Abstract
Angioimmunoblastic T-cell lymphoma (AITL) is a subtype of peripheral T-cell lymphoma (PTCL) strongly correlated with worse clinical outcomes. However, the role of characteristic pathway-related genes in patients with AITL (e.g., subtype typing and pathogenesis) remains unknown. In this study, we intended to understand the potential role and prognostic value of characteristic pathways in AITL and identified a model for subtype identification based on pathway-related functional status. Transcriptomic (RNA-seq) data were obtained from the Gene Expression Omnibus database for three sets of tumor tissues from AITL patients. AITL was divided into three clusters based on the pathway profile of patients and the best clustering k = 3, and differentially expressed genes (DEGs) in the three clusters were analyzed. The top 45 important variables associated with characteristic pathways, such as Huntington's disease, VEGF signaling pathway, nucleotide excision repair, ubiquitin-mediated proteolysis, purine metabolism, olfactory transduction, etc., were used to construct a subtype identification model. The model was experimentally validated and proved to possess good predictive efficacy. In addition, pathway-related subtype typing was significantly associated with different immune cell infiltration in AITL. Further analysis revealed that the drug IC50 values predicted also differed markedly among the different subtypes, thus further identifying some subtype-specific drugs. Our study indicates a potential role of characteristic pathways in AITL staging for the first time, provides novel insights for future research targeting AITL, and points to potential therapeutic options for patients with different subtypes of AITL.
Keywords: Angioimmunoblastic T-cell lymphoma, Pathways, Immune infiltration, Subtype typing model, Specific drugs
1. Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a rare subtype of aggressive peripheral T-cell lymphoma (PTCL) that originates from follicular helper T (TFH) - cells and accounts for 15%–30% of PTCL [1]. It is a highly heterogeneous disease with a variable clinical course, and most clinical manifestations, such as lymph node enlargement, hepatosplenomegaly, bone marrow involvement, and B symptoms, indicate an impaired immune system [2]. The diagnosis of AITL is extremely challenging and requires a constellation of clinical, molecular genetic studies and histopathological findings [1]. Currently, the first-line treatment regimen for ATTL is generally based on a CHOP (cyclophosphamide + doxorubicin + vincristine + prednisone) regimen, supplemented by drugs such as etoposide [1]. However, the 5-year failure-free survival rate for patients ranges from only 13%–20% [3,4]. Relapsed or refractory AITL (R/R AITL) can also be treated with autologous/allogeneic hematopoietic stem cell transplantation (aHSCT), immunotherapy, and targeted drugs, but with limited efficacy [1]. Therefore, a more specific disease subtyping of AITL is needed based on its diverse clinical course. Adopting novel and more effective treatment strategies for patients with AITL is urgently required.
Owing to the critical role of various cellular signaling pathways in the development of organisms and their various organ systems, it appears reasonable to suspect that alterations in these pathways may play a role in the pathogenesis of many malignancies [5]. These enriched pathways in AITL exhibit typical genomic abnormalities, such as DNA damage repair, gene mutations, induction of apoptosis or necrosis, alterations in life metabolism, abnormalities in signal transduction, etc., thereby leading to alterations in the functional status of patients, and impediments to therapy [6,7]. Among them, anabolic signaling pathways (e.g., PI3K/Akt/mTOR and Ras/MEK/ERK) can significantly affect cellular metabolic activity to modulate cell growth, replication, metabolism, motility, etc [7,8]. Abnormalities in the IL-7/JAK/STAT signaling pathway, a key pathway for normal T-cell survival and proliferation, can disrupt the process of T-cell development and result in impaired immune function [7]. In addition, AITL contains a rich tumor microenvironment (TME) component, which includes numerous immune infiltrating cells such as tumor-infiltrating lymphocytes (TILs), macrophages, and mast cells; tumor cells themselves account for only about 10% [9]. Recently, mutational concordance has been observed between malignant cells and immune infiltrating cells within the AITL TME [10]. Infiltration of immune cells is also tightly regulated by signaling pathways. Importantly, T/B-type TILs are key members of the TME and may have important implications for the development and pathological and clinical features of AITL [9].
As mentioned above, alterations in these pathways and their related genes can markedly affect the functional status of patients with AITL, e.g., immunomodulatory processes, which lead to different clinical courses. However, to our knowledge, no reports have focused on the role of characteristic pathways in AITL and the therapeutic value of pathway-related subtype identification in patients with AITL. In this study, we elucidate for the first time the potential role of subtype typing in AITL and develop a model for subtype identification of AITL based on the characteristic pathway. More importantly, we discuss the relationship between AITL typing and drug sensitivity and explore optimal treatment options. Our findings may provide new avenues for further studies on characteristic pathways and individualized treatment of AITL.
2. Materials and methods
2.1. Data collection and preprocessing
Three sets of RNA sequencing (RNA-seq) data (FPKM values) of tumor tissues from AITL patients were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The three datasets, GSE58445, GSE19069, and GSE6338, were based on the same sequencing platform (GPL570) in the GEO database and included 15, 37, and 6 patient samples, respectively. Subsequently, the three sets of data were batch processed using the “ComBat” method in the “sva” R package to be accurate for follow-up analysis. Relevant information on the 186 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and the genes they contain was obtained from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb). The collection of genes corresponding to the 14 functional states of tumor microenvironment (TME) was from the cancerAEA database. Reference data for 22 immune cells were downloaded from CIBERSORTx (https://cibersortx.stanford.edu/) and then assessed for the respective TME immune infiltration in AITL patients using the “cibersort” algorithm. To predict the drug IC50 for AITL individuals, the training set data was downloaded from the GDSC (Genomics of Drug Sensitivity in Cancer) database. Differential IC50 drugs were analyzed and visualized using the “oncoPredict” package in R.
2.2. Collection and functional analysis of patient pathway profiles in AITL
Based on the MSigDB database and scoring of pathways, we obtained patient pathway profiles and clustered patients into three clusters. We further identified 86 characteristic pathways with markedly different activity in the three subtypes using the “agricolae” R package and the “LSD” (least significant difference test) method. We further plotted a heat map of the activity scores for the characteristic pathways between different clusters to illustrate the differences in the three AITL subtypes. To explore the differences in functional status, we calculated Fold Change (FC) values for the activity scores of the characteristic pathways between the two most different subtypes based on the data normalized for extreme differences. Finally, the top 10 pathways with the largest differences in each cluster were selected for visual analysis depending on the FC values. The gene information “KEGG.gmt” was retrieved from the MSigDB database as the background, and differences with a |log2FC| value > 1 and a P-value <0.05 were considered statistically significant.
2.3. Consensus clustering analysis based on patient pathway profiles
Using the “GSVA” R package, we calculated the scores of 186 pathways for each patient in the three datasets of GSE58445, GSE19069, and GSE6338 based on the “single-sample gene set enrichment analysis (ssGSEA)" method and ultimately obtained the patient pathway profiles. The “NbClust” R package was used to calculate 26 different indexes to determine the optimal number of patient clusters. Based on the patient pathway profiles and the clear optimal number of clusters, the AITL samples from the three datasets were subsequently clustered into subgroups using the “kmeans” method in the “NbClust” R package, and the corresponding information was obtained for the different subgroups of patients. The “pca3d-" and “rgl-" R packages were used for principal component analysis to assess sample clustering [11].
2.4. DEGs screening and functional enrichment analysis
The characteristic pathway analysis between different subtypes of AITL samples was performed to elucidate the potential molecular pathways correlated with functional status. Meanwhile, the empirical Bayesian approach of the “limma” R package was used for screening differentially expressed genes (DEGs) between different clusters (|log2FC|>1, p < 0.05) and for gene differential analysis. Next, the DEGs were used to conduct functional enrichment analysis of the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the “clusterProfiler” R package, with a p≤0.05 threshold to identify significantly enriched GO term and KEGG pathways. Finally, the molecular function (MF) enrichment results were visualized. In addition, the functional status of 14 TMEs for each AITL patient in the three subgroups was evaluated using the “ssGSEA” algorithm in the R package “GSVA” with a cut-off value of p≤0.05.
2.5. Immuno-infiltration analysis
To explore the differences in immune cell subtypes among the three subtypes of AITL, the reference data of 22 immune cells were obtained using CIBERSORTx, and the “Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (cibersort)" algorithm was used to assess the relative proportions of the 22 infiltrating immune cells. The R packages “clusterProfiler” and “GSVA” were used for single sample gene set enrichment analysis (ssGSEA) to assess the enrichment fraction of 22 different types of immune cells and 86 different characteristic pathway activities in the three clusters of AITL. Subsequently, we performed a differential analysis of immune cells using the Wilcoxon rank sum test to further identify essential immune cells, and calculated correlations between immune cells and the previously identified DEGs.
2.6. The IC50 prediction and screening of drugs
We explored the differences in drug sensitivity between different subtypes to identify drugs specifically for each subtype. The training set data of patients were downloaded from the GDSC database, and we then predicted the IC50 values of 198 drugs for each individual in the different subtypes using the “oncoPredict” R package. Next, the drugs with differences in IC50 between different clusters were identified using the Wilcoxon rank sum test and the “LSD” method. Finally, based on these results, p-values for the Wilcoxon test were rectified using the Bonferroni method to a corrected cut-off value of p≤0.05 to further narrow the range of drugs with significant differences. The Venn diagram was used to take intersections where differences existed between drug IC50s in different clusters and to visualize them.
2.7. Construction and validation of a model for identification of pathway-related subtypes in AITL
To precisely determine the subtypes of unknown patients for accurate treatment, a machine learning approach was used to develop a model for identifying between different clusters of AITL. The Boruta algorithm is based on the R package “Boruta” and is first used to screen for relevant variables such as characteristic pathways. Subsequently, the 58 AITL samples from the three datasets were randomly (1:1) divided into a training cohort and an internal validation cohort. A subtype typing model was initially constructed using the Random Forest algorithm. Meanwhile, the optimal number of variables was obtained using the training set data to ensure the model's reliability. Next, the training set data and the optimal number of variables screened were used to construct the random forest model, and the model's performance was evaluated using the test set data. The “pROC” R package was used to perform receiver operator characteristic (ROC) curve analysis to confirm the prediction reliability of the model.
2.8. Statistical analysis
All statistical analyses conducted in our study were carried out using R software. Unless specified otherwise, the differences with P < 0.05 were considered statistically significant. Heatmap was fabricated with the “pheatmap” R package, and we performed the boxplot using the “ggpubr” R package.
3. Results
3.1. Processing of sequencing data and pathway-related subtype typing in AITL patients
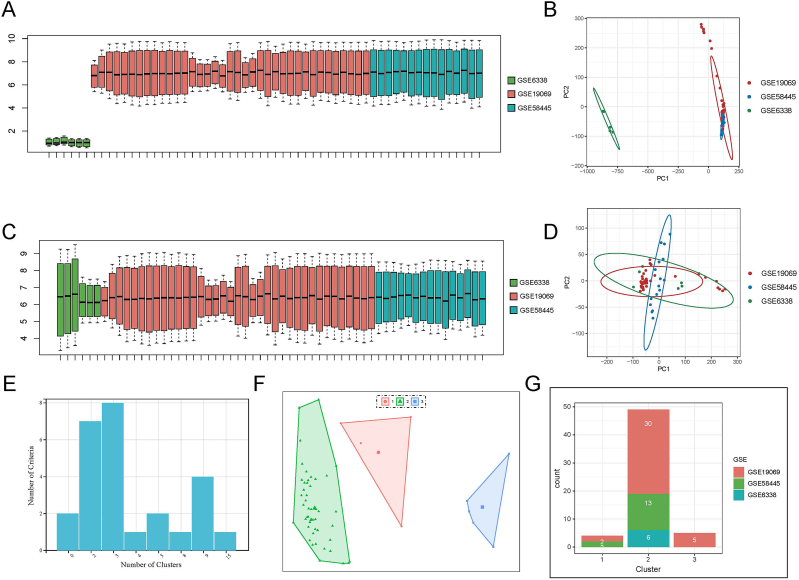
Transcriptomic (RNA-seq) data (sample size: 58) from three sets of tumor tissues in AITL patients were obtained from the GEO database based on the same sequencing platform (GPL570). Before batch processing, the three sets of data from different batches were initially visualized using box plots and principal component analysis (PCA) downscaled plots, which indicated significant batch effects between the three datasets (e.g., different samples and delivery times, different laboratories, etc.) (Fig. 1A and B). Subsequently, we performed batch processing on the three datasets. As shown in Fig. 1C, it could be observed that the batch effect was effectively reduced after batch processing, and the levels of mRNA expression were the same for the three sets of data. There was also no apparent separation between the samples in the three datasets (Fig. 1D).
Fig. 1.
Batch processing and consensus clustering analysis for the three AITL datasets. (A) Box plot of gene expression for untreated batch effects. (B) Principal Component Analysis (PCA) plot for untreated batch effects. (C) Box plot of gene expression after batch effect processing. (D) PCA plot after batch effect processing. (E) Visualization of NbClust results i.e., number of clusters selection. (F): Reduced dimensional visualization of PCA for different clusters. (G) Statistics on the number and origin of samples for the three subtypes.
We obtained the relevant information on the 186 KEGG pathways and the genes they contain. ssGSEA revealed the scoring of the 186 pathways corresponding to each patient and ultimately obtained the patient pathway profile. Consensus clustering analysis was performed on the three datasets, GSE58445, GSE19069, and GSE6338, by calculating 26 different metrics. The clustering algorithm performed the best when more indicators pointed to k = 3, and different clusters had significant treatment choices and prognostic values (Fig. 1E). We then clustered the patients into three subtypes based on their pathway profiles with a clustering number of 3, and obtained the information of the three types of samples corresponding to the patients. Using the pathway profiles, PCA downscaling analysis of the patients showed that the three types of samples were completely separated (Fig. 1F). It indicated that our clustering was very effective, and the three subtypes identified were quite accurate. Meanwhile, the number of patients with AITL and the distribution of their origin were counted in different clusters (subtypes) (Fig. 1G). The results demonstrated that most patients were classified as cluster 2, suggesting that cluster 2 should be the most frequent subtype of AITL, and that patients in all three data sets were partially distributed in cluster 2. Patients in the other two subtypes were not confined to one dataset either. It confirms that the processing of batch effects did not affect the results of our subtype typing, further indicating that our results are more reliable.
3.2. Identification of characteristic pathways in AITL subtypes and functional enrichment analysis
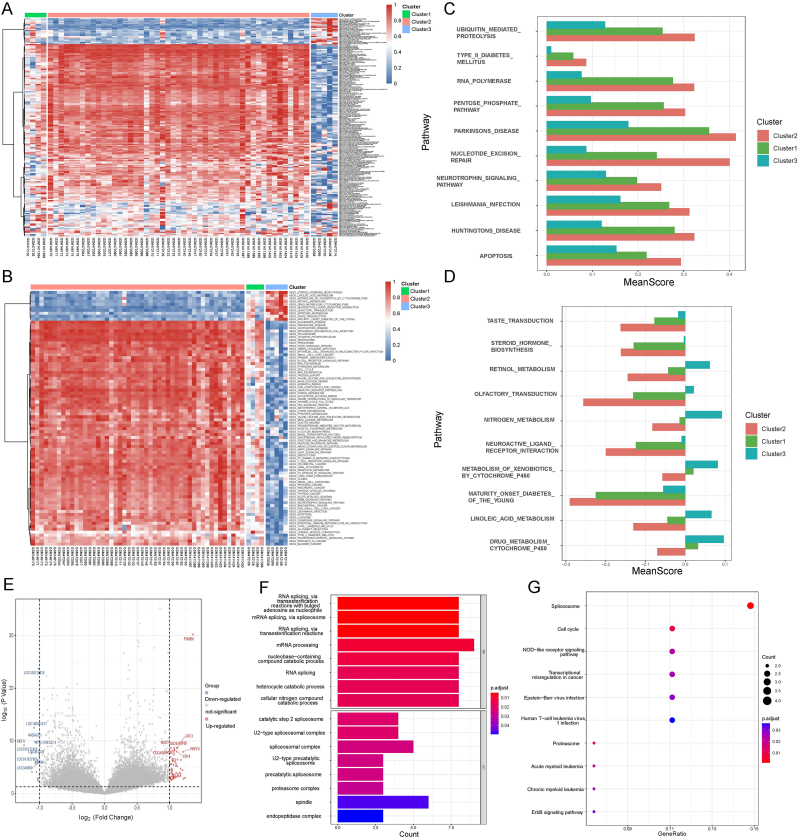
To initially present the differences in the activity of the different pathways in the three subtypes, we sorted the samples by cluster to organize the pathway profiles and plotted the heat map after normalizing the pathway profiles for polar differences on a pathway-by-pathway basis (Fig. 2A). The KEGG pathway enrichment analysis indicated that the different pathways' activity differences in the three types of samples were relatively obvious, especially between cluster 2 and cluster 3. Cluster 2 is mainly enriched in ribosomal bioreactivity, purine metabolism, nucleotide excision repair, and Alzheimer's disease; cluster 3 is significantly enriched in steroid hormone biosynthesis, linoleic acid metabolism, metabolism of xenobiotics by cytochrome p450, and retinol metabolism. In addition, we speculated that cluster 1 might be a transition state isoform between cluster 2 and cluster 3, as shown by the heat map.
Fig. 2.
Pathway differentiation and functional analysis of differentially expressed genes (DEGs) between clusters. (A) The relative activity of all pathways in the three types of samples. (B) The relative activity of characteristic pathways in the three types of samples. (C) Pathways with significantly high activity in cluster 2. (D) Pathways with significantly high activity in cluster 3. Fig. 2C and D shows the mean values of the scores for specific pathways in the three clusters. (E) Volcano map of the differential genes between cluster 2 and cluster 3. Importantly, differences with p≤0.05 & (log2(Fold change [FC])≥1 | log2FC ≤ −1) for t-test were considered statistically significant. (F) GO enrichment analysis of differential genes. (G) KEGG enrichment analysis of differential genes. ***p < 0.001; **p < 0.01; *p < 0.05.
Next, pathways with differential activity in the three subtypes were further identified. We screened the pathways that differed markedly among all three subtypes as characteristic subtype pathways from 186 pathways, and eventually, a total of 86 pathways were identified (Fig. 2B). Fig. 2B indicates that the difference in activity between cluster 2 and cluster 3 is highly remarkable, and cluster1 is definitely between clusters 2 and 3. Combining that with the sample size, we can conclude that cluster 2 is a common AITL and cluster 3 is a rare AITL. Furthermore, there is an intermediate state isoform in between, namely cluster 1.
Based on the results of the above analysis, the differences in functional status between cluster 2 and cluster 3 were then further explored. We calculated Fold change (FC) values for the activity scores of the characteristic pathways between cluster 2 and cluster 3 based on the normalized data with extreme differences. These results found that ten pathways were significantly greater in activity in cluster 3 than in cluster 2; conversely, 76 pathways were notably more active in cluster 2 than in cluster 3. Subsequently, the top 10 pathways with the greatest differences in each of the two categories were selected for visualization (Fig. 2C and D). As illustrated in the figure, cluster 2 was enriched in ubiquitin-mediated protein hydrolysis, RNA polymerase, pentose phosphate pathway, and nucleotide excision repair; cluster 3 was significantly enriched in taste and olfactory transduction, steroid hormone biosynthesis, retinol metabolism, and neuroactive ligand-receptor interactions. Therefore, our findings suggest that differences in the functional status of characteristic pathways may affect AITL biosynthesis and metabolism.
In the molecular level, the differential analysis was performed using t-test to identify differentially expressed genes (DEGs) among patients in cluster 2 and cluster 3. Fig. 2E indicates that 76 DEGs were significantly different between the two subtypes, using p≤0.05 & (FC≥2 | FC≤0.5) as thresholds for the t-test. Subsequently, the DEGs were subjected to GO, KEGG functional enrichment analysis (p≤0.05) (Fig. 2F and G). GO analysis showed that the DEGs were enriched in various processes of RNA splicing, mRNA processing, catabolic processes of nucleotide-containing compounds, and heterocyclic compounds (Fig. 2F). KEGG pathway enrichment analysis (Fig. 2G) indicated that these DEGs mediated spliceosome formation, cell cycle processes, NOD-like receptor signaling pathways, transcriptional misregulation in cancer, Epstein-Barr virus infection, and acute myeloid leukemia. Overall, the DEGs were found to be associated with pathways such as immune response, tumor development and metastasis, and infection, by enrichment analysis and selection of the above characteristic pathways, and that dysfunction of the related pathways may affect the development of AITL and the immune microenvironment. It also highlights that the immune status of the two subtypes is somewhat different, and therefore the following analysis focuses on the immune infiltration of the two subtypes.
3.3. The typing of AITL was correlated with the functional state of the TME
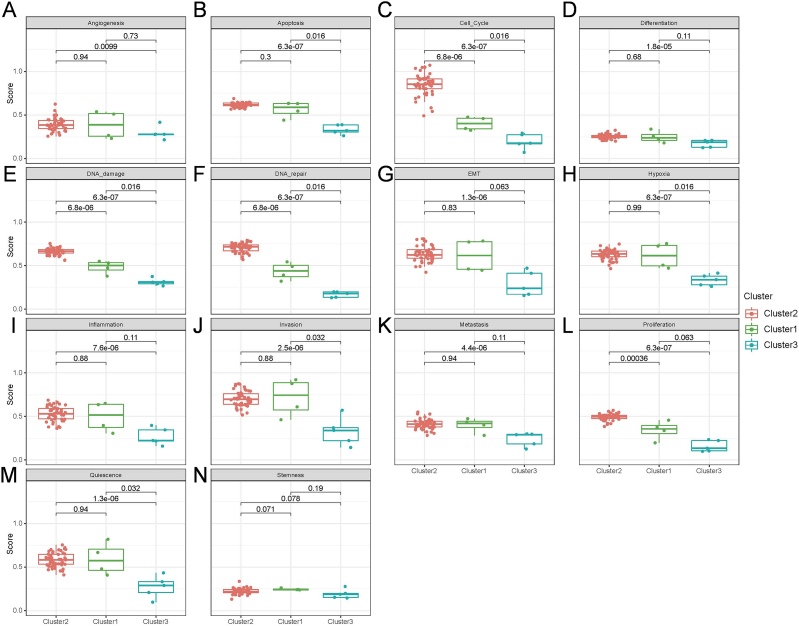
The functional states in the three pathway-related clusters in AITL were analyzed in depth because TME is one of the hot research directions in AITL. We obtained a collection of genes corresponding to the 14 TME functional states based on the cancerAEA database. ssGSEA assessed 14 functional states for each patient, including angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, epithelial-mesenchymal transition (EMT), hypoxia, inflammation, invasion, metastasis, proliferation, quiescence, and stemness (Fig. 3). Fig. 3 further demonstrates the scores of the 14 functional states in the three subtypes of patients with a Wilcoxon rank sum test among groups. The results showed that all 13 functional states were significantly different between the two subtypes of cluster 2 and cluster 3 (p≤0.05), except for stemness.
Fig. 3.
Activity scoring of the 14 functional states in the three clusters of AITL (p < 0.05). (A) Angiogenesis, (B) apoptosis, (C) cell cycle, (D) differentiation, (E) DNA damage, (F) DNA repair, (G) EMT, (H) hypoxia, (I) inflammation, (J) invasion, (K) metastasis, (L) proliferation, (M) quiescence, (N) stemness.
3.4. Immune infiltration of three clusters in AITL
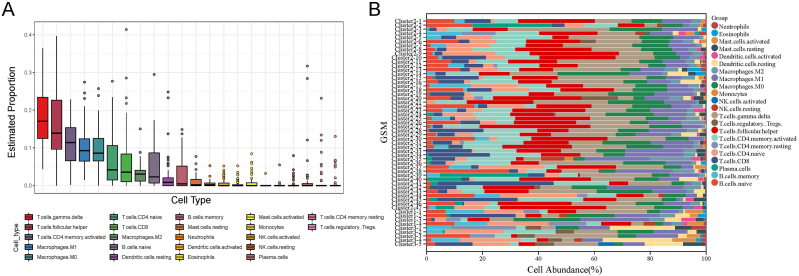
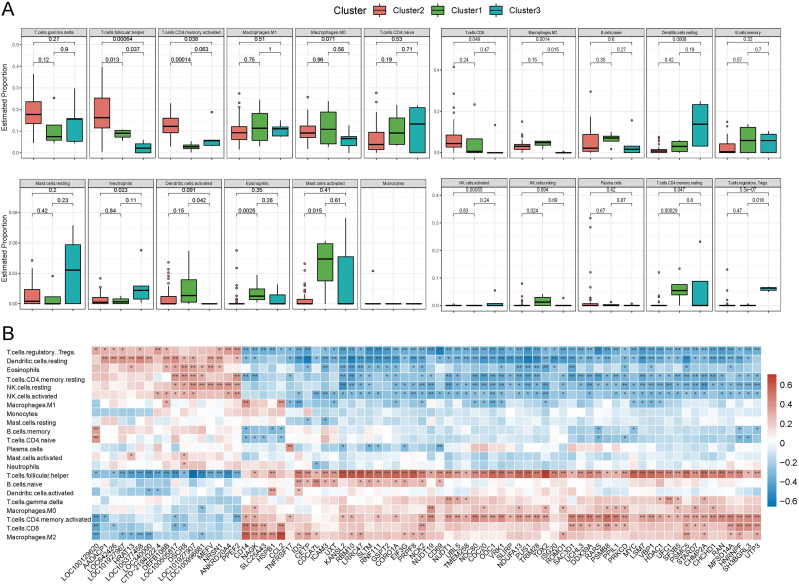
Given the results of the above analysis and the characteristics of AITL, where the three subtypes were significantly related to other immune signatures, we explored the potential relationship between subtype typing of AITL and the infiltration of TME immune cells. The box plots showed the abundance of different infiltrating TME cells as inferred by the cibersort algorithm across all patients and revealed that the infiltration of TME cell types with higher overall abundance in AITL including gamma-delta T cells, follicular helper T cells (TFH), activated memory CD4 T cells, M0 macrophages, M1 macrophages and so on (Fig. 4A). In contrast, the overall abundance of memory-type resting CD4 T cells, regulatory Tregs T cells, activated mast cells, monocytes, activated NK cells, resting NK cells, plasma cells, etc., was significantly lower in AITL patients. Furthermore, Fig. 4B illustrates the proportion of different immune cells in different subtypes of patients. Cluster 2 contains mainly activated memory CD4 T cells, follicular helper T cells, gamma-delta T cells, M0 macrophages, M1 macrophages, M2 macrophages, naive B cells, naive CD4 T cells, CD8 T cells, etc.; cluster 3 is primarily infiltrated with resting dendritic cells, monocytes, M0 macrophages, M1 macrophages, gamma-delta T cells, regulatory Tregs T cells, naive CD4 T cells, activated memory CD4 T cells, and memory B cells; cluster 1 was infiltrated with M0 macrophages, M1 macrophages, M2 macrophages, naive B cells, naive CD4 T cells, and gamma-delta T cells. These findings strongly suggest that the infiltration of TME cells plays a crucial role in the subtype typing of AITL patients.
Fig. 4.
The abundance of immune infiltrating cells. (A) Box plot of the abundance for different immune cells in all patients. (B) Histogram of the proportional stacking for different immune cells in different patients.
Next, we performed a differential analysis of 22 immune infiltrating cells using the Wilcoxon rank sum test to further identify important immune cells (Fig. 5A). Meanwhile, the correlations between these immune cells and the 76 DEGs identified previously were calculated and visualized (Fig. 5B). Based on the above results, we can discriminate some essential immune cells, thereby contributing to our further understanding of the characteristics of the different subtypes. For instance, follicular helper T cells significantly differed between the three subtypes and correlated with almost all of the differential genes, suggesting that it plays a decisive role in subtype typing. The infiltration levels of 10 immune cell types differed markedly in the two subtypes of cluster 2 and cluster 3. Among them, the infiltration levels of follicular helper T cells, activated memory CD4 T cells, CD8 T cells, and M2 macrophages were significantly higher in cluster 2 than in cluster 3, whereas infiltration levels of M1 macrophages, resting dendritic cells, neutrophils, activated NK cells, resting memory CD4 T cells and regulatory Tregs were lower in cluster 2 than in cluster 3. In addition, as previously described, the relatively high expression of 59 DEGs in cluster 2 largely coincided with the type of immune cells they infiltrated (Fig. 5B).
Fig. 5.
The distribution of immune cells in the three subtypes and the relationship with DEGs. (A) Comparison of the abundance levels for immune cells in the 3 subtypes. (B) Correlation between immune cells and differential genes. ***p < 0.001; **p < 0.01; *p < 0.05.
3.5. Subtype-specific drug analysis
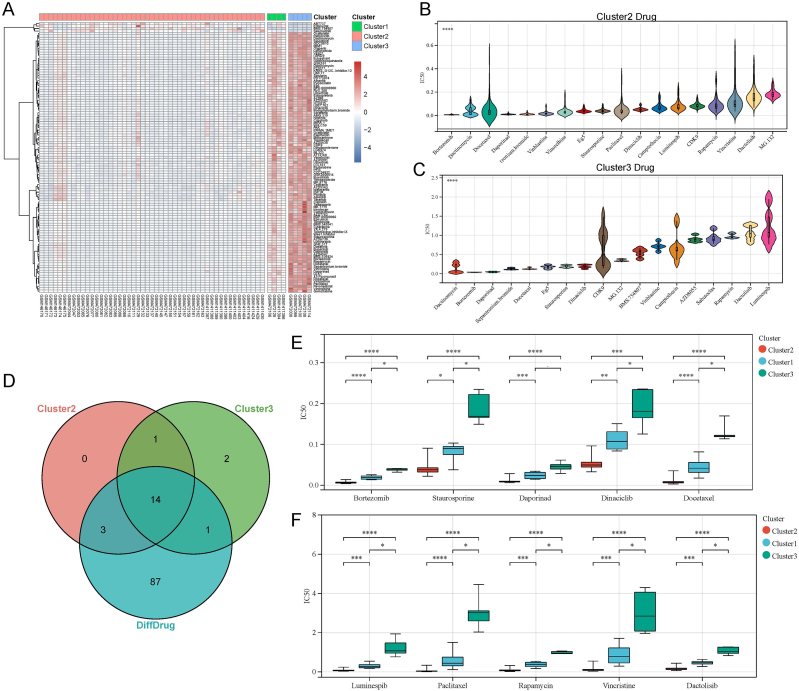
As mentioned above, we can determine that cluster 2 and cluster 3 are subtypes of two states in AITL. Therefore, we then explored the differences in drug-sensitive phenotypes between the two subtypes to identify the drugs specific to each subtype to enhance the effectiveness of clinical treatment. We performed the prediction of drug IC50 for individuals of different subtypes using the training set data and the “LSD” method. Fig. 6A demonstrates that 109 drugs were markedly different between cluster 2 and cluster 3 (corrected p≤0.05). To further ascertain the subtype-specific drugs, we extracted the top 20 drugs with the smallest IC50 for each cluster 2 and cluster 3, ultimately leaving 18 drugs each after de-duplication. The finding shows the names of the 18 drugs selected and their IC50 values in patients (Fig. 6B and C). By analyzing the visualized results, we can also conclude that even for the drugs with the smallest IC50, the IC50 of the corresponding drugs in cluster 3 is commonly higher than that of the drugs in cluster 2. Taken together, it is further indicated that cluster 3 may be the more drug-resistant subtype.
Fig. 6.
Screening of specific drugs for different subtypes of AITL. (A) Heat map for visu-alization of differential IC50 drugs. (B) The 18 drugs with the smallest IC50 in cluster2. (C) The 18 drugs with the smallest IC50 in cluster3. (D) The Venn diagram of drug intersections. (E–F) Box plots of drugs under interest with small IC50. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
Subsequently, we took the intersection of the drugs with the smallest IC50s in clusters 2 and 3 with the drugs that exhibited differences between their IC50s (Fig. 6D). The Venn diagram revealed the existence of three drugs that both differed between cluster 2 and 3 (i.e., significantly smaller IC50 in cluster 2) and were drugs with a smaller IC50 in cluster 2, which we considered to be specific to cluster 2. Similarly, one drug was identified as having a significantly smaller IC50 in cluster 3 and one of the drugs with a smaller IC50 in cluster 3, and we regarded the drug as specific for cluster 3. In addition, we analyzed the results of the different tests for a range of drugs of interest to us. The 10 drugs of interest that intersected with the 20 drugs with the smallest IC50 appearing in cluster 2 or cluster 3 were ultimately filtered out, including bortezomib, staurosporine, daporinad, dinaciclib, docetaxel, luminespib paclitaxel, rapamycin, vincristine, and dactolisib (Fig. 6E and F).
3.6. Identification of subtype recognition models in AITL
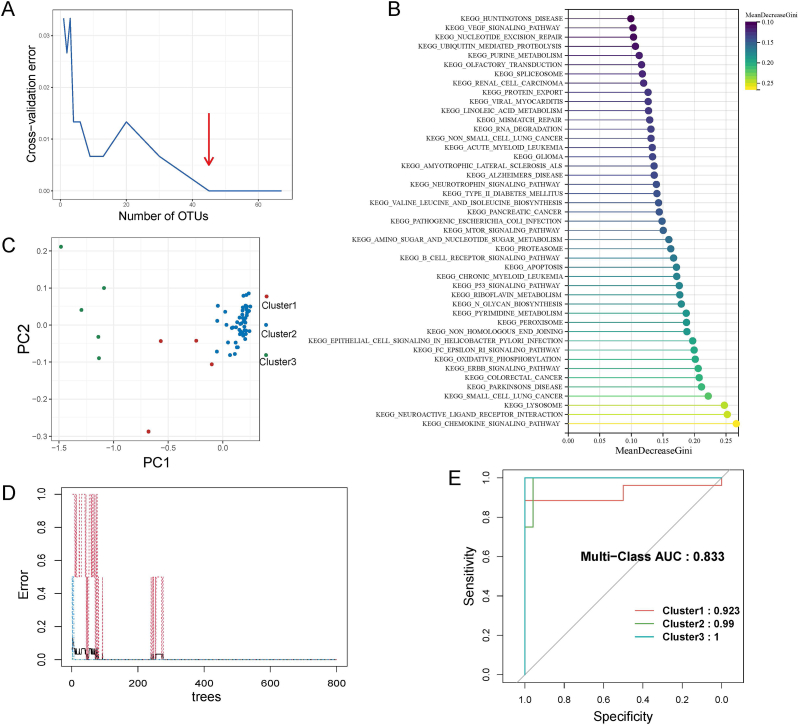
To facilitate clinical diagnosis and improve the efficiency of AITL treatment, we further developed a model of AITL subtype identification based on a machine learning approach for determining unknown patient subtypes. The Boruta algorithm revealed that 67 pathways were more relevant to AITL subtype typing, using the 86 characteristic pathways identified in result 2 as initial variables. Secondly, the samples in each subtype were equally divided into two groups, with one as the training set and the other as the testing set. If it was not possible to divide equally, the training set was assigned one more sample than the testing set. We finally performed a ten-fold cross-validation method to determine the optimal number of variables and the importance of each variable using the training set data and the 67 pathway-related variables (Fig. 7A), thereby further improving the model's predictive performance and avoiding overfitting. As shown in Fig. 7A, the model reached stability when the number of variables was 45, and the top 45 variables in importance were selected (Fig. 7B).
Fig. 7.
Construction and validation of passage-related models. (A) Selection of the number of pathway-related variables. (B) Importance lollipop plot of the 45 significant variables. (C) PCA downscaling analysis after variable screening. (D) Random forest model on the number of trees selected. (E) ROC curves for the testing set, accompanied by AUCs of 0.923, 0.99, and 1, respec-tively.
The PCA downscaling analysis was performed using only 45 critical variables and confirmed that the samples of the three subtypes could still be separated, especially with a clear distinction between cluster 2 and cluster 3 (Fig. 7C). The finding corroborates that the significant variables we screened for exhibit a certain classification ability. Then, the random forest model was constructed using the training set data and the 45 variables filtered. The model showed that it would tend to stabilize after the number of trees was around 300 (Fig. 7D); thus, the ntree was set to 350. The recognition model for the AITL subtype was finally well-trained. The model's performance was evaluated using the testing set data and the area under the ROC curve (AUC) for the model predictions was calculated. The testing cohort showed that the AUC values for the model's prediction accuracy in each cluster 1, 2, and 3 were 0.923, 0.99, and 1, respectively (Fig. 7E), with a blended AUC of 0.833. These findings suggest that our model is highly reliable for identifying AITL subtypes, particularly for assessing the rare subtype cluster 3. Finally, the prediction model was wrapped, and a function was generated: “AITLSubPre”, which is stored in AITLSubPre.R for the convenience of other researchers and clinicians to use the prediction model. The users simply need to call the function and upload the transcriptome data to predict the subtypes of patients, and the model will return the probability that the patient belongs to each subtype.
4. Discussion
Given the complex clinical phenotype and genetic heterogeneity of AITL, subtype classification, prognosis prediction, and precision therapy are particularly challenging missions. With the advent of next-generation sequencing technologies, the mutational landscape of AITL has been identified for over 34 genes, including TET2, DNMT3A, RHOA, IDH2, and TP53 mutations [12,13]. Recently, relatively few studies have indicated that the critical pathways and driver genes of AITL are closely associated with the processes of inflammation and immune response [13], including abnormal T-cell receptor signaling pathways [8,14,15], primary immunodeficiency [16,17], disruption of chemokine signaling pathways [7], DNA damage repair, RNA splicing, infection [13], tumor development and progression, and metastasis [12,18]. Although research on the role of genetic landscapes and their enriched pathways in cancer has progressed rapidly, few studies have focused on the clinical significance of subtype typing for characteristic pathways in AITL. In the current study, we evaluated the therapeutic value of pathway-related subtype identification by dividing AITL patients into three clusters and screening 86 subtype-characterized pathways between clusters. Subsequently, a subtype identification model for AITL patients was constructed and validated based on the characteristic pathways as initial variables. The model could satisfactorily distinguish the functional status of AITL patients with different clusters at the onset of the disease. The present study is the first to predict the functional status (e.g., immune modulation, infection, etc.) of different subtypes in AITL patients using pathway-related subtype typing. Additionally, we systematically reviewed the impact of specific subtyping for AITL in terms of immune cell infiltration, immune-related pathways, and the selection of susceptibility drugs.
Most cellular signaling pathways are differentially expressed with varying degrees of activity in AITL and possess complex regulatory networks. A clustering consensus analysis was initially performed based on an optimal clustering number of k = 3 and the pathway profile of patients and identified three molecular subtypes of AITL related to the pathways. Further analysis revealed 86 subtype-characterized pathways that are differentially expressed in patients with different types of AITL. Importantly, these subtypes resulted in apparently different immune infiltration and treatment options, with the functional status of cluster 1 being an intermediate state subtype between clusters 1 and 2. The 76 DEGs identified in clusters 2 and 3 were primarily enriched in immune response, tumor development and metastasis, and infection pathways by genomic enrichment analysis and selection of the above characteristic pathways. Among them, the volcano plot of DEGs demonstrates that the expression levels of 59 DEGs (e.g., PSMB8, UFC1, NUDT1, HSPB1, CD14, ID3, ODC1, MYC, HNRNPF, RBM10, CHCHD1, NDUFA13, CCL2, ROMO1, RARS, HDAC1, ICAM3) in AITL were up-regulated while the expression levels of 17 DEGs (e.g., LOC100129620, LOC100505817, AADACP1, CTD-2194D22.4, MEFV, LOC105377458, NRSN1, HRAT13, PPEF2, DSCR8) were down-regulated based on the critical criteria (P < 0.05 and |log2FC|>1). This finding indicates that differences in the functional status of characteristic pathways may influence the development of AITL and the immune microenvironment. Interestingly, these DEGs were also highly enriched in hematopoietic regulatory and pro-inflammatory-related pathways [[19], [20], [21], [22], [23]], implying that characteristic pathways may also play a critical role in developing and progressing hematological malignancies and inflammatory diseases.
TME is primarily involved in maintaining cellular homeostasis in the normal tissues, but the components of TME in tumors differ remarkably from those in healthy tissues with respect to cell proportions and cellular status [24]. An increasing body of literature indicated that the TME plays a vital role in the pathogenesis of AITL, suggesting that the biological behaviors of AITL correlate with tumor cells and non-malignant cells in the TME [10]. In fact, the TME of AITL is unique in that it is characterized by a small number of malignant CD4+ TFH cells mixed with infiltration of multiple multispectral immune cells (e.g., T/B lymphocytes, macrophages, etc.) [10]. As described above, the cibersort algorithm, a classical algorithm for immune infiltration analysis, was used to calculate the abundance of various immune infiltrating cell types in tumor samples using transcriptional data from cancer samples. Our study showed the TME cell types predominantly infiltrated in AITL, including gamma-delta T cells, follicular helper T cells, activated memory CD4 T cells, M0 macrophages, M1 macrophages, naive CD4 T cells, etc. Further analysis revealed that the 59 genes previously confirmed to be up-regulated in cluster 2 was found to correspond to the expression profile of this highly infiltrated immune cell population, indicating immune dysfunction, impaired cytotoxicity and alterations in chemokines, and so on. Conversely, cluster 3 was mainly infiltrated by memory resting CD4 T cells, regulatory Tregs T cells, and resting NK cells, whose expression profile contained the 17 DEGs previously described as lowly expressed. It may indicate a lower degree of malignancy in cluster 3. Therefore, we hypothesized that the infiltration of immune cells in TME differs in each AITL subtype, implying that some genes are differentially expressed in different subtypes. Combined with Fig. 3, we found that there are also differences in their functional status in different clusters of AITL, probably due to the effects of differences in the activity of the relevant pathways and their genes described above.
Owing to the absence of a precise therapeutic target, R/R AITL is still linked to a poor clinical prognosis, although with some long-term survivors [13]. Currently, the treatment strategies for AITL mainly include CHOP ± E (etoposide) or BV (brentuximab vedotin) + CHP (without vincristine) [1], rituximab [25], immune checkpoint inhibitors (ICIs) [26], dasatinib [27], ruxolitinib [28], etc. In the study, we ultimately screened for the 10 drugs of interest with the smallest IC50 in different subgroups of AITL, including bortezomib, staurosporine, daporinad, dinaciclib, docetaxel, luminespib, paclitaxel, rapamycin, vincristine, and dactolisib. Bortezomib, a first-generation proteasome inhibitor, is generally used in chemotherapy for multiple myeloma and has potent anti-cancer activity. However, one of its main side effects is to cause severe peripheral neuropathy [29]. Researchers have attempted to use bortezomib in treatment regimens for leukemia and lymphoma in recent years, with good efficacy shown [30,31]. In fact, the chemotherapeutic agent vincristine similarly possesses dose-dependent neurotoxicity, which is a major factor limiting its application [32]. Few studies have focused on the effects of staurosporine. However, as a broadly selective and effective protein kinase inhibitor, it has been hypothesized to play an essential role in treating tumor drug resistance [33]. Its exact role needs to be further defined. The CDK inhibitor dinaciclib and the Hsp90 inhibitor luminespib are widely applied for the treatment of various solid tumors and hematological malignancies (e.g., pancreatic cancer, breast cancer, endometrial cancer, lymphoma, etc.) [[34], [35], [36], [37], [38], [39]]. Similarly, docetaxel has also been proven to be one of the most important chemotherapeutic drugs widely used to treat diverse types of cancer [40]. However, it is mostly administered with other chemotherapeutic agents or targeted drugs rather than alone. Paclitaxel is a well-known anti-cancer agent with a unique mechanism of action to inhibit the cell cycle. In treating lymphoma, we found that paclitaxel is an enhancer [41]. Cao et al. [41] revealed that the combination with paclitaxel significantly augmented the anti-cancer efficacy of CD47-targeted therapy for late-stage non-Hodgkin lymphoma (NHL) by directly evoking the phagocytic capacity of macrophages. Furthermore, rapamycin belongs to a macrolide immunosuppressant that suppresses the mechanistic target of rapamycin (mTOR) protein kinase, thereby controlling the growth and metabolism of tumor cells. Typical rapamycin concentrations (1–8 mg/kg) markedly attenuated tumor burden and improved survival rates in tumor-bearing mice by directly inhibiting tumor cell proliferation [42]. Nevertheless, the mechanism of rapamycin treatment needs to be explored in more detail. The PI3K/mTOR signaling is hyperactivated in Glioblastomas (GBMs) and many additional solid tumors. Hence, multiple tumor-based PI3K inhibitors have been well-studied in various cancers. In a recent study, the researchers found that using the dual PI3K/mTOR inhibitor dactolisib alone induced cytotoxic and pro-apoptotic effects, which are anti-tumor factors [43]. However, Almeida et al. [44] and Civallero et al. [45] also identified that dactolisib is a potential therapeutic route for IL-7R-related cases. Overall, the role of these 10 subtype-sensitive drugs in patients with AITL remains unknown, and more clinical data and experimental studies are needed.
Next, a subtype identification model was constructed in this study based on 45 characteristic pathways as the optimal variables. The model presented a good predictive performance for subtype typing on the training and testing sets. ROC analysis further confirmed the high predictive accuracy of the model and the complete separation of the three types of samples, indicating the clinical applicability of characteristic pathways-based typing models.
Even though the study provides novel avenues for subtype identification in AITL and potential therapeutic targets for individualized treatment of patients, it also contains several limitations. Firstly, although a subtype identification model was constructed to predict the functional status of AITL patients, more prospective studies are needed to assess the accuracy of this model. On the other hand, our findings depend on bioinformatic analysis. More clinical and experimental evidence is needed to explore the exact molecular mechanisms underlying the role of characteristic pathways involved in constructing the model between AITL development and immune microenvironmental features.
In summary, we elucidated the role of characteristic pathways and their related genes in AITL. A subtype identification model for AITL was constructed to determine the potential functional status of different AITL clusters based on 45 important characteristic pathways. Additionally, we not only found that the types and proportions of TME immune infiltrating cells were significantly different between AITL subtypes but also predicted subtype-specific drugs for AITL. Our comprehensive analysis of characteristic pathways brings new insights into the subtype typing of AITL and contributes to the study of targets for the precise treatment of AITL.
Author contributions
HP: study design. ZW: drafting of the manuscript. YZ, HL and CX: critical revision of the manuscript. WG, ZH, and HZ: validating the results. LY, XR, and ZC: curating the data. WG and ZH: managing the software and performing the statistical analysis. SZ: acquisition, assembly, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82070175 and 81600183), the Natural Science Foundation of Hunan Province (2022JJ30830 and 2022JJ70058), the Scientific program of the Health Commission of Hunan Province (20201179) and the Natural Science Foundation of Changsha City (kq2202405).
Data availability statement
The datasets used in this study are available from the corresponding author by request or can be download using the provided hyperlink, full citation, or other persistent identifiers.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Zhihua Wang, Email: wangzhihuazsc@csu.edu.cn.
Hongling Peng, Email: penghongling@csu.edu.cn.
References
- 1.Mohammed Saleh M.F., Kotb A., Abdallah G.E.M., et al. Recent advances in diagnosis and therapy of angioimmunoblastic T cell lymphoma. Curr. Oncol. 2021;28(6):5480–5498. doi: 10.3390/curroncol28060456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lunning M.A., Vose J.M. Angioimmunoblastic T-cell lymphoma: the many-faced lymphoma. Blood. 2017;129(9):1095–1102. doi: 10.1182/blood-2016-09-692541. [DOI] [PubMed] [Google Scholar]
- 3.Deng S., Lin S., Shen J., et al. Comparison of CHOP vs CHOPE for treatment of peripheral T-cell lymphoma: a meta-analysis. OncoTargets Ther. 2019;12:2335–2342. doi: 10.2147/ott.s189825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellin F., LandströM J., Jerkeman M., et al. Real-world data on prognostic factors and treatment in peripheral T-cell lymphomas: a study from the Swedish Lymphoma Registry. Blood. 2014;124(10):1570–1577. doi: 10.1182/blood-2014-04-573089. [DOI] [PubMed] [Google Scholar]
- 5.Lu P.C.W., Shahbaz S., Winn L.M. Benzene and its effects on cell signaling pathways related to hematopoiesis and leukemia. J. Appl. Toxicol. 2020;40(8):1018–1032. doi: 10.1002/jat.3961. [DOI] [PubMed] [Google Scholar]
- 6.Fukumoto K., Nguyen T.B., Chiba S., et al. Review of the biologic and clinical significance of genetic mutations in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018;109(3):490–496. doi: 10.1111/cas.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Liu Z., Mi M., et al. Identification of hub genes and key pathways associated with angioimmunoblastic T-cell lymphoma using weighted gene co-expression network analysis. Cancer Manag. Res. 2019;11:5209–5220. doi: 10.2147/cmar.s185030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heavican T.B., Bouska A., Yu J., et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133(15):1664–1676. doi: 10.1182/blood-2018-09-872549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Leval L., Rickman D.S., Thielen C., et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952–4963. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 10.Pritchett J.C., Yang Z.Z., Kim H.J., et al. High-dimensional and single-cell transcriptome analysis of the tumor microenvironment in angioimmunoblastic T cell lymphoma (AITL) Leukemia. 2022;36(1):165–176. doi: 10.1038/s41375-021-01321-2. [DOI] [PubMed] [Google Scholar]
- 11.Abid A., Zhang M.J., Bagaria V.K., et al. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat. Commun. 2018;9(1):2134. doi: 10.1038/s41467-018-04608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odejide O., Weigert O., Lane A.A., et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiba S., Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia. 2020;34(10):2592–2606. doi: 10.1038/s41375-020-0990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallois D., Dobay M.P., Morin R.D., et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 15.Palomero T., Couronné L., Khiabanian H., et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014;46(2):166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinuesa C.G., Cook M.C., Angelucci C., et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435(7041):452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 17.Ellyard J.I., Chia T., Rodriguez-Pinilla S.M., et al. Heterozygosity for Roquinsan leads to angioimmunoblastic T-cell lymphoma-like tumors in mice. Blood. 2012;120(4):812–821. doi: 10.1182/blood-2011-07-365130. [DOI] [PubMed] [Google Scholar]
- 18.Quivoron C., Couronné L., Della Valle V., et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Liu M., Yang L., Liu X., et al. HNRNPH1 is a novel regulator of cellular proliferation and disease progression in chronic myeloid leukemia. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.682859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wampfler J., Federzoni E.A., Torbett B.E., et al. The RNA binding proteins RBM38 and DND1 are repressed in AML and have a novel function in APL differentiation. Leuk. Res. 2016;41:96–102. doi: 10.1016/j.leukres.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Bahr C., Von Paleske L., Uslu V.V., et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553(7689):515–520. doi: 10.1038/nature25193. [DOI] [PubMed] [Google Scholar]
- 22.Rusmini M., Federici S., Caroli F., et al. Next-generation sequencing and its initial applications for molecular diagnosis of systemic auto-inflammatory diseases. Ann. Rheum. Dis. 2016;75(8):1550–1557. doi: 10.1136/annrheumdis-2015-207701. [DOI] [PubMed] [Google Scholar]
- 23.Fujikawa K., Migita K., Shigemitsu Y., et al. MEFV gene polymorphisms and TNFRSF1A mutation in patients with inflammatory myopathy with abundant macrophages. Clin. Exp. Immunol. 2014;178(2):224–228. doi: 10.1111/cei.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SonugüR F.G., Akbulut H. The role of tumor microenvironment in genomic instability of malignant tumors. Front. Genet. 2019;10:1063. doi: 10.3389/fgene.2019.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delfau-Larue M.H., De Leval L., Joly B., et al. Targeting intratumoral B cells with rituximab in addition to CHOP in angioimmunoblastic T-cell lymphoma. A clinicobiological study of the GELA. Haematologica. 2012;97(10):1594–1602. doi: 10.3324/haematol.2011.061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y., Wu J., Wang Z., et al. Efficacy and safety of geptanolimab (GB226) for relapsed or refractory peripheral T cell lymphoma: an open-label phase 2 study (Gxplore-002) J. Hematol. Oncol. 2021;14(1):12. doi: 10.1186/s13045-021-01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen T.B., Sakata-Yanagimoto M., Fujisawa M., et al. Dasatinib is an effective treatment for angioimmunoblastic T-cell lymphoma. Cancer Res. 2020;80(9):1875–1884. doi: 10.1158/0008-5472.CAN-19-2787. [DOI] [PubMed] [Google Scholar]
- 28.Moskowitz A.J., Ghione P., Jacobsen E., et al. A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood. 2021;138(26):2828–2837. doi: 10.1182/blood.2021013379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto S., Egashira N. Pathological mechanisms of bortezomib-induced peripheral neuropathy. Int. J. Mol. Sci. 2021;22(2) doi: 10.3390/ijms22020888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertaina A., Vinti L., Strocchio L., et al. The combination of bortezomib with chemotherapy to treat relapsed/refractory acute lymphoblastic leukaemia of childhood. Br. J. Haematol. 2017;176(4):629–636. doi: 10.1111/bjh.14505. [DOI] [PubMed] [Google Scholar]
- 31.Arkwright R., Pham T.M., Zonder J.A., et al. The preclinical discovery and development of bortezomib for the treatment of mantle cell lymphoma. Expet Opin. Drug Discov. 2017;12(2):225–235. doi: 10.1080/17460441.2017.1268596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G.Z., Hu Y.H., Li D.Y., et al. Vincristine-induced peripheral neuropathy: a mini-review. Neurotoxicology. 2020;81:161–171. doi: 10.1016/j.neuro.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 33.He Y. Systematic response of staurosporine scaffold-based inhibitors to drug-resistant cancer kinase mutations. Arch. Pharm. (Weinheim) 2020;353(6) doi: 10.1002/ardp.201900320. [DOI] [PubMed] [Google Scholar]
- 34.Howard D., James D., Murphy K., et al. Dinaciclib, a bimodal agent effective against endometrial cancer. Cancers. 2021;13(5) doi: 10.3390/cancers13051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J., Chen P., Liu K., et al. CDK1/2/5 inhibition overcomes IFNG-mediated adaptive immune resistance in pancreatic cancer. Gut. 2021;70(5):890–899. doi: 10.1136/gutjnl-2019-320441. [DOI] [PubMed] [Google Scholar]
- 36.Zhao H., Li S., Wang G., et al. Study of the mechanism by which dinaciclib induces apoptosis and cell cycle arrest of lymphoma Raji cells through a CDK1-involved pathway. Cancer Med. 2019;8(9):4348–4358. doi: 10.1002/cam4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun H.D., Schirm D.K., Felices M., et al. Dinaciclib enhances natural killer cell cytotoxicity against acute myelogenous leukemia. Blood Adv. 2019;3(16):2448–2452. doi: 10.1182/bloodadvances.2019000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piotrowska Z., Costa D.B., Oxnard G.R., et al. Activity of the Hsp90 inhibitor luminespib among non-small-cell lung cancers harboring EGFR exon 20 insertions. Ann. Oncol. 2018;29(10):2092–2097. doi: 10.1093/annonc/mdy336. [DOI] [PubMed] [Google Scholar]
- 39.Tsai H.J., Shih N.Y., Kuo S.H., et al. AUY922 effectively targets against activated B cell subtype of diffuse large B-cell lymphoma and low-grade lymphoma cells harboring genetic alteration-associated nuclear factor-κB activation. Leuk. Lymphoma. 2015;56(9):2674–2682. doi: 10.3109/10428194.2014.995647. [DOI] [PubMed] [Google Scholar]
- 40.Murray Stewart T., Von Hoff D., Fitzgerald M., et al. A Phase Ib multicenter, dose-escalation study of the polyamine analogue PG-11047 in combination with gemcitabine, docetaxel, bevacizumab, erlotinib, cisplatin, 5-fluorouracil, or sunitinib in patients with advanced solid tumors or lymphoma. Cancer Chemother. Pharmacol. 2021;87(1):135–144. doi: 10.1007/s00280-020-04201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao X., Wang Y., Zhang W., et al. Targeting macrophages for enhancing CD47 blockade-elicited lymphoma clearance and overcoming tumor-induced immunosuppression. Blood. 2022;139(22):3290–3302. doi: 10.1182/blood.2021013901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Pandeswara S., Dao V., et al. Biphasic rapamycin effects in lymphoma and carcinoma treatment. Cancer Res. 2017;77(2):520–531. doi: 10.1158/0008-5472.CAN-16-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinzen D., Divé I., Lorenz N.I., et al. Second generation mTOR inhibitors as a double-edged sword in malignant glioma treatment. Int. J. Mol. Sci. 2019;20(18) doi: 10.3390/ijms20184474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almeida A.R.M., Neto J.L., Cachucho A., et al. Interleukin-7 receptor α mutational activation can initiate precursor B-cell acute lymphoblastic leukemia. Nat. Commun. 2021;12(1):7268. doi: 10.1038/s41467-021-27197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Civallero M., Cosenza M., Pozzi S., et al. Activity of BKM120 and BEZ235 against lymphoma cells. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/870918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in this study are available from the corresponding author by request or can be download using the provided hyperlink, full citation, or other persistent identifiers.