Abstract
The sequence data (H. Okamoto et al., Hepatol. Res. 10:1–16, 1998) of a newly discovered single-stranded DNA virus, TT virus (TTV), showed that it did not have the terminal structure typical of a parvovirus. Elucidation of the complete genome structure was necessary to understand the nature of TTV. We obtained a 1.0-kb amplified product from serum samples of four TTV carriers by an inverted, nested long PCR targeted for nucleotides (nt) 3025 to 3739 and 1 to 216 of TTV. The sequence of a clone obtained from serum sample TA278 was compared with those registered in GenBank. The complete circular TTV genome contained a novel sequence of 113 nt (nt 3740 to 3852 [=0]) in between the known 3′- and 5′-end arms, forming a 117-nt GC-rich stretch (GC content, 90.6% at nt 3736 to 3852). We found a 36-nt stretch (nt 3816 to 3851) with an 80.6% similarity to chicken anemia virus (CAV) (nt 2237 to 2272 of M55918), a vertebrate circovirus. A putative SP-1 site was located at nt 3834 to 3839, followed by a TATA box at nt 85 to 90, the first initiation codon of a putative VP2 at nt 107 to 109, the termination codon of a putative VP1 at nt 2899 to 2901, and a poly(A) signal at nt 3073 to 3078. The arrangement was similar to that of CAV. Furthermore, several AP-2 and ATF/CREB binding sites and an NF-κB site were arranged around the GC-rich region in both TTV and CAV. The data suggested that TTV is circular and similar to CAV in its genomic organization, implying that TTV is the first human circovirus.
Recently, the isolation of DNA clones of a new human virus, TT virus (TTV), from a Japanese patient with posttransfusion hepatitis has been reported (11, 13). Epidemiological studies by seminested PCRs showed that TTV is globally widespread among the general population (2, 10, 13, 14). However, the pathogenic nature of this virus is still unclear, even though it seems to be more prevalent in posttransfusion hepatitis patients (2, 5, 13). Okamoto et al. reported that TTV is nonenveloped and has a linear single-stranded DNA (ssDNA) genome (13). They presented a sequence of 3,739 nucleotides (nt) and suggested that TTV may belong to the parvovirus group. Parvovirus has a linear ssDNA genome with a palindromic structure on both ends (1). However, the sequence presented by Okamoto et al. (13) did not contain the palindrome. Here we clarify the full genome structure of TTV.
MATERIALS AND METHODS
Sera.
The serum sample TA278, a key serum for the discovery of TTV, was supplied by M. Mayumi of Jichi Medical School, Tochigi, Japan. We screened the sera of pediatric non-A–G hepatitis patients by a seminested PCR (13). The sera used here included three positive ones (TP88, TP97, and TP110) and a negative one (TP100). These sera were obtained from patients at the Department of Pediatrics, Faculty of Medicine, Tottori University, under informed consent.
DNA extraction.
Each serum sample of 100 μl was treated with 0.5% sodium dodecyl sulfate and 0.12 mg of proteinase K per ml at 37°C for 16 h. The DNA was extracted by phenol, phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform-isoamyl alcohol. The DNA was ethanol precipitated with 20 μg of glycogen (Boehringer, Mannheim, Germany) as a carrier. The DNA was dissolved in 40 μl of 10 mM Tris-HCl (pH 8.3)–0.1 mM EDTA, and it was stored at −20°C until use.
Primers for PCR.
The primers for the seminested short PCR (13) were supplied by M. Mayumi. For the inverted long PCR, we designed nested pairs of inverted primers: TTV2915 (C2915CCCAAACCTTACAACCCTTCC2936) and TTV324 (T347TAGTAGTTTGCAACGTTCTTTG324) for the outer primer pair and TTV3025 (CGCGGATCCG3025GGCCCCCAAGGAAAACCAGTA3046) and TTV195 (AAGGAAAAAAGCGGCCGCA216TTGCCCCTTGACTTCGGTGTG195) for the inner primer pair. For the inner primers, NotI and BamHI tags (indicated in italics) were added for cloning purposes.
PCR.
For TTV genome screening, we utilized the seminested PCR targeted for nt 1915 to 2185 (GenBank no. AB008394). The sensitivity of the PCR was three copies per reaction as determined by a cloned DNA containing nt 946 to 2226 of TTV. To guarantee the sensitivity of each PCR, we added a pair of tubes, each containing six copies of the cloned DNA, as positive controls in the routine screening (7).
For the inverted, nested PCR, we applied a hot-start and long PCR method with minor modifications (7). The first and second PCRs were performed for 35 cycles (each cycle was 94°C for 1 min, 59°C for 1 min, and 72°C for 3 min) with a programmable incubator (API 300; ASTEC, Fukuoka, Japan). The extracted DNA, 10 μl, was used in the first PCR, and a 5-μl aliquot of the first PCR product was used in the second PCR. A reaction cocktail, 50 μl, consisted of 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, a 0.25 mM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 1.25 U of Taq DNA polymerase (Pharmacia Biotech, Uppsala, Sweden), 0.005 U of Deep Vent DNA polymerase (New England Biolabs, Beverly, Mass.), 5% dimethyl sulfoxide, and 2% glycerol. The PCR product was electrophoresed on a 1% agarose S (Nippon Gene, Toyama, Japan) gel, and the gel was stained with ethidium bromide.
Cloning and DNA sequencing.
The 1.0-kb product amplified by the inverted, nested long PCR of DNA derived from TA278 was digested with BamHI and NotI and cloned into a vector, pBluescript SK(−). The BamHI/SacI fragment from one of these clones was recloned into pTZ19U (pTZ27852). From each end of the insert, series of deletion mutants were generated with a Kilo-Sequence Deletion kit (Takara Biomedicals, Kyoto, Japan) (6). In brief, to make a series of deletion mutants from the 5′ end of the insert, pTZ27852 was restricted with BamHI/Sse8387I at the 5′ end of the insert (series BS). Following serial digestions with exonuclease III for up to 10 min, the single-stranded region was digested out with mung bean nuclease. After both ends were repaired by Klenow fragment, the product was self-ligated. Constructs with serial deletions from the 3′ end of the insert were generated similarly with NotI/SacI restriction of pTZ27852 (series NS). At least three of these mutants overlapping in each area were selected for sequencing based on the method of Sanger et al. (15). We used a Thermo Sequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) in an ALFred DNA sequencer (Pharmacia LKB, Uppsala, Sweden). Since the novel GC-rich region was refractory to the Sanger method, we obtained shorter subclones, such as pBS317pma (nt 3706 to 3852 and 1 to 22), from one of the deletion plasmids, pBS317 (nt 3706 to 3852 and 1 to 216). These served in chemical sequencing after the method of Maxam and Gilbert (8) by using [γ-32P]ATP.
Southern blotting.
To confirm the specificity of the newly identified GC-rich region for TTV, PCR products of the inverted PCR were probed by a EcoRI/HindIII insert fragment of pBS317pma; 5′ ends on both of the strands were radiolabeled with [γ-32P]ATP by polynucleotide kinase (Takara Biomedicals). The DNA electrophoresed in a 1% agarose gel was blotted onto a nylon membrane (Hybond N plus; Amersham Pharmacia Biotech) by an alkaline transfer method. Following prehybridization at 65°C for 2 h, the DNA was hybridized with the 32P-probe under a highly stringent condition at 65°C for 16 h. After being washed three times with 40 mM phosphate buffer containing 0.1% sodium dodecyl sulfate (Church’s buffer [3]) at 68°C for 5 min, the membrane was dried and autoradiographed.
Computer work.
Database searches and sequence analyses were performed by FASTA in the DNA Data Bank of Japan (DDBJ) and by Genetyx (version 9.0). For alignment of the DNA, we used Sequencher (version 3.0).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence database with the accession no. AB017911.
RESULTS
PCR.
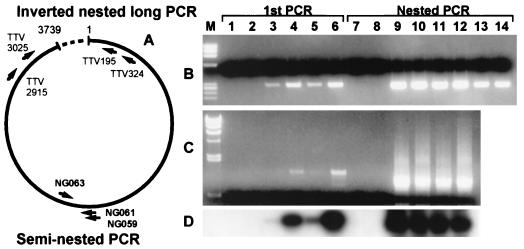
In the seminested short PCR (Fig. 1A), the serum samples TA278, TP88, TP97, and TP110 gave a positive signal with an expected size of 286 nt after the first PCR (Fig. 1B). All of them gave a strong signal with a size of 271 nt after the second PCR. The same set of samples was used in the inverted, nested long PCR (Fig. 1A). Two high-virus-load samples, TP88 and TP110, gave a positive signal at 1.2 kb in the first PCR, and all positive samples gave a strong signal at 1.0 kb after the second PCR (Fig. 1C). The negative control sample, TP100, was consistently negative. The results suggested that this novel sequence is a part of the TTV genome and that its structure is circular instead of linear. Since our inverted, nested PCR was expected to read 931 nt on the reported sequence (nt 3025 to 3739 and 1 to 216), the estimated length of the novel sequence was ∼100 nt.
FIG. 1.
Seminested short PCR and inverted, nested long PCR for TTV. (A) Schematic diagram of the TTV genome and target regions for the seminested and inverted, nested long PCRs. The dotted region in the genome indicates the region identified in this study. Arrows indicate locations and directions of primers. (B) Seminested short PCR. The products were electrophoresed in a 3% agarose gel and stained with ethidium bromide. The expected sizes of the products of the first and second PCRs were 286 and 271 nt, respectively. (C) Inverted, nested long PCR. The products were electrophoresed in a 1% agarose gel and stained with ethidium bromide. The product sizes of the first and second PCRs were 1.2 and 1.0 kb, respectively. (D) Southern blotting of the gel in panel C probed by the insert of pBS317pma (nt 3706 to 3852 and 1 to 22). Note that the autoradiogram for the first PCR is overexposed. Lanes: M, φX174 HaeIII digest for panel B and λ phage HindIII digest for panel C; 1 and 7, template-free control; 2 and 8, TP100 negative serum; 3 to 6 and 9 to 12, TA278, TP88, TP97, and TP110 sera, respectively; 13 and 14, a cloned TTV DNA at six copies per reaction.
To confirm that this novel sequence is a part of the TTV genome, the electrophoresed gel (Fig. 1C) was probed in a Southern blot by the pBS317pma insert encompassing the novel region (nt 3706 to 3852 and 1 to 22). The probe hybridized with the PCR product bands of all positive samples (Fig. 1D, lanes 3 to 6 and 9 to 12) but not with that of TP100 (lanes 2 and 8). This suggested that the novel region is common to all TTV-positive samples.
DNA sequencing.
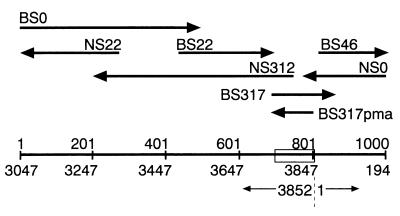
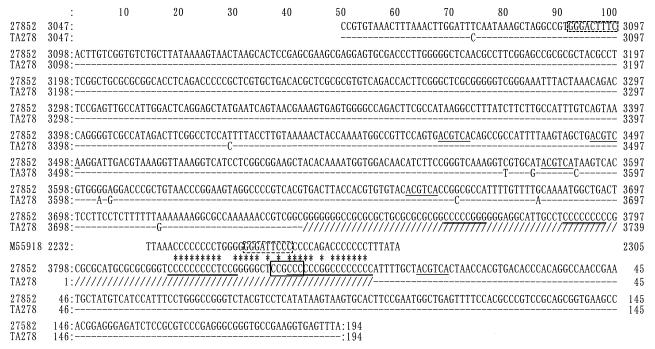
To determine the nucleotide sequence of the novel region, the series of deletion mutants obtained from pTZ27852 from both directions was sequenced (Fig. 2). The enzymatic and chemical sequencing revealed that the 1.0-kb product of the inverted PCR had a total length of 1,000 nt between the inner primers (nt 3047 to 3852 and 1 to 194) (Fig. 3). The similarity of the sequence to that of AB008394 was 98.6% for the 3′-terminal 693 nt (nt 3047 to 3739) and 100% for the 5′-terminal 194 nt (nt 1 to 194). No deletion or addition of nucleotides was found in these two regions. Within the former, 5 of 10 nucleotide changes were located in nt 3577 to 3604. We found a novel region consisting of ∼100 nt between nt 3739 and 1, which was resistant to the Sanger method. By the chemical sequencing, the region was found to be 113 nt long and next to the last C3736GGC3739 of AB008394 and to form a 117-nt-long stretch (nt 3736 to 3852 [=0]) with a GC content of 106/117 (90.6%).
FIG. 2.
Sequence strategy. The insert of pTZ27852, 1.0 kb in size, was sequenced from both directions with two series of deletion mutants: the BS series with 5′-end deletions and the NS series with 3′-end deletions. Shown are the area sequenced (not the size of insert) and the sequence direction of representative clones. Open box on the scale, the novel GC-rich region.
FIG. 3.
Nucleotide sequences of the pTZ27852 insert. The 1,000 nt between the inner primer pairs were compared with those of the previously reported TTV, both from sample TA278 (GenBank no. AB008394). A portion of the CAV genome (M55918) which had an 80.6% similarity to the TTV genome is also included. Dashes, nucleotide matches with AB008394; asterisks, nucleotide matches with M55918; underline, ATF/CREB site; thick underline, AP-2 site; solid box, SP-1 site; dashed box, NF-κB site.
ORF analysis.
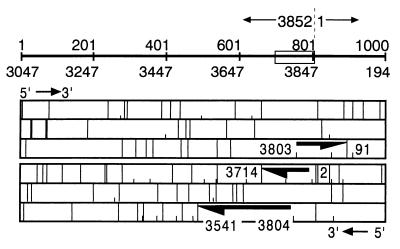
Open reading frame (ORF) analysis revealed three putative ORFs (>100 nt) extending into the novel region. One with 141 nt (nt 3803 to 3852 and 1 to 91) was on the plus strand, capable of coding for 47 amino acids (aa). The other two were on the minus strand with 141 nt (nt 2 to 1 and 3852 to 3714) and 264 nt (nt 3804 to 3541), capable of coding for 47 and 88 aa, respectively (Fig. 4). None of these ORFs were associated with TATA box or poly(A) sites in close proximity.
FIG. 4.
ORF analysis of pTZ27852. The upper panel is for the positive strand, and the lower panel is for the negative strand. The arrows indicate the ORFs found (>100 nt) and their directions. Short separator, initiation codon; long separator, termination codon; box in the scale, the novel GC-rich region.
Computer work.
We searched for some similarities between the sequence of the novel region of the TTV genome (nt 3740 to 3852) and those of the viral genomes registered in GenBank. The 36-nt stretch (nt 3816 to 3851) within the novel region had an 80.6% similarity with a part of the GC-rich region (nt 2237 to 2272) of the chicken anemia virus (CAV) (M55918; 2,319 nt in total) (Fig. 3). We compared the whole genome of TTV with that of CAV (M55918), using the TTV sequence (AB008394; nt 1 to 3739) connected with the novel region of pTZ27852 (nt 3740 to 3852).
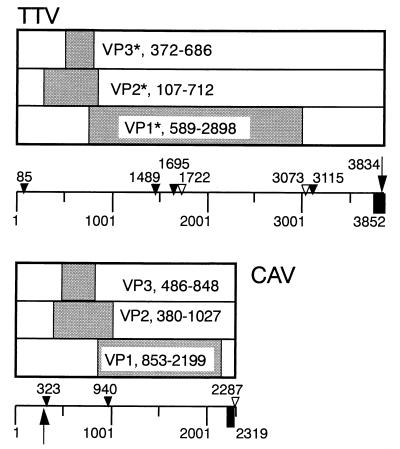
The nucleotide sequence similarities in other regions were insignificant. However, the genomic constructions of these viruses were similar in spite of the differences in their genomic sizes (Fig. 5). CAV has an ORF in each of the three different frames (4, 12). TTV also had a putative ORF in each frame, and the frames’ relative sizes and locations also resembled each other. Following the GC-rich sequence of CAV, there were a SP-1 site, a TATA box, and the first initiation codon of the VP2 ORF (Table 1). TTV had a putative SP-1 site at nt 3834 to 3839, near the end of the GC-rich region, which was followed by a TATA box at nt 85 to 90 and then by the first initiation codon of the putative VP2 ORF at nt 107 to 109. CAV has a poly(A) site after the VP1 ORF. Similarly, the poly(A) site at nt 3073 to 3078 of TTV was located approximately 100 nt downstream from the termination codon of the putative VP1.
FIG. 5.
Comparative scheme of three major ORFs of TTV and CAV. The ORFs of the TTV genome, VP1* through VP3*, are putative. Closed box, the novel GC-rich region; arrow, SP-1 site; open triangle, poly(A) site; closed triangle, TATA box.
TABLE 1.
Comparative genomic organizations of TTV and CAV
| Consensus sequence | Position (length) in:
|
|
|---|---|---|
| TTVa | CAVb | |
| 36-nt stretch | 3816–3851 | 2237–2272 |
| GC-rich region | 3736–3852 (117 nt) | 2206–2272 (67 nt) |
| ORFs | ||
| VP1 (frame 1) | 589–2898 (2,310 nt, 770 aa) | 853–2199 (1,347 nt, 449 aa) |
| VP2 (frame 2) | 107–712 (606 nt, 202 aa) | 380–1027 (648 nt, 216 aa) |
| VP3 (frame 3) | 372–686 (315 nt, 105 aa) | 486–848 (363 nt, 121 aa) |
| SP-1 site (GGGCGG/CCGCCC) | 3834–3839 | 305–310 |
| TATAc (ATATAA, TATATA) | 85–90 | 323–329 |
| Poly(A)c (AATAAA) | 3073–3078 | 2287–2302 |
| AP-2 sitesd (CCC[AC]N[CG][CG][CG]) | 3766–3773, 3788–3795, 3815–3822, 3837–3844 | 2148–2155, 2222–2229, 2237–2244, 2241–2248 |
| ATF/CREB (ACGTCA) | 3465–3470, 3493–3498, 3584–3589, 3659–3664, 9–14 | 157–162, 178–183, 199–204, 232–237, 253–258 |
| NF-κB (GGG[AG][ACT][CT][CT][CT][ACT]) | 3089–3097 | 2249–2258 |
As for the arrangement of putative regulatory sequences around the GC-rich region, the 36-nt stretch of both CAV and TTV contained two areas of putative AP-2 binding sites. In addition, two areas for putative AP-2 sites were found within 100 nt upstream of the 36-nt stretch in both viral genomes. While CAV has five putative ATF/CREB binding sites within 340 nt downstream of the GC-rich region, TTV had five putative ATF/CREB binding sites in the region of nt 3465 to 3852 and 1 to 14. CAV has a putative NF-κB binding site within its 36-nt stretch, which was missing in the 36-nt stretch of TTV. However, we did find one at nt 3089 to 3097.
DISCUSSION
The genomic sequence reported by Okamoto et al. (13) did not conform to a full viral genome of a parvovirus as they suggested, because it lacked terminal repeats. They reported that using an inverted PCR to reveal a circular structure was unsuccessful. We first used a commercial long-PCR kit, which failed. However, by using our nested long-PCR system as reported previously (7), we successfully amplified the novel region by an inverted PCR. The 1.0-kb amplified product contained 693 nt with a 98.6% similarity to the 3′-end region of TTV (AB008394; nt 3047 to 3739) and 194 nt with a 100% similarity to the 5′-end region of TTV (nt 1 to 194). In between, there was a 113-nt GC-rich region (nt 3740 to 3852) following C3736GGC3739, making a 117-nt GC-rich stretch. The results indicated that TTV has a circular genome with 3,852 nt.
Taxonomically, no human virus has been categorized as a circular ssDNA virus. Geminivirus is a plant virus that has one to two circular DNAs as a genome, and Inoviridae and Microviridae are bacterial viruses with a circular genome. Among vertebrate viruses, Circoviridae are known to have a circular ssDNA genome; CAV (4, 12) and the porcine circovirus (9) have been studied most extensively. The former has a 2.3-kb circular ssDNA genome, and the latter has a 1.8-kb one. However, no significant similarity in nucleotide sequences between these two viruses is known. The latter is reported to be more similar to plant circoviruses (9).
The novel region of TTV included a 36-nt stretch (nt 3816 to 3851) which had a 80.6% similarity with one region (nt 2237 to 2270 of M55918) of CAV. However, we could not find any significant nucleotide or amino acid sequence similarity between these two viruses in other regions. Although the TTV genome is ca. 1.5 times larger than that of CAV, both viruses had three major ORFs, one each in three frames. The relative lengths and locations and also the frame usage of the three major ORFs were quite similar (Fig. 5). Furthermore, the relative locations of the GC-rich region, SP-1 site, TATA box, the first initiation codon for VP2, and poly(A) signal following the termination codon of VP1 were also quite similar to each other. The TTV genome (AB008394) also had a second poly(A) signal, at nt 1722 to 1727 in the middle of the putative VP1 ORF. This one may not be significant, since it was not found in a clone we obtained from the same patient (data not shown).
Although there were three putative ORFs extending into the novel region in TTV, we could not find corresponding ORFs in the vicinity of the GC-rich sequence of CAV. These putative ORFs were not associated with nearby TATA box or poly(A) binding sites. These data may suggest that these three ORFs are nonfunctional.
In terms of regulatory sequences for gene expression, CAV has four (D10068) or five (M55918) direct repeats approximately 200 nt downstream from the 36-nt stretch (4, 12). The units are 19 and 21 nt long, respectively, and each contains a region capable of binding ATF/CREB. TTV did not have such long repeats, but it had five putative ATF/CREB binding sites starting from ca. 350 nt upstream to the 36-nt stretch. CAV had two putative AP-2 binding sites within the 36-nt stretch and two additional sites within an area 100 nt upstream. In contrast, TTV had several sites within the 36-nt stretch and two additional sites within an area 100 nt upstream. The CAV 36-nt stretch contained a putative NF-κB binding site, which was not maintained in the TTV 36-nt stretch. However, a putative NF-κB binding site was found at nt 3089 to 3097 of TTV. The putative regulatory element profile provided more evidence that TTV and CAV are related to each other. The results suggested that TTV is the first human virus that belongs to Circoviridae instead of Parvoviridae as previously suggested (13). The exploration of the complete TTV genome will facilitate a better understanding of this virus and its replication.
ACKNOWLEDGMENT
We thank Kenzo Sato for technical advice on chemical sequencing.
REFERENCES
- 1.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2173–2197. [Google Scholar]
- 2.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 3.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claessens J A, Schrier C C, Mockett A P, Jagt E H, Sondermeijer P J. Molecular cloning and sequence analysis of the genome of chicken anaemia agent. J Gen Virol. 1991;72:2003–2006. doi: 10.1099/0022-1317-72-8-2003. [DOI] [PubMed] [Google Scholar]
- 5.Cossart Y. TTV a common virus, but pathogenic? Lancet. 1998;352:164. doi: 10.1016/S0140-6736(05)77802-8. [DOI] [PubMed] [Google Scholar]
- 6.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 7.Kazi A, Miyata H, Kamahora T, Kurokawa K, Katamine S, Hino S. Deleted HTLV-1 provirus in cord-blood samples of babies born to HTLV-1-carrier mothers. Int J Cancer. 1998;77:701–704. doi: 10.1002/(sici)1097-0215(19980831)77:5<701::aid-ijc7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 10.Naoumov N V, Petrova E P, Thomas M G, Williams R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352:195–197. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 12.Noteborn M H M, de Boer G F, van Roozelaar D J, Karreman C, Kranenburg O, Vos J G, Jeurissen S H M, Hoeben R C, Zantema A, Koch G, van Ormondt H, van der Eb A J. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. 1991;65:3131–3139. doi: 10.1128/jvi.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 14.Prescott L E, Simmonds P. Global distribution of transfusion-transmitted virus. N Engl J Med. 1998;339:776–777. doi: 10.1056/NEJM199809103391118. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmonds P, Davidson F, Lycett C, Prescott L, MacDonald D, Ellender J, Yap P, Ludlam C, Haydon G, Gillon J, Jarvis L. Detection of a novel DNA virus (TTV) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]