Abstract
Background and objective
Diabetes can induce diabetic retinopathy (DR), and the blindness caused by this disease is irreversible. The early analysis of mouse retinal images, including the layer and cell segmentation properties of these images, can help to effectively diagnose this disease.
Method
In the study, we design a dilated residual method based on a feature pyramid network (FPN), in which the FPN is adopted as the base network for solving the multiscale segmentation problem concerning mouse retinal images. In the bottom-up encoding pathway, we construct our backbone feature extraction network via the combination of dilated convolution and a residual block, further increasing the range of the receptive field to obtain more context information. At the same time, we integrate a squeeze-and-excitation (SE) attention module into the backbone network to obtain more small object details. In the top-down decoding pathway, we replace the traditional nearest-neighbor upsampling method with the transposed convolution method and add a segmentation head to obtain semantic segmentation results.
Results
The effectiveness of our network model is verified in two segmentation tasks: ganglion cell segmentation and mouse retinal cell and layer segmentation. The outcomes demonstrate that, compared to other supervised segmentation methods based on deep learning, our model attains the utmost precision in both binary segmentation and multiclass semantic segmentation tasks.
Conclusion
The dilated residual FPN is a robust method for mouse retinal image segmentation and it can effectively assist DR diagnosis.
Keywords: Mouse retinal image segmentation, Dilated convolution block, SE, FPN
1. Introduction
Diabetes refers to a cluster of metabolic disorders characterized by elevated blood glucose levels. Hyperglycemia has multiple causes, such as insulin secretion deficiency, impaired biological effects or both.
Long-term diabetes can result in chronic impairment and dysfunction in several tissues, particularly affecting a series of important human organs [1,2]. Numerous prior experiments have substantiated the association between long-term diabetes and the development of diabetic microvasculopathy. Diabetic retinopathy (DR) represents a common form of microangiopathy triggered by persistent hyperglycemia and insulin resistance, leading to impaired vascular endothelial function and damage. In severe cases, DR can have a significant impact on vision and may even result in blindness [3]. In addition, no cure is available for the disease, which can only be alleviated and controlled by certain means.
DR is categorized into nonproliferative and proliferative lesions; the earliest occurring lesions are nonproliferative, and the sites of lesions are mainly distributed in the retinal nerve fiber layer (RNFL), inner retinal layer (IPL), inner nuclear layer (INL), and outer retinal layer (OPL). In comparison to normal mice, diabetic mice exhibit a reduction in thickness in the retinal nerve fiber layer (RNFL), inner plexiform layer (IPL), and inner nuclear layer (INL), alongside a decrease in the number of ganglion cells. Microscopic imaging is an effective approach for observing the above phenomena, and it has been proven that the distinctions between normal and pathological cells or layers in mouse retinal images enable the feasibility of image-based diagnosis, which is essential for quickly and accurately diagnosing potential DR cases [4].
The segmentation of different layers and cells in retinal images is essential for the detection of DR. However, the retinal structure of a mouse is complicated (it includes multiple layers and cells), and manual segmentation of a mouse retinal image is a time-consuming and labor-intensive process [5]. Furthermore, researchers need to master professional knowledge and unify their segmentation criteria. The majority of previous studies, as reported in the literature, have relied on optical coherence tomography (OCT) to facilitate the segmentation of the retinal pigment epithelium and five additional retinal surfaces in mouse retinal images [6,7]. Subsequently, the advancement of computer technology has facilitated the application of machine learning techniques for segmenting mouse retinal images. Srinivasan et al. proposed an automated method, termed S-GTDP (Sparsity-based denoising, Support Vector Machines, Graph Theory, and Dynamic Programming), for segmenting retinal layers in spectral-domain OCT images [8]. With the development of technology, convolutional neural networks (CNNs) have gained significant traction in the field of computer vision due to their ability to automatically learn and extract features from images, ranging from low-level to high-level representations. CNNs provide more robust and extensive object representations than manual functionality. Xiao, Sa et al. employed deep learning-based neural networks to train on a dataset comprising over a thousand segmentations, achieving full automation in the segmentation process of oxygen-induced retinopathy (OIR) images [9]. Kassim et al. introduced a dual pipeline, named “Random Forest (RF) OFB + U-NET,” which integrates deep learning features from U-Net with a low-level image feature filter bank using an RF classifier to achieve vessel segmentation. They made adaptations to the U-Net CNN architecture to generate a foreground vessel regression likelihood map, which was then employed to segment both arteriole and venule blood vessels in mouse dura mater tissues [10].
For practical applications, the automatic segmentation of mouse retinal images faces many challenges, including the following. 1) Many stains and artifacts are present in mouse retinal images, some of which have similar features to ganglion cells, such as stained vacuoles. 2) The edges of some layers are partially or completely blurred. In addition, the adherence and overlap effects of ganglion cells can also be observed in mouse retinal images. The combination of these two factors makes it difficult to correctly divide ganglion cells. 3) Because such images are acquired from an optical microscope, the differences between their illumination and environmental conditions greatly impact the image segmentation results. Moreover, all the above-mentioned approaches have few remarkable effects on mouse retinal image segmentation.
To address the aforementioned challenges, we have transformed the diabetes diagnosis problem using mouse retinal images into an end-to-end segmentation task. Semantic segmentation stands as a crucial problem in the realm of computer vision, allowing us to identify and classify each pixel in the image with respect to its semantic meaning. In recent years, deep learning models have gained widespread use and popularity for semantic segmentation tasks [[11], [12]]. Their ability to automatically learn and extract intricate features from images has proven to be highly effective in achieving accurate and efficient semantic segmentation results [13,14]. Indeed, among the plethora of exceptional deep learning models, one of the most successful and state-of-the-art approaches for semantic segmentation is based on a fully convolutional network (FCN). FCNs are renowned for their ability to process entire images end-to-end, enabling pixelwise predictions, which makes them particularly well-suited for tasks like semantic segmentation [15]. The crux of this method lies in utilizing a CNN as an effective feature extraction tool, wherein the fully connected layer is replaced by convolutional layers. Instead of generating classification scores, spatial feature maps are produced. These maps are subsequently downsampled to yield dense pixel-level outputs, making the approach highly suitable for semantic segmentation. Over time, FCNs have been enhanced and evolved into models such as U-Net and Feature Pyramid Network (FPN), which have demonstrated improved performance and advancements in the field of semantic segmentation [[16], [17], [18], [19]].
Based on the multiscale fusion characteristics of FPN, we use an FPN as our base network to better divide the layers and cells in mouse retinal images. Furthermore, we combine a residual block and dilated convolution to construct a backbone feature extraction network so that the proposed approach can obtain a larger receptive field and further improve its segmentation accuracy for each layer and cell. On this basis, the feature extraction network and feature pyramid are combined to fuse multiscale information, which enables small targets such as the cells and edges of each layer to be accurately segmented. In addition, we add an attention mechanism, which can better distinguish between vacuoles and ganglion cells, to enhance the segmentation accuracy achieved for small targets. In our study, we conducted binary and multiclass semantic segmentation experiments using our proposed model. To evaluate the performance of our model, we employed multiple indicators as metrics. These indicators provide quantitative measures of segmentation accuracy, allowing us to assess the effectiveness and reliability of our approach. Then, we compare the performance of our model with that of unsupervised and supervised state-of-the-art image segmentation methods.
2. Material and methods
In this part, we provide a comprehensive explanation of the detailed overall architecture of the dilation residual-based FPN model proposed in this paper. Additionally, we introduce the constituent units' specific details as proposed in this article.
The architecture proposed in this paper is built upon the FPN model, as depicted in Fig. 1. The network structure of our model has a top-down decoding pathway and a bottom-up encoding pathway, with lateral connections (1 × 1 Conv) linking the corresponding encoding and decoding layers. Next, we discuss the components of our model.
Fig. 1.
The architecture of the dilation residual-based FPN. The dimensions (heights, widths, channels) of the feature maps in each layer are shown above the network components. Our model takes an original image as input and produces a segmentation mask with the same resolution.
2.1. The base network
Many previous networks have used single high-level features. For example, the Faster R–CNN makes use of a convolutional layer with quadruple downsampling [20]. However, this method exhibits an obvious shortcoming: small objects are easily lost and have less pixel information during the downsampling process. To deal with the segmentation problem concerning the obvious size differences among various objects, the classic method applies pyramids for the same image but with different scales to solve this problem, but this approach results in a large number of calculations. Thus, we use an FPN as the base network of the proposed model to segment mouse retinal images, as an FPN can address the problem regarding multiscale changes in object segmentation with a minimal number of calculations.
2.2. Top-down decoding pathway
In the top-down decoding pathway and the lateral connections, the feature map with the strongest semantics from a higher pyramid level is upsampled to obtain features with higher resolutions. The proposed method is similar to a one-sided binary tree, where it merges the feature map acquired from the top layers with a feature map of the corresponding size.
We make use of four transposed convolutional blocks and four normal convolutional blocks to build each branch of this one-sided binary tree. To achieve the upsampling function, we utilize transposed convolutional blocks, comprising a 3 × 3 convolution, group normalization operation, rectified linear unit (ReLU) function, and a transposed convolution. Due to the method's reliance on small batch sizes, we incorporate group normalization in all transposed convolutional blocks. Specifically, the numbers of transposed convolutional blocks in each layer are set as B1 = 3, B2 = 2, and B3 = 1. To enhance the representation and mapping abilities of ourproposed model, we replace the traditional nearest-neighbor upsampling method with the transposed convolution method, where the kernel size is set to 4, and the stride and padding values are set to 2 and 1, respectively. Subsequently, the bottom-up feature map, which goes through a 1 × 1 convolutional layer, and the top-down feature map with the same spatial size are merged by a lateral connection. In addition, we transform the merged feature map of each layer into a map of size 128 × 128 x 128 via a convolution block with a kernel size of 3 × 3 and further merge the output maps. In the final layer, we introduce a segmentation head that upsamples the feature maps to restore the image to its original size. This step ensures that we obtain a semantic segmentation result that aligns with the input size, providing accurate and consistent segmentation outcomes for our model. The structure of the segmentation head includes a convolution with a kernel size of 1 × 1, the nearest-neighbor upsampling method and an identity function. The scale factor is set as 4 to expand the sizes of 128 × 128 x 128 images to the original input size.
2.3. Bottom-up encoding pathway
In the bottom-up encoding pathway, a CNN is generally used to extract features. Previous studies [[21], [22], [23]] have shown that the errors induced by obtaining feature maps mainly originates from two sources: (1) due to limitation regarding neighborhood size, the variance of the estimated value increases; and (2) the convolution layer parameter's error can lead to the deviation of the estimated mean. To mitigate this error, the pooling method effectively reduces both the deviation error induced by the estimated mean and the error caused by the increase in the variance of the estimated value. However, it is essential to note that this pooling approach requires a large number of parameters, which may increase the model's complexity and computational cost. Because pooling layers require separate parameters to store the pooling operations and their associated indices, whereas convolutions with stride = 2 can achieve the same effect by directly downsampling the feature maps. Simultaneously, due to the amount of parameters in a convolutional layer is determined by the kernel and output channels, but not by the size or stride of input feature maps. Moreover, they can learn more complex feature representations compared to pooling, since they are trainable and can adapt to the data being processed. Thus, we use a convolution with a stride size of 2 instead of a convolution pooling layer, which can not only ensure the desired effect but also reduce the number of required parameters.
To better segment the details of small objects, such as the ganglion cells or edges of layers, and efficiently leverage the obtained contextual information, we combine a residual block and dilated convolution to design a backbone network. For constructing the residual block, we utilize the dilated convolution layer instead of the normal convolution layer, thus obtaining a larger receptive field and capturing contextual information with a long scope for lost feature restoration. However, the kernel of dilated convolution is discontinuous, which means that only a portion of the pixels are used for calculation purposes. In this way, similar to a checkerboard, the network loses the continuity of information and causes the gridding effect.
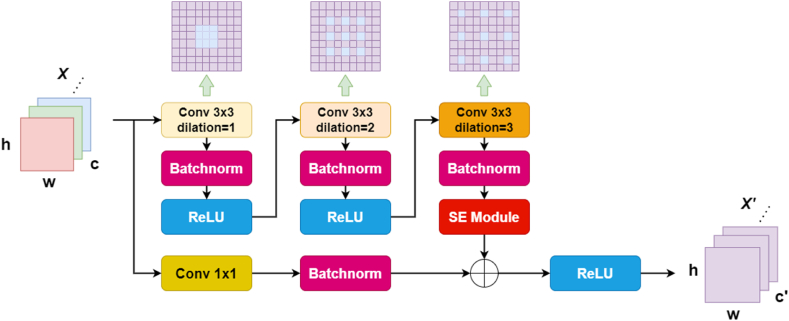
Simultaneously, incorporating a larger dilation rate is advantageous for capturing long-range information, leading to improved segmentation outcomes for larger objects. However, it may result in a relatively weaker effect on smaller objects' segmentation. The selection of an appropriate dilation rate should consider the trade-off between obtaining long-range context and preserving the ability to accurately segment smaller objects. Therefore, we construct a bottom-up encoding pathway with four layers of dilation residual convolutional blocks, and the numbers of dilation residual convolutional blocks in each layer are L1 = 3, L2 = 4, L3 = 6 and L4 = 3. As shown in Fig. 2, we adopt a 3 × 3 kernel in each dilated convolutional layer and set the dilation rates of each layer to 1, 2 and 3 to compose a group of residual blocks, which can prevent the grid effect caused by kernel discontinuity. Furthermore, it should be noted that the initial layer of the dilated convolutional layer has a stride of 2 (“s = 2″ in Fig. 6), whereas all subsequent layers have a stride of 1. Additionally, the circulation of the sawtooth structure between different dilation rates also balances the segmentation effects produced for large and small objects. A normalization layer and a ReLU function are added in each dilated residual convolutional block, which increases the nonlinearity of each block to achieve a better mapping capability.
Fig. 2.
Architecture of the dilation residual module. In the figure, “Conv 3 × 3 dilation” represents a dilated convolution with a kernel size of 3 × 3.
Fig. 6.
Comparison of the segmented images output by each model for the mouse retinal multiclass datasets. (a) Is the original image; (b) is a partial cut of the original image (a); (c) is the ground truth of the original image (a). (d) Our model; (e) the FPN; (f) U-Net; (g) DeepLabV3; (h) the PSPNet.
2.4. Attention mechanism
Furthermore, we integrate the dilation residual block with a squeeze-and-excitation (SE) module, enabling the network to conduct feature recalibration. This approach empowers the network to learn from global information and selectively emphasize feature information while suppressing less relevant ones, leading to further enhancement in object segmentation accuracy [24]. The main architecture of the SE module is shown in Fig. 3, and it consists of a squeeze operation and an excitation operation.
| (1) |
Fig. 3.
The architecture of the squeeze-and-excitation (SE) module consists of a squeeze operation and an excitation operation.
The squeeze operation is completed through the global average pooling process in Eq. (1), which compresses the H x W pixel values of each channel into real numbers, and the excitation operation reduces and restores the dimensionality through two 1 × 1 convolution layers to obtain the relationship between each pair of channels.
3. Experimental methods
Our experiment has been approved by the ethical committee of the Beijing Yizhuang International Biomedical Technology Co., LTD, with the ethical number of 2019S082.
3.1. Data description
Our model is tested on two different groups of mouse retinal datasets to verify its effects on the binary classification problem and the multiclassification problem. The binary classification dataset is collected from the inner nuclear layer, which is used to segment horizontal cells in the inner core layer. The multiclassification dataset is collected from the mouse retina images and consists of multiple layers and ganglion cells. The two datasets are collected from different diabetes mice, with a total of 640 images. The original mouse retinal microscopy images are collected from the Beijing Duan-Dian Pharmaceutical Research and Development Co., Ltd. The original images (with resolutions of 1360 × 1024) are preprocessed by resizing them to 512 × 512. Fig. 4 illustrates the mouse retinal segmentation datasets, where (a) represents the original image for binary segmentation, and (b) corresponds to the ground-truth for binary segmentation; (c) is an original image for multiclass segmentation, and (d) is the corresponding ground truth for multiclass segmentation. One group of datasets contains 316 labels belonging to two categories (background and ganglion cells).
Fig. 4.
The mouse retinal datasets. (a) Is an original image for binary segmentation; (b) is the corresponding ground-truth for binary segmentation; (c) is an original image for multiclass segmentation; (d) is the corresponding ground truth for multiclass segmentation.
Because ganglion cells are easily confused with vacuoles and have different shapes, it is very appropriate to verify the segmentation performance of the model for small objects. The other group of datasets includes 324 labels belonging to seven categories, which are defined as follows: background, retinal nerve fiber layer (RNFL), Inner plexiform layer (IPL), Inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL) and cells in RNFL. Based on these definitions, we can observe that the main differences between different layers and cells lie in their shapes and pixel regions. The pixel area of the layers is large, but the number of layers is low, and the layer edges are blurred in the mouse retinal image. In contrast, the pixel area of the cells is small, but the number of layers is large, and the layer edges are clear in the mouse retinal image. Therefore, it is appropriate to test the segmentation performance of the proposed model for multiscale objects. Additionally, the segmentation regions and labels of the mouse retinal images (forming two categories and seven categories, respectively) are manually annotated by the data provider.
3.2. Experiment setup and environment
The experiments are conducted on the Linux operating system in the Python 3.7 programming language, which is linked with the PyTorch 1.7 deep learning framework. In addition, the experiments are based on the CUDA10.0 GTX 2080Ti graphics card configuration and the CUDNN7.6 basic deep neural network model training library. Our model is trained and evaluated in terms of its performance based on the following experimental settings.
To showcase the effectiveness of our algorithm, we conducted a comparative analysis with four state-of-the-art models: FPN [25], U-Net [26], DeepLabV3 [27], and PSPNet [28]. These models represent some of the most renowned deep learning architectures in the field of semantic segmentation. Widely utilized in diverse scenarios, they have demonstrated impressive segmentation results. As our baseline network, FPN has excellent feature fusion capabilities that enable it to tackle multiple computer vision tasks with great success. U-Net has gained popularity as a widely used architecture for numerous image segmentation tasks, including medical image analysis, cell segmentation, and semantic segmentation of natural images. Its effectiveness in capturing detailed information and preserving spatial context has made it a favored choice in various image segmentation applications. DeepLabV3 is a specialized model architecture explicitly designed for semantic image segmentation. It has also been extended to support panoptic segmentation, which combines instance segmentation and semantic segmentation into a single task. The advantage of PSPNet is that it can capture contextual information on multiple scales, which is important for semantic segmentation tasks where the size and shape of objects vary greatly.
To ensure fairness in the experiments, ResNet 50 was employed as the backbone for all the models. In addition, Kaiming initialization [29] was utilized for initializing the network models, ensuring consistent and reliable results in the comparative experiments. This approach minimizes any potential biases that may arise due to variations in initialization and backbone architecture among the models. During the process of all model training, the initial learning rate is set to 0.0001, and the minibatch size is set to 4. We adopt the Adam optimization method and apply the stepLR algorithm to improve it. The step size is set as 4, gamma is set as 0.92 and the maximum number of iterations is set as 300.
For binary semantic segmentation, the goal is to differentiate between ganglion cells and background pixels in the mouse retinal images. We gain a reliable model training and evaluation by the method of 5-fold cross validation. The original set of 316 mouse retinal images is randomly divided into five subsets. This cross-validation approach allows us to rigorously assess the model's performance by training and testing on different subsets of the dataset, ensuring a comprehensive evaluation of the model's effectiveness and generalization capabilities. In the experiment, we train the model by randomly selecting four subsets, and the remaining subset is used for testing. The loss function we use consists of two parts: the cross-entropy loss () and the dice loss (). The combination of loss functions is represented in Equation (2):
| (2) |
The cross-entropy loss function is used to measure the degree of intersection between the expected output and the actual output, as defined in Equation (3):
| (3) |
where indicates a true label and indicates a predicted segmentation result. The dice loss function is used to calculate the loss of the intersection over union between the prediction result region and the ground-truth region and is defined in Equation (4):
| (4) |
where indicates a ground truth segmentation image and indicates a predicted segmentation image.
Multiclass semantic segmentation: Each layer and the ganglion cells in the mouse retinal images are separated from the background pixels. We implement 5-fold cross-validation to ensure robust model training and evaluation. The initial set of 324 mouse retinal images is randomly partitioned into five subsets, each containing 65, 65, 65, 65, and 66 images, respectively. In each iteration of the cross-validation, we utilize four subsets for model training and withhold the remaining subset for testing and evaluation. The loss functions are the same as those used above for binary classification and segmentation. We train our model with the “freezing-based training” method. The model requires 300 iterations in total. In the first 50 iterations, we freeze the feature extraction module and only train the upsampling module of the model. After that, we unfreeze the feature extraction module and then continue for 250 rounds of iterative training.
3.3. Evaluation indicators
In order to validate and compare the effectiveness of these networks, we adopt four metrics, including mean pixel accuracy (mPA), mean intersection over union (mIoU), sensitivity (SE) and the Dice similarity coefficient (Disc). The mPA represents the average proportion of mouse retinal layers or cells that are correctly classified among all mouse retinal pixels. The mIoU metric calculates the average proportion between the number of correct pixels in each category and the total number of pixel points belonging to this category in the mouse retinal images. SE indicates the proportion of correctly classified samples in each prediction sample category. The Disc demonstrates the correspondence between the ground-truth segmentation label and the predicted segmentation label of the network output. Equations (5)–(8) display the expressions used to compute these metrics.
| (5) |
| (6) |
| (7) |
| (8) |
4. Results and discussion
4.1. Binary semantic segmentation
Table 1 reports the network performance evaluation results achieved for the binary ganglion cell segmentation task in mouse retinal images by five methods (the FPN, U-Net, the PSPNet, DeepLabV3 and our model). The quantitative indicators in the table are computed by averaging the results obtained over the five experiments. The findings indicate that, in relation to all metrics, the proposed model surpasses the other five networks. Additionally, our model produces values that are 0.0371 and 0.0262 higher than the mean IoU (mIoU) scores of the PSPNet and DeepLabV3, respectively. Based on these results, we can draw the conclusion that specific cells in the binary classification mouse retinal datasets are relatively small in size. Consequently, their features cannot be adequately captured by using an atrous spatial pyramid pooling module with a large dilation rate or a pyramid pooling module. This limitation may lead to suboptimal segmentation performance for these smaller cells, highlighting the importance of using an appropriate model architecture that can handle objects of varying sizes effectively. This can also explain why our model performs better, as its attention mechanism and dilated convolution group with different dilation rates can preserve more fine details by selecting the appropriate receptive field for each multiscale object. In Fig. 5, we present a comparison of the segmented images produced by each model in the mouse retina binary dataset. (a) Depicts the original image, (b) shows a partial cut of the original image (a), and (c) represents the corresponding ground truth for (a). The segmented images from the respective models are displayed as follows: (d) Our model, (e) FPN, (f) U-Net, (g) DeepLabV3, and (h) PSPNet. When comparing the segmented images in (e), (f), (g), and (h) with the result of the proposed method (d), it becomes evident that our proposed method achieves more accurate segmentation of ganglion cells, with clearer boundaries. In particular, for the area described by the red box in the figure, we can observe that the cell contour of proposed method is most similar with the true label, and the stained vacuoles are not categorized as ganglion cells. This result further proves that our model is superior to other advanced deep learning methods in terms of small-target segmentation.
Table 1.
Comparison among the indicators of different models.
| mIoU | mPA | Disc | Sensitivity | |
|---|---|---|---|---|
| FPN | 0.8361 | 0.9188 | 0.9107 | 0.8571 |
| U-Net | 0.8345 | 0.9173 | 0.9098 | 0.8614 |
| PSPNet | 0.8093 | 0.9157 | 0.8946 | 0.8587 |
| DeepLabV3 | 0.8202 | 0.9163 | 0.9012 | 0.8638 |
| Our model | 0.8464 | 0.9256 | 0.9168 | 0.8694 |
Fig. 5.
This comparison showcases the segmentation results produced by each model for the mouse retinal binary class datasets.(a) is an original image; (b) is a partial cut of the original image (a); (c) is the ground truth of (a). (d) Our model; (e) the FPN; (f) U-Net; (g) DeepLabV3; (h) the PSPNet.
4.2. Multiclass semantic segmentation
Due to the presence of multiscale objects and obscured boundaries, the segmentation of mouse retinal images becomes challenging. To verify the effectiveness of our model in this article, we carry out a verification experiment on the multiclass datasets and compare it with several famous deep learning models. Mouse retinal image semantic segmentation is performed by the FPN, U-Net, the PSPNet and DeepLabV3 for 7 classes (6 classes of layers, cells and background). Each category and each mIoU result are listed in Table 2. Table 2 illustrates that our model outperforms the other four methods in terms of average mIoUs, with a 0.0152 higher score compared to U-Net. In particular, for Classes 5 and 6, the scores of our model are 0.0372 and 0.0479 higher than that of U-Net, respectively. Through the analysis of the above results, we can conclude that replacing the traditional nearest-neighbor upsampling method with the transposed convolution method yields higher segmentation accuracy in the top-down decoding pathway.
Table 2.
Comparison among the mIoU results of different models.
| Our model | U-Net | PSPNet | DeepLabV3 | FPN | |
|---|---|---|---|---|---|
| Background | 0.9846 | 0.9844 | 0.9798 | 0.9850 | 0.9839 |
| Class 1 | 0.9048 | 0.8943 | 0.8583 | 0.8854 | 0.8996 |
| Class 2 | 0.9345 | 0.9256 | 0.9210 | 0.9339 | 0.9278 |
| Class 3 | 0.9318 | 0.9331 | 0.9016 | 0.9312 | 0.9285 |
| Class 4 | 0.6949 | 0.6926 | 0.6745 | 0.6899 | 0.7084 |
| Class 5 | 0.7047 | 0.6675 | 0.6436 | 0.6774 | 0.6867 |
| Class 6 | 0.6613 | 0.6134 | 0.4951 | 0.5577 | 0.6530 |
| Average | 0.8310 | 0.8158 | 0.7820 | 0.8086 | 0.8269 |
To additionally demonstrate the efficacy of the proposed model, this research performs a generalization assessment on the multi-class mouse retinal datasets. Table 3 demonstrates the superiority of our model over FPN in terms of both mIoU and sensitivity scores, highlighting the effectiveness of our model in multiscale segmentation. At the same time, it can also be seen that the developed structure, which combines dilated convolution and a residual block, is helpful for improving the segmentation performance of the original FPN for multiscale objects. In Fig. 6, we present a comparison of the segmented images generated by each model in a multi-class dataset of mouse retina. (a) represents the original image, (b) shows a partial cut of the original image (a), and (c) displays the corresponding ground truth for (a). The segmented images from the respective models are as follows: (d) Our model, (e) FPN, (f) U-Net, (g) DeepLabV3, and (h) PSPNet. From the white box in Fig. 6, it can be observed that in the deep learning contrast model, FPN (e) has the best visual effect in the segmentation of layers and adhesive cells. However, compared to the label mask (b) in Fig. 6, the output result of FPN is not satisfactory in the details of adhesive cells and layers. From the white box, we can observe that the three independent cells are not completely separated, and the layer segmentation is too smooth, lacking a sense of sawtooth. On the contrary, our model (d) overcomes these two problems and is more similar to the label mask. This also proves that the dilated residual blocks in our model play an important role in the segmentation results. Because dilated residual blocks based on FPN can store more multiscale object details, further enhancing the ability to extract layer and adhesive cell details from mouse retinal images, resulting in more accurate semantic segmentation results.
Table 3.
Comparison among the indicators of different models.
| mPA | mIoU | Disc | Sensitivity | |
|---|---|---|---|---|
| FPN | 0.8868 | 0.8069 | 0.9052 | 0.8764 |
| U-Net | 0.8875 | 0.8158 | 0.8986 | 0.8715 |
| PSPNet | 0.8324 | 0.7820 | 0.8777 | 0.8245 |
| DeepLabV3 | 0.8572 | 0.8086 | 0.8942 | 0.8432 |
| Our model | 0.8984 | 0.8310 | 0.9077 | 0.8873 |
4.3. Ablation study
In this research, we introduced a dilated residual method based on FPN, which demonstrated exceptional performance on the mouse retinal dataset. To assess the model's interpretability, stability, and its ability to effectively segment cells and layers in mouse retinal images, we conducted several ablation experiments on the multiclass mouse retinal dataset. In these experiments, we utilized mIoU as the evaluation metric to quantitatively measure the model's segmentation accuracy and effectiveness.
During the ablation experiments, we established FPN as the baseline model for comparison. By optimizing FPN and incorporating different combinations of the dilated residual module and attention mechanism (AM), we derived three additional models for comparison. To begin, we assessed the performance of the dilated residual module by replacing the standard 3 × 3 convolutional layers in FPN's encoder with the dilated residual module. This substitution allowed for an enlargement of the receptive fields and semantic information without compromising spatial resolution. Through this evaluation, we sought to understand the impact of the dilated residual module on the segmentation results and its potential benefits in handling complex features in the encoder's processing. In Table 4, we can observe that removing the module decreased the model's performance by 1.35%. These findings highlight the significance of the dilated residual module in our proposed model, as it effectively captures more spatial feature information, leading to more accurate segmentation results. In the subsequent experiments, we incorporated the attention mechanism (AM) to modify the encoder module of the baseline model. By integrating the AM, our model can learn to emphasize informative features and suppress weaker ones using global information, enhancing its focus on the contours and intricate details of segmented objects. The experimental results in Table 4 further confirm the effectiveness of the AM, as it improves the mIoU score by 1.06% compared to the baseline model. This improvement demonstrates the importance of attention mechanisms in refining the model's segmentation performance and optimizing feature representation.
Table 4.
Ablation results on the multiclass mouse retinal dataset.
| Index | Baseline | DR | AM | mIoU |
|---|---|---|---|---|
| 1 | ✓ | 0.8069 | ||
| 2 | ✓ | ✓ | 0.8189 | |
| 3 | ✓ | ✓ | 0.8175 | |
| 4 | ✓ | ✓ | ✓ | 0.8310 |
From the results of the ablation study, it can be seen that each module in the proposed semantic segmentation model plays an important role, as verified in Table 4. At the same time, we also found that the segmentation results of the model for certain specific object categories are poor, such as cells attached to layer boundaries and irregular layer boundaries, which may be related to the capacity of the dataset and training strategies, and require further research. In addition, the model's performance was dependent on the image size, with poor performance on small-sized images, which indicates that the model's performance has some dependency on the image size and requires optimization for different image sizes. In conclusion, our ablation study provides important insights into the model's performance and stability, and guides us to further optimize the model.
5. Conclusions
Analyzing mouse retinal images is of great importance for diagnosing DR. In the article, we introduce a deep learning architecture based on an FPN, whose backbone feature extraction network is a combination of dilated convolution and a residual block. On this basis, we integrate an SE attention module into the backbone network to obtain more segmentation object details. In the top-down decoding pathway, we replace the traditional nearest-neighbor upsampling method with the transposed convolution method and add a segmentation head in the last layer to obtain semantic segmentation results that are consistent with the original image sizes. Through an experimental verification, regarding ganglion cell segmentation (binary semantic segmentation), the mPA and mIoU values of our approach reach 0.9732 and 0.9213, respectively; regarding mouse retinal cell and layer segmentation (multiclass semantic segmentation), the mPA and mIoU values reach 0.8984 and 0.8310, respectively. Through a detailed analysis of the results of the ablation experiment, we can observe that the DR module plays a crucial role in obtaining enhanced spatial feature information, resulting in more precise segmentation outcomes. At the same time, the AM module enables our model to selectively emphasize significant information features and suppress weaker features using global information, thereby paying more attention to the contours and details of segmented objects. Therefore, the proposed modules provide the network with better ability to classify multiscale objects and small objects. Overall, we prove that the developed dilation residual FPN is a robust method for mouse retinal image segmentation and DR diagnosis. In addition, we are working on implementing postprocessing steps to further explore the predicted segmentation results. The objective is to obtain detailed statistics, such as cell counts, circumferences, and average areas, which will offer enhanced support for DR diagnosis.
Ethics approval statement
Our experiment has been approved by the ethical committee of the Beijing Yizhuang International Biomedical Technology Co., LTD, with the ethical number of 2019S082.
Author contribution statement
Zhihao Che: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Fukun Bi: Conceived and designed the experiments; Analyzed and interpreted the data.
Yu Sun: Conceived and designed the experiments.
Weiying Xing; Hui Huang; Xinyue Zhang: Contributed reagents, materials, analysis tools or data.
Funding statement
This research was funded by the National Natural Science Foundation of China (Grant No. 61971006).
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Zhihao Che, Email: zhihaoche@163.com.
Fukun Bi, Email: bifukun@163.com.
Yu Sun, Email: yusun@mail.ncut.edu.cn.
Weiying Xing, Email: xwy_1216@163.com.
Hui Huang, Email: huanghui@duan-dian.com.
Xinyue Zhang, Email: zhangxinyue@duan-dian.com.
References
- 1.Jayalakshmi T., Santhakumaran A. 2010. A Novel Classification Method for Diagnosis of Diabetes Mellitus Using Artificial Neural Networks. 02.09. [DOI] [Google Scholar]
- 2.Sumangali K., Geetika B., Ambarkar H. International Conference on Control; 2016. A Classifier Based Approach for Early Detection of Diabetes Mellitus; pp. 389–392. [Google Scholar]
- 3.Duh E.J., Sun J.K., Stitt A.W. Diabetic retinopathy: current understanding, mechanisms,16 and treatment strategies. JCI Insight. 2017;2(14) doi: 10.1172/jci.insight.93751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyoung, Jin, Noh, et al. Scale-space approximated convolutional neural networks for retinal vessel segmentation - ScienceDirect. Comput. Methods Progr. Biomed. 2019;178:237–246. doi: 10.1016/j.cmpb.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 5.F Raz M.M., Remagnino P., Hoppe A., et al. Blood vessel segmentation methodologies in retinal images – a survey. Comput. Methods Progr. Biomed. 2012;108(1):407–433. doi: 10.1016/j.cmpb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Dysli C., Enzmann V., Sznitman R., Zinkernagel M.S., Dysli C., Enzmann V., Sznitman R., Zinkernagel M.S. 2015. Quantitative Analysis of Mouse Retinal Layers Using Automated Segmentation of Spectral Domain Optical Coherence Tomography Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Augustin M., Harper D.J., Merkle C.W., Hitzenberger C.K., Baumann B. 2019. Segmentation of Retinal Layers in OCT Images of the Mouse Eye Utilizing Polarization Contrast. 08.13. [DOI] [Google Scholar]
- 8.Srinivasan P.P., Heflin S.J., Izatt J.A., shavsky V.Y., Farsiu S. Automatic segmentation of up to ten layer boundaries in SD-OCT images of the mouse retina with and without missing layers due to pathology. Biomed. Opt Express. 2014;3(15) doi: 10.1364/BOE.5.000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao S., Bucher F., Wu Y., et al. Web based, fully automated, deep learning segmentation of oxygen induced retinopathy. Invest. Ophthalmol. Vis. Sci. 2018;(9):59. [Google Scholar]
- 10.Kassim Y.M., Glinskii O.V., Glinsky V.V., et al. 2019 IEEE International Conference on Image Processing (ICIP) IEEE; 2019. Deep U-net regression and hand-crafted feature fusion for accurate blood vessel segmentation. [Google Scholar]
- 11.Jc A, Hy A, Kai L B. A review of thyroid gland segmentation and thyroid nodule segmentation methods for medical ultrasound images[J]. Comput. Methods Progr. Biomed., 185. [DOI] [PubMed]
- 12.Bian X., Luo X., Wang C., et al. Optic disc and optic cup segmentation based on anatomy guided cascade network. Comput. Methods Progr. Biomed. 2020:197. doi: 10.1016/j.cmpb.2020.105717. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y., Liu Y., Georgiou T., Lew M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimedia Inf. Retrieval. 2018;7(2):87–93. [Google Scholar]
- 14.Zhu H., Meng F., Cai J., Lu S. Beyond pixels: a comprehensive survey from bottom-up to semantic image segmentation and cosegmentation. J. Vis. Commun. Image Represent. 2016;34:12–27. [Google Scholar]
- 15.Long J., Shelhamer E., Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 16.Ronneberger O., Fischer P., Brox T., U-net . International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015. Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 17.Lin T.-Y., Dollar P., Girshick R., He K., Hariharan B., Belongie S. Feature pyramid networks for object detection. CVPR. 2017;1:4. page. [Google Scholar]
- 18.Kirillov A., He K., Girshick R., Dollar P. A unified architecture for instance and semantic segmentation. http://presentations.cocodataset.org/COCO17-Stuff-FAIR.pdf
- 19.Cui H., Chang Y., Jiang L., et al. Multiscale attention guided U-Net architecture for cardiac segmentation in short-axis MRI images. Comput. Methods Progr. Biomed. 2021;206(1) doi: 10.1016/j.cmpb.2021.106142. [DOI] [PubMed] [Google Scholar]
- 20.Ren S., He K., Girshick R., Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(6):1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 21.Dan C.C., Meier U., Masci J., et al., Flexible, high performance convolutional neural networks for image classification. In Proceedings of the Twenty-Second international joint conference on Artificial Intelligence - Volume Volume Two (IJCAI'11). AAAI Press, 1237–1242..
- 22.Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;25(2) [Google Scholar]
- 23.Ciresan D., Meier U., Schmidhuber J. Computer Vision & Pattern Recognition. IEEE; 2012. Multi-column deep neural networks for image classification. [DOI] [PubMed] [Google Scholar]
- 24.Hu J., Shen L., Sun G. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Squeeze-and-Excitation networks; pp. 7132–7141. [DOI] [Google Scholar]
- 25.Lin T.Y., Dollar P., Girshick R., et al. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE Computer Society; 2017. Feature pyramid networks for object detection. [Google Scholar]
- 26.Ronneberger O., Fischer P., Brox T. Inter-national Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 27.Chen L.C., Papandreou G., Schroff F., et al. 2017. Rethinking Atrous Convolution for Semantic Image Segmentation. [Google Scholar]
- 28.Zhao H., Shi J., Qi X., et al. IEEE Computer Society; 2016. Pyramid Scene Parsing Network. [Google Scholar]
- 29.He K., Zhang X., Ren S., et al. IEEE Computer Society; 2015. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.