Abstract
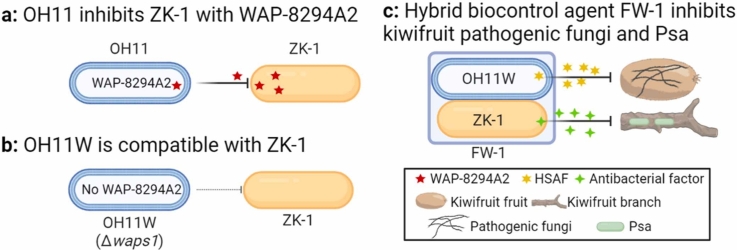
Assembling functional bacterial biocontrol consortia is expected to expand the scope and efficiency of biocontrol agents. Generally, bacterial interspecies interactions lead to incompatibility events, as bacteria can produce antibacterial compounds and/or assemble contact-dependent killing (CDK) devices. Here, we aimed to assemble a bacterial consortium comprising Lysobacter enzymogenes OH11 and Bacillus safensis ZK-1 for the synergistic control of bacterial and fungal diseases of kiwifruit. ZK-1, a native kiwifruit biocontrol bacterium, is effective against Pseudomonas syringae pv. actinidiae (Psa) that causes bacterial kiwifruit canker, but has weak antifungal activity. OH11 is a foreign kiwifruit biocontrol agent with strong antifungal activity. While OH11 was unable to produce anti-Gram-negative metabolites, this strain could utilize type IV secretion system as an antibacterial CDK weapon. We first observed that OH11 could inhibit growth of ZK-1 by generating diffusible anti-Gram-positive antibiotic WAP-8294A2, whereas ZK-1 failed to generate diffusible antibacterial compound to inhibit growth of OH11. To disrupt this interspecies incompatibility, we generated a transgenic OH11-derived strain, OH11W, by deleting the WAP-8294A2 biosynthetic gene and found that OH11W did not kill ZK-1. We further observed that when OH11W and ZK-1 were co-inoculated on agar plates, no CDK effect was observed between them, whereas co-culture of OH11W or ZK-1 with Psa on agar plates resulted in Psa killing, suggesting L. enzymogenes and B. safensis assemble antibacterial CDK weapons against bacterial pathogens, and these CDK weapons did not affect the compatibility between OH11W and ZK-1. Based on these findings, we assembled an OH11W/ZK-1 dependent consortium that was shown to be functional in controlling bacterial canker and several representative fungal diseases of kiwifruit.
Keywords: Lysobacter enzymogenes, Bacillus safensis, Microbial consortium, Kiwifruit disease, Biological control
Graphical Abstract
1. Introduction
Biocontrol bacteria, represented by plant-beneficial members of the genera Bacillus, Pseudomonas, and Lysobacter, are widely applied to control various crop bacterial and fungal diseases [1], [2], [3]. These biocontrol bacteria mainly target and inhibit the growth of pathogens by generating antimicrobial compounds and eliciting plant immune responses, thereby reducing chemical input in agriculture [4], [5], [6]. Although numerous high-performance biocontrol agents have been developed individually as environmental friendly biopesticides, their single use also has limitations in the biocontrol scope and efficiency [7], [8]. To overcome this limitation, bacteriologists have attempted to assemble bacterial consortia by mixing two or more agents together [8], [9]. One notable factor that should be considered in the design of such works is the microbial compatibility of the consortium members.
Biocontrol bacterial interspecies interactions frequently lead to incompatibility events, as most of these members can produce antibacterial compounds and/or assemble contact-dependent killing (CDK) weapons [10], [11]. Diffusible antimicrobial compounds released by biocontrol bacteria can target and damage competitor cell membranes and cell walls, an interaction mode that does not require cell-to-cell contact. Therefore, such antimicrobial compounds can be considered as “long-range” weapons [1], [12]. Meanwhile, bacteria can also assemble CDK weapons represented by type IV (T4SS), type VI (T6SS) and type VII (T7SS) secretion systems that kill neighboring competitors by injecting toxic effector proteins into prey cells, therefore causing their death. These CDK weapons require cell-to-cell contact and are therefore considered as “short-range” weapons whose action does not depend on the presence of “long-range” weapons as defined above [13], [14], [15]. Although it is may be feasible to disrupt “long-range” and “short-range” weapons in biocontrol bacteria through gene manipulation to generate microbial consortia from naturally incompatible to artificially compatible [16], [17], the absence of these weapons, alone or in combination, also has the potential to impair their biocontrol against pathogens and interfere with their competitive fitness in nature [13], [17]. Thus, challenges are raised regarding how to assemble compatible and functional microbial consortia. We attempted to address this crucial issue by choosing representative biocontrol species Lysobacter and Bacillus as working models.
In a recent study, we found that it is difficult to generate compatible combinations between certain Lysobacter and Pseudomonas species, as we found that in most cases, biocontrol Lysobacter species such as L. enzymogenes can use the “short-range” weapon T4SS to kill biocontrol Pseudomonas species, represented by the well-characterized P. fluorescens 2P24 and P. protegens Pf-5 [17]. We also found that certain Bacillus species/strains (i.e. Bacillus subtilis NCD-2) and P. protegens Pf-5 are not naturally compatible after establishing their intercellular contacts, possibly due to the presence of individual or combined “short-range” CDK weapons, represented by the Pseudomonas T6SS and the Bacillus T7SS [17]. Based on these earlier findings, we finally selected the laboratory available Lysobacter enzymogenes OH11 (herein referred to as OH11) and Bacillus safensis ZK-1 (herein referred to ZK-1) to rationally design a possible compatible consortium.
L. enzymogenes OH11 is an antifungal Gram-negative bacterium [18]. It was originally isolated from the pepper rhizosphere and can produce a well-known antibiotic metabolite called heat-stable antifungal factor (HSAF) to target and disrupt the biosynthesis of fungal membrane-associated sphingolipids, thereby exhibiting broad-spectrum antifungal activity [19], [20], [21]. B. safensis ZK-1 is a Gram-positive biocontrol bacterium isolated from kiwifruit fruit [22]. Unlike OH11, ZK-1 efficiently kills Pseudomonas syringae pv. actinidiae (Psa) that causes kiwifruit bacterial canker but has weak antifungal activity, as previously reported [22], [23]. Since the field production of kiwifruit is also severely threatened by several fungal pathogens represented by Diaporthe actinidiae, Colletotrichum gloeosporioides, and Alternaria alternate, which cause kiwifruit soft rot, anthracnose, and brown spot diseases, respectively [24], [25], [26], we therefore aimed to assemble a functional consortium comprising OH11 and ZK-1 to achieve the coordinated control of bacterial and fungal diseases of kiwifruit.
Here, we presented a case study showing how to rationally disrupt certain “long-range” antibacterial weapon possessed by OH11 and assess intercellular contact-dependent compatibility to design a compatible combination with ZK-1, leading to assemble a functional bacterial consortium that could effectively control bacterial and fungal diseases of kiwifruit.
2. Materials and methods
2.1. Plant and microbial strains
Kiwifruit materials were collected from Wuhan Botanical Garden. Details of the bacterial and fungal strains used in this study are described in Table S1. Unless otherwise stated, all bacterial strains were grown in Luria–Bertani (LB) medium at 28 °C and all fungal strains were grown on potato dextrose agar (PDA) medium at 25 °C. Conidia of C. gloeosporioides were induced by inoculation on PDA at 28 °C for 7 days [27], and conidia of A. alternata were induced by inoculating on PDA at 25 °C for 7 days [28].
2.2. Antibacterial activity against Psa
For antagonistic activity against bacteria, 1 mL of an overnight culture of the target bacterium (Psa M228 or Psa C48) was mixed with 25 mL of molten LB agar (LA) medium and poured into a Petri plate. Once solidified, 2 µL of the cell suspensions (OD600, 1.0) of the test bacteria (OH11 or ZK-1) was spot-inoculated on the surface of LA culture plates, each containing the bacteria of interest. 3 plates were used as repetitions in this assay. The plates were incubation at 28 °C for 3 days before photo taking. For contact killing of bacteria, bacteria cells were harvested by centrifugation (6000 rpm for 3 min at room temperature) and washed with sterilized ddH2O. Cells were diluted to an optical density of 1.0 at 600 nm (OD600) and mixed at a ratio of 1:1. The mixtures were cultured at 28 °C for 2 days before observation. A stereoscopic fluorescence microscope (Nikon SMZ25) was used to observe fluorescence signals.
2.3. Antifungal activity against kiwifruit fungal pathogens
For antagonistic activity against fungi, a hyphal plug (5 mm in diameter) of pathogens was transferred to the centre of fresh PDA plates. When the fresh hyphae grew to a suitable size (3 cm in diameter), 5 µL of bacterial cell suspension (washed with sterilized ddH2O and diluted to an OD600 1.0) was inoculated on the edge of the plates. After 3 days of incubation at 25 °C, the antagonistic activity was examined by the inhibition zone around the colonies. For fungal conidia germination inhibition, bacteria strains were grown overnight in LB media. Bacteria cells were harvested by centrifugation (6000 rpm for 3 min at room temperature) and washed with sterilized ddH2O. Cells were diluted to an optical density of 1.0 at 600 nm (OD600). Fungal conidia were induced and filtered through 2 layers of Miracloth to remove mycelia. The conidia were diluted to 200 conidia/µL with sterilized ddH2O. Bacterial cells and fungal conidia were then mixed at a ratio of 1:1 and spotted (2 µL) onto PDA media. Cultures were incubated at 25 °C for 48 h before photo taking.
2.4. Assembly of hybrid biocontrol agent FW-1
The compatibility of OH11 and ZK-1 were examined by antagonistic activity and contact killing between OH11W and ZK-1 with the method described above (2.2). To assemble the hybrid biocontrol agent, OH11W and ZK-1 were cultured overnight separately at 28 °C. Bacteria cells were harvested by centrifugation (6000 rpm for 3 min at room temperature) and washed with sterilized ddH2O. OH11W and ZK-1 cells were diluted to an optical density of 1.0 at 600 nm (OD600) and mixed at different ratios (3:1, 1:1 or 1:3) for further studies. The mixture at the ratio of 1:1 was named as hybrid biocontrol agent FW-1.
2.5. Biocontrol activity against canker and soft rot of kiwifruit
For biocontrol of kiwifruit canker, the wounds on branches of kiwifruit variety “Donghong” were caused by sterilized knife. C48, OH11W, ZK-1 and FW-1 were cultured overnight at 28 °C and diluted to an optical density of 1.0 at 600 nm (OD600). C48 was then mixed with ddH2O, OH11W, ZK-1 and FW-1 at a ratio of 1:1, respectively. The mixtures were inoculated onto the wounds on kiwifruit branches. The wounded branches only inoculated with ddH2O were used as non-inoculated control. The inoculated branches were covered with plastic wrap and cultured in a 16 °C incubator for 20 days before lesion sizes were measured.
For kiwifruit soft rot biocontrol, the kiwifruit fruits were wounded with a sterilized puncher. OH11W, ZK-1 and FW-1 were cultured overnight at 28 °C and diluted to an optical density of 1.0 at 600 nm (OD600). Fresh hyphal plugs (5 mm in diameter) of D. actinidiae were soaked in ddH2O, OH11W, ZK-1 and FW-1 cultures for 20 min, respectively. The treated hyphal plugs were then inoculated onto the wound of the kiwifruits. Inoculated fruits were cultured in a 25 °C incubator for 5 days before measuring their lesion sizes.
3. Results
3.1. Lysobacter enzymogenes OH11 and Bacillus safensis ZK-1 exhibited distinct antimicrobial spectra against bacterial and fungal kiwifruit pathogens
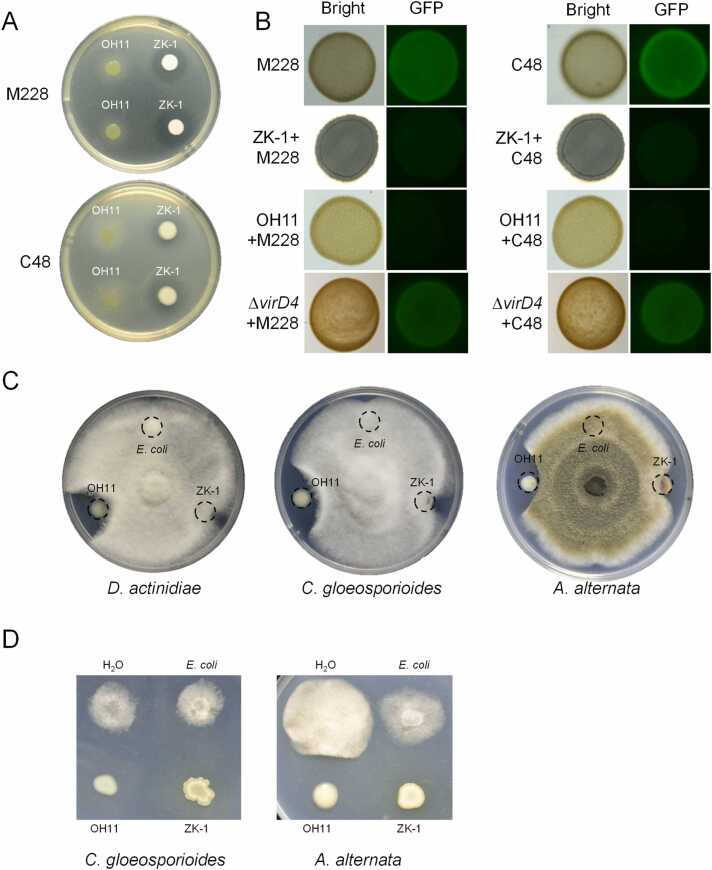
As noted in the Introduction, OH11 is a kiwifruit foreign antifungal bacterium, while ZK-1 is kiwifruit native antibacterial bacterium. To test whether these two strains could be combined into a bacterial consortium for the simultaneous control of the bacterial and fungal kiwifruit pathogens, we first tested the antimicrobial activity of each strain against Psa, D. actinidiae C. gloeosporioides and A. alternata, where the former is the bacterial agent causing kiwifruit canker, and the latter three are the representative fungal pathogens. As shown in Fig. 1A, ZK-1 displayed visible inhibition zones against two Psa strains, M228 and C48 [29], [30], by producing unknown diffusible antibacterial compounds in the culture medium, whereas OH11 failed to do so. However, co-culture of OH11 or ZK-1 with GFP-labelled M228/C48 at a ratio of 1:1 on agar plates, aiming to mimic the cell-to-cell contact between bacterial interspecies described in a recent study in our laboratory [15], led to the discovery that both OH11 and ZK-1 were effective in killing the two tested Psa strains indicated by the intensity of the GFP signal, suggesting that both strains might assemble antibacterial CDK weapons (Fig. 1B). This finding was further validated by the observation that the OH11 T4SS-deficient mutant (ΔvirD4) lost this CDK ability against Psa M228 and C48 under similar testing conditions (Fig. 1B). In the antifungal plate assays, we found that OH11 inhibited the growth of D. actinidiae, C. gloeosporioides and A. alternata, whereas ZK-1 showed very weak (almost nonexistent) antifungal activity (Fig. 1C). In the fungal conidia inhibition assays, we found that the mixing of OH11 with conidia of C. gloeosporioide or A. alternata on agar plates verified that OH11 effectively inhibited the spread of conidia germination to fungal mycelium (Fig. 1D), which was consistent with the reported HSAF effects [19], [20]. Interestingly, we found that co-culture of ZK-1 cells with conidia of D. actinidiae or C. gloeosporioide on agar plates also resulted in a strong inhibition of the spread of conidia germination to the fungal mycelium (Fig. 1D), outlining a possibility that ZK-1 might assemble unidentified antifungal toxins that are induced by contacting with fungi. Nevertheless, these results collectively revealed that OH11 and ZK-1 have distinct antimicrobial spectra, and that both strains could employ antimicrobial compounds as “long-range” weapons to kill fungal or bacterial kiwifruit pathogens, respectively. This finding prompted us to assemble a functional OH11/ZK-1 consortium to simultaneously target and inhibit the growth of bacterial and fungal kiwifruit pathogens with defined “long-range” and/or “short-range” weapons.
Fig. 1.
Lysobacter enzymogenesOH11 andBacillus safensisZK-1 showed different antimicrobial spectra. (A) ZK-1, but not OH11, showed antagonistic activity against P. syringae pv. actinidiae (Psa). M228 and C48 are two Psa strains isolated in China. (B) The killing effect of OH11/ZK-1 co-cultured with Psa on agar plates. When mixed with Psa, both OH11 and ZK-1 inhibited Psa growth, whereas the T4SS-defective mutant of OH11 (ΔvirD4) failed to kill Psa. M228 and C48 were labelled by GFP. Bacteria were mixed at a ratio of 1:1 on LB plates. Fluorescence signals were observed after 2 days of incubation. (C) OH11 showed antagonistic activity against kiwifruit pathogenic fungi D. actinidiae, C. gloeosporioides and A. alternata, whereas ZK-1 showed weak antagonistic activity against these fungal pathogens. (D) Inhibition of fungal conidia germination by co-culturing conidia with OH11/ZK-1 cells on agar plates. When mixed with fungi conidia, both OH11 and ZK-1 inhibited the transfer of conidia to the fungal mycelium of C. gloeosporioides and A. alternata.
3.2. Genetic engineering of OH11 enabled assembly of an artificial consortium compatible with wild-type ZK-1
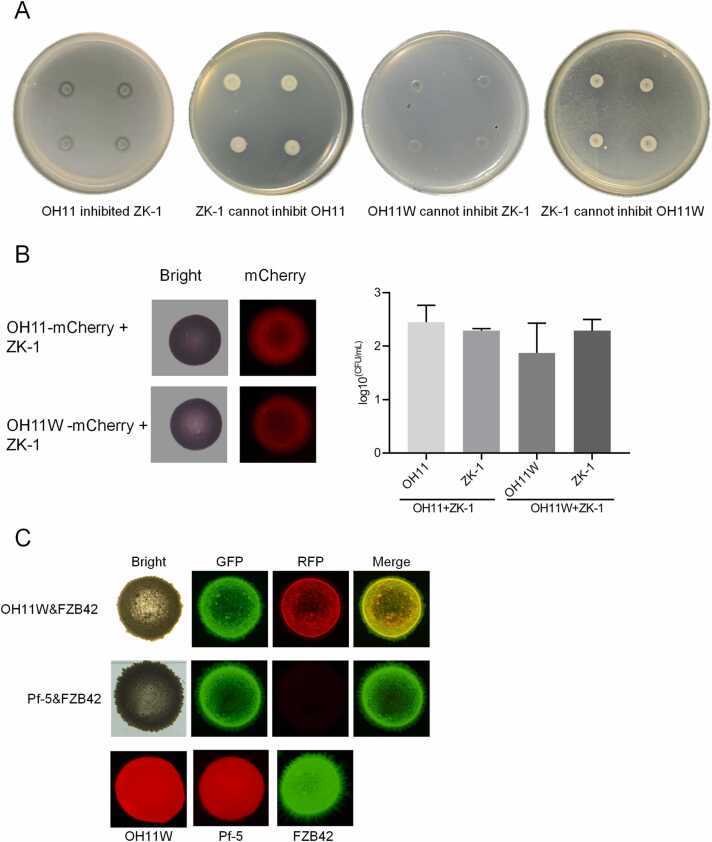
An important factor in the assembly of a functional OH11/ZK-1 consortium is the interspecies compatibility. We first tested this by plate growth inhibition assay based on antimicrobial compounds. By spot inoculation of OH11 on the surface of ZK-1-embedded plates displayed a distinct zone of inhibition (Fig. 2A), suggesting that OH11 was naturally incompatible with ZK-1, because it could kill ZK-1 by producing anti-Gram-positive bacterial factors. We assumed that this growth inhibition was due to the production of anti-Gram-positive bacterial antibiotic WAP-8294A2, a known cyclic depsipeptide, by OH11 [31]. Using a WAP-8294A2-deficient mutant (herein referred to as OH11W) that was previously generated by in-frame deletion of the waps1 gene within the WAP-8294A2 biosynthetic gene cluster in OH11 [31], we found this mutant never produced a zone of growth inhibition against ZK-1 (Fig. 2A). In contrast, ZK-1 failed to suppress the growth of OH11 or OH11W by producing antibacterial compounds (Fig. 2A). We also tested antagonistic activity to OH11W of eight antibacterial Bacillus strains which were isolated from different niches (unpublished data), and found six of them showed inhibition zones to OH11W (Fig. S1). ZK-1 showed strong antagonistic activity to Pseudomonas pathogens, but not to E. coli or OH11 [22], indicating that ZK-1 may produce antimicrobial compounds which specifically inhibits Pseudomonas strains. These findings suggested that disrupting WAP-8294A2 production provided a potentially feasible method for assembling compatible combination comprising OH11W and ZK-1. However, we could not rule out the possibility that OH11W or ZK-1 possessing CDK weapons would hinder their interspecies compatibility. We tested this by co-culturing cells of both strains on agar plates in mixed colonies at an initial 1:1 ratio to create cell-to-cell contact conditions as previously described [15]. Using this protocol, we found that OH11W and ZK-1 are compatible in mixed colonies, because we observed that mixed colonies harboured similar numbers of living cells of each strain after 2 days of co-culture on agar plates (Fig. 2B). To visualize this finding, we further carried out fluorescence microscope assays. However, we were unable to generate fluorescence-labelled ZK-1 due to its difficult genetic manipulation. Alternatively, we selected a well-known GFP-labelled biocontrol bacterium Bacillus amyloliquefaciens FZB42 [32] to test whether the observed interspecies compatibility between Lysobacter and Bacillus species was unique to OH11W and ZK-1 or was common. As shown in Fig. 2C, we did found that co-culture of GFP-labelled FZB42 and the mCherry-labelled OH11W enabled co-observation of GFP and RFP signals in mixed colonies, but co-inoculation of FZB42 and P. protegens Pf-5 at the same ratio in mixed colonies under similar testing conditions resulted in killing of Pf-5 by FZB42 (Fig. 2C). These results raised a strong possibility that genetic modification of OH11 by blocking the production of WAP-8294A2 could facilitate the assembly of an artificially compatible Lysobacter-Bacillus consortium without interspecies killing events.
Fig. 2.
Artificial compatibility between L. enzymogenes OH11 and B. safensis ZK-1 was achieved by inactivating the anti-Gram-positive compound WAP8294A2 produced by OH11. (A) OH11 showed antagonistic activity against ZK-1 on LB plate, while ZK-1 could not inhibit OH11. The WAP-8294A2-deficient mutant OH11W lost its antagonistic activity against ZK-1. (B) OH11 and OH11W could co-exist with ZK-1 when they were mixed. OH11 and OH11W were labelled with mCherry. ZK-1 was then mixed with OH11 or OH11W at a ratio of 1:1 on LB plates. After 2-day culture, the colonies were observed with a microscope and the CFUs of the bacteria were measured. (C) OH11W was also compatible with another biocontrol agent B. amyloliquefaciens FZB42 in mixed colonies. FZB42 was labelled by GFP. mCherry-labelled P. protegens Pf-5 was a control which could be killed by FZB42.
3.3. Establishing an active OH11W/ZK-1 consortium exhibiting inhibitory activities against both bacterial and fungal pathogens of kiwifruit
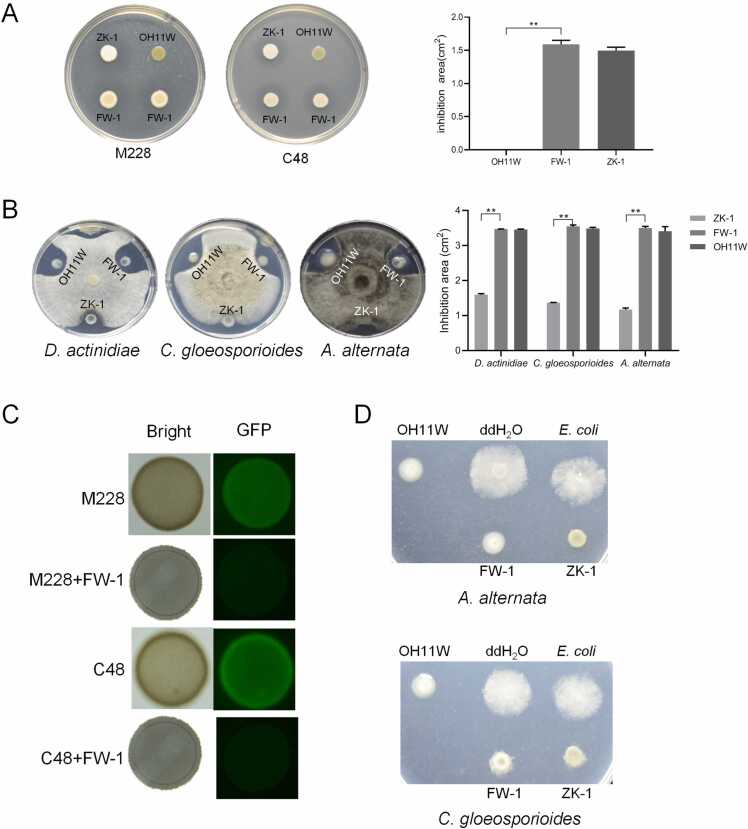
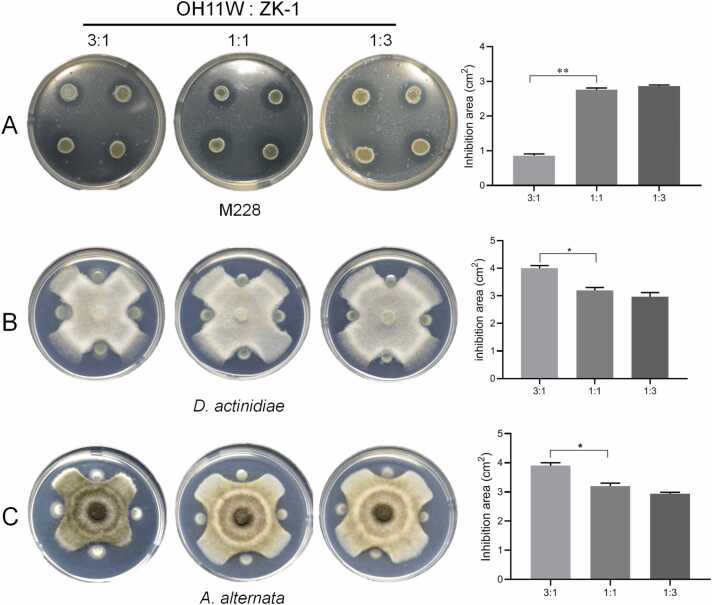
To test whether the artificially assembled OH11W/ZK-1 consortium (herein referred to as FW-1) is functional, we tested its antimicrobial spectrum and activity. As shown in Fig. 3A, we could clearly observe that inoculation of the hybrid FW-1 on the plate surface displayed a visible zone of inhibition against the growth of Psa M228 or C48, similar to the monoculture of ZK-1 itself, but not OH11W. Likewise, the hybrid FW-1 displayed antifungal activity against D. actinidiae, C. gloeosporioide, and A. alternata, similar to OH11W itself, but not ZK-1 (Fig. 3B). By plating cells of FW-1 and Psa M228/C48 together on agar plates to simulate cell-to-cell contact, we clearly observed that FW-1 effectively killed Psa M228 and C48 (Fig. 3C), similar to the situation expressed by OH11 or ZK-1 shown in Fig. 1. FW-1 also retained the ability of OH11/ZK-1 to inhibit the spread of fungal conidia to mycelium, such as C. gloeosporioide or A. alternata (Fig. 3D). These results suggested that the assembled FW-1 consortium retained the antimicrobial ability shared by each consortium member. Next, we showed that varying the 3:1 or 1:3 co-culture ratio between OH11W and ZK-1 did not seem to block the antifungal/antibacterial activity of the consortium (Fig. 4), suggesting that FW-1 was flexible and was promising in application, because it may favor/restrict the growth of one species over the other, resulting in changes in their abundance when it was applied under different native conditions. The consortium seemed to be able to adapt such changes to preserve co-expressed antimicrobial activity.
Fig. 3.
The hybrid biocontrol agent FW-1 exhibited antibacterial and antifungal activity against the kiwifruit pathogens. (A) FW-1 showed similar antibacterial activity as ZK-1. OH11 and ZK-1 were normalized to OD600 = 1.0, and mixed at a ratio of 1:1 to make a hybrid biocontrol agent named as FW-1. FW-1 showed a similar area of inhibition with ZK-1 against Psa on LB plates. Three replicates of each sample were analyzed with a t-test. Asterisks indicate significant differences (P < 0.01). (B) FW-1 showed similar antifungal activity as OH11W. FW-1 showed a similar area of inhibition with OH11W against D. actinidiae, C. gloeosporioides and A. alternata on PDA plates. Three replicates of each sample were analyzed with a t-test. Asterisks indicate significant differences (P < 0.01). (C) Co-culture of FW-1 with GFP-labelled Psa resulted in inhibition of Psa growth by FW-1. (D) FW-1 inhibited conidia germination of C. gloeosporioides and A. alternata during co-cultured with fungi conidia.
Fig. 4.
L. enzymogenesOH11 andB. safensisZK-1 had elastic mixture ratio that balanced antibacterial and antifungal ability. The hybrid biocontrol agents were prepared by mixing OH11W and ZK-1 in different ratios. Antibacterial and antifungal abilities were measured by antagonistic areas against kiwifruit pathogens (A: M228, B: D. actinidiae, C: A. alternata). All the OH11W: ZK-1 ratio showed antibacterial and antifungal ability. The result at ratio of 3:1 (OH11W: ZK-1) showed weak antibacterial activity but strong antifungal ability compared with that at 1:1. Three replicates of each sample were analyzed with a t-test. Asterisks indicate significant differences (**: P < 0.01, *: P < 0.05).
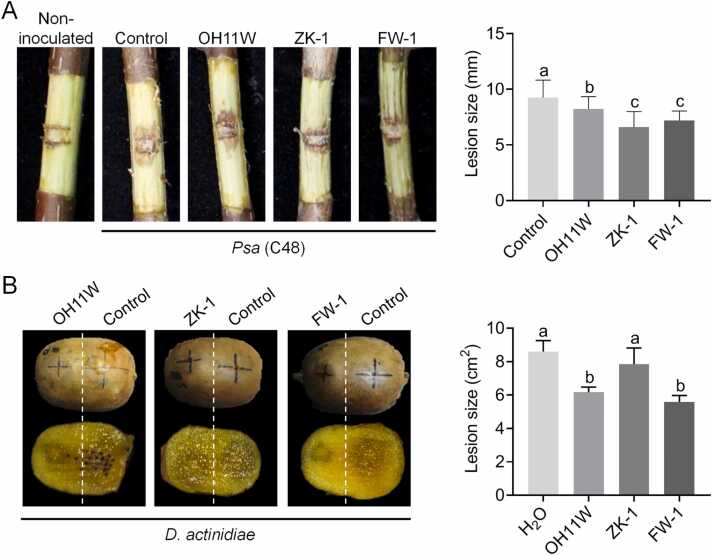
Finally, to investigate whether the assembled OH11W/ZK-1 consortium can effectively control bacterial and fungal diseases of kiwifruit, we conducted respective biocontrol assays in kiwifruit branches and fruits. As shown in Fig. 5A, we found that co-culture of ZK-1 or FW-1 with Psa C48 significantly reduced the development of bacterial canker symptoms compared to negative control (C48 alone). We also observed that OH11W and FW-1 showed biocontrol efficiency in controlling fruit soft rot caused by D. actinidiae (Fig. 5B). These results indicated that FW-1 had both the antibacterial activity of ZK-1 and antifungal activity of OH11, making FW-1 a potential biocontrol agent for the control of bacterial and fungal diseases of kiwifruit.
Fig. 5.
FW-1 showed biocontrol activity against kiwifruit canker and soft rot. In panel A, Psa strain C48 was mixed with sterilized ddH2O (control), or OH11W, ZK-1 and FW-1 at a ratio of 1:1, respectively. The mixtures were then inoculated onto kiwifruit branches. We also included non-inoculated controls that were only inoculated with ddH2O. The inoculated branches were covered with plastic wrap and cultured in an incubator at 16 °C for 20 days before observation. FW-1 showed similar biocontrol ability against kiwifruit canker as ZK-1. Three replicates of each sample were analyzed with a one-way ANOVA test. Different letters indicate the significant differences (P < 0.05). In panel B, hyphal plugs (5 mm in diameter) of D. actinidiae were soaked in ddH2O (control), or cultures of OH11W, ZK-1 and FW-1 for 20 min, respectively. The treated hyphal plugs were then inoculated onto the wounds on kiwifruit which caused by a puncher. The inoculated fruits were cultured in a 25 °C incubator for 5 days before observation. FW-1 showed similar biocontrol ability against kiwifruit soft rot as OH11. Three replicates of each sample were analyzed with a one-way ANOVA test. Different letters indicate the significant differences (P < 0.05).
4. Discussion
Assembling agricultural microbial consortium is an effective means to promote plant health and control pathogen infection [33]. The integration of microbiome profiling and directed isolation of microorganisms is a common and efficient approach in earlier studies [34], [35], [36]. Here, we provided another feasible approach to assemble a functional bacterial consortium comprising Gram-negative (L. enzymogenes OH11) and Gram-positive (B. safensis ZK-1) biocontrol agents by blocking the production of the “long-range” weapons produced by Gram-negative member to design co-existence combinations by assessing their intercellular contact-dependent compatibility. We believe that our strategy appeared to be general and had promising applications in engineering biocontrol microbial consortium, because natural incompatibility between biocontrol species often occurs [10], [37], and the methods employed in our strategy was simple and more feasible.
The most notable finding was the observation of intercellular contact-dependent co-existence between OH11 and ZK-1. As previously reported, OH11 has an active “short-range” CDK weapon, T4SS, that can kill neighbouring bacterial competitors by injecting toxic effectors into prey cells [15]. Interestingly, Bacillus members have also been reported to assemble a similar “short-range” weapon called T7SS, which can kill competitors via cell-to-cell contact [38]. Under our testing conditions, we found that both strains (OH11 and ZK-1) seemed to assemble a potent CDK weapon to kill the phytopathogenic Psa; however, these CDK weapons appear not to “work” when OH11 and ZK-1 cells are mixed together. One possible reason underlying this interesting phenomenon is that, compared to those Gram-negative competitors that were previously targeted and killed by OH11 using the T4SS, such as the plant-beneficial Psedomonas species [15], Gram-positive bacteria have an abundant peptidoglycan layer in their cell envelops, which may act as a physical barrier to block the penetration of T4SS via cell-to-cell contact, leading to a possibly compatible interaction mode between OH11 and Bacillus species. This assumption is partially supported in a recent report where the authors demonstrated that T6SS, another widespread CDK weapon used by Gram-negative bacteria, is relatively difficult to attack Gram-positive bacteria typified by Bacillus [39]. Another possible reason is that the observed coexistence between OH11 and ZK-1 is a killing balancing event, and the two strains in the mixed colony also established contact-dependent competition between cells using their respective CDK weapons. It is also noteworthy that the observed compatibility between OH11 and ZK-1 was consistent with the earlier microbiome analyses, in which the authors reported that application to foreign Bacillus strains to soil would enrich the abundance of native soil Lysobacter species [40]. Since both Lysobacter and Bacillus contain numerous biocontrol members with distinct biocontrol spectra and efficiencies, our findings shed light on the future assembly of diverse functional consortia comprising Lysobacter and Bacillus species.
In a recent study, we showed that the disrupting of bacterial CDK weapons represented by T4SS enable the combination of antifungal OH11 and antibacterial Lysobacter antibioticus OH13 from naturally incompatible to artificially compatible, resulting in the artificially assembled consortium to co-express antifungal and antibacterial activities [17]. In this case, we aimed to use cell-free antimicrobial fermentation broth as the major biocontrol factors, as disrupting those CDK weapons will cause living cells to lose their fitness advantage over their natural microbial competitions [17]. Unlike previous cases, this study searched for “native” interspecies compatibility between OH11 and ZK-1 without genetic manipulation of the respective CDK weapons. It is suggested that this capability retain the natural adaptive advantage conferred by CDK weapons. Therefore, direct application of bacterial cultures is an excellent option for using the assembled OH11W/ZK-1 consortium in the field, and as a case study we demonstrated the biocontrol efficiency of this consortium in the control of major bacterial and fungal diseases of kiwifruit. However, the issue of concern raised in this study was to discuss whether WAP-8294A2-defificient mutant has a wild-type fitness advantage, because if this capacity is impaired, OH11W will have problems with survival and application in the field. Although several studies have demonstrated that antibiotic metabolites produced by Pseudomonas and Bacillus biocontrols have key roles in regulating biofilm formation associated with bacterial colonization [41], [42], [43], we did not found WAP-8294A2 to exert this regulatory role other than its antibacterial effect [31]. One may also ask whether blocking the production of WAP-8294A2 affects the production of the antifungal antibiotic HSAF, and the answer is no, as we previously reported [31]. A third issue is whether WAP-8294A2 blockage is detrimental to OH11 during interspecies competition. In this case, OH11W may have similar competitive ability with OH11 because we found that without WAP-8294A2, OH11 also kills ecologically relevant competitors such as P. fluorescens 2P24 and P. protegens Pf-5 that were previously demonstrated [15]. Therefore, we proposed that OH11 did not seem to have an appreciable negative effect on the bacterial fitness without WAP-8294A2. It is also noteworthy that no Gram-positive bacterial pathogens have been reported to infect kiwifruit. Therefore, the absence of WAP-8294A2 did not appear to affect its application in the control of major kiwifruit diseases caused by Gram-negative Psa and fungal pathogens and importantly, we provided experimental evidence to support this conclusion.
5. Conclusions
This study demonstrated that it was feasible to assemble a functional microbial consortium to create a biocontrol-dependent sustainable kiwifruit industry by designing compatible combination comprising foreign and native kiwifruit biocontrol bacteria, in which the native ZK-1 was expected to have a persistent colonization ability to exert long-term biocontrol effect against Psa, while foreign OH11 was expected to have a rapid killing effect against fungal pathogens through HSAF, similar to chemical fungicides.
CRediT authorship contribution statement
G.Q. and C.Z. conceived the project and designed experiments. L.L., L.L., M.T., Q.W., L.Z., and B.W. performed experiments. L.L., L.L., L.W., and X.S. analyzed data. L.L., L.L., L.W., and X.S. wrote the manuscript. G.Q. and C.Z. revised the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Prof. Huijun Wu (Nanjing Agricultural University) for providing Pf-5 and FZB42 strains used in this study. This study was funded by the Natural Key Research and Development Program (2022YFD1400200 to G.Q.); the Science and Technology Poverty Alleviation Project of Chinese Academy of Sciences (KFJ-FP-202101 to C.Z.); the National Natural Science Foundation of China (U22A20486 and 32072470 to G.Q., 32001955 to L.L., 32272506 to L.L.); Science and Technology Project of Shanxi Branch of China National Tobacco Corporation (KJ-2022-04 to G.Q.); Yangtze River Kiwifruit Industry Technology Research Project (CJZX20210102 to L.L.); Project of CAS Engineering Laboratory for Kiwifruit Industrial Technology (KFJ-PTXM-008 to G.Q.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.021.
Contributor Information
Caihong Zhong, Email: zhongch@wbgcas.cn.
Guoliang Qian, Email: glqian@njau.edu.cn.
Appendix A. Supplementary material
The strains used in this study.
.
Six out of eight antibacterial Bacillus strains showed activity to OH11W.
.
References
- 1.Haas D., Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 2.Fira D., Dimkic I., Beric T., Lozo J., Stankovic S. Biological control of plant pathogens by Bacillus species. J Biotechnol. 2018;285:44–55. doi: 10.1016/j.jbiotec.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 3.Puopolo G., Tomada S., Pertot I. The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J Appl Microbiol. 2018;124:15–27. doi: 10.1111/jam.13607. [DOI] [PubMed] [Google Scholar]
- 4.Ongena M., Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Santoyo G., Orozco-Mosqueda M.D., Govindappa M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci Technol. 2012;22:855–872. [Google Scholar]
- 6.Xie Y., Wright S., Shen Y., Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012;29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fravel D.R. Commercialization and implementation of biocontrol. Annu Rev Phytopathol. 2005;43:337–359. doi: 10.1146/annurev.phyto.43.032904.092924. [DOI] [PubMed] [Google Scholar]
- 8.Massart S., Perazzolli M., Hofte M., Pertot I., Jijakli M.H. Impact of the omic technologies for understanding the modes of action of biological control agents against plant pathogens. Biocontrol. 2015;60:725–746. [Google Scholar]
- 9.Feng S., Jin L., Tang S., Jian Y., Li Z. Combination of rhizosphere bacteria isolated from resistant potato plants for biocontrol of potato late blight. Pest Manag Sci. 2022;78:166–176. doi: 10.1002/ps.6618. [DOI] [PubMed] [Google Scholar]
- 10.Hibbing M.E., Fuqua C., Parsek M.R., Peterson S.B. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein T.A., Ahmad S., Whitney J.C. Contact-dependent interbacterial antagonism mediated by protein secretion machines. Trends Microbiol. 2020;28:387–400. doi: 10.1016/j.tim.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Raaijmakers J.M., De Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 13.Bernal P., Allsopp L.P., Filloux A., Llamas M. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017;11:972–987. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bottai D., Groschel M.I., Brosch R. Type VII secretion systems in Gram-positive bacteria. Curr Top Microbiol Immunol. 2017;404:235–265. doi: 10.1007/82_2015_5015. [DOI] [PubMed] [Google Scholar]
- 15.Shen X., Wang B., Yang N., Zhang L., Shen D., Wu H., et al. Lysobacter enzymogenes antagonizes soilborne bacteria using the type IV secretion system. Environ Microb. 2021;23:4673–4688. doi: 10.1111/1462-2920.15662. [DOI] [PubMed] [Google Scholar]
- 16.Molina-Santiago C., Vela-Corcia D., Petras D., Diaz-Martinez L., Perez-Lorente A.I., Sopena-Torres S., et al. Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Q., Wang B., Shen X., Shen D., Wang B., Guo Q., et al. Unlocking the bacterial contact-dependent antibacterial activity to engineer a biocontrol alliance of two species from natural incompatibility to artificial compatibility. Stress Biol. 2021;1:19. doi: 10.1007/s44154-021-00018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L., Xu K., Shen D., Chou S., Gomelsky M., Qian G. Antifungal weapons of Lysobacter, a mighty biocontrol agent. Environ Microbiol. 2021;23:5704–5715. doi: 10.1111/1462-2920.15674. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Du L., Yuen G., Harris S.D. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Boil Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S., Calvo A.M., Yuen G.Y., Du L., Harris S.D. Induction of cell wall thickening by the antifungal compound dihydromaltophilin disrupts fungal growth and is mediated by sphingolipid biosynthesis. J Eukaryot Microbiol. 2009;56:182–187. doi: 10.1111/j.1550-7408.2008.00384.x. [DOI] [PubMed] [Google Scholar]
- 21.Qian G., Hu B., Jiang Y., Liu F. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric Sci China. 2009;8:68–75. [Google Scholar]
- 22.Wang B., Li L., Lin Y., Shen D., Shao X., Zhong C., Qian G. Targeted isolation of biocontrol agents from plants through phytopathogen co-culture and pathogen enrichment. Phytopathol Res. 2022;4:19. [Google Scholar]
- 23.Pereira C., Costa P., Pinheiro L., Balcao V.M., Almeida A. Kiwifruit bacterial canker: an integrative view focused on biocontrol strategies. Planta. 2021;253:49. doi: 10.1007/s00425-020-03549-1. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Pan H., Liu W., Chen M., Zhong C. First report of Diaporthe actinidiae causing stem-end rot of kiwifruit during post-harvest in China. Plant Dis. 2017;101:1054–1055. [Google Scholar]
- 25.Li L., Pan H., Chen M., Zhang S., Zhong C. First report of anthracnose caused by Colletotrichum gloeosporioides on kiwifruit (Actinidia chinensis) in China. Plant Dis. 2017;101 2151-2151. [Google Scholar]
- 26.Li L., Pan H., Deng L., Wang Z., Li D., Zhang Q., et al. First report of Alternaria tenuissima causing brown spot disease of kiwifruit foliage in China. Plant Dis. 2019;103 582-582. [Google Scholar]
- 27.Nesher I., Minz A., Kokkelink L., Tudzynski P., Sharon A. Regulation of pathogenic spore germination by CgRac1 in the fungal plant pathogen Colletotrichum gloeosporioides. Eukaryot Cell. 2011;10:1122–1130. doi: 10.1128/EC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Yuan S., Sun J., Li Q., Jiang W., Cao J. Ethyl p-coumarate exerts antifungal activity in vitro and in vivo against fruit Alternaria alternata via membrane-targeted mechanism. Int J Food Microbiol. 2018;278:26–35. doi: 10.1016/j.ijfoodmicro.2018.04.024. [DOI] [PubMed] [Google Scholar]
- 29.McCann H., Li L., Liu Y., Li D., Pan H., Zhong C., et al. Origin and evolution of the kiwifruit canker pandemic. Genome Biol Evol. 2017;9:932–944. doi: 10.1093/gbe/evx055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Zhou M., Liu W., Nie J., Huang L. Pseudomonas syringae pv. actinidiae effector HopAU1 interacts with calcium-sensing receptor to activate plant immunity. Int J Mol Sci. 2022;23:508. doi: 10.3390/ijms23010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W., Li Y., Qian G., Wang Y., Chen H., Li Y., et al. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chem. 2011;55(5581–5589) doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chowdhury S.P., Hartmann A., Gao X., Borriss R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42-a review. Front Microbiol. 2015;6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Liu R., You M., Barbetti M.J., Chen Y. Pathogen biocontrol using plant growth-promoting bacteria (PGPR): role of bacterial diversity. Microorganisms. 2021;9:1988. doi: 10.3390/microorganisms9091988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloemberg G., Lugtenberg B.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol. 2001;4:343–350. doi: 10.1016/s1369-5266(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 35.Mendes R., Garbeva P., Raaijmakers J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 36.Mueller U.G., Sachs J.L. Engineering microbiomes to improve plant and animal health. Trends Microbiol. 2015;23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Whipps J.M. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 38.Tassinari M., Doan T., Bellinzoni M., Chabalier M., Ben-Assaya M., Martinez M., et al. The antibacterial Type VII secretion system of Bacillus subtilis: structure and interactions of the pseudokinase YukC/EssB. mBio. 2022;13 doi: 10.1128/mbio.00134-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molina-Santiago C., Pearson J.R., Navarro Y., Berlanga-Clavero M.V., Caraballo-Rodriguez A.M., Petras D., et al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun. 2019;10:1919. doi: 10.1038/s41467-019-09944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong W., Guo S., Jousset A., Zhao Q., Wu H., Li R., et al. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol Biochem. 2017;114:238–247. [Google Scholar]
- 41.Townsley L., Shank E.A. Natural-product antibiotics: cues for modulating bacterial biofilm formation. Trends Microbiol. 2017;25:1016–1026. doi: 10.1016/j.tim.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandin C., Darsonval M., Mayeur C., Le Coq D., Aymerich S., Briandet R. Biofilm formation and synthesis of antimicrobial compounds by the biocontrol agent Bacillus velezensis QST713 in an Agaricus bisporus compost micromodel. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.00327-19. e00327-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zboralski A., Filion M. Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Comput Struct Biotechnol J. 2020;18:3539–3554. doi: 10.1016/j.csbj.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The strains used in this study.
Six out of eight antibacterial Bacillus strains showed activity to OH11W.