Abstract
Cytochrome P450 CYP102A1 is a prototypic biocatalyst that has great potential in chemical synthesis, drug discovery, and biotechnology. CYP102A1 variants engineered by directed evolution and/or rational design are capable of catalyzing the oxidation of a wide range of organic compounds. However, it is difficult to foresee the outcome of engineering CYP102A1 for a compound of interest. Here, we introduce UniDesign as a computational framework for enzyme design and engineering. We tested UniDesign by redesigning CYP102A1 for stereoselective metabolism of omeprazole (OMP), a proton pump inhibitor, starting from an active but nonstereoselective triple mutant (TM: A82F/F87V/L188Q). To shift stereoselectivity toward (R)-OMP, we computationally scanned three active site positions (75, 264, and 328) for mutations that would stabilize the binding of the transition state of (R)-OMP while destabilizing that of (S)-OMP and picked three variants, namely UD1 (TM/L75I), UD2 (TM/A264G), and UD3 (TM/A328V), for experimentation, based on computed energy scores and models. UD1, UD2, and UD3 exhibit high turnover rates of 55 ± 4.7, 84 ± 4.8, and 79 ± 5.7 min−1, respectively, for (R)-OMP hydroxylation, whereas the corresponding rates for (S)-OMP are only 2.2 ± 0.19, 6.0 ± 0.68, and 14 ± 2.8 min−1, yielding an enantiomeric excess value of 92, 87, and 70%, respectively. These results suggest the critical roles of L75I, A264G, and A328V in steering OMP in the optimal orientation for stereoselective oxidation and demonstrate the utility of UniDesign for engineering CYP102A1 to produce drug metabolites of interest. The results are discussed in the context of protein structures.
Keywords: protein design, P450, stereoselectivity, CYP102A1, biocatalyst
Cytochrome P450 CYP102A1 from Bacillus megaterium is a prototypic biocatalyst that has stimulated tremendous interest to harness its catalytic prowess (1, 2). It has been engineered for applications in chemical synthesis, drug discovery, bioremediation, and biotechnology (3, 4). Over the past 2 decades, a large repertoire of CYP102A1 variants has been identified by many laboratories around the world through directed evolution and/or rational design. These variants are capable of metabolizing a wide range of organic compounds, including alkane (5, 6), alkene (7), aromatics (8, 9), and various pharmaceutical drugs such as verapamil and astemizole (10), proton pump inhibitors (11), statins (12), and many more (13). The biochemical, structural, and computational analyses of these variants provide mechanistic insights into the CYP102A1-catalyzed reactions, which further fuel the engineering of CYP102A1 for regioselective and stereoselective oxidation. Some residues and mutations are shown to be important to the catalytic activity and selectivity of CYP102A1 (4, 14, 15, 16, 17). Despite the availability of these variants, it remains challenging to introduce a set of mutations to metabolize a specific compound of interest.
Computational enzyme design offers a solution to enzyme development. Computational enzyme design tools, such as Rosetta (18), PROSS (19), FuncLib (20), and PRODA (21), to name a few, have been successfully used to redesign natural enzymes with improved function (19, 20, 22, 23, 24), screen scaffolding enzymes that are evolved to catalyze target reactions (25), and design de novo enzyme function on inert protein scaffolds taken from nature (26, 27, 28) or artificial intelligence hallucination (29). Notably, Rosetta has been applied to engineer P450s in a few studies (30, 31).
In this work, we introduce UniDesign as a new framework for computational enzyme design in which we feature a systematic approach for generating catalytically relevant small molecule ligand poses. Owing to the generality of the ligand pose sampling method and the physical nature of the UniDesign energy function, we reason that UniDesign may apply to the design of a diversity of small molecule-catalyzing enzymes, given high-quality enzyme scaffold structures and clear catalytic mechanisms. Hereby, we demonstrate UniDesign’s effectiveness by computationally engineering the stereoselectivity of CYP102A1 for hydroxylation of omeprazole (OMP), a proton pump inhibitor, to treat gastric acid hypersecretion disorders. Although prior studies yielded regioselective CYP102A1 variants for hydroxylation of OMP to form 5-hydroxyomeprazole (5OH), they exhibited poor stereoselectivity (11, 32). We experimentally characterized three designer CYP102A1 variants, namely UD1, UD2, and UD3, all of which exhibit high regioselectivity and stereoselectivity for hydroxylation of (R)-OMP. Our results demonstrate the utility of UniDesign for enzyme engineering.
Results
UniDesign for computational enzyme design
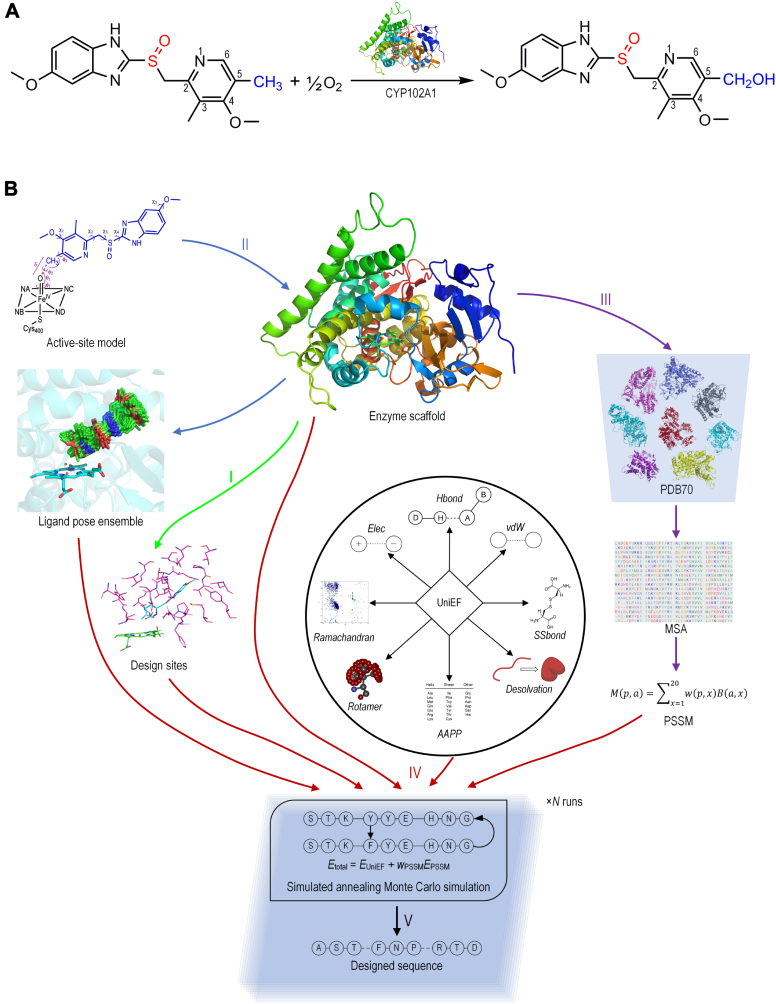
Computational enzyme design generally starts from the construction of an ideal active site model (25, 33) or theozyme (34, 35) that simulates the reaction’s transition state (TS) and searches for optimal active site configurations to stabilize the TS. Alternatively, the TS-mimic near-attack conformation (NAC) (36, 37, 38, 39) model also leads to successful designs (23, 40, 41, 42, 43). We recently presented UniDesign as a computational framework for designing functional protein sequences and showed its effectiveness on protein–nucleic acid interaction design by accurately decoding the CRISPR–Cas PAM recognition (44). In this work, we describe the capability of UniDesign for enzyme design, targeting the CYP102A1-catalyzed hydroxylation of OMP to enhance stereoselectivity (Fig. 1A).
Figure 1.
UniDesign workflow for computational enzyme design.A, hydroxylation of omeprazole (OMP) to produce 5-hydroxyomeprazole (5OH). The position of hydroxylation is colored in blue and the chiral sulphonyl group in red. B, the five main steps, denoted as I−V, are in the UniDesign pipeline. I, selection of design sites. II, generation of ligand poses. III, derivation of the evolutionary position-specific scoring matrix (PSSM) from multiple sequence alignment (MSA) by TM-align search. IV, simulated annealing Monte Carlo simulation for sequence design. V, selection of designer sequences or mutants. Multiple (×N) runs are usually conducted for sequence design due to the Monte Carlo simulation’s stochasticity. TM, triple mutant.
Overall, the UniDesign workflow for enzyme design comprises five steps (Fig. 1B). First, the active site pocket is determined based on the enzyme structure and a suitable set of sites is chosen for design. In the second step, UniDesign employs a “grow-and-check” strategy to generate an ensemble of ligand poses within the active site pocket to represent the TS or NAC states, whereby the pose generation is subject to appropriate geometric constraints. Third, similar to EvoDesign (45), UniDesign employs TM-align (46) to search against a nonredundant Protein Data Bank (PDB) library (PDB70, a subset of PDB with <70% sequence identity) to identify structure analogs to the enzyme scaffold and constructs a position-specific scoring matrix (PSSM) based on the multiple sequence alignment generated by TM-align. Fourth, building on the enzyme scaffold and ligand pose ensemble, UniDesign redesigns the mutable sites to minimize the enzyme–ligand system’s total energy using a composite energy function which is the linear combination of the PSSM and a physics- and knowledge-based energy function (UniDesign energy function, UniEF). The latter is an extension of our protein design energy function EvoEF2 (47) and is utilized to calculate the physical interactions between protein and ligand. An efficient simulated annealing Monte Carlo (SAMC) simulation procedure is used to search the protein sequence space and generate a large number of designer sequences. Finally, after the SAMC simulation, the protein sequence that has the lowest total energy is typically picked as the best design. Due to the SAMC’s stochasticity, it is necessary to run multiple independent simulation trajectories and pool the lowest energy sequence from each run for analysis. In this pipeline, the evolutionary component is set as optional, and the UniEF energy function alone will be used for design if the evolution search is disabled or when the PSSM term is not sufficiently reliable due to very few (e.g., <10) structure analogs available.
Designing CYP102A1 variants for stereoselective hydroxylation of OMP enantiomers
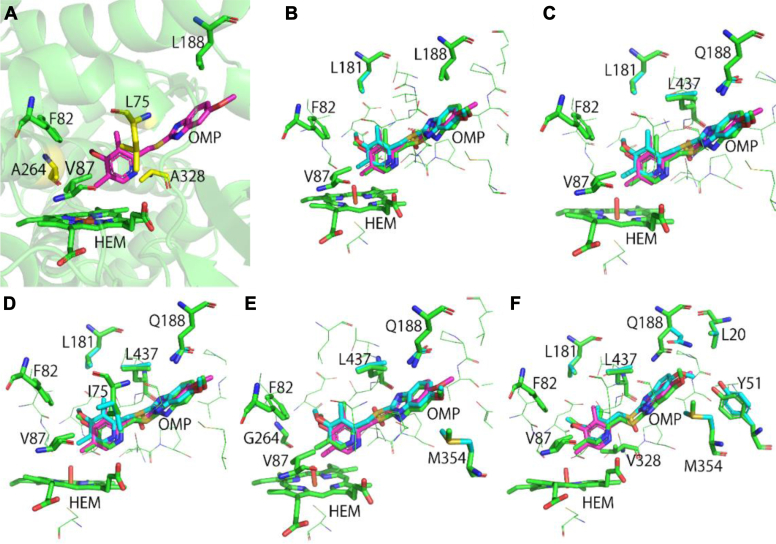
It was reported that the A82F/F87V double mutant (DM) of CYP102A1 is capable of hydroxylating OMP to produce 5OH (11, 16). Since several prior studies showed that L188Q enhances enzyme activity toward non-natural substrates, we also included it in the design (9, 48, 49, 50). We initiated our designs based on this triple mutant (TM: A82F/F87V/L188Q) and selected three residues for redesign, that is, L75, A264, and A328, which are located at the bottom (A264), upper neck (L75), and lower neck (A328) positions in the active site pocket (Fig. 2A); all of them are in direct contact (<4.5 Å) with the pyridinyl ring or the sulfonyl group of OMP (PDB ID: 4KEY). Another 16 surrounding residues were chosen for repacking (for details, see Experimental procedures).
Figure 2.
Computational design and modeling with UniDesign.A, locations of main design sites. Mutable sites L75, A264, and A328 are colored yellow, whereas F82, V87, L188, and HEM are in green. Ligand OMP is colored in magenta. Their coordinates were obtained from the Protein Data Bank (PDB ID: 4KEY). It is of note that the sulfone oxygen is missing from the crystal structure because of weak electron density. B, UniDesign model of the A82F/F87V double mutant (DM). The UniDesign-generated (R)- and (S)-OMP poses are colored green and cyan, respectively. The crystal OMP (in magenta) is depicted for comparison. The main design sites and other sites that adopt different conformations for binding (R)- and (S)-OMP are shown in green and cyan sticks, respectively. The color scheme is the same in B–F. C, UniDesign model of the A82F/F87V/L188Q triple mutant (TM). D, UniDesign model of the UD1 mutant (TM/L75I). E, UniDesign model of the UD2 mutant (TM/A264G). F, UniDesign model of the UD3 mutant (TM/A328V). OMP, omeprazole.
We then used UniDesign to generate ligand poses for (R/S)-OMP modeling in the subsequent design simulations and obtained 7470 and 6998 poses for (R)- and (S)-OMP, respectively, after screening conformation. The details for ligand conformer generation and screening are described in Text S1 in the Supporting information. To examine the accuracy of ligand pose sampling, we performed a repacking simulation for the DM variant. The predicted (R)- and (S)-OMP poses are overall similar to that of the crystal OMP (Fig. 2B), indicative of the high quality of generated conformer ensembles that can cover native-like conformations. In addition, the predicted amino acid side chains are nearly identical for the two conformers, suggesting that DM may favor (R)- and (S)-OMP equally, although their enzyme–ligand interaction energies () differ by ∼4 UniDesign energy units (UEU) (Table S1). Similar results were obtained by repacking the TM mutant, except that two more residues (L437 and Q188) adopt slightly different conformations (Fig. 2C).
Next, we conducted computational saturated mutagenesis on the three design sites to search for point mutations that would shift the enzyme stereoselectivity toward (R)-OMP. For each quadruple mutant (bearing the TM mutations), we carried out design/repacking simulations to minimize the total energy of the whole enzyme–ligand complex system, during which the ligand pose was picked from the aforementioned ensembles.
We hypothesized that enzyme kinetics would enhance for the mutants with favorable enzyme–ligand interaction energy () relative to that of the TM variant (), for example, , and correspondingly, it would reduce for mutants with positive . We further argued that mutants with would be able to shift the enzyme stereoselectivity toward (R)-OMP hydroxylation. According to these hypotheses, we finally chose three mutants, namely UD1 (TM/L75I), UD2 (TM/A264G), and UD3 (TM/A328V), for experimentation (Table 1). These mutants had a negative score and exhibited a relatively small change in enzyme stability (e.g., <5 UEU).
Table 1.
The UniDesign-computed energy changes upon the introduction of mutation relative to the triple variant (TM)
| Variant | Energy changes relative to the TM variant (UniDesign energy units) |
|||
|---|---|---|---|---|
| () | () | () | ||
| TM (A82F/F87V/L188Q) | 0 (−48.86) | 0 (−52.96) | 0 | 0 (−2769.89) |
| UD1 (TM/L75I) | −0.12 (−48.98) | 2.15 (−50.81) | −2.28 | 2.46 (−2767.43) |
| UD2 (TM/A264G) | −0.23 (−49.09) | 1.05 (−51.91) | −1.28 | 4.44 (−2765.45) |
| UD3 (TM/A328V) | −1.27 (−50.13) | 0.52 (−52.44) | −1.79 | 1.46 (−2768.43) |
Computational modeling suggested that the L75I mutation causes a slightly favorable interaction energy change for (R)-OMP ( = −0.12 UEU) and a large interaction energy loss for (S)-OMP ( = 2.15 UEU) (Table 1). Compared to the TM variant, L181, Q188, L437, and the OMP enantiomers choose identical conformations, but the I75 residue adopts different conformations; I75 in the (R)-OMP model forms more favorable contacts (Fig. 2D).
Similar to the L75I case, the A264G mutation led to a moderately favorable interaction energy change for (R)-OMP ( = −0.23 UEU) and a larger interaction energy loss for (S)-OMP ( = 1.05 UEU) (Table 1). Compared to the TM model, the major difference is that the M354 residue adopted different conformations; as shown, M354 in the (R)-OMP model could form closer contact with the ligand (Fig. 2E).
The computational design suggested that the A328V mutation introduced a relatively large favorable interaction with (R)-OMP ( = −1.27 UEU) and a slightly unfavorable interaction with (S)-OMP ( = 0.52 UEU) (Table 1). Compared to the TM, TM/L75I, and TM/A264G models, the OMP enantiomers in TM/A328V adopt different conformations, leading to large conformational variances of the Y51, Q188, and M354 residues (Fig. 2F).
Next, we characterized all these variants to examine if the three designer mutants could shift the stereoselectivity profiles for OMP hydroxylation.
NADPH oxidation rates of CYP102A1 variants
To evaluate the effects of mutations on catalytic activity, we first determined the rates of NADPH oxidation under steady-state conditions in the presence of racemic OMP. Among all variants, DM exhibits the highest kcat of 1306 min−1, comparable to 1500 min−1 reported by Dr Munro’s group (16). TM has a reduced kcat of 943 min−1. All three designer variants exhibit relatively high kcat values for NADPH oxidation, ranging from 662 to 1276 min−1 (Table 2). Among them, UD2 is the most active with a kcat of 1276 min−1, whereas UD1 is the least active with a kcat of 662 min−1. Compared to DM and TM, designer variants exhibit elevated Michaelis constants (KM), resulting in a decrease in catalytic efficiency in terms of NADPH oxidation.
Table 2.
Rates of NADPH oxidation by CYP102A1 variants
| Variant | kcat (min−1) | KM (μM) | kcat/KM (μM−1 min−1) | Coupling (%) |
|---|---|---|---|---|
| DM (A82F/F87V) | 1306.0 ± 45.3 | 28.8 ± 6.8 | 45.3 | 3.0 |
| TM (DM/L188Q) | 943.1 ± 26.2 | 24.4 ± 4.8 | 38.6 | 5.2 |
| UD1 (TM/L75I) | 662.2 ± 28.1 | 86.6 ± 11.8 | 7.6 | 6.5 |
| UD2 (TM/A264G) | 1276.0 ± 96.8 | 131.3 ± 26.7 | 9.7 | 5.5 |
| UD3 (TM/A328V) | 1144.0 ± 57.2 | 59.5 ± 15.8 | 19.2 | 5.9 |
The rates were determined in 1× PBS solution at 25 °C as described in Experimental procedures.
Regioselectivity of CYP102A1 variants for OMP hydroxylation
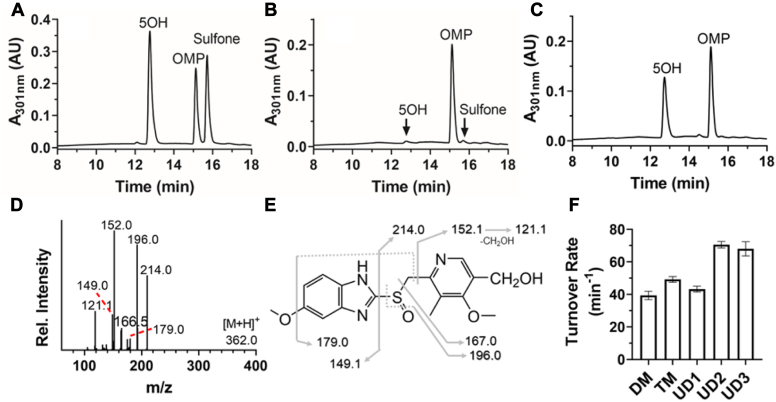
To confirm the sites of monooxygenation, OMP metabolites produced by the CYP102A1 variants were analyzed by liquid chromatography-tandem mass spectrometry. Authentic OMP, 5OH, and sulfone metabolites elute at 12.7, 15.1, and 15.8 min, respectively (Fig. 3A). As expected, the metabolism of OMP by human liver microsomes produced low levels of 5OH and sulfone metabolites (Fig. 3B). In marked contrast, the metabolism of OMP by the UniDesign variants produces predominantly a metabolite that elutes at 12.7 min as the authentic 5OH (Figs. 3C and S1). We compared the MS2 spectrum of this monooxygenated metabolite obtained at m/z 362 to further investigate its identity. As shown, the MS2 fragments for the UD3 variants were observed at m/z 121.1, 149.0, 152.0, 179.0, 196.0, and 214.0 (Fig. 3D), which are identical to those of the authentic 5OH metabolite (Fig. S1). The fragmentation pattern shown in Figure 3E is in agreement with those recently reported by Shin et al. (51). The consistency in retention time and MS2 fragmentation patterns suggest that the UniDesign variants hydroxylate OMP to predominately produce the 5OH metabolite, indicative of high regioselectivity. Under steady-state turnover conditions, hydroxylation of OMP by UD2 and UD3 is enhanced to produce 5OH at a rate of 71 ± 2.0 and 68 ± 4.3 min−1, respectively (Fig. 3F), compared with the DM variant at 39.4 ± 2.6 min−1 and the TM variant at 49.7 ± 1.7 min−1. The turnover rate for UD1 is 43.2 ± 1.8 min−1, similar to the DM variant. Together, our data show that UD2 and UD3 have increased activity for the OMP → 5OH transformation. The enhancement of production activity may be a result of the synergistic effect of the relatively high NADPH oxidation rates and coupling efficiencies (Table 2).
Figure 3.
Regioselective hydroxylation of racemic OMP by CYP102A1 variants. Hydroxylation of racemic OMP was performed as described in Experimental procedures, and metabolites were analyzed by LC-MS/MS to confirm identities of major metabolites. A, elution of authentic OMP, 5OH, and sulfone metabolites; (B) elution of OMP metabolites by HLM; (C) elution of OMP metabolites by UD3; (D) MS2 spectrum of the major metabolite produced by UD3 at 12.7 min in C; (E) assignments of MS2 peaks observed in D; (F) steady-state turnover rates for production of 5OH metabolite. 5OH, 5-hydroxyomeprazole; HLM, human liver microsome; LC-MS/MS, liquid chromatography-tandem mass spectrometry; OMP, omeprazole.
Stereoselectivity of CYP102A1 variants for OMP hydroxylation
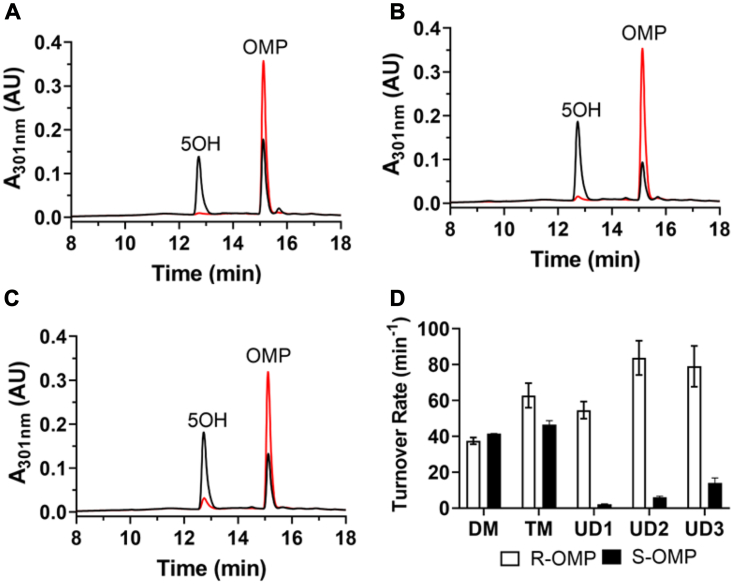
To examine the stereoselective hydroxylation of OMP, we further determined the turnover rates in the presence of either (R)- or (S)-OMP under steady-state conditions. In the presence of (R)-OMP, all three UniDesign variants produce 5OH as the predominant metabolite as observed at 12.7 min (Fig. 4, A–C, black traces). In marked contrast, (S)-OMP is barely metabolized by the three variants as evidenced by the minor peaks at 12.7 min (Fig. 4, A–C, red traces). Among all variants, UD2 exhibits the highest turnover rate for (R)-OMP at 84 ± 4.8 min−1, followed by UD1 and UD3 at 55 ± 4.7 and 79 ± 5.7 min−1, respectively (Fig. 4D). In the case of (S)-OMP, the turnover rates decrease to 2.2 ± 0.19, 6.0 ± 0.68, and 14 ± 2.8 min−1 for UD1, UD2, and UD3, respectively. The shifted stereoselectivity profiles lead to an enantiomeric excess (ee) value of 92, 87, and 70% for UD1, UD2, and UD3, respectively. By contrast, the ee values for DM and TM are −5 and 15%, respectively. Thus, the UniDesign predictions suggested highly stereoselective CYP102A1 variants for (R)-OMP hydroxylation.
Figure 4.
Stereoselective hydroxylation of OMP enantiomers by CYP102A1 variants.A–C, represent HPLC elution profiles of metabolites produced by UD1, UD2, and UD3, respectively, in the presence of either (R)-OMP (black trace) or (S)-OMP (red trace). D, steady-state turnover rates for the 5OH metabolite. The rates of DM and TM are included for comparison. 5OH, 5-hydroxyomeprazole; DM, double mutant; OMP, omeprazole; TM, triple mutant.
Total turnover number for (R)-5OH production
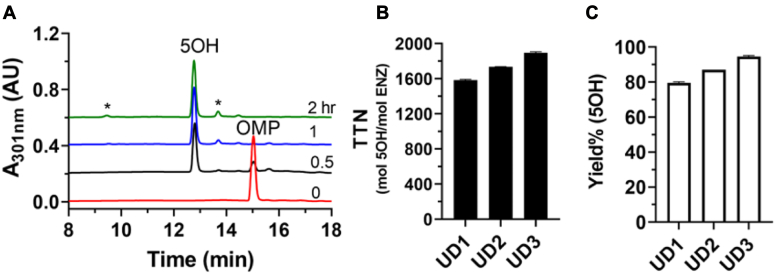
Next, we determined the total turnover numbers (TTNs) for the production of (R)-5OH to examine the utility of the designer variants for meaningful biosynthesis. Figure 5A shows a representative HPLC elution profile for the UD1 variant at different reaction times. In the control sample (t = 0), only substrate (R)-OMP was observed at 13.2 min (Fig. 5A, red trace). At 30 min, the amount of (R)-OMP is decreased by ∼80% with a concomitant increase in the amount of (R)-5OH observed at 12.7 min (Fig. 5A, black trace). Further incubation led to the complete depletion of (R)-OMP and the formation of minor byproducts (Fig. 5A). The TTNs calculated at the maximal yields of (R)-5OH are 1586 ± 8.5, 1734 ± 2.8, and 1894 ± 14.1 (mol 5OH/mol P450) for UD1, UD2, and UD3, respectively (Fig. 5B), corresponding to a 5OH production yield of 80, 87, and 94%, respectively (Fig. 5C).
Figure 5.
Total turnover numbers of the three UniDesign variants for production of the (R)-5OH metabolite. Hydroxylation of (R)-OMP was carried out at 25 °C in 1× PBS buffer containing 0.5 mM (R)-OMP, 0.5 mM NADP+, 0.25 μM variant enzyme, and 50 μg/ml glucose dehydrogenase. Product (R)-5OH was quantified at 15 min intervals by HPLC until depletion of (R)-OMP. A, representative HPLC traces for the UD1 variant at 0, 0.5, 1, and 2 h intervals. Minor byproducts are marked with asterisks. B, TTNs of the three UniDesign variants. C, yields of the three UniDesign variants for production of the (R)-5OH metabolite. 5OH, 5-hydroxyomeprazole; OMP, omeprazole; TTN, total turnover number.
Discussion
Many computational approaches have been developed and applied to engineer enzymes with varying degrees of success. The most widely used Rosetta (18) lacks an integrated ligand pose sampling method and relies on third-party tools like OpenEye OMEGA (52) for generating conformers. The PROSS tool (19), which is specialized in stability design, excludes the redesign of active site residues and evaluates only the combination of potential mutations from phylogenetic analysis. PROSS does not model ligands. FuncLib (20), an enzyme function design tool, considers the combination of phylogeny-suggested mutations within the active site pocket, and it either ignores the ligand or fixes the crystal ligand. As a result, FuncLib lacks the capability of designing activities specifically for substrates of interest. PRODA (21) employs a similar ligand pose generation strategy to UniDesign, and it can theoretically serve as another general-purpose enzyme design tool. A notable weakness of PRODA is that it requires intensive human intervention and manual efforts, making it difficult for nonexperts to use. Compared to the aforementioned tools, UniDesign not only provides a flexible ligand pose generation and screening strategy, allowing for function design toward specified substrates, but also reduces manual efforts to a minimal, controllable extent.
As demonstrated in this study, we have used UniDesign to successfully design highly stereoselective CYP102A1 mutants for the hydroxylation of (R)-OMP. The simple , , and metrics overall correlate well with the experimental outcomes. The predicted positive values match the reduced turnover rates in the correct order for all three UniDesign variants (Table 1 and Fig. 4). The predicted negative scores correspond to the elevated turnover rates for the metabolism of (R)-OMP by UD2 and UD3, whereas the turnover rates of UD1 and TM are comparable, presumably due to a negligible level of (−0.12 UEU) (Table 1 and Fig. 4). The most negative for UD1 also matches the highest stereoselectivity with an ee value of 92%. Together, these metrics work well with the UniDesign models, although they seem simple. We argue that these metrics may also work for other enzyme systems for engineering stereoselectivity.
The consistency in prediction and experimental results highlights the important roles of residues I75, G264, and V328 in differentiating (R)- from (S)-OMP to allow stereoselective oxidation. The reactive intermediate responsible for the monooxygenation of inert C-H bonds by P450 enzymes is a high-valent iron (IV)-oxo (or ferryl) species, which is highly oxidizing (53). Thus, OMP must be oriented in the correct pose in the vicinity of the ferryl intermediate for monooxygenation to ensure regioselective and stereoselective oxidation. Since the three UniDesign variants maintain the regioselectivity, the stereoselectivity likely arises from differential interaction energy change in favor of (R)-OMP (Table 1). Positions 75 and 328 are located in two well-studied regions that are known to affect substrate specificity due to their proximity to the bound substrate (Fig. 2); position 75 is in the helix B/C region and position 328 is in the loop between helix K and sheet β3. It is conceivable that the changes in active site geometry and/or volume arising from L75I and A328V mutations may alter OMP binding in favor of (R)-OMP. It is shown that the single A328V mutation dramatically enhances substrate binding and the production formation rate (14). The L75I mutation was also observed in a few stereoselective CYP102A1 variants that were screened by directed evolution (10, 54). Position 264 is located in helix I which contains the critical T268 residue for proton delivery required for catalysis (55, 56). The A264G mutation exerts no adverse effect on proton delivery since the UD2 variant exhibits high rates for both NADPH oxidation and product turnover. It is not intuitive how the A264G mutation, which should result in slight enlargement of the active site volume, would increase stereoselective by itself. However, a comparison of the structure models for TM and TM/A264G suggested that A264G led to an observable conformational change of M354 (Fig. 2, C and E), which may help differentiate (R)- from (S)-OMP. Nonetheless, the three designer variants achieve higher turnover rates and stereoselectivity for (R)-OMP than a triple variant R47L/F87V/L188Q as reported previously (32). The amount of (R)-5OH produced by the UD3 variant is 59-fold more than that produced by human liver microsomes under conventional conditions (Figs. 3B and 5).
We also wonder if the UniDesign energies and/or models could explain the poor stereoselectivity of DM and TM. It should be noted that the absolute interaction energy () cannot explain the preference for (R)- or (S)-OMP. For example, all five variants (DM, TM, UD1, UD2, and UD3) have more negative (Table S1), which would indicate a preference for (S)-OMP. However, the experimental outcome is that DM shows nearly no preference, while the remaining four prefer (R)-OMP to some extent. We reason that the inherent interaction energy bias toward (S)-OMP could be in part due to the independent parameterization of two enantiomers. Thus, we further argue the rationality of using relative energy (or energy change relative to a reference), rather than the absolute value, to make predictions. Although the energy scores cannot explain the stereoselectivity of DM and TM, the UniDesign models provide some hints. In the repacked DM and TM models, the (R/S)-OMP–binding residues adopt almost identical poses (Fig. 2, B and C), suggesting that the two variants may not have a strong preference for (R)- or (S)-OMP.
Like any other computational enzyme design tool, UniDesign has potential limitations. One notable limitation is that UniDesign cannot predict active site-distant mutations that enhance enzyme activity. This is a common problem for almost all computational enzyme design methods. Such mutations are usually discovered through directed evolution (57, 58) rather than physical energy functions with a short distance cutoff. The combination of UniDesign and directed evolution will serve as a strengthened solution to enzyme engineering. Another limitation is that UniDesign considers only the fixed-backbone protein design simulations at the current stage, which has the advantage of alleviating sampling complexity. While this is somewhat reasonable when the mutations take place in the well-ordered regions, it has been shown that mutations in the loop regions can induce large backbone conformational changes (59, 60). Modeling the protein backbone flexibility explicitly would be beneficial to enzyme variant generation and selection.
Conclusions
In this work, we introduce UniDesign as a computational framework for enzyme design and engineering. We have demonstrated its effectiveness by successfully redesigning the CYP102A1 enzyme to substantially enhance its stereoselectivity for (R)-OMP hydroxylation. Computational modeling suggests that L75I, A264G, and A328V are critical mutations that facilitate the shift of stereoselectivity profiles toward (R)-OMP oxidation. UniDesign should be broadly applicable in protein design and engineering.
Experimental procedures
Development of UniDesign for small molecule–catalyzing enzyme design
We recently reported UniDesign for designing protein–nucleic acid interactions (44). The UniEF is a score function optimized for protein sequence design; it is an extension of our former protein design score function EvoEF2 (47), which is based on the CHARMM19 (61) residue topologies and atom parameters but not the CHARMM force field energy function. UniEF is a linear combination of nine energy terms including van der Waals, hydrogen bonding, electrostatics, desolvation, disulfide bonding, amino acid propensity, Ramachandran, rotamer preference, and amino acid reference energy (see Fig. 1). UniEF has different energy terms and equations compared to traditional molecular mechanics force fields such as CHARMM (61) that do not have an explicit hydrogen-bonding term. The overall methodology of UniDesign for enzyme design is similar to that for protein–nucleic acid interaction design (44). The major differences lie in the generation of atomic parameters and residue topologies and the conformation and pose sampling for small molecule ligands in enzyme design. We parameterized ligand atoms based on their chemical similarity to the atoms in amino acid groups. The small molecule residue topology that describes the bond connections and internal coordinates is generated from its atomic Cartesian coordinates, starting from three heavy atoms that are most relevantly involved in the catalytic constraints with a protein or cofactor residue. One grand challenge for computational enzyme design is to adequately sample the ligand poses in the TS or NAC states. To address this, we developed a small molecule placement approach (termed “grow-and-check”) to generate the TS or NAC ligand poses by extending a similar protocol in PRODA (21). Briefly, given an enzyme structure, UniDesign truncates all its amino acids into glycines and then starts to “grow” ligand poses by varying the catalytic geometric restraints between the ligand and the anchor residue and sampling the torsional angles of rotatable bonds within the ligand. By progressively growing the ligand atoms, UniDesign will “check” the catalytic constraints between a ligand pose and other catalytic residues as well as the pose’s internal energy (termed “Internal” energy) and external energy with protein backbone (termed “Backbone” energy). The repulsive van der Waals term of UniEF is used to calculate the two energies. Ligand poses that satisfy all the catalytic constraints with both energies below the given thresholds are generated. Depending on the flexibility of the ligand, the ligand pose ensemble can be huge (e.g., >100 K poses). In this case, the poses with both internal and backbone energy ranked in the top (e.g., 20%) are retained for design. Finally, the “ProteinDesign” command in UniDesign is used for enzyme sequence design or mutant evaluation. A step-by-step protocol is provided in Text S1 including Tables S3–S13 and Figs. S2–S7 for enzyme design with UniDesign, using CYP102A1 redesign as an example.
Design and selection of stereoselective CYP102A1 variants
We sought to redesign the CYP102A1 TM variant to enhance its stereoselectivity for the hydroxylation of (R)-OMP. By analyzing the crystal structure of the CYP102A1 heme domain (PDB ID: 4KEY) in complex with OMP (named 1C6), we chose three sites for mutagenesis, that is, L75, A264, and A328, and another 16 surrounding residues for side chain repacking, including L20, P25, V26, L29, Y51, S72, L181, I263, E267, T268, T327, P329, M354, T436, L437, and T438. Each mutable site was substituted into any other 19 amino acids, resulting in 57 quadruple mutants. The ligand poses were generated and sampled through the small molecule placement procedure as described above, and 7470 and 6998 poses were retained for (R)- and (S)-OMP, respectively (Text S1). We then used UniDesign to repack the enzyme–ligand complex for each variant to minimize the system’s total energy through SAMC simulations, with (R)- or (S)-OMP poses sampled from the two ensembles and amino acid side chain conformations taken from the Dunbrack2010 backbone-dependent rotamer library (62). After each simulation, the (R/S)-OMP ligand pose was determined with protein residue rotamers simultaneously reintroduced, and the total energy and the enzyme–ligand interaction energy were calculated. Five independent SAMC simulation trajectories were conducted for each quadruple mutant, producing five designs, among which the one with the lowest total energy was taken as the optimal design. The relative enzyme–OMP interaction energy () together with the UniDesign model was used to guide mutant selection.
Chemicals
All chemicals used are of the highest purity available. Authentic OMP, 5OH OMP, and OMP sulfone were purchased from Cayman Chemicals. NADPH, NADP+, and D-glucose were purchased from Sigma-Aldrich. Carbon monoxide gas was from Cryogenic Gas.
Construction, expression, and purification of CYP102A1 variants
Mutations were introduced to template plasmid complementary DNA (cDNA) using Quikchange site-directed mutagenesis kit according to the manufacturer’s instructions (Agilent Technologies). The mutagenic primers used to construct the DM, TM, and three UniDesign variants are provided in Table S2. We first constructed the DM mutant by introducing the F87V mutation into the plasmid cDNA of pCW-CYP102A1A82F6×His that we prepared previously (63). Then the L188Q mutation was introduced to DM to prepare the TM mutant. Three UniDesign mutants were prepared using the TM plasmid as a template and a pair of mutagenic primers. All mutants were confirmed by cDNA sequencing at the University of Michigan DNA Sequencing Core Facility. A 6× His tag was inserted after the start codon ATG to facilitate purification. CYP102A1 variants were overexpressed in bacterial Escherichia coli C41(DE3) cells and purified on a Histrap HP affinity column (Cytiva Life Sciences) as previously described (63).
Determination of NADPH oxidation rates
The rate of NADPH oxidation was determined under steady-state conditions as previously described (63). In brief, CYP102A1 variants (0.1 μM) and various concentrations of racemic OMP (0–0.2 mM) were preincubated in 0.5 ml of 1× PBS buffer for 5 min at 25 °C. The reaction was then initiated by the addition of 0.2 mM NADPH and monitored at 340 nm for 30 s on a UV-visible spectrophotometer (Shimadzu PC 2043 UV). The kinetic parameters of KM and kcat were obtained by fitting the initial velocities at a variety of OMP concentrations to a Michalis–Menten equation.
Identification of major OMP metabolites by CYP102A1 variants
Major metabolites of OMP were identified using liquid chromatography-tandem mass spectrometry on a Thermo TSQ Ultra AM mass spectrometer equipped with a Shimadzu binary HPLC system. The retention time and MS2 spectra of the metabolites were compared with authentic OMP standards that were chromatogramed on a reverse phase C18 column (Zobax C18, 3 × 150 mm, Agilent Technologies). The binary mobile phase consists of 60 mM ammonium acetate (pH 5.8) (A) and acetonitrile (B). The flow rate was 0.4 ml/min and the gradient was as follows: 10% B for 2 min, then 30% B in 10 min, 70% B in 15 min, and 90% B in 16 min and held for 4 min. The MS2 spectra were recorded with the following parameters: spray voltage of 3.5 kV, vapor temperature of 300 °C, capillary temperature of 290 °C, and collision energy of 35%.
Determination of turnover rates for OMP hydroxylation
The turnover rate was determined in 0.2 ml of 1× PBS buffer containing 0.2 mM OMP, 5 mM glucose, 0.2 mM NADP+, and 50 μg/ml glucose dehydrogenase. After equilibrating at 25 °C for 5 min, the CYP102A1 variant was added to 0.1 μM to initiate the reaction. The reaction was terminated in 10 min by the addition of 0.2 ml acetonitrile. After centrifuging at 16,000g for 10 min, the samples were analyzed to quantify the amount of product 5OH on a Shimadzu HPLC system equipped with a photodioarray detector and autosampler. Elution of 5OH was monitored at 301 nm, and the area under the curve was used for quantification based on a calibration with a known amount of authentic 5OH. We determined the coupling efficiency, defined as the percentage of NADPH used to produce 5OH, by calculating the ratio of the turnover rate over the rate of NADPH oxidation. To determine the ee value, metabolism of (R)- or (S)-enantiomer was carried out under the same conditions as described above. The molar ratio of (R)- versus (S)-5OH was used to calculate the ee values on the condition that the sum of (R)- and (S)-5OH equals 100%.
Determination of TTN
The TTNs were determined in 1× PBS buffer containing 0.5 mM (R)-OMP, 10 mM glucose, 0.5 mM NADP+, and 50 μg/ml glucose dehydrogenase. After equilibrating at 25 °C for 5 min, the CYP102A1 variant was added to 0.25 μM to initiate the reaction. Aliquots of the reaction mixture were terminated at an interval of 30 min, and 5OH was quantified as aforementioned. TTN was expressed as mol 5OH/mol P450.
Data availability
The UniDesign is available at https://github.com/tommyhuangthu/UniDesign. All computational data and scripts in this work are available at https://doi.org/10.5281/zenodo.7881449.
Supporting information
This article contains supporting information (4, 16, 53).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The computational research in this work was supported through the computational resources and services provided by Advanced Research Computing at the University of Michigan.
Author contributions
X. H., Y. E. C., and H. Z. conceptualization; X. H. methodology; X. H., Y. S., and H. Z. writing–original draft; X. H., Y. O., Y. E. C., and H. Z. writing–review and editing; X. H., Y. S., and H. Z. investigation; Y. O., Y. E. C., and H. Z. resources; X. H., Y. E. C., and H. Z. project administration; Y. E. C. and H. Z. supervision.
Funding and additional information
This work was supported by grants from the National Institutes of Health (GM149016 to X. H., GM077430 to Y. O., HL159900, HL147527, HL159871 to Y. E. C., and ES030791 to H. Z.) and the UM Pandemic Research Recovery fund (to H. Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Contributor Information
Xiaoqiang Huang, Email: xiaoqiah@umich.edu.
Haoming Zhang, Email: haom@umich.edu.
Supporting information
References
- 1.Dunham N.P., Arnold F.H. Nature's machinery, repurposed: expanding the repertoire of iron-dependent oxygenases. ACS Catal. 2020;10:12239–12255. doi: 10.1021/acscatal.0c03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munro A.W., Leys D.G., McLean K.J., Marshall K.R., Ost T.W., Daff S., et al. P450 BM3: the very model of a modern flavocytochrome. Trends Biochem. Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 3.Yun C.H., Kim K.H., Kim D.H., Jung H.C., Pan J.G. The bacterial P450 BM3: a prototype for a biocatalyst with human P450 activities. Trends Biotechnol. 2007;25:289–298. doi: 10.1016/j.tibtech.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Whitehouse C.J., Bell S.G., Wong L.L. P450(BM3) (CYP102A1): connecting the dots. Chem. Soc. Rev. 2012;41:1218–1260. doi: 10.1039/c1cs15192d. [DOI] [PubMed] [Google Scholar]
- 5.Fasan R., Chen M.M., Crook N.C., Arnold F.H. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. Int. Ed Engl. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 6.Meinhold P., Peters M.W., Chen M.M., Takahashi K., Arnold F.H. Direct conversion of ethane to ethanol by engineered cytochrome P450 BM3. ChemBioChem. 2005;6:1765–1768. doi: 10.1002/cbic.200500261. [DOI] [PubMed] [Google Scholar]
- 7.Kubo T., Peters M.W., Meinhold P., Arnold F.H. Enantioselective epoxidation of terminal alkenes to (R)- and (S)-epoxides by engineered cytochromes P450 BM-3. Chemistry. 2006;12:1216–1220. doi: 10.1002/chem.200500584. [DOI] [PubMed] [Google Scholar]
- 8.Carmichael A.B., Wong L.L. Protein engineering of Bacillus megaterium CYP102. The oxidation of polycyclic aromatic hydrocarbons. Eur. J. Biochem. 2001;268:3117–3125. doi: 10.1046/j.1432-1327.2001.02212.x. [DOI] [PubMed] [Google Scholar]
- 9.Li Q.S., Ogawa J., Schmid R.D., Shimizu S. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2001;67:5735–5739. doi: 10.1128/AEM.67.12.5735-5739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawayama A.M., Chen M.M., Kulanthaivel P., Kuo M.S., Hemmerle H., Arnold F.H. A panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chemistry. 2009;15:11723–11729. doi: 10.1002/chem.200900643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler C.F., Peet C., McLean K.J., Baynham M.T., Blankley R.T., Fisher K., et al. Human P450-like oxidation of diverse proton pump inhibitor drugs by 'gatekeeper' mutants of flavocytochrome P450 BM3. Biochem. J. 2014;460:247–259. doi: 10.1042/BJ20140030. [DOI] [PubMed] [Google Scholar]
- 12.Kim K.H., Kang J.Y., Kim D.H., Park S.H., Park S.H., Kim D., et al. Generation of human chiral metabolites of simvastatin and lovastatin by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2011;39:140–150. doi: 10.1124/dmd.110.036392. [DOI] [PubMed] [Google Scholar]
- 13.Reinen J., van Leeuwen J.S., Li Y., Sun L., Grootenhuis P.D., Decker C.J., et al. Efficient screening of cytochrome P450 BM3 mutants for their metabolic activity and diversity toward a wide set of drug-like molecules in chemical space. Drug Metab. Dispos. 2011;39:1568–1576. doi: 10.1124/dmd.111.039461. [DOI] [PubMed] [Google Scholar]
- 14.Haines D.C., Hegde A., Chen B., Zhao W., Bondlela M., Humphreys J.M., et al. A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation. Biochemistry. 2011;50:8333–8341. doi: 10.1021/bi201099j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raner G.M., Hatchell A.J., Morton P.E., Ballou D.P., Coon M.J. Stopped-flow spectrophotometric analysis of intermediates in the peroxo-dependent inactivation of cytochrome P450 by aldehydes. J. Inorg. Biochem. 2000;81:153–160. doi: 10.1016/s0162-0134(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 16.Butler C.F., Peet C., Mason A.E., Voice M.W., Leys D., Munro A.W. Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 2013;288:25387–25399. doi: 10.1074/jbc.M113.479717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasan R., Meharenna Y.T., Snow C.D., Poulos T.L., Arnold F.H. Evolutionary history of a specialized P450 propane monooxygenase. J. Mol. Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alford R.F., Leaver-Fay A., Jeliazkov J.R., O'Meara M.J., DiMaio F.P., Park H., et al. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenzweig A., Goldsmith M., Hill S.E., Gertman O., Laurino P., Ashani Y., et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell. 2016;63:337–346. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khersonsky O., Lipsh R., Avizemer Z., Ashani Y., Goldsmith M., Leader H., et al. Automated design of efficient and functionally diverse enzyme repertoires. Mol. Cell. 2018;72:178–186.e175. doi: 10.1016/j.molcel.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X., Han K., Zhu Y. Systematic optimization model and algorithm for binding sequence selection in computational enzyme design. Protein Sci. 2013;22:929–941. doi: 10.1002/pro.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hettiaratchi M.H., O'Meara M.J., O'Meara T.R., Pickering A.J., Letko-Khait N., Shoichet M.S. Reengineering biocatalysts: computational redesign of chondroitinase ABC improves efficacy and stability. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J., Huang X., Xue J., Zhu Y. Computational redesign of penicillin acylase for cephradine synthesis with high kinetic selectivity. Green Chem. 2018;20:5484–5490. [Google Scholar]
- 24.Tian Y., Huang X., Li Q., Zhu Y. Computational design of variants for cephalosporin C acylase from Pseudomonas strain N176 with improved stability and activity. Appl. Microbiol. Biotechnol. 2017;101:621–632. doi: 10.1007/s00253-016-7796-x. [DOI] [PubMed] [Google Scholar]
- 25.Huang X., Xue J., Zhu Y. Computational design of cephradine synthase in a new scaffold identified from structural databases. Chem. Commun. 2017;53:7604–7607. doi: 10.1039/c7cc02270k. [DOI] [PubMed] [Google Scholar]
- 26.Jiang L., Althoff E.A., Clemente F.R., Doyle L., Rothlisberger D., Zanghellini A., et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothlisberger D., Khersonsky O., Wollacott A.M., Jiang L., DeChancie J., Betker J., et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 28.Siegel J.B., Zanghellini A., Lovick H.M., Kiss G., Lambert A.R., St Clair J.L., et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh A.H., Norn C., Kipnis Y., Tischer D., Pellock S.J., Evans D., et al. De novo design of luciferases using deep learning. Nature. 2023;614:774–780. doi: 10.1038/s41586-023-05696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashworth M.A., Bombino E., de Jong R.M., Wijma H.J., Janssen D.B., McLean K.J., et al. Computation-aided engineering of cytochrome P450 for the production of Pravastatin. ACS Catal. 2022;12:15028–15044. doi: 10.1021/acscatal.2c03974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D., Ma Y., Zhou Y., Gou J., Zhong Y., Zhao L., et al. A structural and data-driven approach to engineering a plant cytochrome P450 enzyme. Sci. China Life Sci. 2019;62:873–882. doi: 10.1007/s11427-019-9538-3. [DOI] [PubMed] [Google Scholar]
- 32.Ryu S.H., Park B.Y., Kim S.Y., Park S.H., Jung H.J., Park M., et al. Regioselective hydroxylation of omeprazole enantiomers by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2014;42:1493–1497. doi: 10.1124/dmd.114.058636. [DOI] [PubMed] [Google Scholar]
- 33.Huang X., Xue J., Lin M., Zhu Y. Use of an improved matching algorithm to select scaffolds for enzyme design based on a complex active site model. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss G., Celebi-Olcum N., Moretti R., Baker D., Houk K.N. Computational enzyme design. Angew. Chem. Int. Ed Engl. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 35.Zanghellini A. De novo computational enzyme design. Curr. Opin. Biotechnol. 2014;29:132–138. doi: 10.1016/j.copbio.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Bruice T.C. A view at the millennium: the efficiency of enzymatic catalysis. Acc. Chem. Res. 2002;35:139–148. doi: 10.1021/ar0001665. [DOI] [PubMed] [Google Scholar]
- 37.Bruice T.C., Lightstone F.C. Ground state and transition state contributions to the rates of intramolecular and enzymatic reactions. Acc. Chem. Res. 1999;32:127–136. [Google Scholar]
- 38.Bruice T.C., Benkovic S.J. Chemical basis for enzyme catalysis. Biochemistry. 2000;39:6267–6274. doi: 10.1021/bi0003689. [DOI] [PubMed] [Google Scholar]
- 39.Hur S., Bruice T.C. The mechanism of catalysis of the chorismate to prephenate reaction by the Escherichia coli mutase enzyme. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1176–1181. doi: 10.1073/pnas.022628599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R., Wijma H.J., Song L., Cui Y., Otzen M., Tian Y., et al. Computational redesign of enzymes for regio- and enantioselective hydroamination. Nat. Chem. Biol. 2018;14:664–670. doi: 10.1038/s41589-018-0053-0. [DOI] [PubMed] [Google Scholar]
- 41.Cui Y., Wang Y., Tian W., Bu Y., Li T., Cui X., et al. Development of a versatile and efficient C–N lyase platform for asymmetric hydroamination via computational enzyme redesign. Nat. Catal. 2021;4:364–373. [Google Scholar]
- 42.Lin M., Wang F., Zhu Y. Modeled structure-based computational redesign of a glycosyltransferase for the synthesis of rebaudioside D from rebaudioside A. Biochem. Eng. J. 2020;159 [Google Scholar]
- 43.Wang P., Zhang S., Zhang J., Zhu Y. Computational design of penicillin acylase variants with improved kinetic selectivity for the enzymatic synthesis of cefazolin. Biochem. Eng. J. 2021;175 [Google Scholar]
- 44.Huang X., Zhou J., Yang D., Zhang J., Xia X., Chen Y.E., et al. Decoding CRISPR–Cas PAM recognition with UniDesign. Brief Bioinform. 2023;24:bbad133. doi: 10.1093/bib/bbad133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearce R., Huang X., Setiawan D., Zhang Y. EvoDesign: designing protein-protein binding interactions using evolutionary interface profiles in conjunction with an optimized physical energy function. J. Mol. Biol. 2019;431:2467–2476. doi: 10.1016/j.jmb.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang X., Pearce R., Zhang Y. EvoEF2: accurate and fast energy function for computational protein design. Bioinformatics. 2020;36:1135–1142. doi: 10.1093/bioinformatics/btz740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q.-S., Schwaneberg U., Fischer P., Schmid R.D. Directed evolution of the Fatty-acid Hydroxylase P450 BM-3 into an Indole-hydroxylating catalyst. Chemistry. 2000;6:1531–1536. doi: 10.1002/(sici)1521-3765(20000502)6:9<1531::aid-chem1531>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 49.van Vugt-Lussenburg B.M., Damsten M.C., Maasdijk D.M., Vermeulen N.P., Commandeur J.N. Heterotropic and homotropic cooperativity by a drug-metabolising mutant of cytochrome P450 BM3. Biochem. Biophys. Res. Commun. 2006;346:810–818. doi: 10.1016/j.bbrc.2006.05.179. [DOI] [PubMed] [Google Scholar]
- 50.van Vugt-Lussenburg B.M., Stjernschantz E., Lastdrager J., Oostenbrink C., Vermeulen N.P., Commandeur J.N. Identification of critical residues in novel drug metabolizing mutants of cytochrome P450 BM3 using random mutagenesis. J. Med. Chem. 2007;50:455–461. doi: 10.1021/jm0609061. [DOI] [PubMed] [Google Scholar]
- 51.Shin S.H., Park Y., Park M.H., Byeon J.J., Lee B.I., Choi J., et al. Profiling and Identification of omeprazole metabolites in mouse brain and plasma by isotope ratio-monitoring liquid chromatography-mass spectrometric method. Life (Basel) 2020;10:115. doi: 10.3390/life10070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boström J., Greenwood J.R., Gottfries J. Assessing the performance of OMEGA with respect to retrieving bioactive conformations. J. Mol. Graphics Model. 2003;21:449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 53.Rittle J., Green M.T. Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 54.Meinhold P., Peters M.W., Hartwick A., Hernandez A.R., Arnold F.H. Engineering cytochrome P450 BM3 for terminal alkane hydroxylation. Adv. Synth. Catal. 2006;348:763–772. [Google Scholar]
- 55.Yeom H., Sligar S.G., Li H., Poulos T.L., Fulco A.J. The role of Thr268 in oxygen activation of cytochrome P450BM-3. Biochemistry. 1995;34:14733–14740. doi: 10.1021/bi00045a014. [DOI] [PubMed] [Google Scholar]
- 56.Clark J.P., Miles C.S., Mowat C.G., Walkinshaw M.D., Reid G.A., Daff S.N., et al. The role of Thr268 and Phe393 in cytochrome P450 BM3. J. Inorg. Biochem. 2006;100:1075–1090. doi: 10.1016/j.jinorgbio.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 57.Arnold F.H. Design by directed evolution. Acc. Chem. Res. 1998;31:125–131. [Google Scholar]
- 58.Romero P.A., Arnold F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kellogg E.H., Leaver-Fay A., Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joyce M.G., Girvan H.M., Munro A.W., Leys D. A single mutation in cytochrome P450 BM3 induces the conformational rearrangement seen upon substrate binding in the wild-type enzyme. J. Biol. Chem. 2004;279:23287–23293. doi: 10.1074/jbc.M401717200. [DOI] [PubMed] [Google Scholar]
- 61.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 62.Shapovalov M.V., Dunbrack R.L., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su M., Chakraborty S., Osawa Y., Zhang H. Cryo-EM reveals the architecture of the dimeric cytochrome P450 CYP102A1 enzyme and conformational changes required for redox partner recognition. J. Biol. Chem. 2020;295:1637–1645. doi: 10.1074/jbc.RA119.011305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UniDesign is available at https://github.com/tommyhuangthu/UniDesign. All computational data and scripts in this work are available at https://doi.org/10.5281/zenodo.7881449.