Highlights
-
•
LINC00482 participated in the development of prostate cancer with bone metastasis.
-
•
LINC00482 acted as a competing endogenous RNA by sponging miR-2467-3p in prostate cancer.
-
•
LINC00482 activate Wnt/β-catenin signaling pathway by sponging miR-2467-3p.
Keywords: LINC00482, miR-2467-3p, Wnt/β-catenin, Prostate cancer, Bone metastasis
Abstract
This study was designed to investigate the biological functions of LINC00482 in prostate cancer (PCa) with bone metastasis. TCGA dataset of PCa was applied for LINC00482 expression analysis and real time PCR was used to verify the expression level of LINC00482 in PCa tissues as well as PCa bone metastatic tissues. To detect the biological functions of LINC00482 in vitro, various assays were used including CCK-8, EdU, colony formation and transwell assays. The biological functions of LINC00482 were also identified in vivo by inoculating PCa cells into the left cardiac ventricle of mice, followed by evaluating the osteolytic lesions and osteolytic score. In addition, Starbase and Lncbase databases were applied for predicting the potential target miRNA of LINC00482, while TargetScan and Starbase databases were used for predicting the potential target of miRNA. The luciferase reporter assay was utilized to determine the interactions among these molecules and western blotting was employed to verified the targeted proteins. Results showed that high expression level of LINC00482 was observed in bone metastatic PCa tissues and associated with PCa progression. Silencing of LINC00482 inhibited cell proliferation, migration and invasion in PCa. Furthermore, LINC00482 was proved to act as a competing endogenous RNA by sponging miR-2467-3p to activate Wnt/β-catenin signaling pathway, which may be a promising therapeutic target for PCa with bone metastasis.
1. Introduction
Prostate cancer (PCa) is the most common non-skin malignancy for men and remains the second leading cause of cancer-related deaths.[1], [2], [3] Although there are some improvements in early screening strategies and development of clinical therapies in PCa, an increasing rate of aggressive PCa has been observed and bone metastasis is the leading cause of PCa deaths.[4], [5], [6] The development of bone metastasis contains multiple steps, including colonization, dormancy, reactivation and development, reconstruction.[7] To better develop effective therapies, it’s urgently needed to obtaining a comprehensive understanding of the mechanism in prostate cancer with bone metastasis.
Long non-coding RNAs (lncRNAs), defined as non-protein-coding RNAs longer than 200 nucleotides, has played important roles not only on the regulation between protein and DNA but also cellular functions. The dysregulation of lncRNAs may induce many human diseases, especially cancers.[8], [9] Abundance evidences demonstrated that lncRNAs could participate in the network of competing endogenous RNA (ceRNA) by sponging microRNAs (miRNA) to modulate target genes expression, resulting in regulation on cellular functions, including cell proliferation, differentiation, apoptosis, cell cycle and cell invasion.[10], [11], [12], [13], [14] In PCa, several lncRNAs has contributed to tumor progression as ceRNAs. For example, lncRNA AC245100.4 has been proved to enhance PCa tumorigenesis through miRNA-145-5p/RBBP5 axis.[15] Additionally, lncRNA TUG1 was able to promote PCa progression via miRNA-128-3p/YES1 axis.[16] Accordingly, other lcnRNA may also be involved in PCa tumorigenesis acting as ceRNA. Downregulation of LINC00482 in bladder cancer has been proved to suppress tumor-associated inflammation as well as angiogenesis by downregulating MMP-15 via FOXA1.[17] However, the role of LINC00482 was still unclear in PCa.
The Wnt/β-catenin signaling pathway has been involved in many crucial cellular functions including stem cell renewal, organ formation and cell survival.[18] Activation of Wnt has been occurred in lung, breast and hematopoietic malignancies and resulting in tumor recurrence.[19] Interestingly, it’s reported that lncRNA SNHG1 could regulate cell proliferation, autophagy and apoptosis in PCa cells by modulating the PI3K/AKT/mTOR and Wnt/β-catenin signaling pathways.[20] Accordingly, we hypothesized that LINC00482 may regulate PCa progression via regulating Wnt/β-catenin signaling pathway.
Collectively, in this study, high LINC00482 expression has been identified as a characteristic molecular change in PCa and the biological role of LINC00482 on PCa progression has been investigated. Furthermore, LINC00482 has been found to promote bone metastasis of PCa through activating Wnt/β-catenin signaling pathway by sponging miR-2467-3p. The LINC00482/miR-2467-3p/Wnt/β-catenin axis might be a potential therapeutic target for bone metastatic PCa.
2. Methods
2.1. Patients and tumor tissues
A total of 79 archived PCa tissues, including 43 non-bone metastatic and 36 bone metastatic PCa tissues as well as 30 paired adjacent normal tissues (ANT) and PCa tissues were obtained from Zhejiang Provincial People’s Hospital. All subjects have written informed consent and approval from the Zhejiang Provincial People’s Hospital were obtained. The expression level of LINC00482 and clinical profile of PCa dataset were obtained from The Cancer Genome Atlas (TCGA).
2.2. Cell culture
The human primary PCa cell (22RV1), lymph node metastatic cell (LNCaP), bone metastatic PCa cell (C4-2B, PC-3 and VCaP), brain metastatic cell (DU145) and normal prostate epithelial cell (RWPE-1) were obtained from the Shanghai Chinese Academy of Sciences Cell Bank (China). RWPE-1 cells were cultured in defined keratinocyte-SFM (Invitrogen, US), while 22RV1, PC-3 and LNCaP were grown in RPMI-1640 medium (Life Technologies, US) with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS, Life Technologies, US). VCaP and DU145 were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, US) with 10% FBS. C4-2B cells were cultured in T-medium (Invitrogen, US) with 10% FBS. All cell lines were incubated at a humidified atmosphere of 5% CO2 at 37 °C.
2.3. RNA interference and vectors
The two shRNA plasmids targeting LINC00482 and corresponding negative control plasmid were purchased from Genechem (Shanghai, China) and cloned into the pLKO.1-puro lentivirus vector (Addgene, Watertown, MA, USA). The RNA interference sequences were listed as follows: shControl, CCGG TGGTTTACATGTTGTGTGAAGCTCGAGCTTCACACAACATGTAAACCATTTTTG; LINC00482 shRNA#1, CCGGGCAGACAGGCCAGGAAGAGCGCTCGAGCGCTCTTCCTG; GCCTGTCTGCTTTTTG, LINC00482 shRNA#2 (CCGGGCCAGGAAGAGCGCCTGCAAGCTCGAGCTTGCAGGCGCTCTTCCTGGCTTTTTG). The PCa cells in the control group were transfected with the corresponding negative control plasmid. Stable cell lines transfected with two shRNAs and corresponding negative control plasmids were selected by using puromycin (Solarbio, China). Transfection of miRNA and plasmids was conducted using Lipofectamine 3000 (Invitrogen, US).
2.4. Real time PCR
The total RNA from PCa cell lines or tissues were extracted by using Trizol (Invitrogen, US) according to the manufacturer’s instructions. LINC00482 reverse transcription was performed by using M−MLV reverse transcriptase (Invitrogen, US). The expression levels of LINC00482, FZD3, FZD4 and FZD5 were detected by using SYBR Green (TaKaRa, China) in ABI 7900HT Real Time PCR System (Applied Biosystems, US). GAPDH was served as the internal control. miR-2467-3p expression level was detected by TaqMan MicroRNA (Applied Biosystems, US) with U6 as an internal control. All the primers were listed as follows: LINC00482 (F: 5′-AGGGGTAACCTACCGGGAAA-3′, R: 5′-CTTGGCCAGAGCTCCAGAAG-3′), miR-2467-3p (F-5′-GCCGAGGGACAGGCACCTGA-3′; R-5′-CTCAACTGGTGTCGTGGA-3′), FZD3 (F: 5′- GGCTCTCATAGTTGGCATTCCC-3′, R: 5′- TGGAGTACCTGTCGGCTCTCAT-3′), FZD4 (F: 5′- TTCACACCGCTCATCCAGTACG −3′, R: 5′- ACGGGTTCACAGCGTCTCTTGA −3′), FZD5 (F: 5′- TGGAACGCTTCCGCTATCCTGA −3′, R: 5′-GGTCTCGTAGTGGATGTGGTTG-3′), GAPDH (F-5′-TCCCATCACCATCTTCCAGG-3′; R-5′-GATGACCCTTTTGGCTCCC-3′), U6 (F-5′-AACGAGACGACGACAGAC-3′; R-5′-GCAAATTCGTGAAGCGTTCCATA-3′).
2.5. Cell proliferation, migration and invasion analysis
The Cell Counting Kit 8 (CCK8) (Dojindo, Japan) was used for PCa cell proliferation analysis. The spectrophotometric absorbance at 450 nm was detected by using a spectrophotometer (Molecular Devices, US). The DNA synthesis rate was measued by the 5-ethynyl-20-deoxyuridine (EdU) assay kit (Ribobio, China) according to the manufacturer’s instruction. For the colony formation, cells cultured for 14 days were fixed for 15 min with 30% formaldehyde and then stained with 0.1% crystal violet. The transwell assay was applied to evaluate cell migration and BioCat Matrigel Invasion Chamber (BD Biosciences, US) was used for cell invasion assay. The number of migrated and invasive cells were counted in three random fields.
2.6. Animal study
All mouse experiments were approved by the Ethics Committee of Zhejiang Provincial People’s Hospital. 24 of 6-week-old BALB/c-nu male mice were divided into three groups. The LINC00482 shRNA#1 and #2 transfected or control PC-3 cells were inoculated into the left cardiac ventricle of the anaesthetized mice. All mice were sacrificed at the 6th week after modeling and their hind limbs were removed for analysis. Osteolytic lesions were observed on radiographs. The osteolytic area was measured using ImageJ.
2.7. Nuclear-cytoplasmic fractionation
Nuclear-cytoplasmic fractionation was performed by using the Protein and RNA Isolation System (Ambion, US). GAPDH was served as a cytoplasmic control, while U6 was served as a nuclear control.
2.8. Osteoclastogenesis assay
RAW264.7 cells (1×104) were cultured on 96-well plates and grown in the conditioned media (CM) from indicated PCa cells. Media were changed every day. Osteoclasts were counted on day 5. The osteoclasts were fixed with 4% paraformaldehyde/PBS, and TRAP expression was examined by staining with a kit (G1050; Servicebio). Osteoclasts were defined as TRAP-positive multinucleated cells containing more than 3 nuclei.
2.9. Luciferase reporter assay
Cells were transfected with LINC00482 wt luciferase or LINC00482 mutant luciferase plasmid, together with miR-2467-3p mimics or inhibitors using Lipofectamine 3000 (Invitrogen, US) according to the manufacturer’s instruction. After 24 h incubation, cells were harvested for luciferase measurement by using a Dual Luciferase Reporter Assay Kit (Promega, US).
2.10. RNA immunoprecipitation (RIP) assay
The Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, US) was applied for RIP assay to exploring the binding relationship between LINC00482 and miR-2467-3p in PCa cell lines according to the instruction. Cell lysates lysed by RIP lysis buffer were incubated in RIP buffer with magnetic beads conjugated with a negative control IgG (Millipore, US) or human anti-Argonaute 2 (Ago2) antibody (Millipore, US), followed by incubating with proteinase K and precipitation. Purified RNA was used for real time PCR analysis.
2.11. Western blotting analysis
Proteins were extracted with RIPA buffer and then run on the polyacrylamide gel, followed by transferring to polyvinylidene difluoride membranes (Millipore, US). The membranes were blocked for 1 h by 5% non-fat dry milk in PBS with 0.1% Tween 20. Then the membranes were incubated with primary antibodies, β-catenin (1:1000, Santa Cruz Biotech, CA), FZD4 (1:1000, Santa Cruz Biotech, CA), FZD5 (1:1000, Santa Cruz Biotech, CA) and GAPDH (1:1000, Santa Cruz Biotech, CA), followed by HRP-conjugated secondary antibodies (1: 1000, Santa Cruz Biotech, CA) incubation for 1 h at room temperature. ECL western blotting kit (Amersham Biosciences, UK) was applied for the detection.
2.12. Statistical analysis
The data were shown as mean ± SD. An unpaired two-tailed t test was applied for comparison between two groups of the in vitro data. The Mann-Whitney or Chi-square test was used to for animal study. Spearman-Pearson correlation was used for statistical correlation. Survival curve was obtained by the Kaplan-Meier method with a log-rank test. All experiments were repeated three times and data analyses were conducted by GraphPad Prism (GraphPad Software, US). p < 0.05 was considered to be statistically significant.
3. Results
3.1. LINC00482 was upregulated in bone metastatic PCa tissues and associated with PCa progression
Based on the lncRNA sequencing data of PCa from TCGA, LINC00482 was upregulated in primary PCa tissues compared with that in normal tissues (Fig. 1A and B). Higher expression level was also observed in the dead events of PCa patients as well as in PCa patients with higher Gleason score (Fig. 1C and D). To further verified the expression level of LINC00482 in PCa tissues, 79 archived PCa tissues, including 43 non-bone metastatic and 36 bone metastatic PCa tissues as well as 30 paired adjacent normal tissues (ANT) and PCa tissues were applied for real time PCR analysis. As shown in Fig. 1E, increased LINC00482 expression level was observed in PCa tissue compared to ANT. Furthermore, LINC00482 expression was upregulated in bone metastatic PCa tissue compared to that in non-bone metastatic PCa tissues (Fig. 1F) as well as in PCa patients with higher Gleason score (Fig. 1G). Consistently, LINC00482 expression was significantly upregulated in different types of PCa cells (22RV1, LNCaP, C4-2B, PC-3, DU145 and VCaP) compared to normal prostate epithelial cells (RWPE-1) and the highest expression level of LINC00482 was observed in bone metastatic C4-2B cells (Fig. 1H). Kaplan-Meier survival analysis indicated that high expression level of LINC00482 was associated with poor overall survival (p < 0.05, Fig. 1I) as well as bone metastasis-free survival (p < 0.05, Fig. 1J). high expression level of LINC00482 was also associated with poor progression free interval from the TCGA database (p = 0.01, Fig. 1K). These results indicated that LINC00482 was associated with PCa metastasis.
Fig. 1.
LINC00482 was upregulated in bone metastatic PCa tissues and associated with progression and prognosis in PCa patients. (A) Upregulated LINC00482 expression levels in 52 paired PCa tissues compared with the matching normal tissues by analyzing PCa lncRNA sequencing dataset from TCGA. (B) Increased LINC00482 expression levels in PCa tissues compared with normal tissues by analyzing PCa lncRNA sequencing dataset from TCGA. (C) Higher LINC00482 expression levels in dead events of PCa patients by analyzing PCa lncRNA sequencing dataset from TCGA. (D) Increased LINC00482 expression levels in PCa tissues with higher Gleason score by analyzing PCa lncRNA sequencing dataset from TCGA. (E) Real time PCR analysis of LINC00482 expression in 30 paired PCa tissues and ANT. (F) Real time PCR analysis of LINC00482 expression in 43 non-bone metastatic and 36 bone metastatic PCa samples. (G) Real time PCR analysis of LINC00482 expression in PCa patients with different Gleason score. (H) Real time PCR analysis of LINC00482 expression in normal prostate epithelial cell (RWPE-1), primary PCa cell (22RV1), lymph node metastatic cell (LNCaP), bone metastatic PCa cell (C4-2B, PC-3 and VCaP) and brain metastatic cell (DU145). (I) Kaplan-Meier analysis of overall survival of PCa patients with high LINC00482 expression versus low LINC00482 expression. (J) Kaplan-Meier analysis of bone metastatic-free survival of PCa patients with high LINC00482 expression versus low LINC00482 expression. (K) Kaplan-Meier analysis of progression free survival of PCa patients with high LINC00482 expression versus low LINC00482 expression by analyzing PCa lncRNA sequencing dataset from TCGA.
3.2. Silencing of LINC00482 inhibited cell proliferation, migration and invasion in PCa
The specific biological role of LINC00482 in PCa was further analyzed by gene set enrichment analysis (GSEA) based on the lncRNA sequencing data from TCGA. The result showed that high expression of LINC00482 positively and significantly coorelated with proliferation and metastasis (Fig. 2A). To determine whether silencing of LINC00482 could regulate biological activities in PCa cells, LINC00482 expression was successfully downregulated in PC3 and C4-2B cells by using shRNAs compared with the control plasmid (Fig. 2B). The CCK-8, EdU and colony formation assay indicated that silencing of LINC00482 could attenuate the proliferation of PC3 and C4-2B cells (Fig. 2C – F). Furthermore, the migratory and invasive abilities were also inhibited after silencing of LINC00482 in PCa cells (Fig. 3A and B). Meanwhile, to explore the effect of LINC00482 on osteoclastogenesis, we performed osteoclastogenesis assays and found that CM from PCa cells with LINC00482 knockdown significantly inhibited the osteoclastogenesis of RAW264.7 cells compared to the control PCa cells (Fig. 3C and D). To investigate the role of LINC00482 in bone metastasis of PCa in vivo, PC-3 cells transfected with LINC00482 shRNA#1 were inoculated into cardiac ventricle of mice. As shown in Fig. 3E – H and Fig. S1, silencing of LINC00482 in mice showed less bone metastasis ability by X-ray and decreased bone metastasis site as well as osteolytic area compared to the control group. Collectively, these results demonstrated that silencing of LINC00482 could inhibit bone metastasis in vitro and in vivo.
Fig. 2.
Downregulation of LINC00482 inhibited proliferation in PCa cells. (A) GSEA showed high expression of LINC00482 significantly and positively correlated with proliferation signature and metastatic signature. (B) The expression of LINC00482 in PCa cell lines transfected with LINC00482 shRNAs and corresponding negative control plasmid. (C) Silencing of LINC00482 inhibited cell proliferation in PC-3 cell line by CCK-8 assay. (D) Silencing of LINC00482 inhibited cell proliferation in C4-2B cell line by CCK-8 assay. (E) Silencing of LINC00482 inhibited DNA damage and cell death in PCa cell lines by EdU assay. (F) Silencing of LINC00482 inhibited colony formation in PCa cell lines.
Fig. 3.
Downregulation of LINC00482 inhibits bone metastasis in PCa. (A) Silencing of LINC00482 suppressed PCa cells migration ability. (B) Silencing of LINC00482 suppressed PCa cells invasion ability. (C) Osteoclast differentiation assays by TRAP staining in the presence of CM from indicated cells. (D) Quantification of the number of TRAP+ multinuclear osteoclasts. (E) Representative radiographic and HE images of bone metastasis in mice. Arrows indicated osteolytic lesions. T, tumor tissue; N, normal tissue. (F) The number of bone metastatic mice in the indicated groups. (G) Bone metastasis site per mouse in the indicated groups. (H) Osteolytic area per mouse in the indicated groups.
3.3. LINC00482 acted as a ceRNA by sponging miR-2467-3p and regulated cell metastasis
To identify the subcellular localization of LINC00482 in PC-3 and C4-2B cells, cellular fractionation was performed and the result showed that LINC00482 was primarily localized in the cell cytoplasm (Fig. 4A). Both Starbase and Lncbase databases were applied to predict the potential miRNA targets of LINC00482 and miR-2467-3p may act as a biological target of LINC00482 (Fig. 4B). Then the expression level of miR-2467-3p was found to be upregulated after the silencing of LINC00482 in PCa cells (Fig. 4C). Additionally, less expression of miR-2467-3p was observed in PCa tissue compared to that in ANT. LINC00482 was negatively correlated with miR-2467-3p (p = 0.0025, Fig. 4E). Based on the bioinformatic database Starbase, the potential binding site (Fig. 4F) and luciferase reporter assay was performed to validate the binding of miR-2467-3p with LINC00482. In Fig. 4G, miR-2467-3p mimics significantly decreased luciferase activity in LINC00482-wild type but not in LINC00482-mutant in both PC-3 and C4-2B cells. To further verify the binding between LINC00482 and miR-2467-3p, anti-Ago2 RIP assay was applied. The result showed that the endogenous LINC00482 was specifically enriched in PCa cells treated with miR-2467-3p mimics (Fig. 4H), indicating that LINC00482 directly targeted miR-2467-3p. Furthermore, miR-2467-3p inhibitor could notably reverse the inhibitory effect of PCa cells transfected with LINC00482 shRNA#1 on proliferation, migration and invasion (Fig. 4I and J).
Fig. 4.
LINC00482 acted as a ceRNA by sponging miR-2467-3p and regulated cell metastasis. (A) Cellular localization of LINC00482 in PCa cell line. GAPDH served as a cytoplasmic localization marker and U6 served as nuclear localization marker. (B) miR-2467-3p was predicted to be interacted with LINC00482 by Starbase and Lncbase databases. (C) Relative expression level of miR-2467-3p in PCa cell lines treated with LINC00482 shRNAs. (D) Relative expression level of miR-2467-3p in 30 paired PCa tissues and ANT. (E) Spearman-Pearson correlation between LINC00482 and miR-2467-3p. (F) Predicted LINC00482 targeting sequence and mutant sequences in 3′UTR of miR-2467-3p. (G) Luciferase assay reporter assay showed the luciferase activities in each group of PCa cell lines. (H) RIP was performed in PCa cell lines transfected with miR-2467-3p mimics. (I) The percentages of EdU positive cells in PCa cell lines transfected with LINC00482 shRNA#1 or miR-2467-3p inhibitor. (J) Migration and invasion ability of PCa cell lines transfected with LINC00482 shRNA#1 or miR-2467-3p inhibitor.
3.4. LINC00482 regulated Wnt signaling pathway by sponging miR-2467-3p
GSEA analysis has further revealed that high expression of LINC00482 positively correlated with Wnt signaling pathway (Fig. 5A). In Fig. 5B and C, both silencing of LINC00482 and miR-2467-3p mimics could decrease luciferase activity of Wnt/β-catenin signaling pathway and the protein expression level of β-catenin in the nuclei in PCa, indicating that LINC00482 may regulate Wnt/β-catenin signaling pathway by sponging miR-2467-3p. To identify the potential target of miR-2467-3p, both Starbase and TargetScan databases were used and three targets (FZD3, FZD4 and FZD5) were predicted to be the targets of miR-2467-3p (Fig. 6A). Then the expression levels of FZD4 and FZD5 were significantly downregulated in PCa cells treated with miR-2467-3p mimics, except for FZD3 (Fig. 6B). As shown in Figure D-E, luciferase assay revealed that miR-2467-3p mimics significantly decreased luciferase activity in FZD4-wild type and FZD5-wild type but not in FZD4-mutant and FZD5-mutant in both PC-3 and C4-2B cells, indicating that both FZD4 and FZD5 were the targets of miR-2467-3p. Furthermore, miR-2467-3p inhibitor was able to reverse the inhibitory effect of PCa cells transfected with LINC00482 shRNA#1 on the mRNA and protein expression levels of FZD4 and FZD5 (Fig. 6F – H). Collectively, these results indicated that LINC00482 may activate Wnt signaling pathway by sponging miR-2467-3p.
Fig. 5.
LINC00482 induced WNT signaling pathway. (A) GSEA showed high expression of LINC00482 significantly and positively correlated with Wnt signaling pathway. (B) miR-2467-3p or silencing of LINC00482 significantly reduced luciferase activity in PCa cell lines. (C) Western blotting analysis of β-catenin in PCa cell lines treated with miR-2467-3p, LINC00482 shRNA#1 and corresponding negative control.
Fig. 6.
LINC00482 regulated FZD4 and FZD5 by sponging miR-2467-3p. (A) Predicted targets of miR-2467-3p in TargetScan and Starbase. (B) Real time PCR analysis of FZD3, FZD4 and FZD5 in PCa cell lines treated with miR-2467-3p mimics. (C) Predicted miR-2467-3p targeting sequence and mutant sequences in the 3′UTRs of FZD4 and FZD5. (D) miR-2467-3p significantly reduced luciferase activity in PCa cell lines transfected with reporter of FZD4 and FZD5 in PC-3 cells. (E) miR-2467-3p significantly reduced luciferase activity in PCa cell lines transfected with reporter of FZD4 and FZD5 in C4-2B cells. (F) The expression levels of FZD4 and FZD5 in PC-3 cells treated with LINC00482 shRNA#1 and miR-2467-3p inhibitor. (G) The expression levels of FZD4 and FZD5 in PC-3 cells treated with LINC00482 shRNA#1 and miR-2467-3p inhibitor. (H) Western blotting analysis of FZD4 and FZD5 in PCa cell lines treated with LINC00482 shRNA#1 and miR-2467-3p.
3.5. The correlation between LINC00482 and FZD4, FZD5 or β-catenin in PCa
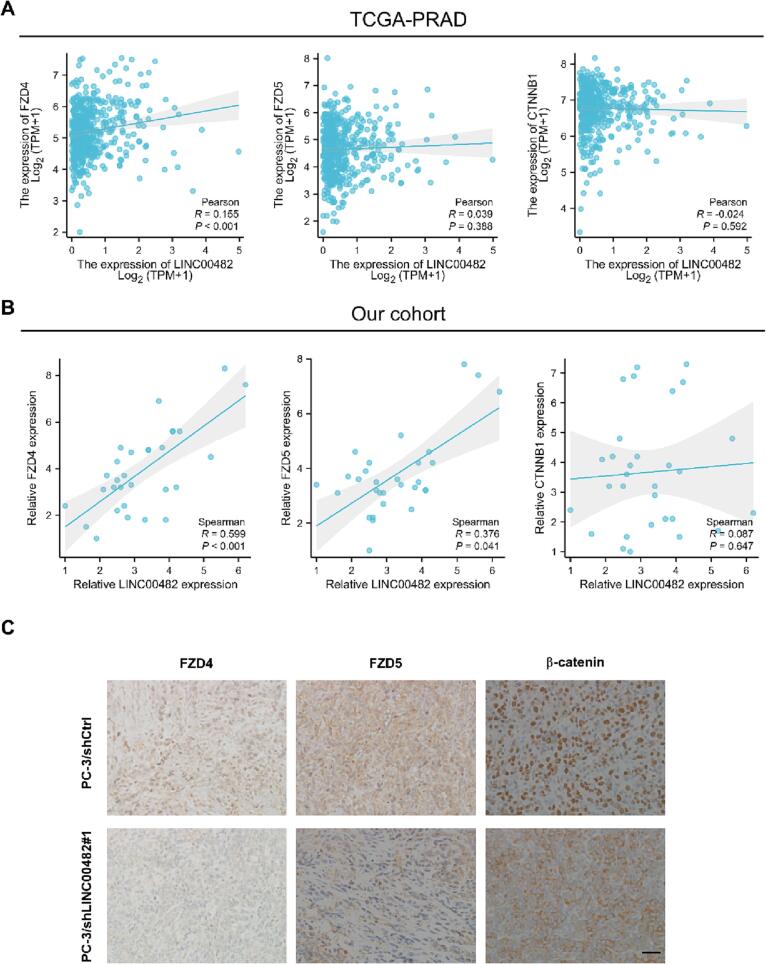
Next, we investigated the correlation between LINC00482 and FZD4, FZD5 or β-catenin in PCa. In TCGA-PRAD database, the LINC00482 expression was positively correlated with FZD4 expression, but not FZD5 and CTNNB1 (which encodes β-catenin) (Fig. 7A). In our cohort, we found that the LINC00482 expression was positively correlated with FZD4 and FZD5 expression, but not CTNNB1 (Fig. 7B). The different result of the correlation between LINC00482 and FZD5 in TCGA-PRAD cohort and our cohort may be due to that there are other more powerful factors which regulates FZD5 expression in TCGA-PRAD cohort. No correlation between LINC00482 and CTNNB1 supported our findings, which is that LINC00482 upregulated FZD4/5 expression, further promoting nuclear transport of β-catenin instead of CTNNB1 mRNA level. Moreover, we also analyzed FZD4/FZD5 expression and β-catenin location in bone metastases section from Fig. 2. The results indicated that compared with the control group, FZD4/FZD5 expression and nuclear β-catenin expression were significantly decreased in the shLINC00482#1 group (Fig. 7C).
Fig. 7.
The correlation between LINC00482 and FZD4, FZD5 or β-catenin in PCa (A-B) The correlation between LINC00482 and FZD4, FZD5 or CTNNB1 in PCa in TCGA-PRAD cohort (A) and our cohort (B). (C) Immunohistochemistry analysis of FZD4/FZD5 expression and β-catenin location in bone metastases section from Fig. 2.
4. Discussion
For decades, PCa has remained a life-threatening disease worldwide in men.[21] A majority of PCa-related mortality caused by metastatic disease, such as lung, liver, lymph nodes and bone metastasis. Bone metastasis has been the most common in PCa with osteolytic and osteoblastic lesions. [22] However, the exact mechanisms underlying PCa bone metastasis remain unclear. It’s urgently necessary to further explore this area, which may provide references for finding potential therapeutic targets on PCa with bone metastasis.
In this study, LINC00482, a novel lncRNA, was identified to be associated with PCa bone metastasis and poor overall survival, which acted as an oncogenic role on PCa cell proliferation, migration and invasion. LncRNAs could act as miRNA sponges resulting in the release of target genes in the downstream, which is a canonical way to investigate the molecular mechanisms of lncRNAs in cancers.[23], [24] A series of lncRNA has been proved to be responsible for PCa progression, including lncRNA HOTAIRM1, lncRNA PRNCR1, lncRNA LINC01006, lncRNA PART1 and so on.[25], [26], [27], [28] However, the mechanisms of how lncRNA regulate bone metastasis in PCa are still unclear and few study has revealed the mechanisms. In a previous study, it’s reported that lncRNA NORAD was able to induce bone metastasis in PCa by enhancing the PCa extracellular vesicle release through miRNA-541-3p/PKM2 axis.[29] Another study also demonstrated that lncRNA HCG18 and MCM3AP-AS1 were associated with bone metastasis.[30] For LINC00482, it’s reported that silencing of LINC00482 was able to suppress angiogenesis and tumor-associated inflammation in bladder cancer through modulating MMP-15 via FOXA1.[17] Consistently, silencing of LINC00482 was able to inhibit bone metastasis in PCa in vivo.
To better understanding the mechanism of LINC00482 on bone metastasis of PCa, we further investigated the LINC00482-involved ceRNA network. In the ceRNA networks, miRNAs are crucial mediators which could serve roles in the development of PCa.[31], [32] The target miRNA of LINC00482 was predicted and verified as miR-2467-3p which had the complementary binding site. Studies have revealed that miR-2467-3p could inhibit tumor progression in various types of cancers, including cervical, colorectal, non-small cell lung cancer.[33], [34], [35] Xiao et al. reported that LINC01224 promotes progression and cisplatin resistance in non-small lung cancer by sponging miR-2467.[36] In colorectal cancer, LINC01224 promotes cell proliferation, migration and invasion by sponging miR-2467.[37] In cervical cancer, LINC01410 promotes tumor progression via targeting miR-2467-3p/VOPP1 axis.[33] Therefore, we analyzed the expression of LINC01224 and LINC01410 in TCGA-PRAD database and found that LINC01224 and LINC01410 were decreased in prostate cancer compared with normal tissues (Fig. S2A and C). However, survival analysis indicated that two lncRNAs’ expression were not associated with poor progression-free survival (Fig. S2B and D). Therefore, the biological role of LINC01224 and LINC01410 should be further explored in the future study. The ceRNA network also contains a target gene of miRNA for the regulatory axis.[38] The target mRNAs of miR-2467-3p were found to be FZD4 and FZD5. Previous studies showed that Wnt receptor FZD4 and FZD5 were responsible for PCa migration to some extent.[39], [40] The abnormal regulation of Wnt/β-catenin signaling pathway could enhance cell proliferation and differentiation, as well as renewal of cancer stem cell, resulting in tumorigenesis and therapy response.[41] In this study, LINC00482 could act as the sponge for miR-2467-3p to regulate PCa progression via Wnt/β-catenin signaling pathway, which may be a promising therapeutic target for PCa with bone metastasis.
5. Conclusion
In conclusion, this study revealed that LINC00482 was an up-regulated lncRNA in PCa with bone metastasis and associated with poor overall and bone metastatic-free survival in PCa patients, which could regulate cell proliferation, migration and invasion. We also demonstrated that LINC00482 could act as miRNA sponges to target miR-2467-3p/Wnt/β-catenin axis resulting in PCa progression inhibition. These findings might provide a promising strategy for clinical therapies on PCa with bone metastasis.
CRediT authorship contribution statement
Shiyao Liao: Validation, Data curation, Writing – original draft. Xuemei Fang: Data curation, Formal analysis. Kai Zhou: Data curation, Formal analysis. Tingxiao Zhao: . Lichen Ji: Visualization, Investigation. Wei Zhang: Investigation. Xugang Zhong: Data curation, Formal analysis, Investigation. Fabo Feng: Data curation, Formal analysis, Investigation. Jun Lv: . Yao Kang: Conceptualization, Writing – review & editing. Danjie Zhu: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Danjie Zhu reports financial support was provided by Zhejiang Provincial Health Bureau Science Foundation of China.
Funding
This study was supported by grants from Zhejiang Provincial Health Bureau Science Foundation of China, NO. 2022ky034, Natural Science Foundation of Zhejiang Province, NO. LQ19H160014, Medical Health Science and Technology Project of Zhejiang Provincial Health Commission, NO. 2019KY320.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2023.100494.
Contributor Information
Jun Lv, Email: 13858010120@163.com.
Yao Kang, Email: kyc0730@163.com.
Danjie Zhu, Email: zhudj@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
Supplementary Fig. 2.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Lowrance W.T., Breau R.H., Chou R., Chapin B.F., Crispino T., Dreicer R., Jarrard D.F., Kibel A.S., Morgan T.M., Morgans A.K., Oh W.K., Resnick M.J., Zietman A.L., Cookson M.S. Advanced Prostate Cancer: AUA/ASTRO/SUO Guideline PART I. J Urol. 2021;205(1):14–21. doi: 10.1097/JU.0000000000001375. [DOI] [PubMed] [Google Scholar]
- 3.M.F. Monn, A.J. Tatem, L. Cheng. Prevalence and management of prostate cancer among East Asian men: Current trends and future perspectives. Urol Oncol. 2016. 34. 58. e51-59. [DOI] [PubMed]
- 4.Bhagirath D., Yang T.L., Dahiya R., et al. MicroRNAs as Regulators of Prostate Cancer Metastasis. Adv Exp Med Biol. 2018;1095:83–100. doi: 10.1007/978-3-319-95693-0_5. [DOI] [PubMed] [Google Scholar]
- 5.Dalela D., Sun M., Diaz M., Karabon P., Seisen T., Trinh Q.-D., Menon M., Abdollah F. Contemporary Trends in the Incidence of Metastatic Prostate Cancer Among US Men: Results from Nationwide Analyses. Eur Urol Focus. 2019;5(1):77–80. doi: 10.1016/j.euf.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X. Interactions between cancer cells and bone microenvironment promote bone metastasis in prostate cancer. Cancer Commun (Lond) 2019;39:76. doi: 10.1186/s40880-019-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croucher P.I., McDonald M.M., Martin T.J. Bone metastasis: the importance of the neighbourhood. Nat Rev Cancer. 2016;16(6):373–386. doi: 10.1038/nrc.2016.44. [DOI] [PubMed] [Google Scholar]
- 8.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller Jörg, Hofacker I.L., Bell I., Cheung E., Drenkow J., Dumais E., Patel S., Helt G., Ganesh M., Ghosh S., Piccolboni A., Sementchenko V., Tammana H., Gingeras T.R. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 10.Z Xing, S Li, Z Liu, et al. The long non-coding RNA LINC00473 contributes to cell proliferation via JAK-STAT3 signaling pathway by regulating miR-195-5p/SEPT2 axis in prostate cancer. Biosci Rep. 2020. 40. [DOI] [PMC free article] [PubMed]
- 11.Qu J., Li M., Zhong W., et al. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8:17110–17116. [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X., Zhang D.-H., Wu N., Xiao J.-H., Wang X., Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 13.Hu C.H., Yang X.J., Yu L., et al. Long non-coding RNA LINC00173 serves as sponge for miR-338-3p to promote prostate cancer progression via regulating Rab25. Eur Rev Med Pharmacol Sci. 2020;24:9290–9302. doi: 10.26355/eurrev_202009_23011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Li Y. Long non-coding RNA NORAD contributes to the proliferation, invasion and EMT progression of prostate cancer via the miR-30a-5p/RAB11A/WNT/beta-catenin pathway. Cancer Cell Int. 2020;20:571. doi: 10.1186/s12935-020-01665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie H., Zhao J., Wan J., Zhao J., Wang Q., Yang X.u., Yang W., Lin P., Yu X. Long noncoding RNA AC245100.4 promotes prostate cancer tumorigenesis via the microRNA1455p/RBBP5 axis. Oncol Rep. 2021;45(2):619–629. doi: 10.3892/or.2020.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao S.D., Ma J.X., Liu Y., et al. Long non-coding TUG1 accelerates prostate cancer progression through regulating miR-128-3p/YES1 axis. Eur Rev Med Pharmacol Sci. 2020;24:619–632. doi: 10.26355/eurrev_202001_20038. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Zhang L., Wei N.a., Sun Y., Pan W., Chen Y. Silencing LINC00482 inhibits tumor-associated inflammation and angiogenesis through down-regulation of MMP-15 via FOXA1 in bladder cancer. Aging (Albany NY) 2021;13(2):2264–2278. doi: 10.18632/aging.202247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croce J.C., McClay D.R. Evolution of the Wnt pathways. Methods Mol Biol. 2008;469:3–18. doi: 10.1007/978-1-60327-469-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Wang F., Xu H., et al. Long Non-Coding RNA SNHG1 Regulates the Wnt/beta-Catenin and PI3K/AKT/mTOR Signaling Pathways via EZH2 to Affect the Proliferation, Apoptosis, and Autophagy of Prostate Cancer Cell. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.552907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akoto T., Saini S. Role of Exosomes in Prostate Cancer Metastasis. Int J Mol Sci. 2021;22(7):3528. doi: 10.3390/ijms22073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller E.T., Brown J. Prostate cancer bone metastases promote both osteolytic and osteoblastic activity. J Cell Biochem. 2004;91(4):718–729. doi: 10.1002/jcb.10662. [DOI] [PubMed] [Google Scholar]
- 23.Liu X.-H., Sun M., Nie F.-q., Ge Y.-B., Zhang E.-B., Yin D.-D., Kong R., Xia R., Lu K.-H., Li J.-H., De W., Wang K.-M., Wang Z.-X. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13(1) doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang C., Qiu S., Sun F., Li W., Wang Z., Yue B., Wu X., Yan D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Wang L., Wang L., Wang Q., Yosefi B., Wei S., Wang X., Shen D. The function of long noncoding RNA HOTAIRM1 in the progression of prostate cancer cells. Andrologia. 2021;53(2):e13897. doi: 10.1111/and.13897. [DOI] [PubMed] [Google Scholar]
- 26.Bardhan A., Banerjee A., Basu K., Pal D.K., Ghosh A. PRNCR1: a long non-coding RNA with a pivotal oncogenic role in cancer. Hum Genet. 2022;141(1):15–29. doi: 10.1007/s00439-021-02396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L., Han F., Yang J., Ming X., Chen L. Long noncoding RNA LINC01006 exhibits oncogenic properties in cervical cancer by functioning as a molecular sponge for microRNA285p and increasing PAK2 expression. Int J Mol Med. 2021;47(4) doi: 10.3892/ijmm.2021.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X., Zhang L., Tian J., Ma J. Long non-coding RNA PART1 predicts a poor prognosis and promotes the malignant progression of pancreatic cancer by sponging miR-122. World J Surg Oncol. 2021;19(1) doi: 10.1186/s12957-021-02232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu C.-y., Chen J., Qin X.-H., You P., Ma J., Zhang J., Zhang H.e., Xu J.-D. Long non-coding RNA NORAD promotes the prostate cancer cell extracellular vesicle release via microRNA-541-3p-regulated PKM2 to induce bone metastasis of prostate cancer. J Exp Clin Cancer Res. 2021;40(1) doi: 10.1186/s13046-021-01891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y Chen, Z Chen, J Mo, et al. Identification of HCG18 and MCM3AP-AS1 That Associate With Bone Metastasis, Poor Prognosis and Increased Abundance of M2 Macrophage Infiltration in Prostate Cancer. Technol Cancer Res Treat. 2021. 20. 1533033821990064. [DOI] [PMC free article] [PubMed]
- 31.Cochetti G., Poli G., Guelfi G., et al. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016;9:7545–7553. doi: 10.2147/OTT.S119027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guelfi G., Cochetti G., Stefanetti V., Zampini D., Diverio S., Boni A., Mearini E. Next Generation Sequencing of urine exfoliated cells: an approach of prostate cancer microRNAs research. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-24236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F., Wen C. LINC01410 Knockdown Suppresses Cervical Cancer Growth and Invasion via Targeting miR-2467-3p/VOPP1 Axis. Cancer Manag Res. 2020;12:855–861. doi: 10.2147/CMAR.S236832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao H., Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2020;235(3):2881–2890. doi: 10.1002/jcp.29193. [DOI] [PubMed] [Google Scholar]
- 35.Chen H., Tan X., Ding Y.i. Knockdown SNHG20 Suppresses Nonsmall Cell Lung Cancer Development by Repressing Proliferation, Migration and Invasion, and Inducing Apoptosis by Regulating miR-2467-3p/E2F3. Cancer Biother Radiopharm. 2021;36(4):360–370. doi: 10.1089/cbr.2019.3430. [DOI] [PubMed] [Google Scholar]
- 36.Xiao S., Sun L., Ruan B., Li J., Chen J., Xiong J., Jiang Y., Song Z. Long non-coding RNA LINC01224 promotes progression and cisplatin resistance in non-small lung cancer by sponging miR-2467. Pulm Pharmacol Ther. 2021;70:102070. doi: 10.1016/j.pupt.2021.102070. [DOI] [PubMed] [Google Scholar]
- 37.Chen L., Chen W., Zhao C., Jiang Q.i. LINC01224 Promotes Colorectal Cancer Progression by Sponging miR-2467. Cancer Manag Res. 2021;Volume 13:733–742. doi: 10.2147/CMAR.S281625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan Z., Zielske S.P., Ibrahim K.G., et al. Wnt and beta-Catenin Signaling in the Bone Metastasis of Prostate Cancer. Life (Basel) 2021;11 doi: 10.3390/life11101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiele S., Zimmer A., Göbel A., Rachner T.D., Rother S., Fuessel S., Froehner M., Wirth M.P., Muders M.H., Baretton G.B., Jakob F., Rauner M., Hofbauer L.C. Role of WNT5A receptors FZD5 and RYK in prostate cancer cells. Oncotarget. 2018;9(43):27293–27304. doi: 10.18632/oncotarget.25551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165. doi: 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]