Abstract
The α-chemokine SDF-1 binds CXCR4, a coreceptor for human immunodeficiency virus type 1 (HIV-1), and inhibits viral entry mediated by this receptor. Since chemokines are potent chemoattractants and activators of leukocytes, we examined whether the stimulation of HIV target cells by SDF-1 affects the replication of virus with different tropisms. We observed that SDF-1 inhibited the entry of X4 strains and increased the infectivity of particles bearing either a CCR5-tropic HIV-1 envelope or a vesicular stomatitis virus G envelope. In contrast to the inhibitory effect of SDF-1 on X4 strains, which is at the level of entry, the stimulatory effect does not involve envelope-receptor interactions or proviral DNA synthesis. Rather, we observed an increased ability of Tat to transactivate the HIV-1 long terminal repeat in the presence of the chemokine. Therefore, the effects of SDF-1 on the HIV-1 life cycle can be multiple and opposite, including both an inhibition of viral entry and a stimulation of proviral gene expression.
Chemokines are peptides of 70 to 100 amino acids that activate and induce the migration of leukocytes (5, 46). They bind to seven transmembrane-spanning, G protein-coupled receptors. At least two subfamilies, CXC (or α) and CC (or β) chemokines, are distinguished with respect to the position of the first two cysteine residues. SDF-1 (stromal cell-derived factor 1) is a CXC chemokine secreted constitutively by several types of cells (5). Functional variants of SDF-1 are generated by differential splicing of mRNAs. SDF-1α is four amino acids shorter than SDF-1β at the carboxy terminus. Both species bind with high affinity to the CXCR4 receptor, which is expressed in many tissues. SDF-1 stimulates CD34+ cells, pre-B cells, monocytes, neutrophils, and peripheral blood lymphocytes (PBLs), as indicated by intracellular [Ca2+] changes and chemotaxis (1, 5, 26, 36). SDF-1 can also induce apoptosis of primary CD8+ T cells (25). SDF-1 is involved in the homing of T cells and monocytes (9) and plays a key role in the development of the embryo. The knockout of the SDF-1 or of the CXCR4 gene is lethal in mouse embryos, leading to serious developmental defects of the immune, hematopoietic, circulatory, cardiac, and central nervous systems (35, 45, 47).
The roles played by chemokines in human immunodeficiency virus (HIV)-induced pathology are poorly understood. Several chemokine receptors serve as coreceptors for HIV entry into target cells, with a major role played by CCR5 and CXCR4 (12, 34). The interaction of gp120, the surface envelope glycoprotein of the virion, with both CD4 and the coreceptor triggers conformational changes of the envelope which expose fusogenic epitopes. These events lead to fusion of the viral and plasma membranes (13, 32). Macrophage-tropic strains of HIV (now classified as R5 viruses [7]) replicate in macrophages and in primary CD4+ T cells and use the CCR5 receptor. T-tropic HIV type 1 (HIV-1) isolates (referred to as X4 viruses) replicate in primary or established CD4+ lymphocytes and use the CXCR4 receptor. Viruses isolated in the initial stages of infection use primarily CCR5, while those isolated from patients with advanced disease use CXCR4 in addition to, or in place of, CCR5 (16). This observation suggests that the use of a particular chemokine receptor may have important consequences for HIV pathogenesis. The CCR5 ligands (the β-chemokines RANTES, MIP-1α, and MIP-1β) and the CXCR4 ligand SDF-1 inhibit the entry of R5 and X4 viruses, respectively (8, 12, 14, 38). In view of these inhibitory effects, it has been proposed that an increased production of β-chemokines may protect against HIV infection and disease progression (14). The inhibition of HIV entry by chemokines is independent of G protein signalling pathways and relies both on the occupancy and on the ligand-induced internalization of the coreceptors (2, 3, 23, 43). However, chemokines may exert a more complex action on HIV infection, since the β-chemokine RANTES has been reported to increase viral replication in monocytes or macrophages (29, 39). The molecular mechanisms of this enhancement are not fully understood. It is sensitive to pertussis toxin and therefore involves signalling through Gi proteins (29, 31). It has also been reported that MIP-1α, MIP-1β, and RANTES increase replication of X4 strains in PBLs by stimulating the transcription of the CXCR4 gene (19) or by increasing cell surface colocalization of CD4 and CXCR4 (31). Whether the inhibiting and stimulating effects of β-chemokines, observed in vitro on replicating HIV, have any impact on the outcome of the in vivo infection is not known.
We have examined the effect of SDF-1 on the in vitro replication of HIV-1 viruses with various tropisms. We observed that the chemokine exerts opposite effects on HIV-1 replication, depending on which receptor is used for entry. SDF-1 inhibited the entry of X4 strains and increased the infectivity of R5 strains and of vesicular stomatitis virus G (VSV-G) pseudotypes. In contrast to the inhibitory effect of SDF-1 on X4 strains, which takes place at the entry level, the stimulatory effect occurred later in the viral cycle by increasing the ability of Tat to transactivate the HIV-1 long terminal repeat (LTR). Therefore, the effects of SDF-1 on the HIV-1 life cycle can be multiple and opposite, including both the inhibition of viral entry and the stimulation of provirus transcription.
MATERIALS AND METHODS
Viruses, cells, and reagents.
HIV-1 (NL43, JRCSF, and NLAD8 strains) and HIV(VSV), a defective HIV-1 with the env gene deleted and pseudotyped with the VSV-G envelope, were produced, and infections were performed as described previously (33, 38, 41). The HIV-GFP reporter virus, which encodes the GFP marker in the place of Nef and has the env gene deleted, was a kind gift from D. Gabuzda (Dana-Farber Cancer Institute, Boston, Mass.) (24). HIV-GFP(VSV) was generated like HIV(VSV). The NLAD8 provirus was constructed by introducing a portion of the env gene from the R5-tropic clone of AD8 into pNL43 and was a kind gift from E. Freed (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). P4 cells (HeLa CD4+ CXCR4+ LTR-lacZ) and P4C5 cells (HeLa CD4+ CXCR4+ CCR5+ LTR-lacZ) (3, 33) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. CEMX174 and Jurkat T-cell lines and U937 and HL60 monocytic cell lines were grown in RPMI 1640 supplemented with 10% fetal calf serum. PBLs were isolated from healthy, HIV-negative volunteers, stimulated for 3 days with phytohemagglutinin (PHA) (1 μg/ml; Wellcome), and grown in the presence of interleukin-2 (IL-2) (3 ng/ml; PeproTech, Inc.). SDF-1α, referred to in this report as SDF-1, was chemically synthesized (20) and was a kind gift of F. Baleux (Institut Pasteur, Paris, France). Bordetella pertussis toxin was purchased from Calbiochem (San Diego, Calif.).
Measurement of HIV infection in P4C5 cells. (i) Single-cycle assay.
P4C5 cells (104 per well) in 96-well plates were cultured for 24 h before infection with 200 μl of viral supernatants (in triplicate) in the absence or in the presence of SDF-1 and with 20 ng of DEAE dextran/ml and 20 mM HEPES buffer (pH 7.3) as described previously (41). After the indicated periods, the cells were washed to remove unbound virus. After 24 h, β-gal activity was measured. The cells were lysed in 100 μl of 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 2.5 mM EDTA, 50 mM β-mercaptoethanol, and 0.125% Nonidet P-40. The lysates were mixed with 100 μl of 80 mM NaPO4 (pH 7.4), 10 mM MgCl2, 50 mM β-mercaptoethanol, and 6 mM chlorophenol red–β-galactopyranoside monoNa salt and incubated at 37°C before the optical density at 540 nm was measured. The values given are means ± standard deviations of triplicates. Variations for each point were less than 10%.
(ii) Study of HIV replication in P4C5 cells.
P4C5 cells (104 per well) in 96-well plates were cultured for 24 h before infection with 200 μl of viral supernatants (in triplicate) in the absence or in the presence of SDF-1 and with 20 ng of DEAE dextran/ml and 20 mM HEPES buffer (pH 7.3) as described previously (41). The viral inoculum corresponded to 2 and 2.5 ng of p24 for NL43 and JRCSF viruses, respectively. After viral exposure (2 h at 37°C) and every following day, the supernatants were harvested and replaced with fresh medium containing or not containing SDF-1. Gag p24 concentrations were measured in each supernatant by enzyme-linked immunosorbent assay (ELISA) (Dupont).
Measurement of HIV infection in other cell lines.
For infections with the HIV-GFP(VSV) reporter virus, 4 × 105 target cells were exposed overnight, with 5 ng of DEAE dextran/ml, to a viral dose corresponding to 100 ng of p24, in the absence or in the presence of SDF-1. The cells were washed to remove unbound virus and further incubated with or without SDF-1 for 8 h. The cells were then fixed and analyzed by flow cytometry with a Facscalibur (Becton Dickinson, Mountain View, Calif.) with CellQuest software. For infection of PBLs, 4 × 105 PBLs per well in 96-well plates were exposed to 200 μl of viral supernatants (in duplicate) in the absence or in the presence of SDF-1 and 20 mM HEPES buffer (pH 7.3). The viral inoculum corresponded to 1 ng of p24 of NLAD8 virus. After 4 h at 37°C, the cells were washed and resuspended in 200 μl of fresh medium containing 3 ng of IL-2/ml. At each cell passage, the supernatants were harvested and replaced with fresh medium containing or not containing SDF-1. Gag p24 concentrations were measured in each supernatant by ELISA.
Analysis of viral DNA synthesis.
P4 cells (6 × 106) were exposed to the indicated strains of HIV-1 (750 ng of p24), in the presence or in the absence of SDF-1. Seventeen hours later, low-molecular-weight DNA was prepared by Hirt extraction, EcoRI digested, and analyzed by Southern blotting with a 1.9-kb fragment from the pol region of pNL43 as a probe, as described previously (40).
Measurement of cytosolic Gag p24 amounts.
P4 cells were preincubated for 20 min in the absence or in the presence of SDF-1 and exposed to the indicated strains of HIV-1 (300 ng of p24) for 1 h at 37°C in the absence or in the presence of the chemokine. Cytosolic fractions were prepared as described previously (33). Briefly, to remove virus adsorbed at the cell surface, the cells were incubated for 10 min on ice with 1 ml of pronase (7 mg/ml, freshly prepared in DMEM with 20 mM HEPES). The cells were resuspended with 1 ml of ice-cold DMEM containing 10% fetal calf serum, washed in ice-cold phosphate-buffered saline, and resuspended in 2 ml of swelling buffer (Tris-HCl [pH8], 10 mM; KCl, 10 mM; EDTA, 1 mM) for 15 min at 4°C. The cells were then subjected to mechanical disruption by 15 strokes of a 7-ml β-pestle Dounce homogenizer. Nuclei and unbroken cells were pelleted at 3,000 rpm for 3 min at 4°C in a Heraeus Varifuge centrifuge. The resulting postnuclear supernatant was spun at 60,000 rpm in a TL-100 Beckman centrifuge for 10 min at 4°C to separate the membrane- and vesicle-rich pellet from the cytosolic supernatant. The supernatant was adjusted to 0.5% Triton X-100 and analyzed for p24 content by ELISA.
Analysis of LTR activity.
P4 cells were seeded at 8 × 104 per well of a 24-well plate 24 h before transfection by the Ca phosphate coprecipitation technique. The cells were transfected with 1 μg of the pLTRX-Luc plasmid, encoding the luciferase reporter gene driven by the HIV-1 LTR (42), and with or without 30 ng of the pCMV-Tat plasmid, carrying the tat gene driven by the cytomegalovirus immediate-early promoter (42). This ratio of pCMV-Tat versus pLTRX-Luc plasmid was chosen in order to stay in the linear part of the dose-response curve of Tat activity (42). Six hours after transfection, the cells were incubated, when stated, with SDF-1 and/or with pertussis toxin and lysed 18 h later as previously described (42). Luciferase activity contained in cytoplasmic extracts was measured with a luminometer (Lumat LB9501; Bertold). The results are expressed as relative luciferase units (RLU) per μg of cellular protein and were obtained by subtracting background signal (from nontransfected cells) from each value and dividing the figure by the amount of cytoplasmic protein contained in the sample. Experiments were performed in triplicate, and variations for each point were less than 10%.
RESULTS
Dual effects of SDF-1 on HIV infection.
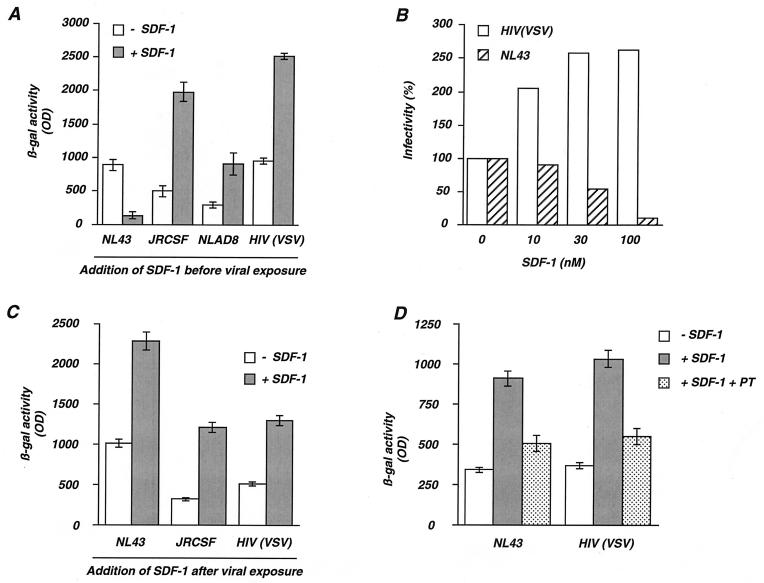
We examined the effects of SDF-1 on the replication of HIV in a single-cycle assay by using the P4C5 indicator cells. P4C5 cells are HeLa cells expressing the CD4, CXCR4, and CCR5 receptors, and they harbor an integrated lacZ gene driven by the HIV-1 LTR, which is inducible by the viral transactivator Tat (3, 33). Upon infection with X4 or R5 strains of HIV, P4C5 cells produce Tat, and viral replication can be quantified by using a β-galactosidase (β-gal)-based colorimetric assay. P4C5 cells were preincubated with SDF-1 (300 nM) for 20 min and exposed either to the X4 strain NL43 or the R5 strains JRCSF and NLAD8. As expected, β-gal activity, measured 24 h after exposure to NL43, was 10-fold lower in the presence of SDF-1 (Fig. 1A), confirming the inhibitory activity of the chemokine on the entry of X4 strains (8, 38). By contrast, β-gal activity induced by exposure to JRCSF and NLAD8 strains was three- to four-fold higher in the presence of SDF-1 (Fig. 1A). Infections were also performed with HIV-1 particles devoid of gp120 or gp41 and coated with the VSV-G envelope glycoprotein [HIV(VSV)]. VSV-G binds membrane phospholipids, allowing virus entry independently of the CD4 and chemokine receptors (11). Infection of P4C5 cells with HIV(VSV) was also increased by SDF-1 (Fig. 1A). Thus, stimulation of HIV infection by SDF-1 is independent of gp120- gp41-mediated binding and entry.
FIG. 1.
Opposite effects of SDF-1 on HIV replication. (A). Effect of SDF-1 added before virus exposure. P4C5 indicator cells (HeLa CD4+ CXCR4+ CCR5+ LTR-lacZ+) were preincubated for 20 min in the absence (− SDF-1) or in the presence (+ SDF-1) of SDF-1 (300 nM) and exposed to either NL43 (X4 strain), JRCSF and NLAD8 (R5 strains), or HIV(VSV), a defective HIV-1 with the env gene deleted and pseudotyped with the VSV-G envelope. Virus doses were 5 ng of p24 for NL43, JRCSF, or NLAD8 and 1 ng of p24 for HIV(VSV), which is more infectious (33). After 24 h, the cells were lysed and β-gal activity was measured in the cell extracts. The values are the means ± standard deviations of triplicates. (B) Dose-response analysis of the effect of SDF-1. P4C5 cells were preincubated with or without SDF-1 and exposed to NL43 or to HIV(VSV). After 24 h, the cells were lysed and β-gal activity was measured. For each indicated concentration of SDF-1, the values are expressed as percent infectivity, with 100% corresponding to the β-gal activity induced by each virus in the absence of SDF-1. (C) Effect of SDF-1 added after virus exposure. P4C5 cells were exposed for 2 h to NL43, JRCSF, or HIV(VSV), and unbound virus was removed. After 16 h, the cells were incubated with SDF-1 (100 nM), and β-gal activity was measured 9 h later. The values are means ± standard deviations of triplicates. (D) The stimulatory effect of SDF-1 is inhibited by pertussis toxin. P4 cells were exposed for 2 h to NL43 or HIV(VSV), and unbound virus was removed. After 16 h, the cells were incubated for 1 h with or without pertussis toxin (PT) (5 μg/ml). SDF-1 (200 nM) was then added as indicated, and β-gal activity was measured 9 h later. The values are means ± standard deviations of triplicates. Similar results were observed with P4C5 cells or when smaller amounts of pertussis toxin (50 ng/ml) were used (not shown). The data are representative of at least three independent experiments. OD, optical density.
The extent of the stimulation of viral replication induced by SDF-1 was examined for various concentrations of the chemokine. The increase of HIV(VSV) (Fig. 1B) and of R5 strain (not shown) infectivity was dose dependent and was observed at concentrations as low as 3 to 10 nM. These nanomolar concentrations are consistent with those inducing the migration and activation of leukocytes (1, 5, 9, 26) and the inhibition of X4 strain infection (Fig. 1B).
Since stimulation of HIV replication by SDF-1 is independent of gp120- or gp41-mediated entry, we examined at which step of the viral cycle the chemokine acts. SDF-1 was added 16 h after virus exposure. At this period of the cycle, incoming virions have performed reverse transcription and proviral DNA has been integrated in the host chromosome. P4C5 cells were exposed for 2 h to NL43, JRCSF, or HIV(VSV); unbound virus was removed; and 16 h later, the cells were incubated with SDF-1 (100 nM). β-Gal activity was measured 9 h later (Fig. 1C). The chemokine induced a two- to fourfold increase of β-gal activity upon infection with JRCSF or HIV(VSV), indicating that SDF-1 stimulates viral replication when added at a late step of the viral cycle. Since HIV(VSV) is a defective pseudotype that performs only a single cycle of replication, stimulation was not due to an effect of SDF-1 at an early step of a second viral cycle. Interestingly, under these experimental conditions, infection with the X4 strain NL43 was also increased by SDF-1 (Fig. 1C). Therefore, SDF-1 exerts opposite effects on the replication of X4 strains: the chemokine inhibits viral infection when added before or during virus exposure, whereas it increases the efficiency of infection when added at later steps of the viral cycle. The stimulatory effect of SDF-1 on HIV replication was sensitive to pertussis toxin (Fig. 1D), implicating Gi-linked signal transduction pathways in the activity of the chemokine.
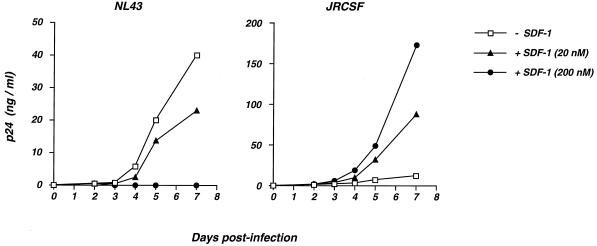
We then examined whether the enhancing effect of SDF-1 could be observed on multiple rounds of viral replication. P4C5 cells were exposed either to the X4 strain NL43 or the R5 strain JRCSF (with a low viral inoculum, corresponding to 2 ng of p24) in the absence or in the presence of SDF-1 (at 20 or 200 nM) which was maintained throughout the study. Viral replication was assessed by measuring p24 release in cell supernatants (Fig. 2). The NL43 strain led to a concentration of 40 ng of p24/ml at day 7 postinfection (p.i.). As expected, SDF-1 totally inhibited the replication of the X4 strain at a concentration of 200 nM. The inhibition was only partial when the chemokine was used at 20 nM (Fig. 2). The JRCSF strain replicated less efficiently than NL43 in P4C5 cells, and p24 concentrations reached 12 ng/ml at day 7 p.i. The presence of SDF-1 induced a dramatic increase of replication of the R5 strain, with p24 reaching 88 and 173 ng/ml at day 7 p.i. for 20 and 200 nM SDF-1, respectively (Fig. 2). Therefore, the stimulatory effect of SDF-1 on the infectivity of R5 strains could be observed on both single-cycle and multiple-round infection assays.
FIG. 2.
Effects of SDF-1 on a multiple-round infection assay of HIV replication. P4C5 cells were exposed either to the X4 strain NL43 (right panel) or the R5 strain JRCSF (left panel), in the absence (− SDF-1) or in the presence (+ SDF-1) of SDF-1 (at 20 or 200 nM). The viral inoculum corresponded to 2 and 2.5 ng of p24 for NL43 and JRCSF, respectively. Viral replication was assessed by measuring p24 release in cell supernatants at the indicated days p.i. SDF-1 was added to the cultures after each harvesting in order to maintain the indicated concentrations of the chemokine. The data are means of triplicates, and the standard deviation of each experimental point was below 10% (not shown). The data are representative of two independent experiments.
SDF-1 does not affect the entry of R5 strains.
We examined the effect of SDF-1 on the entry of incoming particles by measuring cytosolic Gag p24 content. We previously reported that the measurement of cytosolic p24 soon after virus exposure is a reliable assay for the assessment of virus entry events leading to authentic cell infection (33). P4C5 cells were incubated for 1 h at 37°C with the NL43, NLAD8, or JRCSF strain in the absence or the presence of SDF-1 (200 nM). The chemokine was added 20 min before virus exposure in order to allow binding to CXCR4. Cytosolic Gag p24 contents were measured after virus exposure (Fig. 3A). SDF-1 induced a fourfold decrease in cytosolic p24 content detected upon exposure to the X4 strain NL43 (Fig. 3A). This observation confirmed that the chemokine inhibits the entry of the X4 strain. In contrast, SDF-1 did not significantly affect the cytosolic p24 contents after exposure to the R5 strains NLAD8 and JRCSF (Fig. 3A). Therefore, the enhanced infection with R5 strains in the presence of SDF-1 is not a consequence of a more efficient entry.
FIG. 3.
Analysis of the effect of SDF-1 on cytosolic Gag p24 content and proviral DNA synthesis in target cells and on LTR activity. (A) Cytosolic Gag p24 content. P4C5 cells were preincubated for 20 min with (+ SDF-1) or without (− SDF-1) SDF-1 (200 nM) and exposed to the indicated viruses (with a dose of 300 ng of p24) for 1 h at 37°C. The cells were treated with pronase, to eliminate virus adsorbed at the cell surface, and lysed. Postnuclear supernatants were separated into cytosol and pellet fractions. The pellet fraction corresponds to cellular membranes and vesicles. Gag p24 content was measured in the cytosolic fraction. (B) Synthesis of proviral DNA in newly infected cells. P4C5 cells were preincubated for 30 min with (+) or without (−) SDF-1 (200 nM) and exposed to the indicated viruses (with a dose of 750 ng of p24). Seventeen hours later, low-molecular-weight DNA was extracted and analyzed by Southern blotting with an HIV probe from the pol region. Samples were digested with EcoRI, which produced, for NL43, diagnostic fragments with sizes of 5.7 and 9.1 kb from viral linear (L) and one LTR circular (C) DNA, respectively. The HIV(VSV) pseudotype generated two fragments: a 5.7-kb linear species and a 7.8-kb circular species. JRCSF, whose genome does not contain an EcoRI site, yielded a 9.7-kb signal corresponding to full-length proviral DNA. Samples were also hybridized with a probe corresponding to the mitochondrial gene coding for cytochrome b to verify that similar amounts of low-molecular-weight DNA were loaded in each lane (not shown). (C) Effect of SDF-1 on HIV-1 LTR activity. P4 cells were transfected with 1 μg of pLTRX-Luc, with or without 30 ng of pCMV-Tat DNA. After 6 h, the cells were incubated in the absence or in the presence of SDF-1 (200 nM), and 18 h later luciferase activities were measured in cell extracts. Where indicated, pertussis toxin (PT) (50 ng/ml) was added simultaneously with SDF-1. Similar results were observed at higher doses of pertussis toxin (5 μg/ml [not shown]). The data are representative of three independent experiments.
SDF-1 does not affect the efficiency of reverse transcription.
The amount of newly reverse-transcribed proviral DNA was monitored. P4C5 cells were preincubated for 30 min at 37°C with or without SDF-1 (200 nM) and exposed to either the NL43, HIV(VSV), or JRCSF viral strain, in the presence or in the absence of the chemokine. Nonintegrated viral DNA was extracted 17 h later and analyzed by Southern blotting with a probe from the pol region (Fig. 3B). Samples were digested with EcoRI, which, in the case of NL43, produced diagnostic fragments of 5.7 and 9.1 kb for viral linear (L) DNA and one LTR circular (C) DNA, respectively (40). The HIV(VSV) pseudotype generated two fragments: a 5.7-kb linear species and a 7.8-kb circular species, shorter than that of NL43 due to the deletion in the env gene. JRCSF, whose genome does not contain an EcoRI site, yielded a 9.7-kb signal corresponding to full-length proviral DNA. No signal was detected when target cells were treated with zidovudine, indicating that the hybridizing DNA was actually de novo synthesized during the infection period (not shown). The detection of the NL43-specific DNA signal was greatly reduced in the presence of SDF-1 (Fig. 3B), confirming the block of X4 strain entry by the chemokine (8, 38). In contrast, SDF-1 did not significantly affect proviral DNA synthesis of the HIV(VSV) or JRCSF strains. Viral DNA molecules undergoing circularization after their transport to the nucleus were used as markers to monitor nuclear import of preintegration complexes (10). For HIV(VSV), the ratios of circular DNA to total viral DNA were equivalent with and without SDF-1. Thus, SDF-1 did not significantly affect the efficiency of proviral DNA synthesis and subsequent nuclear import of preintegration complexes. This observation was confirmed in three independent experiments and by quantitative analysis of hybridization signal intensity by phosphorimager scanning (not shown). Increased HIV infectivity by SDF-1 is therefore not due to an enhancement of viral entry or of the reverse transcription process.
SDF-1 increases Tat-mediated provirus transcription.
We investigated whether SDF-1 acts at a late step of the viral cycle by increasing Tat-mediated transactivation of the HIV-1 LTR. HeLa-CD4 cells were transiently cotransfected with the pLTRX-Luc plasmid, which contains the luciferase reporter gene driven by the HIV-1 LTR, and with a Tat expression vector (pCMV-Tat). Six hours after the addition of DNA, the cells were incubated in the presence or in the absence of SDF-1 (200 nM), and luciferase activity was measured 18 h later (Fig. 3C). In the absence of SDF-1, the LTR promoter yielded a baseline luciferase activity of 6 RLU, which rose to 690 RLU upon transfection of the pCMV-Tat DNA. The baseline activity of the LTR (without Tat) was not significantly increased by SDF-1 and reached 9 RLU. In the presence of Tat, luciferase activity was significantly higher in SDF-1-treated cells and rose to 2,000 RLU (Fig. 3C). This stimulation corresponded, in four independent experiments, to a mean increase of luciferase activity of (3.1 ± 0.5)-fold in the presence of SDF-1 (not shown). We then examined whether pertussis toxin inhibited the effect of SDF-1 on Tat transactivation. In the absence of SDF-1, equivalent levels of luciferase activity were measured with and without pertussis toxin (Fig. 3C), indicating that the compound did not affect the ability of the LTR to be transactivated by Tat. However, the toxin abrogated the stimulatory effects of SDF-1 (Fig. 3C). Altogether, these experiments indicated that SDF-1 stimulates Tat-mediated transcription through a mechanism involving Gi-linked signal transduction pathways.
Stimulatory effects of SDF-1 in monocytic and lymphoid cells.
With the aim of analyzing the effect of SDF-1 on a single round of viral infection in monocytic and lymphoid cell lines, experiments were performed with a VSV-G pseudotype of the HIV-GFP reporter virus. This defective recombinant virus contains a deletion in the env gene and encodes the GFP marker protein in place of Nef (24). Infected cells can be detected by measuring GFP expression by flow cytometry. HeLa CD4 cells, U937 and HL60 monocytic cell lines, and CEMX174 and Jurkat lymphoid cells were exposed to equivalent viral inocula (100 ng of p24). The proportion of GFP-expressing cells, measured 24 h later, varied from 12 to 23% (Fig. 4). In contrast, the HUT78 lymphoid cell line and PBLs stimulated by PHA and grown in the presence of IL-2 were poorly infected by HIV-GFP(VSV) (<1% GFP-expressing cells [not shown]). This experiment showed that susceptibility to HIV-GFP(VSV) infection varies from one cell line to another. The effect of SDF-1 on HIV replication was then studied in susceptible cell lines. In HeLa cells, in U937 and HL60 monocytic cells, and in CEMX174 lymphoid cells, the proportion of GFP-expressing cells was higher when infections were performed in the presence of SDF-1 (Fig. 4). No effect was observed in Jurkat cells (Fig. 4). In MT4 lymphoid cells, exposure to HIV-GFP(VSV) led to 85% GFP+ cells, and this proportion was not increased by the chemokine (not shown). These experiments showed that SDF-1 increases the efficiency of HIV infection in a variety of cell lines, including HeLa CD4, U937, HL60, and CEMX174, whereas others, such as Jurkat and MT4, are not sensitive to the chemokine.
FIG. 4.
Analysis of HIV-1 infection in the presence of SDF-1 in various cell lines. HeLa CD4 cells (clone P4), U937 and HL60 monocytic cells, and CEMX174 and Jurkat lymphoid cell lines were infected with the HIV-GFP defective virus pseudotyped with the VSV-G envelope, which contains, in place of nef, the gene coding for GFP. Following integration and proviral expression, the cells synthetize GFP, which can be detected by flow cytometry. The cell lines were exposed to HIV-GFP(VSV) (100 ng of p24) with (+ SDF-1) or without (− SDF-1) SDF-1 (200 nM). After an overnight incubation, the cells were washed to remove unbound virus and further incubated with or without SDF-1 for 8 h. Infection was then revealed by flow cytometry analysis on gated living cells. The values are presented as two-color dot blots, with the green fluorescence (GFP) on the abscissa and the red fluorescence on the ordinate. The percentage of GFP+ cells is indicated in each panel. In the absence of viral infection, these percentages were <1% (not shown). The data are representative of three independent experiments.
Replication-competent virus was then used to study the effect of SDF-1 on HIV replication in PBLs. PHA-stimulated PBLs from six different donors were exposed to the R5 strain NLAD8 in the presence or in the absence of SDF-1. Viral replication was monitored by a time course measurement of p24 concentrations in cell supernatants. In two donors, we observed a two- to threefold increase of p24 levels when infection was performed in the presence of SDF-1. No significant effect of SDF-1 was observed in PBLs from four other donors. Representative examples of responsive and unresponsive donors is shown Fig. 5 (donors A and B, respectively).
FIG. 5.
Analysis of HIV-1 replication in the presence of SDF-1 in PBLs. PHA-stimulated PBLs (from six different donors) were exposed to the R5 strain NLAD8 (viral inoculum corresponding to 1 ng of p24) in the presence (+ SDF-1) or in the absence (− SDF-1) of SDF-1 (200 nM). Viral replication was assessed by measuring p24 release in cell supernatants at the indicated days p.i. SDF-1 was added to the cultures after each harvesting in order to maintain the indicated concentrations of the chemokine. The values are means ± standard deviations of duplicates. In two of the donors, we observed a two- to threefold increase of p24 levels when infection was performed in the presence of SDF-1. A representative example of a responsive donor is provided in the left panel (donor A). No significant effect of SDF-1 was observed in PBLs from four other donors. An example of an unresponsive donor is donor B (right panel).
DISCUSSION
By binding to its receptor, CXCR4, the α-chemokine SDF-1 interferes with the entry of X4 HIV strains into target cells (8, 38) (Fig. 1 and 3). We show here that this chemokine exerts an additional and opposite effect on infection by both X4 and R5 viruses by enhancing Tat-mediated transcriptional activity of the HIV LTR.
Several lines of evidence indicate that the enhancement of HIV-1 infection by SDF-1 takes place at a late stage of the viral cycle rather than at the level of entry. First, the chemokine increased to the same extent the infectivities of R5 strains and viruses pseudotyped with the VSV-G envelope, indicating that enhancement is independent of gp120- or gp41-mediated entry. Second, time course experiments showed that SDF-1 was equally efficient when added 16 h after virus exposure, confirming that the positive effect of SDF-1 is not due to a modification of the number or of the localization of coreceptors at the cell surface. Moreover, under these experimental conditions, infection with both X4 and R5 strains was increased by SDF-1. Third, a direct analysis of entry events showed that SDF-1 inhibited cytosolic Gag p24 uptake and proviral DNA synthesis of X4 strains when added before virus exposure but had no significant effects on R5 strains and on VSV-G-pseudotyped viruses. Lastly, evidence was obtained for an increased transactivation by Tat of the HIV LTR in the presence of SDF-1. Interestingly, the extent of the stimulation of transcriptional activity of the viral promoter (three- to fourfold) was in the same range as that of viral infectivity. This suggests that the positive effect of SDF-1 on HIV infection is mainly, if not only, due to stimulation of proviral gene expression.
Other cytokines have dichotomous effects on the viral cycle. Tumor necrosis factor alpha (TNF-α), transforming growth factor β (TGF-β), gamma interferon (IFN-γ), IL-4, IL-10, and IL-13 also exert positive and negative effects on HIV infection (15, 37, 44). Some of these effects vary according to the timing of HIV infection and include cellular activation and upregulation of coreceptor expression. The β-chemokines RANTES, MIP-1α, and MIP-1β can increase HIV replication in monocytes, macrophages, and lymphocytes, although their major effect is to inhibit the entry of R5 strains (19, 29, 31, 39). The positive effects of β-chemokines are pertussis toxin sensitive (29, 31). Dolei et al. suggested that β-chemokines may increase the entry of X4 strains into PBLs as a consequence of an accumulation of CXCR4 transcripts (19). However, no evidence of modified CXCR4 surface levels was provided in that study. Recently, Kinter et al. reported that β-chemokines have no effect on CXCR4 cell surface levels but increase surface colocalization of CD4 and CXCR4 (31). The mechanisms producing positive effects of β-chemokines on HIV replication are therefore not yet fully understood.
We show here that increases of HIV infectivity and of LTR activity induced by SDF-1 were sensitive to pertussis toxin, implying that cellular Gi-linked signal transduction pathways mediate the action of the chemokine. The signaling pathways activated by chemokine receptors are multiple and not fully understood (5, 22, 46). Chemokine receptors couple to members of the Gi or Gq family of G proteins. SDF-1, like other chemokines, induces phosphorylation of its receptor and stimulates G protein activation, inositol phosphate generation, and calcium elevation (22, 23, 46). Downstream events include the tyrosine kinases Pyk2 and mitogen-activated protein kinase and phosphatidylinositol 3-kinase (18, 28, 46). Furthermore, chemokines may upregulate transcription factors, such as nuclear transcription factor κB, which may, in turn, augment HIV transcription (22, 30). By using an HIV-GFP(VSV) reporter virus, we show here that the positive effect of SDF-1 on HIV infection is observed not only in HeLa-derived indicator cells but also in U937 and HL60 monocytic cells and in CEMX174 lymphoid cells. In contrast, cell lines like Jurkat or MT4 were not susceptible, although they express surface CXCR4 molecules fully functional for HIV entry. However, there are several indications that a dissociation exists between HIV binding and signalling through chemokine receptors. First, deletion of the C-terminal cytoplasmic domain of CXCR4 or of CCR5 does not affect HIV entry, although it alters chemokine-induced receptor downregulation (2, 3, 23, 43). Second, SDF-1 analogs, which act as CXCR4 antagonists, do not promote Ca2+ intracellular changes although they retain their anti-HIV activity (17, 27). Third, the function of CCR5 as an HIV-1 coreceptor is not related to its ability to signal or to be desensitized upon ligand binding (4, 21). It is therefore conceivable that in Jurkat or MT4 T cells, CXCR4 allows virus entry but does not activate signalling pathways leading to an enhancement of Tat activity.
We also studied the effect of SDF-1 in primary lymphocytes. A significantly increased replication of R5 strains was observed in cells from two of the six analyzed donors, whereas no effect was detected in the others (Fig. 5). This inconsistent effect of SDF-1 does not argue against the relevance of the phenomenon in the natural target cells of HIV replication. Indeed, primary cell populations are heterogeneous, possibly containing various proportions of cells susceptible to the effect of SDF-1 on HIV infection. This could mask the effects observed in homogeneous, immortalized cell clones. Crude supernatants from PBL cultures also contain variable levels of numerous cytokines which are either stimulators or inhibitors of HIV replication (15). Stimulatory cytokines include IL-1β, IL-2, IL-3, IL-6, IL-12, TNF-α and TNF-β, and the macrophage and granulocyte-macrophage colony-stimulating factors. IFN-α and IFN-β suppress HIV replication, whereas TGF-β, IL-4, IL-10, IL-13, and IFN-γ either induce or suppress HIV expression, depending on the culture system (15). Inconsistent effects in PBLs may therefore reflect variations in the relative levels of endogenous cytokines or other unidentified factors. Interestingly, Kinter et al. also observed donor-to-donor variability with regard to the stimulatory activity of β-chemokines on HIV replication in primary CD4+ T cells (31).
The in vivo relevance of the positive effect of SDF-1 on HIV replication is a matter for speculation. The pathogenesis of HIV is multifactorial and is influenced by both viral and host factors (15). The multifactorial nature of HIV pathogenesis is reflected in the highly variable rates of disease progression which are observed in infected individuals. Since a huge number of replication cycles is required for disease progression, even a three- to fourfold increase in the efficiency of the viral cycle could have significant effects on virus loads. Disease progression is associated with a shift in chemokine receptor usage, from CCR5 to CXCR4 (16). Whether this shift is induced by the chemokines themselves and/or by other host or viral factors is not known. The relative influence of the positive and negative effects of the β-chemokines and of SDF-1 on HIV replication in vivo remains unknown. A delicate balance among a wide array of host positive and negative factors, including chemokines, likely determines the net rate of viral replication in HIV-infected individuals (15). The stimulating activity of chemokines should also be taken into account when considering the use of chemokine analogs as anti-HIV agents (6, 12).
ACKNOWLEDGMENTS
We thank Susan Michelson for critical reading of the manuscript and F. Baleux, D. Gabuzda, E. Freed, N. Landau, and A. Miyanohara for the kind gift of reagents.
This work was supported by grants from the Agence Nationale de Recherche sur le SIDA (ANRS) and the Pasteur Institute.
REFERENCES
- 1.Aiuti A, Webb I J, Bleul C, Springer T, Gutierrez-Ramos J C. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitor to peripheral blood. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Locati M, Kennedy P E, Murphy P M, Berger E A. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 3.Amara A, Le Gall S, Schwartz O, Salamero J, Montes M, Loetscher P, Baggioloni M, Virelizier J L, Arenzana-Seisdedos F. HIV co-receptor down-regulation as anti-viral principle. SDF-1α dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aramori I, Ferguson S S, Bieniasz P D, Zhang J, Cullen B, Cullen M G. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 1997;16:4606–4616. doi: 10.1093/emboj/16.15.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini M, Moser B. Blocking chemokine receptors. J Exp Med. 1997;186:1189–1191. doi: 10.1084/jem.186.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clarck-Lewis Y, Sodroski J, Springer T A. The lymphocyte chemo-attractant SDF-1 is a ligand for lestr/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 11.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairns J S, D’Souza M P. Chemokines and HIV-1 second receptors: the therapeutic connection. Nat Med. 1998;4:563–568. doi: 10.1038/nm0598-563. [DOI] [PubMed] [Google Scholar]
- 13.Clapham P R. HIV and chemokines: ligands sharing cell-surface receptors. Trends Cell Biol. 1997;7:264–268. doi: 10.1016/S0962-8924(97)01075-1. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 15.Cohen O J, Kinter A, Fauci A S. Host factors in the pathogenesis of HIV disease. Immunol Rev. 1997;159:31–48. doi: 10.1111/j.1600-065x.1997.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump M P, Gong J H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J L, Baggiolini M, Sykes B D, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolei A, Biolchini A, Serra C, Curreli S, Gomes E, Dianzani F. Increased replication of T-cell tropic HIV strains and CXCR4 induction in T cells treated with MIP-1α, MIP-1β and RANTES β chemokines. AIDS. 1998;12:183–190. doi: 10.1097/00002030-199802000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein 1beta-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 22.Ganju R K, Brubaker S A, Meyer J, Dutt P, Yang Y M, Qin S X, Newman W, Groopman J E. The alpha-chemokine, stromal cell-derived factor-1 alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J Biol Chem. 1998;273:23169–23175. doi: 10.1074/jbc.273.36.23169. [DOI] [PubMed] [Google Scholar]
- 23.Haribabu B, Richardson R M, Fisher I, Sozzani S, Peiper S C, Horuk R, Ali H, Snyderman R. Regulation of human chemokine receptors CXCR4. Role of phosphorylation in desensitization and internalization. J Biol Chem. 1997;272:28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- 24.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newmann W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 25.Herbein G, Mahlknecht U, Batliwalla F, Gregersen P, Pappas T, Butler J, O’Brien W A, Verdin E. Apoptosis of CD8(+) T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature. 1998;395:189–194. doi: 10.1038/26026. [DOI] [PubMed] [Google Scholar]
- 26.Hesselgesser J, Liang M, Hoxie J, Greenberg M, Brass L F, Orsini M J, Taub D, Horuk R. Identification and characterization of the CXCR4 chemokine receptor in human T cell lines: ligand binding, biological activity, and HIV-1 inefectivity. J Immunol. 1998;160:877–883. [PubMed] [Google Scholar]
- 27.Heveker N, Montes M, Germeroth L, Amara A, Trautmann A, Alizon M, Schneider-Mergener J. Dissociation of the signalling and antiviral properties of SDF-1-derived small peptides. Curr Biol. 1998;8:369–376. doi: 10.1016/s0960-9822(98)70155-1. [DOI] [PubMed] [Google Scholar]
- 28.Jones S A, Moser B, Thelen M. A comparison of post-receptor signal transduction events in Jurkat cells transfected with either IL-8R1 or IL-8R2. Chemokine mediated activation of p42/p44 MAP-kinase (ERK-2) FEBS Lett. 1995;364:211–214. doi: 10.1016/0014-5793(95)00397-r. [DOI] [PubMed] [Google Scholar]
- 29.Kelly M D, Naif H M, Adams S L, Cunningham A L, Lloyd A R. Dichotomous effects of β-chemokines on HIV replication in monocytes and monocyte-derived macrophages. J Immunol. 1998;160:3091–3095. [PubMed] [Google Scholar]
- 30.Kingsman S M, Kingsman A J. The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem. 1996;240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 31.Kinter A, Catanzaro A, Monaco J, Ruiz M, Justement J, Moir S, Arthos J, Oliva A, Ehler L, Mizell S, Jackson R, Ostrowski M, Hoxie J, Offord R, Fauci A S. CC-chemokines enhance the replication of T-tropic strains in CD4+ T cells: Role of signal transduction. Proc Natl Acad Sci USA. 1998;95:11880–11885. doi: 10.1073/pnas.95.20.11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong P D, Wyatt R, Robinson J, Sweet R, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maréchal V, Clavel F, Heard J M, Schwartz O. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J Virol. 1998;72:2208–2212. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 35.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 36.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B cell growth-stimulating factor. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naif H M, Li S, Ho-Shon M, Mathijs J M, Williamson P, Cunningham A L. The state of maturation of monocytes into macrophages determines the effects of IL-4 and IL-13 on HIV replication. J Immunol. 1997;158:501–506. [PubMed] [Google Scholar]
- 38.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine, stromal cell derived factor 1 (SDF-1), is the ligand for LESTR/fusin and prevents infection by lymphocyte-tropic HIV-1 syncytium-inducing strains. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 39.Schmidtmayerova H, Sherry B, Bukrinsky M. Chemokines and HIV replication. Nature. 1996;382:767. doi: 10.1038/382767a0. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz O, Maréchal V, Danos O, Heard J M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J M. Antiviral activity of the proteasome on incoming HIV-1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz O, Virelizier J L, Montagnier L, Hazan U. A microtransfection method using luciferase gene for the study of human immunodeficiency virus long terminal repeat activity. Gene. 1990;88:197–205. doi: 10.1016/0378-1119(90)90032-m. [DOI] [PubMed] [Google Scholar]
- 43.Signoret N, Oldridge J, Pelchen-Matthews A, Klasse P J, Tran T, Brass L F, Rosenkilde M M, Schwartz T W, Holmes W, Dallas W, Luther M A, Wells T N C, Hoxie J A, Marsh M. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sozzani S, Ghezzi S, Iannolo G, Luini W, Borsatti A, Polentarutti N, Sica A, Locati M, Mackay C, Wells T N, Biswas P, Vicenzi E, Poli G, Mantovani A. Interleukin 10 increases CCR5 expression and HIV infection in human monocytes. J Exp Med. 1998;187:439–444. doi: 10.1084/jem.187.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 46.Ward S G, Bacon K, Westwick J. Chemokines and T lymphocytes: more than an attraction. Immunity. 1998;9:1–11. doi: 10.1016/s1074-7613(00)80583-x. [DOI] [PubMed] [Google Scholar]
- 47.Zou Y, Kottmann A, Kuroda M, Taniuchi I, Littman D R. Function of the chemokine receptor CXCR4 in hematopoiesis and in cerebellar development. Nature. 1998;391:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]