites distinct from the sites for heparin, acetylated low-density lipoprotein, dermatan sulfate, and CS-E. Glycobiology. 2008;18:638–648. doi: 10.1093/glycob/cwn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown C.R., Gupta S., Qin J., Racie T., He G., Lentini S., Malone R., Yu M., Matsuda S., Shulga-Morskaya S., et al. Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res. 2020;48:11827–11844. doi: 10.1093/nar/gkaa670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He C., Migawa M.T., Chen K., Weston T.A., Tanowitz M., Song W., Guagliardo P., Iyer K.S., Bennett C.F., Fong L.G., et al. High-resolution visualization and quantification of nucleic acid-based therapeutics in cells and tissues using Nanoscale secondary ion mass spectrometry (NanoSIMS) Nucleic Acids Res. 2021;49:1–14. doi: 10.1093/nar/gkaa1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juliano R.L. Chemical Manipulation of the Endosome Trafficking Machinery: Implications for Oligonucleotide Delivery. Biomedicines. 2021;9:512. doi: 10.3390/biomedicines9050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer S.R. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller C.M., Wan W.B., Seth P.P., Harris E.N. Endosomal Escape of Antisense Oligonucleotides Internalized by Stabilin Receptors Is Regulated by Rab5C and EEA1 During Endosomal Maturation. Nucleic Acid Therapeut. 2018;28:86–96. doi: 10.1089/nat.2017.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Rietz H., Hedlund H., Wilhelmson S., Nordenfelt P., Wittrup A. Imaging small molecule-induced endosomal escape of siRNA. Nat. Commun. 2020;11:1809. doi: 10.1038/s41467-020-15300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munson M.J., O'Driscoll G., Silva A.M., Lázaro-Ibáñez E., Gallud A., Wilson J.T., Collén A., Esbjörner E.K., Sabirsh A. A high-throughput Galectin-9 imaging assay for quantifying nanoparticle uptake, endosomal escape and functional RNA delivery. Commun. Biol. 2021;4:211. doi: 10.1038/s42003-021-01728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborn M.F., Alterman J.F., Nikan M., Cao H., Didiot M.C., Hassler M.R., Coles A.H., Khvorova A. Guanabenz (Wytensin) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res. 2015;43:8664–8672. doi: 10.1093/nar/gkv942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou F.C., Chen H.Y., Kuo C.C., Sytwu H.K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018;19:430. doi: 10.3390/ijms19020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F.T., Patterson R.J., Wang J.L. Intracellular functions of galectins. Biochim. Biophys. Acta. 2002;1572:263–273. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., Meng H., Xu S., Qi X. Galectins for Diagnosis and Prognostic Assessment of Human Diseases: An Overview of Meta-Analyses. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2020;26:e923901. doi: 10.12659/MSM.923901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An Y., Xu S., Liu Y., Xu X., Philips C.A., Chen J., Méndez-Sánchez N., Guo X., Qi X. Role of Galectins in the Liver Diseases: A Systematic Review and Meta-Analysis. Front. Med. 2021;8:744518. doi: 10.3389/fmed.2021.744518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 32.Mayor S., Pagano R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannes L., Wunder C. Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic. 2011;12:956–962. doi: 10.1111/j.1600-0854.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald E., Savage B., Zech T. Connecting the dots: combined control of endocytic recycling and degradation. Biochem. Soc. Trans. 2020;48:2377–2386. doi: 10.1042/BST20180255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kzhyshkowska J., Gratchev A., Martens J.H., Pervushina O., Mamidi S., Johansson S., Schledzewski K., Hansen B., He X., Tang J., et al. Stabilin-1 localizes to endosomes and the trans-Golgi network in human macrophages and interacts with GGA adaptors. J. Leukoc. Biol. 2004;76:1151–1161. doi: 10.1189/jlb.0504300. [DOI] [PubMed] [Google Scholar]
- 37.Hansen B., Longati P., Elvevold K., Nedredal G.I., Schledzewski K., Olsen R., Falkowski M., Kzhyshkowska J., Carlsson F., Johansson S., et al. Stabilin-1 and stabilin-2 are both directed into the early endocytic pathway in hepatic sinusoidal endothelium via interactions with clathrin/AP-2, independent of ligand binding. Exp. Cell Res. 2005;303:160–173. doi: 10.1016/j.yexcr.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Bus T., Traeger A., Schubert U.S. The great escape: how cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B. 2018;6:6904–6918. doi: 10.1039/c8tb00967h. [DOI] [PubMed] [Google Scholar]
- 39.Maxfield F.R. Weak bases and ionophores rapidly and reversibly raise the pH of endocytic vesicles in cultured mouse fibroblasts. J. Cell Biol. 1982;95:676–681. doi: 10.1083/jcb.95.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daussy C.F., Wodrich H. "Repair Me if You Can": Membrane Damage, Response, and Control from the Viral Perspective. Cells. 2020;9 doi: 10.3390/cells9092042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dings R.P.M., Miller M.C., Nesmelova I., Astorgues-Xerri L., Kumar N., Serova M., Chen X., Raymond E., Hoye T.R., Mayo K.H. Antitumor agent calixarene 0118 targets human galectin-1 as an allosteric inhibitor of carbohydrate binding. J. Med. Chem. 2012;55:5121–5129. doi: 10.1021/jm300014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corral J.M., Puerto-Nevado L.D., Cedeño M., Río-Vilariño A., Mahillo-Fernández I., Galeano C., Baños N., García-Foncillas J., Dómine M., Cebrián A. Galectin-1, a novel promising target for outcome prediction and treatment in SCLC. Biomed. Pharmacother. 2022;156:113987. doi: 10.1016/j.biopha.2022.113987. [DOI] [PubMed] [Google Scholar]
- 44.Scheiter A., Evert K., Reibenspies L., Cigliano A., Annweiler K., Müller K., Pöhmerer L.M.G., Xu H., Cui G., Itzel T., et al. RASSF1A independence and early galectin-1 upregulation in PIK3CA-induced hepatocarcinogenesis: new therapeutic venues. Mol. Oncol. 2022;16:1091–1118. doi: 10.1002/1878-0261.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johannes L., Jacob R., Leffler H. Galectins at a glance. J. Cell Sci. 2018;131:jcs208884. doi: 10.1242/jcs.208884. [DOI] [PubMed] [Google Scholar]
- 46.Juliano R.L., Ming X., Carver K., Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Therapeut. 2014;24:101–113. doi: 10.1089/nat.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Juliano R.L., Carver K., Cao C., Ming X. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target. 2013;21:27–43. doi: 10.3109/1061186X.2012.740674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 50.Roth C.M. Molecular and cellular barriers limiting the effectiveness of antisense oligonucleotides. Biophys. J. 2005;89:2286–2295. doi: 10.1529/biophysj.104.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen K.D., Kopecková P., Kopecek J. Antisense oligonucleotides delivered to the lysosome escape and actively inhibit the hepatitis B virus. Bioconjugate Chem. 2002;13:975–984. doi: 10.1021/bc025559y. [DOI] [PubMed] [Google Scholar]
- 52.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang X.H., Sun H., Nichols J.G., Allen N., Wang S., Vickers T.A., Shen W., Hsu C.W., Crooke S.T. COPII vesicles can affect the activity of antisense oligonucleotides by facilitating the release of oligonucleotides from endocytic pathways. Nucleic Acids Res. 2018;46:10225–10245. doi: 10.1093/nar/gky841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang X.H., Sun H., Hsu C.W., Nichols J.G., Vickers T.A., De Hoyos C.L., Crooke S.T. Golgi-endosome transport mediated by M6PR facilitates release of antisense oligonucleotides from endosomes. Nucleic Acids Res. 2020;48:1372–1391. doi: 10.1093/nar/gkz1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowdy S.F., Setten R.L., Cui X.S., Jadhav S.G. Delivery of RNA Therapeutics: The Great Endosomal Escape. Nucleic Acid Therapeut. 2022;32:361–368. doi: 10.1089/nat.2022.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattarai S.R., Muthuswamy E., Wani A., Brichacek M., Castañeda A.L., Brock S.L., Oupicky D. Enhanced gene and siRNA delivery by polycation-modified mesoporous silica nanoparticles loaded with chloroquine. Pharm. Res. (N. Y.) 2010;27:2556–2568. doi: 10.1007/s11095-010-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lönn P., Kacsinta A.D., Cui X.S., Hamil A.S., Kaulich M., Gogoi K., Dowdy S.F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016;6:32301. doi: 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng J., Zeidan R., Mishra S., Liu A., Pun S.H., Kulkarni R.P., Jensen G.S., Bellocq N.C., Davis M.E. Structure-function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J. Med. Chem. 2006;49:6522–6531. doi: 10.1021/jm060736s. [DOI] [PubMed] [Google Scholar]
- 59.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016;44:7314–7330. doi: 10.1093/nar/gkw595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang X.H., Nichols J.G., De Hoyos C.L., Sun H., Zhang L., Crooke S.T. Golgi-58K can re-localize to late endosomes upon cellular uptake of PS-ASOs and facilitates endosomal release of ASOs. Nucleic Acids Res. 2021;49:8277–8293. doi: 10.1093/nar/gkab599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pempe E.H., Xu Y., Gopalakrishnan S., Liu J., Harris E.N. Probing structural selectivity of synthetic heparin binding to stabilin protein receptors. J. Biol. Chem. 2012;287:20774–20783. doi: 10.1074/jbc.M111.320069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandey M.S., Weigel P.H. A hyaluronan receptor for endocytosis (HARE) link domain N-glycan is required for extracellular signal-regulated kinase (ERK) and nuclear factor-kappaB (NF-kappaB) signaling in response to the uptake of hyaluronan but not heparin, dermatan sulfate, or acetylated low density lipoprotein (LDL) J. Biol. Chem. 2014;289:21807–21817. doi: 10.1074/jbc.M114.565846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou B., Weigel J.A., Fauss L., Weigel P.H. Identification of the hyaluronan receptor for endocytosis (HARE) J. Biol. Chem. 2000;275:37733–37741. doi: 10.1074/jbc.M003030200. [DOI] [PubMed] [Google Scholar]
- 64.Gheysen L., Soumoy L., Trelcat A., Verset L., Journe F., Saussez S. New Treatment Strategy Targeting Galectin-1 against Thyroid Cancer. Cells. 2021;10 doi: 10.3390/cells10051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cabral F., Miller C.M., Kudrna K.M., Hass B.E., Daubendiek J.G., Kellar B.M., Harris E.N. Purification of hepatocytes and sinusoidal endothelial cell from mouse liver perfusion. J. Vis. Exp. 2018;13:56993. doi: 10.3791/56993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available by contacting Prof. Edward Harris at eharris5@unl.edu.