Abstract
The presence of low-pathogenic H7 avian influenza virus (AIV), which is associated with live-bird markets (LBM) in the Northeast United States, was first detected in 1994 and, despite efforts to eradicate the virus, surveillance of these markets has resulted in numerous isolations of H7 AIVs from several states from 1994 through 1998. The hemagglutinin, nonstructural, and matrix genes from representative H7 isolates from the LBM and elsewhere were sequenced, and the sequences were compared phylogenetically. The hemagglutinin gene of most LBM isolates examined appeared to have been the result of a single introduction of the hemagglutinin gene. Evidence for evolutionary changes were observed with three definable steps. The first isolate from 1994 had the amino acid threonine at the −2 position of the hemagglutinin cleavage site, which is the most commonly observed amino acid at this site for North American H7 AIVs. In January 1995 a new genotype with a proline at the −2 position was detected, and this genotype eventually became the predominant virus isolate. A third viral genotype, detected in November 1996, had an eight-amino-acid deletion within the putative receptor binding site. This viral genotype appeared to be the predominant isolate, although isolates with proline at the −2 position without the deletion were still observed in viruses from the last sampling date. Evidence for reassortment of multiple viral genes was evident. The combination of possible adaptive evolution of the virus and reassortment with different influenza virus genes makes it difficult to determine the risk of pathogenesis of this group of H7 AIVs.
The natural host and reservoir for influenza virus is believed to be wild waterfowl, gulls, and shorebirds (11, 20, 23). Poultry are not considered to be a normal host for the virus (5, 6, 9, 10). However, avian influenza viruses (AIVs) appear to routinely cross over from the wild-bird reservoir to poultry, including chickens, turkeys, gamebirds, domestic ducks, ratites, and other commercially raised birds. AIV in poultry may cause asymptomatic infections or a range of disease symptoms from mild respiratory disease to severe systemic infection with high mortality (6). When AIV does infect poultry, control measures are often instituted to prevent the spread of the virus because of the potential for a virulence shift causing a serious disease outbreak. Particular emphasis is placed on the H5 or H7 hemagglutinin subtypes of AIV isolates because these are the only subtypes clearly shown to cause highly pathogenic avian influenza in poultry (6). Although most isolates of H5 and H7 AIV are considered to be of low pathogenicity, the high mutation rate of AIV is thought to allow these subtypes of viruses to change to a highly pathogenic AIV with an alarming frequency (6, 8).
Currently, an ongoing outbreak of H7 AIV has been observed since 1994, primarily in the live-bird markets (LBMs) in the Northeast United States (16, 18). These LBMs, often catering to specific ethnic groups, provide a variety of live poultry, including chickens, turkeys, gamebirds, and ducks, that can be slaughtered on site or sold live to the consumer (29). As part of an ongoing state and federal surveillance program, many different subtypes of AIV have been isolated from these LBMs. Epidemiological investigations to try to determine the origins of a specific outbreak (trace-back testing) and eradication procedures initiated to control the outbreak have not kept the markets free of AIV (16, 28). For the most recent reporting period of October 1996 to September 1997, a total of 36 LBMs in three states have been found to be positive by virus isolation of H7N2 AIV (18). This can possibly be related to several factors: the infected premises are not thoroughly cleaned, trace-back testing efforts are not able to find all of the infected flocks, or multiple introductions of AIV are occurring on poultry farms that allow new outbreaks to occur (28, 29). H7 AIVs have also spread from the LBMs to larger commercial chicken and turkey operations in Pennsylvania, resulting in quarantine measures and eradication efforts in the infected flocks (28). The disease has remained of low pathogenicity based on pathogenicity testing and the observation that no additions of basic amino acids at the cleavage site of the hemagglutinin gene have occurred (18). However, the H7 viruses do appear to be contributing to increased mortality and to production losses in the large commercial flocks (31).
This study was designed to study several important questions concerning the H7 outbreak in the LBMs. First, we sought to determine whether the outbreak was the result of a single introduction of virus or multiple introductions of virus. Second, we sought to determine whether reassortment of different gene segments is occurring between the multiple hemagglutinin subtypes of AIV circulating in the LBMs. Third, we wanted to determine whether the H7 viruses are evolving in poultry in the LBMs. The HA1 segment of the hemagglutinin gene and the complete coding sequence of the nonstructural and matrix genes were sequenced from selected H7 LBM isolates and compared phylogenetically with other AIV isolates. Finally, this study provides a unique opportunity to follow the H7 outbreak over several years to see whether AIVs evolve in the poultry host and to continue to monitor these viruses for changes in pathogenicity.
MATERIALS AND METHODS
Virus.
All virus isolates sequenced for this study were obtained from the National Veterinary Services Laboratories in Ames, Iowa. Viruses were received in allantoic fluid after passage in embryonated chicken eggs. Isolates were passaged one additional time at the Southeast Poultry Research Laboratory to make working stocks of the virus. The isolates and accession numbers for the sequences used here are given in Table 1.
TABLE 1.
AIV examined in this study
| Isolate | Subtype | Date isolated (mo-yr or yr) | Cleavage site sequence | Deletion in HA1a | GenBank accession nos. (hemagglutinin, NS, and M genes) |
|---|---|---|---|---|---|
| NA H7 LBM | |||||
| CK/NJ/15086-3/94 | H7N3NSA | Feb-94 | PENPKTR/G | No | AF072383, AF074267, AF073180 |
| CK/NY/13142-5/94 | H7N2NSB | Feb-94 | PENPKTR/G | No | AF072384, AF001409, AF073181 |
| CK/NY/4447-7/94 | H7N2NSA | Oct-94 | PENPKTR/G | No | AF072385, AF074268, AF073182 |
| TK/NY/4450-5/94 | H7N2NSB | Oct-94 | PENPKTR/G | No | AF072386, AF074269, AF073183 |
| GF/NY/13820-3/95 | H7N2NSB | Jan-95 | PENPKTR/G | No | AF072387, AF074270, AF073184 |
| CK/NY/13833-7/95 | H7N2NSB | Jan-95 | PENPKPR/G | No | AF072388, AF074271, AF073185 |
| CK/NY/19542-5/95 | H7N2NSB | Mar-95 | PENPKTR/G | No | AF072389, AF074272, AF073186 |
| CK/NY/3112-1/95 | H7N2NSB | Oct-95 | PENPKPR/G | No | AF072390, AF074273, AF073187 |
| CK/RI/4328/95 | H7N2NSB | Nov-95 | PENPKPR/G | No | AF072391, AF074274, AF073188 |
| CK/NY/3202-7/96 | H7N2NSB | Nov-96 | PENPKPR/G | Yes | AF072392, AF001410, AF073189 |
| CK/NY/8030-2/96 | H7N2NSB | Dec-96 | PENPKPR/G | No | AF072393, AF074275, AF073190 |
| GF/PA/7777-1/96 | H7N2NSB | Dec-96 | PENPKPR/G | Yes | AF072394, AF074276, AF073191 |
| CK/PA/11767-1/97 | H7N2NSB | Feb-97 | PENPKPR/G | Yes | AF072395, AF074277, AF073192 |
| CK/NY/6777-3/97 | H7N2NSB | Dec-97 | PENPKPR/G | Yes | AF072396, AF074278, AF073193 |
| TK/PA/7975/97 | H7N2NSB | Dec-97 | PENPKPR/G | Yes | AF072397, AF074279, AF073194 |
| CK/PA/13552-1/98 | H7N2NSB | Feb-98 | PENPKPR/G | Yes | AF072398, AF074280, AF073195 |
| Quail/NY/13989-51/98 | H7N2NSA | Feb-98 | PENPKPR/G | No | AF072399, AF074281, AF073196 |
| NA H7 non-LBM | |||||
| TK/OR/71 | H7N3NSB | 1971 | PENPKTR/G | No | M31689b, U96740, AF073197 |
| Seal/MA/1/80 | H7N7 | 1980 | PENPKTR/G | No | K00429b |
| TK/CO/13356/91 | H7N3NSA | Feb-91 | PENPKTR/G | No | AF072400, AF074282, AF073198 |
| Rhea/NC/39482/93 | H7N1NSA | Jul-93 | PENPKTR/G | No | U20468b, AF007036, AF073199 |
| Quail/AR/16309-7/94 | H7N3NSA | 1994 | PENPKTR/G | No | AF072401, AF074283, AF073200 |
| TK/UT/24721-10/95 | H7N3NSA | Apr-95 | PENPKTR/G | No | AF072402, AF074284, AF073201 |
| Eurasian H7 isolates | |||||
| TK/England/63c | H7N3 | 1963 | PETPKRRR/G | No | U20462b, None, None |
| CK/Jena/1816/87 | H7N7 | 1987 | PEIPKGR/G | No | U20469b, None, None |
| FPV/Rostock/34 | H7N1NSA | 1934 | PEPSKKREKR/G | No | J02111b, J02118b, X05905b |
| NA H5d | |||||
| Mallard/WI/169/75 | H5N3NSA | 1975 | PQRETR/G | No | U85375b, AF073202 |
| CK/PA/1370/83c | H5N2NSA | 1983 | PQKKKR/G | No | U96739b, X13413b |
| CK/PA/13609/93 | H5N2NSB | 1993 | PQRKTR/G | No | U85383b, AF073203 |
| Emu/TX/39924/93 | H5N2NSA | 1993 | PQRKTR/G | No | U85384b, AF073204 |
| CK/Mexico/31382-7/94 | H5N2NSB | 1994 | PQRETR/G | No | U85385b, AF073205 |
Presence of a 24-nucleotide deletion in the HA1 segment of the H7 isolate.
Sequence obtained from GenBank.
Highly pathogenic AIV.
For the NA H5 isolates, only the GenBank accession numbers for the NS and M genes are given.
Molecular cloning and sequencing of influenza virus genes.
RNA from the isolates was extracted with Purescript RNA extraction kit (Gentra, Minneapolis, Minn.) from infected allantoic fluid prior to reverse transcriptase PCR (RT-PCR) amplification. For the nonstructural (NS) and matrix (M) gene segments, RNA was reverse transcribed by using Superscript II (Life Technologies) RT enzyme with incubation at 42°C for 1 h. PCR was performed at a 51°C annealing temperature for 31 cycles. Primers were to the conserved 12 and 13 bp present on the 5′ and 3′ ends of each viral segment. The PCR product was electrophoresed in an agarose gel, and the DNA, corresponding in size to the gene segment of interest, was extracted with the Agarose Gel DNA extraction kit (Boehringer Mannheim, Indianapolis, Ind.). The DNA was cloned into the pAmp1 (Life Technologies) plasmid vector by using a ligation-independent cloning system. Colonies were screened by PCR with internal primers, positive cultures were grown overnight, and plasmid was extracted by using the High Pure Plasmid Isolation Kit (Boehringer Mannheim). Plasmids were sequenced by using the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer, Foster City, Calif.) run on a 373A automated sequencer (Perkin-Elmer). The H7 gene was also similarly amplified by RT-PCR, but the PCR product was directly sequenced. All primer sequences are available upon request.
Nucleotide and amino acid sequence phylogenetic analysis.
Assembly of sequencing contigs, translation of the nucleotide sequence into the protein sequence, and initial multiple sequence alignments were performed with the Lasergene (DNASTAR, Madison, Wis.) group of programs. Phylogenetic trees for each gene were generated by using the maximum parsimony method with 100 bootstrap replicates in a heuristic search with the PAUP 3.1 software program (25). Midpoint rooting was used for all trees.
RESULTS
The HA1 coding sequence and the complete coding sequence for the NS and M gene segments from 17 H7 AIV isolates from the Northeast United States were sequenced. The HA1 sequence data were compared to an additional six North American (NA) H7 isolates and three Eurasian (EA) H7 isolates, and the NS and M data were compared to five NA H7, one EA H7, and four NA H5 isolates.
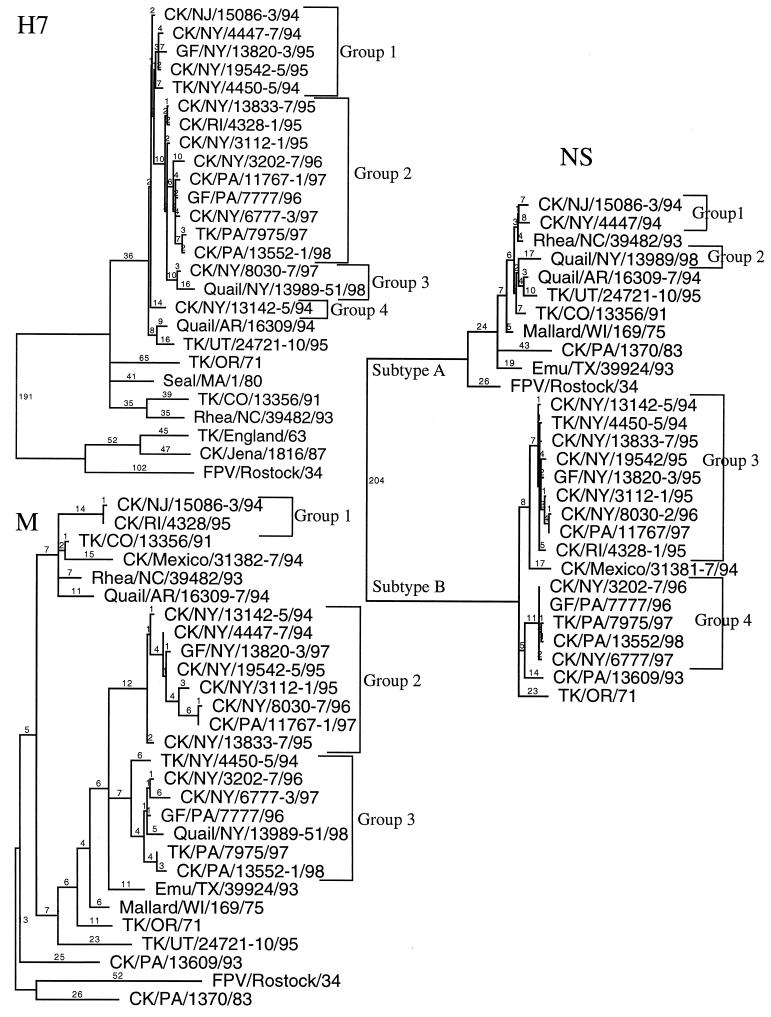
Phylogenetic trees developed by using parsimony were created for each gene based on nucleotide sequence data. The HA1 coding sequence had 981 nucleotides for most of the isolates examined. One exception was the non-LBM H7 isolate A/Quail/Arkansas/16309-7/94, which had an additional amino acid, serine (Fig. 1). Also several EA AIVs had additional basic amino acids near the hemagglutinin cleavage site, an important factor in these isolates being highly pathogenic. A previously unrecognized deletion of 24 nucleotides, eight amino acids, was present in the HA1 gene of six of the most recent LBM isolates (Fig. 1). The HA1 phylogenetic tree demonstrated a close relationship between all H7 LBM isolates and several other NA H7 AIV isolates, but four distinct clusters of H7 LBM strains were observed: five isolates were in group 1, nine in group 2, two in group 3, and one in group 4 (Fig. 2, H7). A group was defined when at least 15 nucleotides were different between clusters of isolates. Groups 3 and 4 had only a few members, and it is unclear whether these isolates represent separate introductions of H7 virus or are just outliers from the remaining isolates. The HA1 coding sequence of 14 isolates from groups 1 and 2 had two unusual changes that became the predominant H7 isolates circulating in the LBMs (Table 2). The first H7 AIVs isolated in 1994 and 1995 had a threonine at the −2 position of the HA1-HA2 cleavage site. In January 1995 the first isolate with a proline at the −2 position was observed, and all the H7 isolates obtained after March 1995 had the proline at this position. In November 1996, the first isolate with a deletion of 24 nucleotides, eight amino acids, was observed. Isolates with this deletion became predominant, with six of eight isolates sequenced from November 1996 to February 1998 having the deletion. This deletion was not observed in any other H7 isolates examined.
FIG. 1.
Amino acid sequence comparison of the HA1 coding region of three North American H7 isolates selected to display unique structural features of the HA1 coding region. The amino acids which have been shown by molecular modeling to correspond to the receptor binding site identified in the H3 structure are in boldface and are underlined. The serine insertion in the quail isolate between positions 136 and 137 is indicated by the dash at that position in the two 1996 isolates. The amino acid numbering system is based upon the consensus H7 amino acid sequence. The cleavage site separating the HA1 and HA2 is indicated by the gap.
FIG. 2.
Phylogenetic trees from the H7 hemagglutinin (H7) subtype, NS subtypes (groups A and B), and the M gene from H7 isolates from the LBMs in the Northeast United States and other representative AIV isolates are presented. All trees were generated with PAUP 3.1 computer program, are the result of 100 bootstrap replicates, and are midpoint rooted. Branch lengths are included on each tree. The H7 LBM isolates are grouped (1 to 4 for the H7 and NS trees and 1 to 3 for the M tree) according to their associations on the tree. Abbreviations: CK, chicken; TK, turkey; GF, guinea fowl; and FPV, Fowl plague virus. Standard two-letter abbreviations are used for states in the United States.
TABLE 2.
Sequence characteristics and comparisons of H7 AIVs from the LBM Isolates in the Northeast United States
| H7 LBM isolate | Neuramini-dase and NS subtypes | Date of isolation (mo-yr) | Cleavage site aa at −2 positiona | Deletion in HA1 geneb | Numerical codec for:
|
No. of nucleotide changesd | Discriminat-ing sitese | ||
|---|---|---|---|---|---|---|---|---|---|
| H | NS | M | |||||||
| CK/NJ/15086-3/94 | N3NSA | Feb-94 | T | No | 1 | 1 | 1 | 0 | 0 |
| CK/NY/13142-5/94 | N2NSB | Feb-94 | T | No | 4 | 3 | 2 | 15 | 0 |
| CK/NY/4447-7/94 | N2NSA | Oct-94 | T | No | 1 | 1 | 2 | 11 | 0 |
| TK/NY/4450-5/94 | N2NSB | Oct-94 | T | No | 1 | 3 | 3 | 11 | 0 |
| GF/NY/13820-3/95 | N2NSB | Jan-95 | T | No | 1 | 3 | 2 | 14 | 0 |
| CK/NY/13833-7/95 | N2NSB | Jan-95 | P | No | 2 | 3 | 2 | 16 | 10 |
| CK/NY/19542-5/95 | N2NSB | Mar-95 | T | No | 1 | 3 | 2 | 9 | 0 |
| CK/NY/3112-1/95 | N2NSB | Oct-95 | P | No | 2 | 3 | 2 | 17 | 12 |
| CK/RI/4328/95 | N2NSB | Nov-95 | P | No | 2 | 3 | 1 | 17 | 11 |
| CK/NY/3202-7/96 | N2NSB | Nov-96 | P | Yes | 2 | 4 | 3 | 31 | 16 |
| GF/PA/7777-1/96 | N2NSB | Dec-96 | P | Yes | 2 | 4 | 3 | 25 | 20 |
| CK/NY/8030-2/96 | N2NSB | Dec-96 | P | No | 3 | 3 | 2 | 28 | 12 |
| CK/PA/11767-1/97 | N2NSB | Feb-97 | P | Yes | 2 | 3 | 2 | 27 | 20 |
| CK/NY/6777-3/97 | N2NSB | Dec-97 | P | Yes | 2 | 4 | 3 | 27 | 20 |
| TK/PA/7975/97 | N2NSB | Dec-97 | P | Yes | 2 | 4 | 3 | 33 | 20 |
| CK/PA/13552-1/98 | N2NSB | Feb-98 | P | Yes | 2 | 4 | 3 | 32 | 20 |
| Quail/NY/13989-51/98 | N2NSA | Feb-98 | P | No | 3 | 2 | 3 | 41 | 12 |
Amino acid (aa) present in the −2 position of the hemagglutinin cleavage site.
Presence of a 24-nucleotide deletion in the HA1 segment of the H7 isolate.
Numerical codes for hemagglutinin (H), NS, and M phylogenetic groups as determined in Fig. 2.
Number of nucleotide changes in HA1 gene from index virus A/CK/NJ/15086-3/94.
Number of nucleotide changes in HA1 gene at 20 discriminative sites from the index virus A/CK/NJ/15086-3/94.
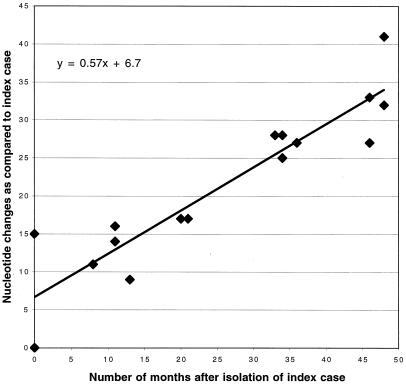
The 14 isolates from groups 1 and 2 are likely from the same lineage, and the nucleotide differences between them appear to be the result of mutations accumulating over time. The HA1 sequences were also compared at 20 phylogenetically discriminative nucleotide sites spread throughout the gene (Table 3). Discriminative changes are nucleotide differences that allow a phylogenetic tree to be determined and can be nucleotide changes shared by as few as two viral isolates. However, the 20 selected nucleotide sites in Table 3 include nucleotide differences that were observed between groups with more than two isolates per group and nucleotide changes that primarily varied according to the date of virus isolation. These 20 changes appear to be from a progressive accumulation of point mutations from CK/NJ/15086-3/94 to CK/PA/13552-1/98. These data, along with the total number of nucleotide changes with CK/NJ/15086-3/94 as the isolate closest to the progenitor virus, are presented in Table 2. Both the total number of nucleotide changes and the changes at the 20 discriminating sites increased over time, suggesting a single lineage of virus. The total number of nucleotide changes was also plotted against the date of isolation to help calculate an observed mutation rate for this gene (Fig. 3).
TABLE 3.
Nucleotide sequence at discriminative sites of the HA1 protein
| H7 LBM isolate | Nucleotide sequence at discriminative site:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 84a | 126 | 218b | 258 | 270 | 310 | 326 | 342 | 389 | 468 | 530 | 543 | 586 | 732 | 765 | 876 | 894 | 900 | 942 | 958 | |
| CK/NJ/15086-3/94 | T | C | A | G | A | G | A | T | G | C | G | G | G | T | G | A | A | C | C | A |
| CK/NY/13142-5/94 | T | C | A | G | A | G | A | T | G | C | G | G | G | T | G | A | A | C | C | A |
| CK/NY/4447-7/94 | T | C | A | G | A | G | A | T | G | C | G | G | G | T | G | A | A | C | C | A |
| TK/NY/4450-5/94 | T | C | A | G | A | G | A | T | G | C | T | G | G | T | G | A | A | C | C | A |
| GF/NY/13820-3/95 | T | C | A | G | A | G | A | T | G | C | G | G | G | T | G | A | A | C | C | A |
| CK/NY/13833-7/95 | C | T | A | G | T | G | A | T | A | A | G | G | G | C | A | A | G | C | T | C |
| CK/NY/19542-5/95 | T | C | A | G | A | G | A | T | G | C | G | G | G | T | G | A | A | C | C | A |
| CK/NY/3112-1/95 | C | T | C | G | T | G | A | T | A | A | G | G | G | C | A | T | G | C | T | C |
| CK/RI/4328/95 | C | T | A | G | T | G | A | T | A | A | G | G | G | C | A | T | G | C | T | C |
| CK/NY/3202-7/96 | T | A | C | A | T | A | G | C | A | A | A | G | A | C | A | T | G | C | T | C |
| GF/PA/7777-1/96 | C | T | C | A | T | A | G | C | A | A | A | A | A | C | A | T | G | T | T | C |
| CK/NY/8030-2/96 | C | T | C | G | T | G | A | T | A | A | T | G | G | C | A | T | G | C | T | C |
| CK/PA/11767-1/97 | C | T | C | A | T | A | G | C | A | A | A | A | A | C | A | T | G | T | T | C |
| CK/NY/6777-3/97 | C | T | C | A | T | A | G | C | A | A | A | A | A | C | A | T | G | T | T | C |
| TK/PA/7975/97 | C | T | C | A | T | A | G | C | A | A | A | A | A | C | A | T | G | T | T | C |
| CK/PA/13552-1/98 | C | T | C | A | T | A | G | C | A | A | A | A | A | C | A | T | G | T | T | C |
| Quail/NY/13989-51/98 | C | T | C | G | T | G | A | T | A | A | T | G | G | C | A | T | G | C | T | C |
Nucleotide position based on the HA1 nucleotide sequence of consensus H7 sequence.
Nucleotide sequence positions in boldface represent nonsynonymous nucleotide changes.
FIG. 3.
Comparison of the evolutionary rate of the HA1 segment of the hemagglutinin gene from H7 AIV isolates from the LBMs of the Northeastern United States. The number of nucleotide changes from the earliest H7 isolate, A/CK/NJ/15086-3/94, are given on the y axis, and the number of months after each virus was isolated compared to the index case is given on the x axis. Linear regression analysis was performed to determine the evolutionary rate, and the equation for the line is indicated.
The M and NS genes were also sequenced and compared phylogenetically. By using 15 nucleotide differences as a dividing point between groups of isolates, the NS gene had four defined groups and the M gene had three defined groups (Fig. 2). To facilitate the comparison of isolates, the numbers of the defined groups for the HA1, NS, and M genes were tabulated to give a code for each virus (Table 2). For the 17 isolates examined, 10 unique codes were assigned to the different isolates. Five isolates with the 2-4-3 code had the proline at the −2 position and the deletion in the HA1 gene. Otherwise no code had more than two members.
DISCUSSION
An important question that needed to be addressed in this study was whether infections in poultry in the LBMs in the Northeast United States were the result of a single introduction or multiple introductions of H7 virus. The data clearly suggest that 14 of the 17 isolates had hemagglutinin sequences consistent with a single introduction of the hemagglutinin gene. Three other isolates had a large number of nucleotide changes that made it unclear if they were outliers to the other isolates or were unique introductions of virus. Examination of the 20 discriminating mutations would suggest they were outliers from the main group and not unique introductions of the virus because they had no unique nucleotide differences at these 20 sites.
A second question was whether these H7 viruses were reassorting with other subtypes of AIV present in the LBMs. Neuraminidase subtyping of H7 isolates had already documented the presence of N2 and N3 isolates, and both group A and group B NS subtypes were present (Table 1). The phylogenetic data from both the NS and M genes strongly suggest that reassortment is occurring with regularity among H7 LBM isolates. This contrasts with the extended H5 outbreak in Mexico that occurred from 1993 to the present in which no evidence of reassortment was observed when the HA and NS genes from representative isolates were examined (9). The difference in the Mexican H5 outbreak and the LBM H7 outbreak is that no other hemagglutinin subtypes of AIV were circulating in Mexico but many subtypes of AIV are known to be circulating in the LBMs in the Northeast United States (18). Because of the pool of influenza genes in the LBMs and the apparent routine reassortment between viruses, H7 AIV isolates have many unique constellations of viral genes. The diversity in the constellation of genes for each isolate makes it difficult to predict which isolates may present a greater risk for becoming highly pathogenic. The pathogenicity of AIV is known to be controlled by multiple genes, with the hemagglutinin gene being the most important factor known (21, 26, 27). Only the H5 and H7 hemagglutinin subtypes are currently associated with the highly pathogenic phenotype of AIV, but differences in pathogenicity can also be observed with viruses classified as having low pathogenicity. The virulence shift from a low-pathogenicity to a highly pathogenic AIV can occur quickly by the accumulation of basic amino acids at the hemagglutinin cleavage site either by insertion or substitution events or by glycosylation changes that allow the cleavage site to be more accessible to endogenous proteases (3, 13). Because of the high mutation rate inherent in influenza viruses, increased vigilance in the surveillance of H5 and H7 viruses is warranted.
A third important question to be addressed was whether the H7 lineage of viruses is evolving in the LBM poultry or whether it remains evolutionarily static as has been previously proposed for AIVs (12, 17). The data strongly suggest that nonsynonymous mutations and even deletions are becoming fixed in the circulating population and that certain genotypes of virus are being lost. It is not clear whether these changes are completely the result of the viruses becoming better adapted to the poultry host or whether past efforts to eradicate the virus may have inadvertently selected for one genotype of the virus over another. The earliest isolates from the LBMs had a threonine at the −2 position of the cleavage site, a finding which is consistent with what has been observed in cleavage site sequencing of 18 other wild-bird and poultry H7 AIV isolates of North American origin (19). To examine the mutation rates for the H7 LBM isolates, the isolates from 1994 were compared to determine which one had the nucleotide sequence relationship closest to all the other H7 LBM isolates. This isolate would be assumed to be the virus closest to the progenitor virus and would be used as a point of comparison for pairwise analysis of the other isolates in the study. A/CK/NJ/13142-5/94, which was one of the first H7 viruses isolated from the LBM system, was selected as the index case and served as a reference point for determining the mutation rate and for quantifying nucleotide changes in the hemagglutinin gene. The mutation rate, estimated by using linear regression analysis, for the H7 LBM isolates was 7.0 × 10−3 nucleotide substitutions per site per year (Fig. 3). This mutation rate is similar to mutation rates previously described for H3 human influenza virus isolates (6.7 × 10−3 nucleotide substitutions per site per year) (7) and is higher than that reported for H3 equine influenza virus isolates (3.1 × 10−3 nucleotide substitutions per site per year) (4). However, the estimated H7 AIV mutation rate is less than that previously described for the Mexican H5 outbreak (9). The Mexican H5 outbreak and the Northeast LBM H7 outbreak provide the only two examples of an avian influenza outbreak that has been repeatedly sampled over several years. Both outbreaks show that AIVs adapt to their new poultry hosts, and both have a measurable rate of evolutionary change. The y-intercept value of 6.7 suggests that the isolate A/CK/NJ/13142-5/94 was probably not the earliest isolate in this outbreak, and that H7 viruses may have been circulating in the LBMs for over a year before the first H7 isolation of AIV.
Two additional H7 isolates, including a 1994 Arkansas quail isolate and a 1995 Utah turkey isolate, were sequenced to determine whether these isolates were related to the LBM outbreaks (14, 16). The HA1 genes of both isolates were more closely related to the LBM H7 genes than to the other H7 HA1 genes in this study, but the isolates were different enough so that it appeared that both outbreaks were the result of separate introductions of the virus. Also, both isolates had threonine at the −2 position of the cleavage site. Multiple sublineages of H7 HA1 genes appear to be present in North America as has been described for the H5 HA1 gene (9).
Circulating AIVs, in addition to the risk of increased pathogenesis, also have the potential of crossing species barriers to infect humans or other mammals. This perceived risk of AIVs being spread from poultry to humans has greatly increased after the recent crossover of an H5 AIV in Hong Kong in 1997 (24). The H7 hemagglutinin subtype has also previously been implicated in two different incidences of crossover to humans, although in both cases only mild disease was observed (1, 30). It is not known whether the H7 hemagglutinin protein predisposes the virus to easily cross species barriers. As previously indicated, the rampant reassortment of AIVs in the LBMs could increase the risk of species crossover because it would increase the chances of the occurrence of the correct constellation of genes to create a virus that replicates efficiently in mammals (2, 15, 22).
The H7 LBM outbreak in the Northeast United States has provided a unique opportunity to observe how an AIV can infect several different kinds of poultry and presumably make adaptive changes to the new host. The evolution of the virus also seems to favor certain lineages of virus, and other lineages appear to be lost. However, this experiment in evolution does not come without risk. The main concern is that the H7 viruses may have mutations that allow them to become highly pathogenic, but we must also be concerned with the public health implications of a new hemagglutinin subtype crossing over to humans, causing a new pandemic of avian influenza. Continued efforts to eradicate the viruses from the LBMs and associated production facilities would be the most prudent control measure.
ACKNOWLEDGMENTS
We thank Suzanne DeBlois and Joan Beck for technical support.
This work was supported by USDA/ARS Cris Project number 6612-32000-016.
REFERENCES
- 1.Banks J, Speidel E, Alexander D J. Characterization of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. doi: 10.1007/s007050050329. [DOI] [PubMed] [Google Scholar]
- 2.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 3.Bosch F X, Garten W, Klenk H D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 4.Daly J M, Lai A C, Binns M M, Chambers T M, Barrandeguy M, Mumford J A. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J Gen Virol. 1996;77:661–671. doi: 10.1099/0022-1317-77-4-661. [DOI] [PubMed] [Google Scholar]
- 5.Davidson W R, Yoder H W, Brugh M, Nettles V F. Serological monitoring of Eastern Wild Turkeys for antibodies to Mycoplasma spp. and avian influenza viruses. J Wildl Dis. 1988;24:348–351. doi: 10.7589/0090-3558-24.2.348. [DOI] [PubMed] [Google Scholar]
- 6.Easterday B C, Hinshaw V S, Halvorson D A. Influenza. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. Ames, Iowa: Iowa State University Press; 1997. pp. 583–605. [Google Scholar]
- 7.Fitch W M. The variety of human virus evolution. Mol Phylogenet Evol. 1996;5:247–258. doi: 10.1006/mpev.1996.0018. [DOI] [PubMed] [Google Scholar]
- 8.Garcia M, Crawford J M, Latimer J W, Rivera-Cruz M V Z E, Perdue M L. Heterogeneity in the hemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J Gen Virol. 1996;77:1493–1504. doi: 10.1099/0022-1317-77-7-1493. [DOI] [PubMed] [Google Scholar]
- 9.Garcia M, Suarez D L, Crawford J M, Latimer J W, Slemons R D, Swayne D E, Perdue M L. Evolution of H5 subtype avian influenza A viruses in North America. Virus Res. 1997;51:115–124. doi: 10.1016/s0168-1702(97)00087-7. [DOI] [PubMed] [Google Scholar]
- 10.Hopkins B A, Skeeles J K, Houghten G E, Slagle D, Gardner K. A survey of infectious diseases in wild turkeys (Meleagridis gallopavo silvestris) from Arkansas. J Wildl Dis. 1990;26:468–472. doi: 10.7589/0090-3558-26.4.468. [DOI] [PubMed] [Google Scholar]
- 11.Kawaoka Y, Chambers T M, Sladen W L, Webster R G. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247–250. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 12.Kawaoka Y, Gorman O T, Ito T, Wells K, Donis R O, Castrucci M R, Donatelli I, Webster R G. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res. 1998;55:143–156. doi: 10.1016/s0168-1702(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 13.Kawaoka Y, Naeve C W, Webster R G. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology. 1984;139:303–316. doi: 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 14.Kleven S H. Proceedings of the U.S. Animal Health Association 99th Annual Meeting. U.S. Richmond, Va: Animal Health Association; 1995. Report of the committee on transmissible diseases of poultry and other avian diseases; pp. 550–588. [Google Scholar]
- 15.Murphy B R, Hinshaw V S, Sly D L, London W T, Hosier N T, Wood F T, Webster R G, Chanock R M. Virulence of avian influenza A viruses for squirrel monkeys. Infect Immun. 1982;37:1119–1126. doi: 10.1128/iai.37.3.1119-1126.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomeroy B S E. Proceedings of U.S. Animal Health Association 98th Annual Meeting. U.S. Richmond, Va: Animal Health Association; 1994. Avian influenza; pp. 512–523. [Google Scholar]
- 17.Scholtissek C. Molecular evolution of influenza viruses. Virus Genes. 1995;11:209–215. doi: 10.1007/BF01728660. [DOI] [PubMed] [Google Scholar]
- 18.Senne D. Proceedings of the U.S. Animal Health Association 101st Annual Meeting. U.S. Richmond, Va: Animal Health Association; 1997. NVSL report on avian influenza; pp. 497–498. [Google Scholar]
- 19.Senne D A, Panigrahy B, Kawaoka Y, Pearson J E, Suss J, Lipkind M, Kida H, Webster R G. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425–437. [PubMed] [Google Scholar]
- 20.Slemons R D, Johnson D C, Osborn J S, Hayes F. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 1974;18:119–124. [PubMed] [Google Scholar]
- 21.Snyder M H, Buckler-White A J, London W T, Tierney E L, Murphy B R. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987;61:2857–2863. doi: 10.1128/jvi.61.9.2857-2863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder M H, Clements M L, Herrington D, London W T, Tierney E L, Murphy B R. Comparison by studies in squirrel monkeys, chimpanzees, and adult humans of avian-human influenza A virus reassortants derived from different avian influenza virus donors. J Clin Microbiol. 1986;24:467–469. doi: 10.1128/jcm.24.3.467-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stallknecht D E. Ecology and epidemiology of avian influenza viruses in wild bird populations: waterfowl, shorebirds, pelicans, cormorants, etc. In: Swayne D E, Slemons R D, editors. Proceedings of the Fourth International Symposium on Avian Influenza. U.S. Richmond, Va: Animal Health Association; 1998. pp. 61–69. [Google Scholar]
- 24.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford D. Phylogenetic analysis using parsimony, version 3. Champaign, Ill: Illinois Natural History Survey; 1997. [Google Scholar]
- 26.Tian S F, Buckler White A J, London W T, Reck L J, Chanock R M, Murphy B R. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/NY/78 virus and its reassortants in squirrel monkey respiratory tract. J Virol. 1985;53:771–775. doi: 10.1128/jvi.53.3.771-775.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treanor J J, Snyder M H, London W T, Murphy B R. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology. 1989;171:1–9. doi: 10.1016/0042-6822(89)90504-7. [DOI] [PubMed] [Google Scholar]
- 28.Trock S, Eckroade R J, Davison S, Ziegler A F. Proceedings of the U.S. Animal Health Association 101st Annual Meeting. U.S. Richmond, Va: Animal Health Association; 1997. Chronology of events on the recent AI outbreaks in Pennsylvania; pp. 510–513. [Google Scholar]
- 29.Trock S C. Epidemiology of influenza in live bird markets and ratite farms. In: Swayne D E, Slemons R D, editors. Proceedings of the Fourth International Symposium on Avian Influenza. U.S. Richmond, Va: Animal Health Association; 1998. pp. 77–79. [Google Scholar]
- 30.Webster R G, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med. 1981;304:911. doi: 10.1056/NEJM198104093041515. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.Ziegler A F, Eckroade R J, Davison S. Proceedings of the 47th Western Poultry Disease Conference. Conference and Event Services, University of California—Davis, Davis, Calif. 1998. Nonpathogenic H7N2 avian influenza in Pennsylvania in 1997; pp. 68–69. [Google Scholar]