Abstract
Hematology analysis, a common clinical test for screening various diseases, has conventionally required a chemical staining process that is time-consuming and labor-intensive. To reduce the costs of chemical staining, label-free imaging can be utilized in hematology analysis. In this work, we exploit optical diffraction tomography and the fully convolutional one-stage object detector or FCOS, a deep learning architecture for object detection, to develop a label-free hematology analysis framework. Detected cells are classified into four groups: red blood cell, abnormal red blood cell, platelet, and white blood cell. In the results, the trained object detection model showed superior detection performance for blood cells in refractive index tomograms (0.977 mAP) and also showed high accuracy in the four-class classification of blood cells (0.9708 weighted F1 score, 0.9712 total accuracy). For further verification, mean corpuscular volume (MCV) and mean corpuscular hemoglobin (MCH) were compared with values obtained from reference hematology equipment, with our results showing reasonable correlation in both MCV (0.905) and MCH (0.889). This study provides a successful demonstration of the proposed framework in detecting and classifying blood cells using optical diffraction tomography for label-free hematology analysis.
Keywords: Label-free imaging, Optical diffraction tomography, Hematology analysis, Deep learning, Object detection
Highlights
-
•
Blood cell detection and classification is shown optical diffraction tomography.
-
•
Superior classification and detection performance is obtained through an FCOS backbone.
-
•
High computational efficiency is achieved.
-
•
Quantitative analysis of cell indices is possible from refractive index distributions.
-
•
Feasible correlation with reference hematology analysis equipment is found.
1. Introduction
Hematology analysis is one of the most common clinical tests used to look for various disorders such as sepsis [1], [2], [3], infection [4], [5], [6], anemia [7], [8], [9], and blood cancer [10], [11], [12], [13]. Chemical staining has traditionally been used for blood analysis, with requirements for complex equipment, numerous chemical reagents, difficult system calibration and procedures, and highly skilled personnel, factors that can significantly impact the status of cells during the process [14], [15]. Additionally, the staining process itself is time-consuming and difficult; it is generally estimated that it takes approximately 45 min for specialists to stain cells in accordance with strict staining protocols and to assess the staining quality under a microscope [15].
To solve this problem, various methods using label-free imaging modalities in hematology analysis have been studied. For instance, Raman microscopy [16], [17], [18], hyperspectral imaging [19], [20], [21], and defocusing phase-contrast imaging [22], [23] have been exploited for hematology analysis. These methods use the endogenous imaging contrast of samples themselves without using any staining agent, but require complex optical systems and long data acquisition times. Recently, investigations have adopted quantitative phase imaging (QPI) to enable reasonably quick and simple label-free imaging of biological samples [24], [25]. The morphological and biochemical characteristics of a cell can be observed concurrently using QPI by measuring the phase delay of light passing through the sample and reconstructing the refractive index (RI) of the sample through the relationship between the scattered light and the sample.
Recent rapid advances in artificial intelligence (AI) have led to the application of AI to the QPI of various types of biological samples in a wide range of tasks including classification [26], [27], [28], [29], [30], segmentation [31], [32], [33], and inference [34], [35], [36]. In the field of blood cell identification, QPI and AI demonstrate a good synergy since they can simultaneously check morphological and biological properties in addition to their label-free nature and uniform data quality that is not affected by staining quality [37], [28], [29].
In this study, we utilize optical diffraction tomography (ODT) [38], a label-free and three-dimensional (3D) QPI modality that measures the RI distribution of a sample, and a deep learning technology for the hematological analysis of blood samples. Details of each step can be found in Fig. 1. To develop an object detection model for blood cells, the fully convolutional one-stage object detector (FCOS) [39], an anchor-free multitask deep learning model, is implemented and trained. With this framework, detected cells are classified into four groups: red blood cell (RBC), abnormal red blood cell (ARBC), platelet (PLT), and white blood cell (WBC). We demonstrate that the trained model is able to detect most of the cells, showing high accuracy in the four-class classification of blood cells. In addition, morphological and biochemical quantities are calculated, namely mean corpuscular volume (MCV) and mean corpuscular hemoglobin (MCH), for RBC and ARBC groups based on their RI distributions. Our quantitative analysis results also show highly acceptable agreement with commercial hematology analysis equipment.
Figure 1.
Schematic diagram of hematological analysis using ODT. (a) A 3D RI tomogram of a blood sample is acquired by partially coherent ODT. (b) The deep learning model automatically detects blood cells and classifies them into four types: RBC, ARBC, PLT, and WBC. See Section 2.2 for a detailed explanation of the object detection model implementation. (c) For RBCs and ARBCs, morphological and biochemical quantities such as MCV and MCH are measured by their RI distribution.
2. Materials and methods
2.1. Dataset preparation
A total of 171 blood samples were collected from Chungnam National University Hospital (CNUH, Republic of Korea), and ODT was exploited to obtain RI tomograms of the blood samples. (This study was approved by the institutional review board at CNUH.) For each blood sample, 27 measurements were carried out because single-measurement coverage was restricted to 234 μm× 234 μmdue to field-of-view limitations. Each measured 3D RI tomogram had a lateral resolution of 0.162 μm/pixel and axial resolution of 0.731 μm/pixel.
Bounding boxes and cell types for all blood cell instances were annotated for 1,224 3D RI tomograms by three experts. The blood cells were categorized into four types: RBC, ARBC meaning a morphologically irregular RBC, PLT, and WBC. The criteria for morphological irregularity was determined by annotators. A total of 115,143 cell instances were annotated to train and evaluate the object detection model (Table 1). The whole dataset was divided into a 7:2:1 ratio for training, validation, and testing.
Table 1.
Distribution of cell types in the dataset.
| Type of dataset | RBC | PLT | WBC | ARBC |
|---|---|---|---|---|
| Train | 67,595 | 6,391 | 87 | 6,435 |
| Validation | 19,604 | 1,608 | 19 | 1,567 |
| Test | 9,661 | 1,143 | 18 | 1,015 |
| Total | 96,860 | 9,142 | 124 | 9,017 |
2.2. Training details of object detection models
In previous studies [40], [41], YOLOv3 [42] or its variant model has been widely used and has shown reasonable performance. However, detection failures of PLTs were observed due to their small size compared to RBCs or WBCs. Hence, for the detection and identification of blood cells of various sizes, the FCOS, or fully connected one-stage object detector, [39], was exploited in this work, which is an anchor-free one-stage object detection framework optimized for detecting objects of various sizes at once.
For model implementation, the MMDetection library [43] was utilized, which is a PyTorch-based open-source object detection toolbox. FCOS detects objects by performing classification, regression, and centerness calculations on feature maps extracted from the backbone. In our experiments, we employed focal loss (FL, (1)), generalized intersection over union (GIoU, (2)) loss, and binary cross entropy (BCE, (3)) loss to evaluate classification, regression, and centerness prediction. These loss functions are the same as the improvement methods described in the original FCOS paper [39]. The mathematical formulations for these loss functions are defined as follows:
| (1) |
| (2) |
| (3) |
In this paper, we applied all the enhancements presented in the work of Tian et al. [39] to the FCOS framework.
For preprocessing, min-max normalization was applied to the RI images. An augmentation ratio of 0.5 was applied to the training dataset using the horizontal flip operation. In the training phase, the batch size was set to 4. The AdamW optimizer was used, and the learning rate was applied as 0.0005. In addition, mixed precision and gradient clipping were applied, and the number of epochs was 30.
We tested three backbone networks for the FCOS framework, namely ResNet-18, ResNet-50, and the Swin Transformer, to find the architecture with the best performance [44], [45]. Since ResNet, one of the most famous convolutional neural networks, is known as a good feature extractor in various vision tasks, two variants of ResNet were tested for the backbone of our network. Recently, backbones based on transformers [46] have also shown high performance in various vision tasks. First achieving state-of-the-art performance in object detection tasks in 2021 [45], the Swin Transformer has a characteristic that it performs self-attention only among patches within each window. Our data has the feature that similarly shaped and sized cells are evenly distributed throughout the entire image; thus, to investigate whether the features of the Swin Transformer's architecture can be advantageous for our images, we chose the Swin Transformer as one of the backbones. In the case of ResNet-18, the Feature Pyramid Networks (FPNs) to Path Aggregation Feature Pyramid Network (PAFPNs) were adjusted according to the model output, while for the Swin Transformer, the numbers of output channels and FPN input channels were adjusted.
Our network was implemented in PyTorch 1.12 using a graphics processing unit (GPU) server (Tesla V100 32GB). The GPU server environment was as follows: Python 3.8.12, CUDA 11.6, CUDNN 8.3.3, MMCV 1.5.1, and MMDetection 2.24.1.
2.3. Evaluation of the trained object detection model
A total of five metrics—mean average precision (mAP), recall (7), precision (8), F1 score (9), and total accuracy (10)—were calculated to evaluate the performance of the trained object detection model. In the case of mAP, the definition in the PASCAL VOC dataset was applied (average of 11-point interpolated AP, (4), (5), (6)). For an IoU threshold of 0.5, true and false cases were distinguished, and the average of the area under the precision-recall curve (PR curve) was used for each category. When calculating the base area, recall was divided into 0.1 units and interpolated with the highest precision value in the relevant section. Each metric is defined as follows:
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
2.4. Comparison of blood cell indices
Refractive index, which is used as an imaging contrast in ODT, is a biochemical quantity related to the number of molecules per unit volume. With a 3D RI tomogram, RI can be used to calculate not only morphological quantities such as corpuscular volume (CV) but also biochemical quantities like corpuscular hemoglobin (CH). Therefore, hematological analysis using ODT opens the door to quantitative analysis beyond simple blood cell counting and classification. First, a cell contour is generated by a simple thresholding-based contouring algorithm. In this work, the threshold was set to 0.005, which means the cell contour was set based on the voxels where RI exceeds the medium RI + 0.005. Then CV is measured by equation (11).
| (11) |
where denotes the number of voxels in a cell contour and is the volume of a single voxel, which is 0.0192 μm3 for the device used in this study. CH is calculated as equation (12):
| (12) |
where is the sum of RI values for all voxels in the cell contour, denotes the RI of the medium (1.337 for water), and RII means refractive index increment [47], [48], [49], [50], [51], 0.0029 for this study.
By averaging the above quantities of each RBC instance in the same blood sample, the MCV and MCH of the blood sample are obtained. In this study, MCV and MCH were calculated for RBCs detected through the trained FCOS model for 1,539 3D RI tomograms, which were not included in the dataset for object detection model training and evaluation. The calculated values were compared with those measured from reference hematology analysis equipment (DxH-800 and LH-780, Beckman Coulter).
3. Results and discussion
3.1. Evaluation of the trained object detection model
The cell detection performance of each of the three backbones (ResNet-18, ResNet-50, and Swin Transformer) was first compared. Among the models used as the FCOS backbone, the Swin Transformer showed the best performance by a very narrow margin (Table 2). The highest performance was 0.977 mAP when the backbone network was the Swin Transformer, but it also had the largest number of model parameters, about 1.7 times that of ResNet-18. ResNet-18 and ResNet-50 showed similar performance.
Table 2.
Comparison of the detection performance of each FCOS backbone.
| Backbone | mAP | Number of model parameters |
|---|---|---|
| ResNet-18 | 0.970 | 21.45 M |
| ResNet-50 | 0.970 | 32.12 M |
| Swin Transformer | 0.977 | 35.56 M |
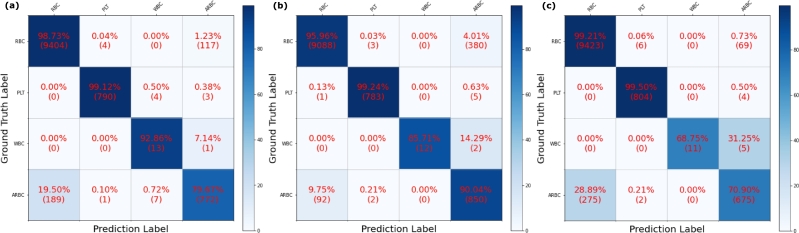
The classification performance of the three backbone networks was also compared. ResNet-18 showed the highest classification performance with 0.9708 weighted F1 score and 0.9712 total accuracy. While the difference between these results and those from the Swin Transformer is relatively small (about 0.03), when compared to ResNet-50, ResNet-18 had higher scores of more than 0.1 (Table 3) Detailed classification results of each backbone network can be found in Fig. 2. Considering RBCs and ARBCs only, ResNet-18 failed 306 cases, while ResNet-50 and Swin Transformer failed 472 and 344 cases, respectively. Because of the severe data imbalance for the WBC class (only 124 cases in 115,143 cells), the WBC classification performance of each model was neglected.
Table 3.
Comparison of the classification performance of each FCOS backbone.
| Backbone | Class | Recall | Precision | F1 score | Weighted F1 score | Total Accuracy |
|---|---|---|---|---|---|---|
| ResNet-18 | RBC | 0.9873 | 0.9803 | 0.9838 | 0.9708 | 0.9712 |
| PLT | 0.9912 | 0.9937 | 0.9925 | |||
| WBC | 0.9286 | 0.5417 | 0.6842 | |||
| ARBC | 0.7967 | 0.8645 | 0.8292 | |||
| ResNet-50 | RBC | 0.9596 | 0.9899 | 0.9745 | 0.9593 | 0.9568 |
| PLT | 0.9924 | 0.9937 | 0.9930 | |||
| WBC | 0.8571 | 1.0000 | 0.9231 | |||
| ARBC | 0.9004 | 0.6871 | 0.7795 | |||
| Swin | RBC | 0.9921 | 0.9716 | 0.9818 | 0.9663 | 0.9680 |
| Transformer | PLT | 0.9950 | 0.9901 | 0.9926 | ||
| WBC | 0.6875 | 1.0000 | 0.8148 | |||
| ARBC | 0.7090 | 0.8964 | 0.7918 | |||
Figure 2.
Normalized confusion matrix results for each FCOS backbone: (a) ResNet-18, (b) ResNet-50, and (c) Swin Transformer. ResNet-18 showed the best performance in classifying RBCs and ARBCs.
In summary, ResNet-18 performed the best in terms of classification while achieving competitive detection performance despite having the smallest number of model parameters. Therefore, ResNet-18 was chosen to be utilized as the backbone of our hematology analysis framework.
Compared with the results of a previous related study [41], our model improved mAP by about 0.23, and the accuracy for the RBC, WBC, and PLT groups improved by more than 3%. Since our imaging modality is relatively new, it should be noted that there are no previous experimental results using the same type of modality.
Although our model achieved an F1 score over 0.8 for ARBC classification, it should be noted that the criteria for the ARBC/RBC boundary are ambiguous. Hence, there is a limit to determine the model's performance in identifying ARBCs through its accuracy with the label, since the standard of morphological irregularity for an ARBC depends on the annotator. To investigate ARBC classification performance in more detail, t-SNE [52] analysis, which is a method of reducing high-dimensional data to low-dimensional data, was performed using extracted features from the trained FCOS model. T-sne result can be found in Fig. 3 (a), and Fig. 3 (b) shows representative sample RI images. For the feature extraction, cells detected in the RBC and ARBC channels were used, meaning that features were able to be extracted for both classes, as FCOS utilizes multiple binary classifiers for multi-class identification.
Figure 3.
t-SNE result of the classification model: (a) t-SNE plot and (b) representative sample RI images for each legend entry.
By projecting high-dimensional features extracted from the trained FCOS model onto a two-dimensional plane, we could see that the features were largely divided into two clusters with a gray area between them. As a result of visualizing numerous points corresponding to each prediction/label result shown in the figure legend, the morphological characteristics of a clear circular shape for RBCs and a clear irregular shape for ARBCs were observed at the ends, while cells of a shape difficult to assign to either class were observed in the middle area. These results demonstrate that our FCOS model distinguished RBCs and ARBCs based on their degree of morphological irregularity.
3.2. Comparison of blood cell indices
In terms of MCV and MCH, our results showed acceptable agreement with the reference equipment. Fig. 4 (a), (b) and (c) show MVC data comparison, and (d), (e), and (f) show MCH comparison. The Pearson correlation coefficients for MCV and MCH were 0.905 and 0.889, respectively, while the gradients of Passing–Bablok regression were 1.04 and 0.97, respectively.
Figure 4.
Comparison of RBC indices, (a–c) MCV and (d–f) MCH, obtained from our model and the reference device. CBA denotes our result, and Coulter means the reference device.
The indices of the RBC and ARBC classes were also compared. Fig. 5 (a) shows cell indices comparison of MCV, and Fig. 5 (b) shows MCH results. There was a clear difference between the physical quantity distribution of RBCs and ARBCs. The MCV and MCH of the ARBCs were clearly lower than those of the RBCs. A larger divergence of the ARBC MCV and MCH was also observed, caused by the irregularity of the cell shape.
Figure 5.
Comparison of cell indices, (a) MCV and (b) MCH, for RBC and ARBC classes.
Discussion about the issue of cell contouring is required in this section. For an accurate quantitative analysis, an appropriate cell contour is necessary. Because of the low axial resolution caused by the missing cone problem [35], the RI distribution changes depending on the shape and angle of the cell. This causes distortion of the cell contour and consequently affects quantitative analysis. To solve this issue, recognition of the lying angle of each cell and corresponding compensation will be studied in follow-up research.
4. Conclusions
This study showed that the detection and classification of blood cells using ODT technology is feasible for label-free hematology analysis and also that physical quantities can be calculated with QPI equipment that uses refractive index as the imaging contrast. We applied FCOS, an anchor-free one-stage detection framework, for hematological analysis in 3D RI tomograms of blood samples. Our model achieved 0.977 mAP for blood cell detection, and achieved 0.9708 weighted F1 score and 0.9712 total accuracy for four-class blood cell classification (RBC, ARBC, PLT, and WBC). The RBC indices from the RI distribution in each cell were also calculated and compared with those obtained from reference equipment. The result showed reasonable correlation in MCV (0.905) and MCH (0.889) with the reference equipment.
While previous study [29] has conducted hematological analysis on single red blood cells (RBCs) using 3D QPI, the significance of our research lies in detecting blood cells and calculating the physical quantity of blood cells from the same type of blood sample used in actual clinical settings. Our proposed framework has high potential to be extended to more various types of hematology analysis, such as WBC subtype classification, through additional data acquisition.
Ethics approval statement
This study was approved by the institutional review board at Chungnam National University Hospital, Republic of Korea. (CNUH 2020-110914) As a retrospective study, waiver of informed consent was approved.
Funding
This work was supported in part by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (HI21C1161), in part by an Institute of Information and communications Technology Planning and Evaluation (IITP) grant funded by the Ministry of Science and ICT (MSIT), Republic of Korea (No.2020-0-01336, Artificial Intelligence Graduate School Program (UNIST)), and in part by a grant of the National Research Foundation of Korea (NRF) funded by the MSIT, Republic of Korea (NRF-2021R1F1A1057818).
CRediT authorship contribution statement
Dongmin Ryu; Taeyoung Bak: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Daewoong Ahn: Analyzed and interpreted the data.
Hayoung Kang; Sanggeun Oh: Contributed reagents, materials, analysis tools or data.
Hyun-seok Min: Conceived and designed the experiments; Analyzed and interpreted the data.
Sumin Lee; Jimin Lee: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Declaration of Competing Interest
Dongmin Ryu, Daewoong Ahn, Hayoung Kang, Sangguen Oh, Hyun-seok Min, and Sumin Lee have financial interests in Tomocube Inc., a company that commercializes optical diffraction tomography and quantitative phase imaging instruments. The other authors declare no competing financial interests.
Contributor Information
Sumin Lee, Email: slee@tomocube.com.
Jimin Lee, Email: jiminlee@unist.ac.kr.
Data availability
The authors do not have permission to share data.
References
- 1.Murphy K., Weiner J. Use of leukocyte counts in evaluation of early-onset neonatal sepsis. Pediatr. Infect. Dis. J. 2012;31(1):16–19. doi: 10.1097/INF.0b013e31822ffc17. [DOI] [PubMed] [Google Scholar]
- 2.Chandramohanadas R., Park Y., Lui L., Li A., Quinn D., Liew K., Diez-Silva M., Sung Y., Dao M., Lim C.T., et al. Biophysics of malarial parasite exit from infected erythrocytes. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0020869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskurt O.K., Gelmont D., Meiselman H.J. Red blood cell deformability in sepsis. Am. J. Respir. Crit. Care Med. 1998;157(2):421–427. doi: 10.1164/ajrccm.157.2.9611103. [DOI] [PubMed] [Google Scholar]
- 4.Klinger M.H., Jelkmann W. Role of blood platelets in infection and inflammation. J. Interferon Cytokine Res. 2002;22(9):913–922. doi: 10.1089/10799900260286623. [DOI] [PubMed] [Google Scholar]
- 5.Al-Gwaiz L.A., Babay H.H. The diagnostic value of absolute neutrophil count, band count and morphologic changes of neutrophils in predicting bacterial infections. Med. Princ. Pract. 2007;16(5):344–347. doi: 10.1159/000104806. [DOI] [PubMed] [Google Scholar]
- 6.Honda T., Uehara T., Matsumoto G., Arai S., Sugano M. Neutrophil left shift and white blood cell count as markers of bacterial infection. Clin. Chim. Acta. 2016;457:46–53. doi: 10.1016/j.cca.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 7.de Haan K., Ceylan Koydemir H., Rivenson Y., Tseng D., Van Dyne E., Bakic L., Karinca D., Liang K., Ilango M., Gumustekin E., et al. Automated screening of sickle cells using a smartphone-based microscope and deep learning. npj Digit. Med. 2020;3(1):1–9. doi: 10.1038/s41746-020-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson C.I., Hopkins P.L., Cabello-Inchausti B., Melnick S.J., Robinson M.J. The peripheral blood smear in patients with sickle cell trait: a morphologic observation. Lab. Med. 2000;31(8):445–447. [Google Scholar]
- 9.Dhaliwal G., Cornett P.A., Tierney L.M., Jr Hemolytic anemia. Am. Fam. Phys. 2004;69(11):2599–2606. [PubMed] [Google Scholar]
- 10.Nowakowski G.S., Hoyer J.D., Shanafelt T.D., Zent C.S., Call T.G., Bone N.D., LaPlant B., Dewald G.W., Tschumper R.C., Jelinek D.F., et al. Percentage of smudge cells on routine blood smear predicts survival in chronic lymphocytic leukemia. J. Clin. Oncol. 2009;27(11):1844. doi: 10.1200/JCO.2008.17.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S.S., Aasen G.A., Dolan M.M., Linden M.A., McKenna R.W., Rudrapatna V.K., Trottier B.J., Drawz S.M. Early diagnosis of intravascular large b-cell lymphoma: clues from routine blood smear morphologic findings. Lab. Med. 2014;45(3):248–252. doi: 10.1309/LMSVEOKLN18M5XTV. [DOI] [PubMed] [Google Scholar]
- 12.Khvastunova A.N., Kuznetsova S.A., Al-Radi L.S., Vylegzhanina A.V., Zakirova A.O., Fedyanina O.S., Filatov A.V., Vorobjev I.A., Ataullakhanov F. Anti-cd antibody microarray for human leukocyte morphology examination allows analyzing rare cell populations and suggesting preliminary diagnosis in leukemia. Sci. Rep. 2015;5(1):1–13. doi: 10.1038/srep12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sant M., Allemani C., Tereanu C., De Angelis R., Capocaccia R., Visser O., Marcos-Gragera R., Maynadié M., Simonetti A., Lutz J.-M., et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the haemacare project. Blood. 2010;116(19):3724–3734. doi: 10.1182/blood-2010-05-282632. [DOI] [PubMed] [Google Scholar]
- 14.Doan M., Sebastian J.A., Pinto R.N., McQuin C., Goodman A., Wolkenhauer O., Parsons M.J., Acker J.P., Rees P., Hennig H., et al. Label-free assessment of red blood cell storage lesions by deep learning. BioRxiv. 2018 [Google Scholar]
- 15.Ojaghi A., Carrazana G., Caruso C., Abbas A., Myers D.R., Lam W.A., Robles F.E. Label-free hematology analysis using deep-ultraviolet microscopy. Proc. Natl. Acad. Sci. 2020;117(26):14779–14789. doi: 10.1073/pnas.2001404117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramoji A., Neugebauer U., Bocklitz T., Foerster M., Kiehntopf M., Bauer M., Popp J. Toward a spectroscopic hemogram: Raman spectroscopic differentiation of the two most abundant leukocytes from peripheral blood. Anal. Chem. 2012;84(12):5335–5342. doi: 10.1021/ac3007363. [DOI] [PubMed] [Google Scholar]
- 17.Atkins C.G., Buckley K., Blades M.W., Turner R.F. Raman spectroscopy of blood and blood components. Appl. Spectrosc. 2017;71(5):767–793. doi: 10.1177/0003702816686593. [DOI] [PubMed] [Google Scholar]
- 18.Jones R.R., Hooper D.C., Zhang L., Wolverson D., Valev V.K. Raman techniques: fundamentals and frontiers. Nanoscale Res. Lett. 2019;14(1):1–34. doi: 10.1186/s11671-019-3039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verebes G.S., Melchiorre M., Garcia-Leis A., Ferreri C., Marzetti C., Torreggiani A. Hyperspectral enhanced dark field microscopy for imaging blood cells. J. Biophotonics. 2013;6(11–12):960–967. doi: 10.1002/jbio.201300067. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q., Wang J., Zhou M., Li Q., Wang Y. Spectral-spatial feature-based neural network method for acute lymphoblastic leukemia cell identification via microscopic hyperspectral imaging technology. Biomed. Opt. Express. 2017;8(6):3017–3028. doi: 10.1364/BOE.8.003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Zhou M., Qiu S., Sun L., Liu H., Li Q., Wang Y. Adaptive and automatic red blood cell counting method based on microscopic hyperspectral imaging technology. J. Opt. 2017;19(12) [Google Scholar]
- 22.Roma P.M.S., Siman L., Hissa B., Agero U., Braga E.M., Mesquita O.N. Profiling of individual human red blood cells under osmotic stress using defocusing microscopy. J. Biomed. Opt. 2016;21(9) doi: 10.1117/1.JBO.21.9.090505. [DOI] [PubMed] [Google Scholar]
- 23.Roma P., Siman L., Amaral F., Agero U., Mesquita O. Total three-dimensional imaging of phase objects using defocusing microscopy: application to red blood cells. Appl. Phys. Lett. 2014;104(25) [Google Scholar]
- 24.Chhaniwal V., Singh A.S., Leitgeb R.A., Javidi B., Anand A. Quantitative phase-contrast imaging with compact digital holographic microscope employing Lloyd's mirror. Opt. Lett. 2012;37(24):5127–5129. doi: 10.1364/OL.37.005127. [DOI] [PubMed] [Google Scholar]
- 25.Lee S., Park H., Kim K., Sohn Y., Jang S., Park Y. Refractive index tomograms and dynamic membrane fluctuations of red blood cells from patients with diabetes mellitus. Sci. Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-01036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozaki Y., Yamada H., Kikuchi H., Hirotsu A., Murakami T., Matsumoto T., Kawabata T., Hiramatsu Y., Kamiya K., Yamauchi T., et al. Label-free classification of cells based on supervised machine learning of subcellular structures. PLoS ONE. 2019;14(1) doi: 10.1371/journal.pone.0211347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G., Ahn D., Kang M., Park J., Ryu D., Jo Y., Song J., Ryu J.S., Choi G., Chung H.J., et al. Rapid species identification of pathogenic bacteria from a minute quantity exploiting three-dimensional quantitative phase imaging and artificial neural network. Light: Sci. Appl. 2022;11(1):1–12. doi: 10.1038/s41377-022-00881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu D., Kim J., Lim D., Min H.-S., Yoo I.Y., Cho D., Park Y. BME Frontiers; 2021. Label-Free White Blood Cell Classification Using Refractive Index Tomography and Deep Learning; p. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim G., Jo Y., Cho H., Min H.-s., Park Y. Learning-based screening of hematologic disorders using quantitative phase imaging of individual red blood cells. Biosens. Bioelectron. 2019;123:69–76. doi: 10.1016/j.bios.2018.09.068. [DOI] [PubMed] [Google Scholar]
- 30.Go T., Kim J.H., Byeon H., Lee S.J. Machine learning-based in-line holographic sensing of unstained malaria-infected red blood cells. J. Biophotonics. 2018;11(9) doi: 10.1002/jbio.201800101. [DOI] [PubMed] [Google Scholar]
- 31.Choi J., Kim H.-J., Sim G., Lee S., Park W.S., Park J.H., Kang H.-Y., Lee M., Do Heo W., Choo J., et al. Label-free three-dimensional analyses of live cells with deep-learning-based segmentation exploiting refractive index distributions. BioRxiv. 2021 [Google Scholar]
- 32.Lee J., Kim H., Cho H., Jo Y., Song Y., Ahn D., Lee K., Park Y., Ye S.-J. Deep-learning-based label-free segmentation of cell nuclei in time-lapse refractive index tomograms. IEEE Access. 2019;7:83449–83460. [Google Scholar]
- 33.Lee M., Lee Y.-H., Song J., Kim G., Jo Y., Min H., Kim C.H., Park Y. Deep-learning-based three-dimensional label-free tracking and analysis of immunological synapses of car-t cells. eLife. 2020;9 doi: 10.7554/eLife.49023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo Y., Cho H., Park W.S., Kim G., Ryu D., Kim Y.S., Lee M., Joo H., Jo H., Lee S., et al. Data-driven multiplexed microtomography of endogenous subcellular dynamics. BioRxiv. 2020 doi: 10.1038/s41556-021-00802-x. [DOI] [PubMed] [Google Scholar]
- 35.Ryu D., Ryu D., Baek Y., Cho H., Kim G., Kim Y.S., Lee Y., Kim Y., Ye J.C., Min H.-S., et al. Deepregularizer: rapid resolution enhancement of tomographic imaging using deep learning. IEEE Trans. Med. Imaging. 2021;40(5):1508–1518. doi: 10.1109/TMI.2021.3058373. [DOI] [PubMed] [Google Scholar]
- 36.Rivenson Y., Zhang Y., Günaydın H., Teng D., Ozcan A. Phase recovery and holographic image reconstruction using deep learning in neural networks. Light: Sci. Appl. 2018;7(2):17141. doi: 10.1038/lsa.2017.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C.L., Mahjoubfar A., Tai L.-C., Blaby I.K., Huang A., Niazi K.R., Jalali B. Deep learning in label-free cell classification. Sci. Rep. 2016;6(1):1–16. doi: 10.1038/srep21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hugonnet H., Lee M., Park Y. Optimizing illumination in three-dimensional deconvolution microscopy for accurate refractive index tomography. Opt. Express. 2021;29(5):6293–6301. doi: 10.1364/OE.412510. [DOI] [PubMed] [Google Scholar]
- 39.Tian Z., Shen C., Chen H., He T. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. Fcos: fully convolutional one-stage object detection; pp. 9627–9636. [Google Scholar]
- 40.Liang Y., Pan C., Sun W., Liu Q., Du Y. Global context-aware cervical cell detection with soft scale anchor matching. Comput. Methods Programs Biomed. 2021;204 doi: 10.1016/j.cmpb.2021.106061. [DOI] [PubMed] [Google Scholar]
- 41.Alam M.M., Islam M.T. Machine learning approach of automatic identification and counting of blood cells. Healthc. Technol. Lett. 2019;6(4):103–108. doi: 10.1049/htl.2018.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redmon J., Farhadi A. Yolov3: an incremental improvement. 2018. arXiv:1804.02767 arXiv preprint.
- 43.Chen K., Wang J., Pang J., Cao Y., Xiong Y., Li X., Sun S., Feng W., Liu Z., Xu J., Zhang Z., Cheng D., Zhu C., Cheng T., Zhao Q., Li B., Lu X., Zhu R., Wu Y., Dai J., Wang J., Shi J., Ouyang W., Loy C.C., Lin D. MMDetection: open mmlab detection toolbox and benchmark, arXiv preprint. 2019. arXiv:1906.07155
- 44.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 45.Liu Z., Lin Y., Cao Y., Hu H., Wei Y., Zhang Z., Lin S., Guo B. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021. Swin transformer: hierarchical vision transformer using shifted windows; pp. 10012–10022. [Google Scholar]
- 46.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017;30 [Google Scholar]
- 47.Barer R. Interference microscopy and mass determination. Nature. 1952;169(4296):366–367. doi: 10.1038/169366b0. [DOI] [PubMed] [Google Scholar]
- 48.Popescu G., Park Y., Lue N., Best-Popescu C., Deflores L., Dasari R.R., Feld M.S., Badizadegan K. Optical imaging of cell mass and growth dynamics. Am. J. Physiol., Cell Physiol. 2008;295(2):C538–C544. doi: 10.1152/ajpcell.00121.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhernovaya O., Sydoruk O., Tuchin V., Douplik A. The refractive index of human hemoglobin in the visible range. Phys. Med. Biol. 2011;56(13):4013. doi: 10.1088/0031-9155/56/13/017. [DOI] [PubMed] [Google Scholar]
- 50.Sung Y., Tzur A., Oh S., Choi W., Li V., Dasari R.R., Yaqoob Z., Kirschner M.W. Size homeostasis in adherent cells studied by synthetic phase microscopy. Proc. Natl. Acad. Sci. 2013;110(41):16687–16692. doi: 10.1073/pnas.1315290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mashaghi A., Swann M., Popplewell J., Textor M., Reimhult E. Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics. Anal. Chem. 2008;80(10):3666–3676. doi: 10.1021/ac800027s. [DOI] [PubMed] [Google Scholar]
- 52.Van der Maaten L., Hinton G. Visualizing data using t-sne. J. Mach. Learn. Res. 2008;9(11) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.