Abstract
Centrioles are critical for fundamental cellular processes, including signaling, motility, and division. The extent to which centrioles are present after cell cycle exit in a developing organism is not known. The stereotypical lineage of Caenorhabditis elegans makes it uniquely well-suited to investigate this question. Using notably lattice light-sheet microscopy, correlative light electron microscopy, and lineage assignment, we found that ~88% of cells lose centrioles during embryogenesis. Our analysis reveals that centriole elimination is stereotyped, occurring invariably at a given time in a given cell type. Moreover, we established that experimentally altering cell fate results in corresponding changes in centriole fate. Overall, we uncovered the existence of an extensive centriole elimination program, which we anticipate to be paradigmatic for a broad understanding of centriole fate regulation.
Extensive and stereotyped centriole elimination during C. elegans development is controlled by cell fate.
INTRODUCTION
Centrioles are evolutionarily conserved microtubule-based organelles fundamental for critical cellular and developmental processes (1). Centrioles recruit the pericentriolar material (PCM) and thus assemble centrosomes in animal cells (2). Moreover, centrioles template axoneme formation in primary cilia and flagella (3). Despite such fundamental roles, centrioles are absent from some differentiating cells, including some tissue types in Drosophila, as well as vertebrate myotubes (4–10). Whether the absence of centrioles is a frequent occurrence has not been investigated in any system. Moreover, whether centrioles vanish stochastically or else in a stereotyped manner has not been addressed in a comprehensive manner.
We set out to systematically monitor centriole fate in a developing organism. Because of its invariant lineage (11, 12), Caenorhabditis elegans is uniquely well-suited to this end. Furthermore, the components essential for centriole assembly have been identified in a systematic manner in C. elegans (13), and the molecular architecture of worm centrioles has been resolved (14). Centrioles in C. elegans embryos are ~175 nm by 120 nm in dimensions and comprise ninefold radially symmetric microtubule singlets. In addition to microtubules, worm centrioles harbor notably SAS-6, which is located centrally and seeds organelle assembly, as well as SAS-4, which localizes in the vicinity of the microtubules, whereas SAS-7 localizes more peripherally (Fig. 1A) (14). Both SAS-6 and SAS-4 are incorporated in the forming organelle and undergo little if any exchange with the cytoplasmic pool of proteins thereafter (15–19). Moreover, worms lack motile cilia and harbor only ciliated sensory neurons (20), so that centriole fate can be investigated in this system without potentially confounding constraints from these derived structures.
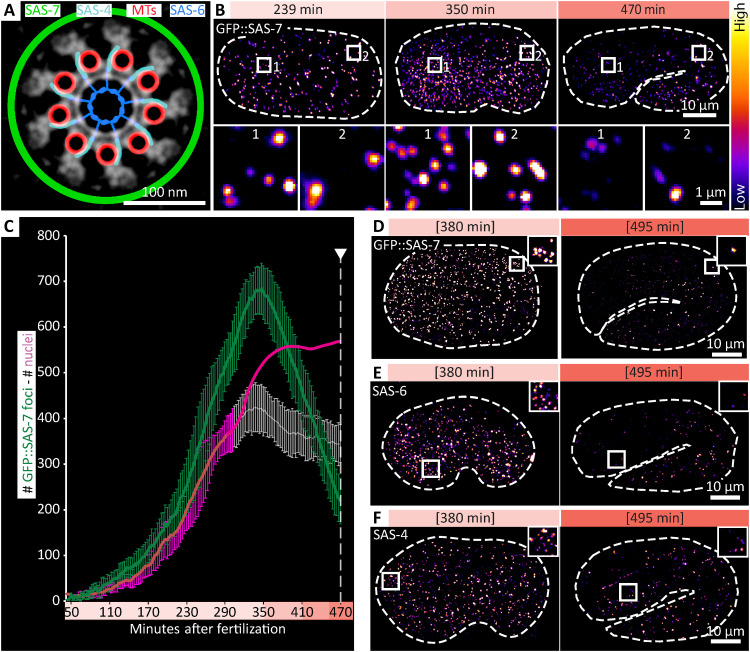
Fig. 1. Extensive centriole elimination during C. elegans embryogenesis.
(A) Localization of proteins used in this study represented schematically on a cross section of an inverted electron microscopy (EM) image modified from (14). MTs, microtubules (B) Images from lattice light-sheet time-lapse microscopy at the indicated times after fertilization, illustrating increase (compare 239 to 350 min) and then decrease (compare 350 to 470 min) of green fluorescent protein (GFP)::SAS-7 foci number. High magnification insets of an anterior and posterior region are shown below. Embryos also expressed mCherry (mCh)::HIS-58 [not shown, but see (C)]. (C) Quantification of GFP::SAS-7 foci (green) over time from lattice light-sheet data (n = 11 embryos; average and SD). Corresponding nuclear values are represented in magenta. Actual average counts of mCh::HIS-58 are shown until ~300 min after fertilization (dark line, with SD), after which the nuclear detection pipeline identified less nuclei than expected (gray line, with SD), primarily because nuclei are very crowded at that time. Note that such an underestimate was not observed for the smaller and sparser centriolar foci (see also fig. S1B). From this point onward, actual nuclear counts (12) are provided as well (magenta line). Here and throughout the manuscript, arrowhead and dashed line indicate twitching onset. Note that because of the resolution limit, one focus of GFP::SAS-7 might correspond to one, two, or sometimes four centrioles. (D to F) Bean (~380 min) and twofold stage (~495 min) embryos expressing GFP::SAS-7 immunostained for GFP (D), as well as wild-type embryos at the same stages immunostained for SAS-6 (E) and SAS-4 (F). Squared brackets indicate estimated time based on morphology. Brightness and contrast was set differently for (D) compared to (E) and (F). Actual number of foci at 495 min: 120 GFP::SAS-7; 113 SAS-6; 189 SAS-4.
Immunostaining experiments indicated that select centriolar markers are absent from some cells toward the end of C. elegans embryogenesis (15, 21). Whether these observations reflect selectively impaired antibody accessibility, loss of some centriolar components in a subset of cells or else bona fide centriole elimination is unknown. Moreover, the centrioles that template the ciliary axoneme of sensory neurons degenerate toward the end of embryogenesis (21, 22). Some centriolar markers also disappear during larval development from select cells, including terminally differentiated intestinal and seam cells (23). Although such initial observations indicate that centrioles are eliminated from some cells, comprehensive mapping of centriole fate onto the C. elegans lineage has not been conducted, precluding an assessment of the magnitude and potential stereotypy of this process.
RESULTS
We set out to determine potential changes in overall centriole number during worm embryogenesis. C. elegans embryos exhibit a proliferation phase until ~350 min after fertilization, by which time most of the 558 cells present at hatching have been generated (fig. S1A) (12). In the morphogenesis phase that follows, embryos are successively in the bean, comma, and 1.5-fold stages, which precedes the twofold stage, when muscle activity leads to twitching (fig. S1A). During morphogenesis, most cells exit the cell cycle and terminally differentiate, whereas a few others merely arrest cell cycle progression, resuming proliferation during the larval stages (hereafter referred to as “proliferative cells”) (11, 24). To monitor centrioles, we used a modified lattice light-sheet microscope to image embryos expressing centriolar green fluorescent protein (GFP)::SAS-7, which exhibited the brightest signal among centriolar markers tested, and nuclear mCherry::HIS-58 (Fig. 1B and movie S1) (25). The resulting number of GFP::SAS-7 foci was determined at each time point by a faithful machine learning algorithm (fig. S1B). As expected, GFP::SAS-7 foci numbers increased regularly during the proliferation phase, peaking at ~680 (±60 SD) counts (Fig. 1C). This number is higher than that of cells because there is more than one centriole per cell, although neighboring centrioles within a cell are sometimes detected as single entities given their proximity (see legend of Fig. 1C). Strikingly, we found that GFP::SAS-7 foci number began decreasing after the proliferation phase, reaching ~220 (±50) at the onset of twitching, beyond which monitoring with live imaging cannot be conducted (Fig. 1C). To extend these observations beyond twitching, we analyzed fixed embryos expressing GFP::SAS-7, finding similar numbers at the 1.5-fold stage, followed by a further diminution (Fig. 1D and fig. S1, C and E). To test whether this drastic diminution extends to other centriolar markers, we immunostained staged wild-type embryos for the core centriolar proteins SAS-6 and SAS-4, with an analogous outcome (Fig. 1, E and F, and fig. S1, D, F, and G). Overall, we conclude that the number of foci bearing GFP::SAS-7, SAS-6, and SAS-4 diminishes in a dramatic manner during the morphogenesis phase of C. elegans embryogenesis.
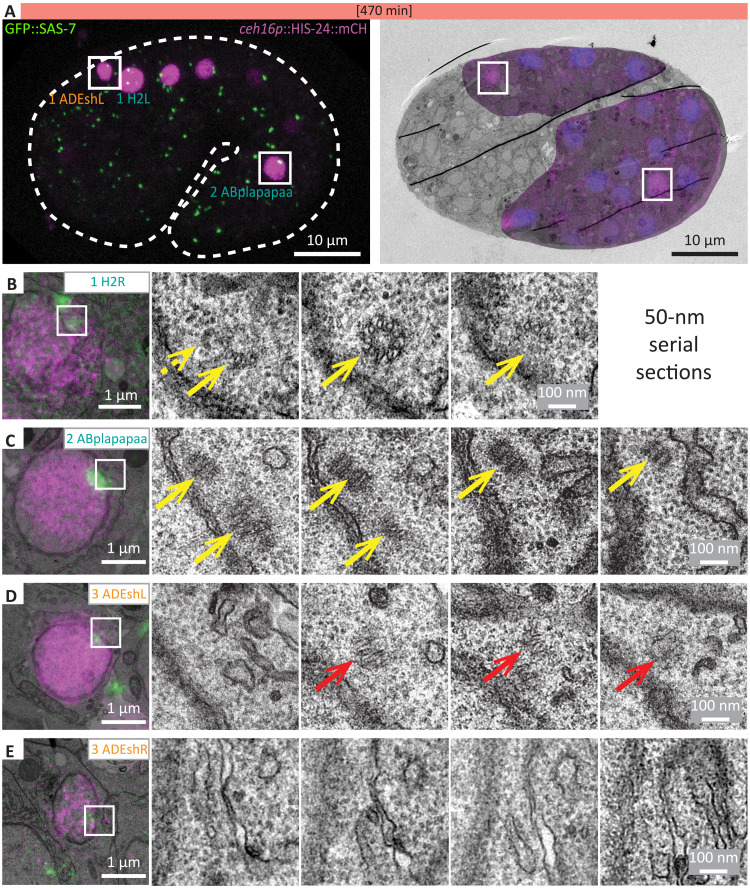
We next addressed whether the diminution of foci harboring centriolar proteins is mirrored by a likewise loss of the signature centriolar microtubules at the ultrastructural level. To aid the recognition of cells in the densely packed 1.5-fold embryo, we focused on a subset of cells expressing HIS-24::mCherry driven by the ceh-16 promoter, corresponding to cells with different GFP::SAS-7 focal intensities. These included the proliferating cells ABpl/rapapaa, which harbored a bright focus of GFP::SAS-7, and H2L/R, where the foci were less bright, as well as the nonproliferating cells ADEshL/R, in which GFP::SAS-7 was weaker still (Fig. 2A and fig. S1, H and I). We used correlative light and electron microscopy (CLEM) to identify nuclei and centrioles in these cells, which were then analyzed by serial-section electron microscopy (EM). We found centrioles with characteristic microtubule blades in all six proliferating cells analyzed (Fig. 2, B and C). In the three nonproliferating cells, by contrast, no structure with organized microtubules was found, except that, in one case, a structure suggestive of a centriole remnant was observed (Fig. 2, D and E). Thus, bright GFP::SAS-7 foci correspond to centrioles, whereas at least some of the cells in which GFP::SAS-7 foci intensity are diminishing undergo centriole elimination. Together, these findings indicate that loss of GFP::SAS-7 foci can be used as a proxy for centriole elimination.
Fig. 2. CLEM reveals centriole elimination in cells with weak GFP::SAS-7 foci.
(A) Overlay of single-plane dual-color confocal microscopy of a 1.5-fold embryo expressing ceh-16p::HIS-24::mCh and GFP::SAS-7 with corresponding EM image. In the overlay EM image, additional nucei are stained using Hoechst in blue. Cells analyzed by correlative light and electron microscopy (CLEM) are indicated (left), with insets showing regions magnified in (C) and (D). (B to E) Consecutive 50-nm sections of the proliferating seam progenitor cell H2R (B) (n = 3), ABplapapaa (n = 3) (C), and two examples of the nonproliferating ADEshL/R cell [(D) from the cell shown in (A); (E) from another embryo]; note that one such cell harbored what appeared like a centriole remnant (red arrow) (n = 1) (D), whereas no visible centriolar structure was detected in the second cell (E) (n = 2). Note also that no higher magnification EM images were acquired for (E), explaining the more pixelated appearance of these images compared to the other ones. Yellow arrows point to centrioles, dashed yellow arrow to a potential centriole, and red arrows to centriole remnants. Gray levels of the GFP::SAS-7 signal were differently adjusted for (B) and (E). Note that, in (B), no clear centriolar structure appeared to be associated with the second GFP::SAS-7 focus that is visible on the top left in the overlay image.
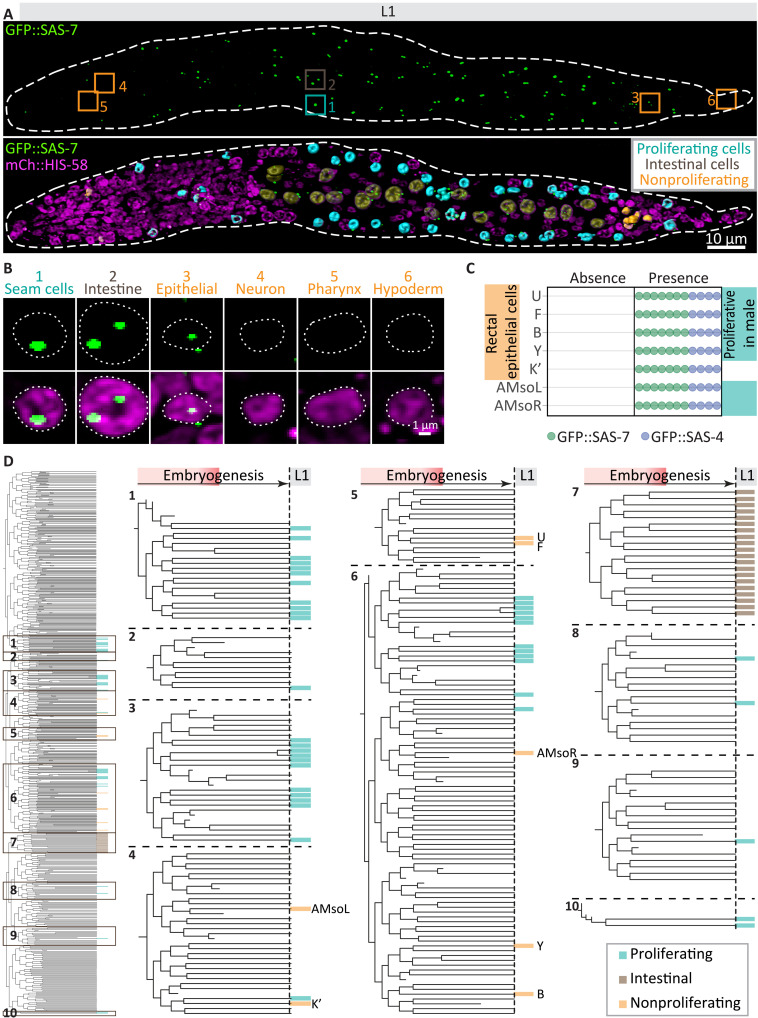
To what extent do GFP::SAS-7 foci numbers diminish until the end of embryogenesis, and is centriole elimination stereotyped? Because the small size and tight packing of cells preclude addressing this question during the second part of embryogenesis, we turned to the L1 larval stage that immediately follows. Here, most cells are further apart from one another and their identification is aided by clear anatomical landmarks, cell type–specific nuclear characteristics, and detailed atlases (11, 26). Strikingly, this analysis revealed that, of the 558 cells present at the end of embryogenesis, 68 invariably maintained foci harboring both GFP::SAS-7 and GFP::SAS-4 (Fig. 3 and fig. S2). As anticipated from the requirement of centrioles for centrosome assembly in C. elegans (13, 27), we found that all 41 proliferating cells maintained centrioles (Fig. 3, A and B, and figs. S2 and S3A). Moreover, all 20 intestinal cells, which later endoreduplicate, with some undergoing endomitosis (28, 29), maintained centrioles in L1 larvae, although they undergo elimination later in development (figs. S2 and S3A) (23). We found that 7 of the 497 cells that have terminally exited the cell cycle nevertheless invariably retained GFP::SAS-7 and GFP::SAS-4 foci: five cells located in the rectal region (U, F, B, Y, and K′) and two amphid socket cells located in the head region (AMsoR/L) (Fig. 3, A to C, and figs. S2 and S3A). We confirmed the identity of these seven cells using cell type–specific markers (fig. S3, B to D). Six of these seven cells are proliferative in the male and are the only male blast cells that are nonproliferative in the hermaphrodite (11, 30). We observed that nonproliferative cells generally, but not always, harbored a weaker GFP::SAS-7 focus than proliferative cells (Fig. 3B and fig. S4, A and B). Such differences may stem from nonproliferating cells having two centrioles compared to four in proliferating cells, as well as from different GFP::SAS-7 levels depending on cell cycle stage or cell type. Regardless, lower intensities of GFP::SAS-7 in the L1 stage do not seem to reflect ongoing elimination, since such foci generally remain present in later larval stages, 20 hours or more after the L1 larval stage (fig. S4, C to E). By way of comparison, the weak GFP::SAS-7 focus present in ADEshL/R in the embryo despite the absence of detectable centriolar microtubules vanishes entirely within approximately 5 hours. Together, these observations reveal the existence of a centriole elimination program that results in stereotyped organelle removal in ~88% of cells during C. elegans embryogenesis (Fig. 3D).
Fig. 3. Centriole elimination is stereotyped.
(A) Three-dimensional rendition of lattice light-sheet imaging of paralyzed L1 larva expressing GFP::SAS-7 (top) and the pan-nuclear marker mCherry::HIS-58 (magenta), with segmentation of centriole-maintaining proliferating (cyan), intestinal (brown), and nonproliferating (orange) cells overlaid (bottom). Boxes indicate cells magnified in (B). (B) Maximum z-projections of exemplary cells with centrioles (seam, V1L; intestine, int2V; epithelial, B), or without centriole (neuron, M4; pharyngeal, PM3L; hypodermal, hyp10). (C) Scoring of individual worms expressing either GFP::SAS-7 (green) or GFP::SAS-4 (blue) for the presence or absence of foci in the indicated nonproliferating cells (n = 11). The turquoise highlight marks cells that are proliferative in the male. (D) Entire embryonic lineage tree (left) and 10 magnified parts thereof (right) showing proliferating (cyan), intestinal (brown), and nonproliferating (orange) cells maintaining centrioles (n = 11; see also fig. S2). Note that, apart from the intestinal E lineage, cells maintaining centrioles are not highly related to each other.
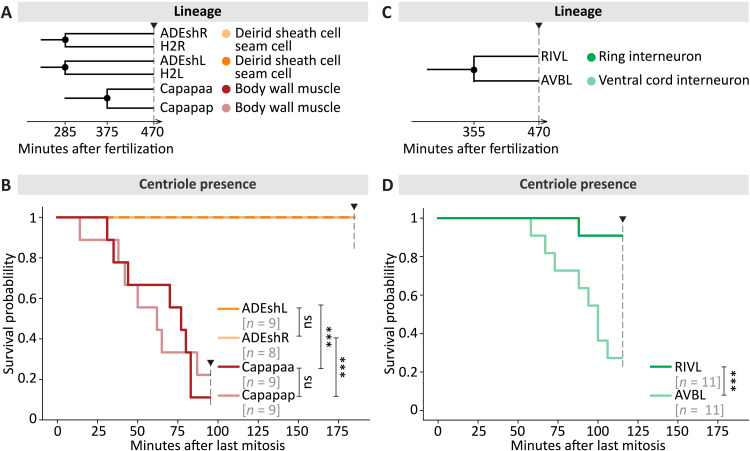
We set out to test whether centrioles are eliminated invariably at a given time following cell cycle exit or, alternatively, with cell type–specific kinetics. To distinguish between these possibilities, we estimated the time point of centriole elimination after the last mitotic division by monitoring GFP::SAS-7 foci using the modified lattice light-sheet microscope and a custom analysis pipeline (fig. S5). We analyzed 80 specific cells in several embryos in this manner, including 65 cells undergoing terminal differentiation. We found that, in general, mirror cells—equivalent cells on the left and right sides of the animal—and sister cells exhibited similar elimination kinetics after the last mitosis (Fig. 4, A and B, and fig. S6, A and B). Thus, the timing of GFP::SAS-7 focus loss was analogous between ADEshL and ADEshR, as well as between Capapaa and Capapap (Fig. 4B). Strikingly, in addition, we found that the timing of GFP::SAS-7 foci disappearance differed between cell types. For instance, GFP::SAS-7 foci were still detected 185 min after the last mitosis in ADEshL (9/9 cells), whereas they were already lacking by 95 min in Capapaa (eight of nine cells) (Fig. 4, A and B), with similar outcomes for the mirror cell ADEshR and the sister cell Capapaa, respectively. Moreover, we noted that elimination timing differed between sister cells in some cases (Fig. 4, C and D, and fig. S6C). This is exemplified by RIVL, in which GFP::SAS-7 was usually still detected 115 min after mitosis (10 of 11 cells), and its sister AVBL, where this was rarely the case (3 of 11 cells; Fig. 4, C and D). RIVL and AVBL give rise to neurons with distinct physiology, whereas most other sister cells analyzed give rise to body wall muscle cells with identical fate and a likewise timing of GFP::SAS-7 focus disappearance (fig. S6). Together, these findings establish that centriole elimination kinetics are cell type–specific.
Fig. 4. Timing of centriole elimination following mitosis is cell type–dependent.
(A and C) Lineage relationship and fate of cells analyzed for centriole maintenance in (B) and (D). Black disks on the lineage tree indicate the time point of mitosis. (B and D) Kaplan-Meier curves reporting the presence of GFP::SAS-7 foci adjacent to nuclei in indicated cells. Sister cells are shown in related colors. The number of nuclei analyzed is shown in gray. Pairwise log-rank test was performed. ns, not significant (P > 0.1); ***P < 0.005.
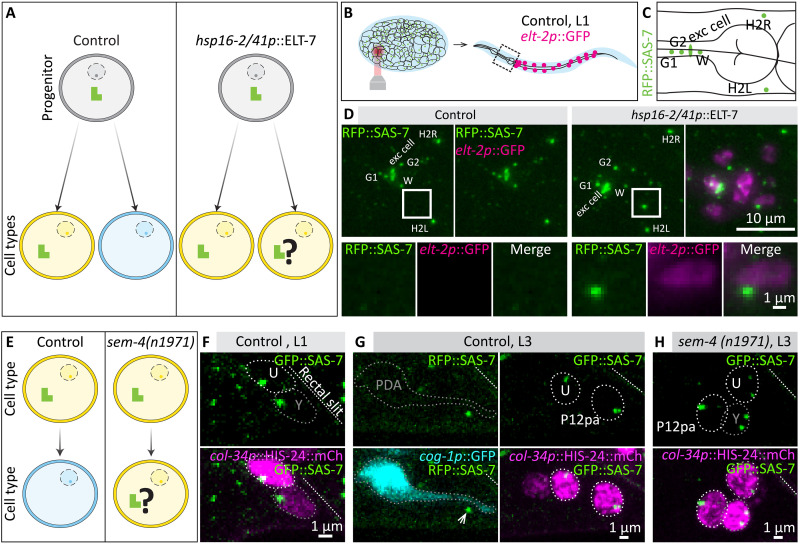
We investigated further the relationship between cell fate and centriole fate. In principle, centriole elimination could be imparted either by cell fate or by a centriole-intrinsic program. To distinguish between these possibilities, we sought to change the fate of progenitor cells that normally yield daughter cells without centrioles into that of cells that normally maintain centrioles (Fig. 5A). To this end, we utilized ELT-7, a transcription factor that can transdetermine progenitor cell fate, with ELT-7 expression being sufficient to turn pharyngeal progenitors, which normally yield cells lacking centrioles, into intestinal progenitors, which normally yield cells maintaining them (31). We used a strain in which ELT-7 expression can be induced with heat shock, monitoring excess intestinal cells with elt-2p::GFP (31); moreover, these animals expressed red fluorescent protein (RFP)::SAS-7 to monitor centrioles. Global ELT-7 overexpression before morphogenesis resulted in embryonic arrest, precluding rigorous analysis of a potential impact on centriole elimination (fig. S7, A to D). To circumvent this issue, we overexpressed ELT-7 in merely a few cells by directing an infrared laser onto a small region in the embryo anterior, thereby generating a local heat shock and transdetermining only a few cells (Fig. 5B). We scored the pharyngeal region of the resulting L1 larvae for the presence of elt-2p::GFP-positive cells (Fig. 5, B and C). As shown in Fig. 5D and fig. S7 (E to G), we found that ~63% of such cells maintained RFP::SAS-7 foci, with the remainder presumably reflecting cells that adopted the intestinal fate following centriole elimination onset (see legend of fig. S7G). Together, these findings establish that centriole fate can be altered following a change in progenitor cell fate.
Fig. 5. Cell fate imparts centriole fate.
(A) Schematic of transdetermination experiment. In control conditions, a progenitor (gray) may give rise to a cell type that maintains centrioles (yellow) and one that eliminates them (blue). Following transdetermination of a pharyngeal progenitor into an intestinal progenitor using hsp16-2/41::ELT-7, both daughter cells adopt an intestinal fate. (B and C) Schematics of local heat shock and region analyzed in the L1 (B), as well as of cells that normally maintain centrioles in part of the pharyngeal region (C), corresponding to (D). (D) Maximum z-projection of live wide-field microscopy of control L1 larva expressing red fluorescent protein (RFP)::SAS-7 and elt-2p::GFP (left), and of L1 larva expressing hsp16-2/41p::ELT-7 in addition (right). Both larvae have been locally heat-shocked during the late proliferation phase. Boxes point to regions of interest magnified below. (E) Schematic of transdifferentiation inhibition experiment. In control conditions, the Y cell, which harbors centrioles in the L1, transdifferentiates into PDA, which is usually devoid of centrioles in the L3. Transdifferentiation fails in sem-4(n1971) mutants. (F and G) Maximum confocal microscopy z-projections of live paralyzed L1 and L3 control worms expressing GFP::SAS-7 (F) or RFP::SAS-7 (G). In L1, the Y cell marked by the nuclear marker col-34p::HIS-24:mCh is positioned close to the rectal slit (slanted dashed line), and always maintains a GFP::SAS-7 focus (n = 17). Y then transdifferentiates into a PDA neuron marked by cog-1p::GFP by the L3 stage (G); RFP::SAS-7 foci were usually absent in PDA neurons (n = 17/21), despite clear foci being detected in the adjacent rectal epithelial cells U and P12pa (arrow). Weak (one cell) or very weak foci (three cells) were found in the other cases. (H) Maximum confocal microscopy z-projections of live paralyzed L3 sem-4(n1971) mutant worms expressing GFP::SAS-7 and col-34p::HIS-24::mCherry; all 12 such animals analyzed harbored a strong focus of GFP::SAS-7 in the Y cell.
We investigated also whether centriole fate can be altered when the fate of a differentiated cell is changed, using as a model the differentiated rectal epithelial cell Y, which harbors centrioles in the L1 larva (Fig. 5, E and F). In the wild-type L1 to L2 transition, the Y cell migrates away from the rectum and transdifferentiates into a PDA neuron (11, 32, 33) (Fig. 5, F and G). In the subsequent L3 larval stage, PDA neurons typically did not harbor a focus and rarely a very weak focus of RFP::SAS-7 (Fig. 5G). Transdifferentiation of the Y cell into a PDA neuron fails in sem-4(n1971) mutant animals, in which the Y cell remains at its original location (33). We found that, in all cases where this configuration was observed in sem-4(n1971) mutant L3 larvae, GFP::SAS-7 foci were present (Fig. 5H). We conclude that centriole fate is instructed by cell fate also in differentiated cells.
DISCUSSION
We report that centrioles are eliminated in most cells upon terminal differentiation during C. elegans embryogenesis, such that merely ~12% of cells harbor centrioles by the time of hatching. We found that centriole fate is stereotyped, with centriole elimination kinetics being characteristic of a given cell type. Moreover, we established that seven terminally differentiated cells retain foci harboring both GFP::SAS-7 and GFP::SAS-4. In addition, our analysis uncovered that centriole fate is governed by cell fate.
Serial-section EM analysis of a few cells established that the disappearance of foci bearing GFP::SAS-7 reflects bona fide centriole loss. In our larger-scale analysis, we used the presence of both GFP::SAS-7 foci and GFP::SAS-4 foci as a proxy for centrioles in embryos and L1 larvae. We note, however, that GFP::SAS-4 foci are sometimes lost before GFP::SAS-7 foci at later stages of development (see fig. S4, C to E), illustrating that elimination kinetics can differ between markers in some cases. Therefore, systematic serial-section EM would be required to determine with absolute certainty whether all foci bearing GFP::SAS-7 and GFP::SAS-4 correspond to bona fide centrioles.
Centriole elimination is not restricted to C. elegans embryos (30, 31). In Drosophila, for instance, centrioles are lost in the developing eye (4) and in a number of different tissue types upon polyploidization (7–10). Centriole elimination also occurs during oogenesis across metazoan organisms (34–38). Mechanistic insight regarding oogenesis centriole elimination was uncovered in Drosophila (39). In the wild-type, the departure of the Polo kinase from the PCM results in centriole elimination, whereas artificial centrosomal tethering of Polo prevents organelle removal (39). In the starfish, however, inhibiting Plk1 kinase activity does not lead to precocious centriole elimination (40), such that the extent to which a PLK-based mechanism operates across species and cell types to modulate centriole persistence remains to be determined. The stereotyped centriole fate map unveiled here provides a powerful means to identify further mechanisms, including through forward genetic and functional genomic screens. This would also enable investigation of the importance of extensive centriole elimination for worm physiology.
Whether widespread centriole elimination is likewise present in other organisms remains to be determined. Perhaps centrioles in the worm are essential after terminal differentiation only in ciliated sensory neurons, although centrioles are removed from the ciliary base once formed (21, 41, 42). Alternatively, centrioles might be prone to elimination in C. elegans because of their unusual architecture, since they are ~4 times shorter than their vertebrate counterpart and contain microtubule singlets instead of the usual triplets/doublets (43–46). Human cells lacking δ- or ε-tubulin, which are absent in C. elegans, generate centrioles with microtubule singlets that are unstable (47). By extrapolation, it might be that C. elegans centrioles are inherently more labile because they bear singlets.
We found that all cells that will later proliferate maintain centrioles during embryogenesis, as expected from the requirement of centrioles for bipolar spindle assembly in C. elegans. A monopolar spindle assembles in early embryos devoid of centrioles, for example, after fertilization by such-1(t1668) mutant sperm (48). This is in contrast to Drosophila or vertebrate cells, where bipolar spindle assembly occurs despite the absence of centrioles, through a centrosome-independent spindle assembly route (49–51). Moreover, de novo centriole formation does not appear to operate either in C. elegans, since embryos fertilized by such-1(t1668) mutant sperm devoid of centrioles go through the first two cell cycles without making new centrioles (48).
In summary, we established the first spatially resolved map of centriole fate in a developing organism, thereby discovering extensive centriole elimination during C. elegans embryogenesis. We established that centriole elimination is stereotyped and directed by cell fate, leading us to refer to the process as programmed centriole elimination. Just like programmed cell death was later found to be widespread and of critical importance in other systems (52), we anticipate that programmed centriole elimination will be uncovered in other physiological contexts where centriole fate must be carefully regulated. Furthermore, it appears reasonable to speculate that such a program may go awry in disease settings, and thereby perturb the fundamental functions normally exerted by centrioles.
MATERIALS AND METHODS
Strains
C. elegans strains were cultured according to standard procedures (53) and are listed in table S1.
Sample preparation for live imaging
For lattice light-sheet microscopy, embryos were dissected and deposited on a poly-l-lysine–coated 25-mm-diameter coverslip using a mouth pipette. Embryos were kept in M9 during imaging in a water bath of the sample chamber.
For other imaging modalities, embryos were dissected in M9, transferred onto a 2% agarose pad, covered with a coverslip, and sealed with VaLaP (1:1:1 mixture of petroleum:jelly:lanolin:paraffin wax).
For L1 larva analysis, gravid adults were bleached in a bleaching solution (71% 1 M NaOH and 29% NaOCl), washed four times with M9, and the embryos were allowed to hatch overnight in M9. L1 larvae were anesthetized using 100 mM NaN3 in M9 on an agar pad and imaged within 1 hour.
Microscopy and analysis
To achieve minimal phototoxicity and excellent temporal resolution, we used a modified lattice light-sheet microscope (25, 54) at the Advanced Imaging Center at Janelia Research Campus (USA). The microscope is equipped with a Thorlabs numerical aperture (NA) 0.6, 5.5-nm working distance water-dipping lens (TL20X-MPL) for light-sheet excitation, and a Zeiss 1.0 NA water-dipping objective with 2.2-mm working distance (421452–9800) for light-sheet detection, as well as two Hamamatsu Orca Flash 4.0 scientific complementary metal-oxide semiconductor (sCMOS) cameras for simultaneous multiple color imaging. The total system magnification is ×63. A square lattice pattern (inner NA: 0.34; outer NA: 0.4; envelope: 3; crop: 10) was used for generating the lattice light-sheet. Volumes were acquired in the “Z galvo & DO XZ stage” mode, wherein the sample stage is moved directly in line with the optical axis of the detection objective. Excitation was performed with 488- and 560-nm laser lines with an exposure time of 10 ms. The voxel size was 0.108 × 0.108 × 0.25 μm. The emission light was split to the two cameras using a 561-nm long-pass filter (Semrock, Di03-R561-t3-32x40). Furthermore, emission was filtered by a 617-nm bandpass filter (Semrock, FF02-617/73-25) or a 520-nm bandpass filter (Semrock, FF01-520/35-25). A system correction for the excitation and detection paths was performed as previously described (54). Images from the lattice light-sheet were deconvolved before further processing with 10 iterations of Richardson-Lucy deconvolution using experimentally measured point spread functions for each excitation wavelength (https://github.com/aicjanelia/LLSM). Imaging was performed at 22° to 24°C.
Wide-field microscopy was performed on a Zeiss Axio Observer D.1 inverted microscope, equipped with a 63× Plan-Apochromat (NA 1.4) objective connected to an Andor Zyla 4.2 sCMOS camera and a light-emitting diode (LED) light source (Lumencor SOLA II), controlled by the open-source μManager software (55). Pixels were binned 2 × 2. Confocal images were acquired on an upright Leica SP8 with two hybrid photon counting detectors (HyD) and a transmission photomultiplier tube for bright field, and equipped with a 63× HC Plan-Apochromat (NA 1.4), using a 405-, a 488-, and a 552-nm solid-state laser light for excitation and a DFC 7000 GT (B/W) camera. For CLEM, fluorescence images were acquired on an inverted confocal Leica SP8 microscope with HyD detectors and a 60× HC Plan-Apochromat (NA 1.3) glycerol immersion objective. All confocal microscopes were set to 8-kHz resonance scanning mode, 8× line averaging, and 2× frame accumulation with a pixel size of 81 nm, and the pinhole was set to 1 Airy unit. Notch filters were used.
Single time point images of L1 larvae were acquired in 0.5-μm steps. For the analysis of adults, focal planes were acquired in 0.7-μm steps. Images were rotated, z-projected, and brightness/contrast adjusted for each channel using Fiji (ImageJ). Unless otherwise indicated, brightness/contrast was kept the same for a specific channel within an experiment.
Fixation
For obtaining large quantities of embryos, gravid worms were bleached for 3 to 4 min in a bleaching solution and subsequently washed four times in M9. Paraformaldehyde fixation was performed as described (56). Briefly, worms were incubated in 3.8% paraformaldehyde in cytoskeleton buffer [10 mM MES (pH 6.1), 138 mM KCl, 2 mM MgCl2, and 2 mM EGTA] and 8% sucrose at room temperature for 10 min. Embryos were washed for 12 min in phosphate-buffered saline (PBS) and then resuspended in mounting solution (0.189 M n-propyl gallate, 90% glycerol, and 10% PBS).
Methanol fixation was performed similarly as described (57), with slight modifications. Briefly, worms were dissected on poly-l-lysine–coated slides, frozen on metal blocks deposited on dry ice, and subsequently freeze-cracked. Slides were then fixed in ice-cold methanol at −20°C for 2 min. After two washes in PBS, slides were blocked in 2% BSA in PBS for 20 min. After one wash with PBS + 0.05% Tween 20 (PBS-T) and one with PBS, slides were incubated with primary antibodies overnight at 4°C. After PBS-T and PBS washes, slides were incubated with secondary antibodies at room temperature for 45 min. Thereafter, slides were incubated in PBS-T and Hoechst 33258 (1 μg/ml) (1:1000) for 5 min, washed again with PBS-T and PBS, and lastly mounted in mounting solution. The following rabbit primary antibodies were used: 1:1000 SAS-6 (18); 1:800 SAS-4 (17). Secondary antibodies were goat anti-rabbit Alexa Fluor 568 (1:1000) (Thermo Fisher Scientific, A11011).
Counting overall number of centrioles and nuclei in the embryo
For determining the overall numbers of GFP::SAS-7 foci, the lattice light-sheet data were converted to Imaris files and the centriole channel was median-filtered (kernel size = 3 × 3 × 1 pixel). Surfaces were segmented using LabKit (58) by classifying the image into background and centriole signals. In Imaris, close centriolar foci were separated using the proprietary machine learning algorithm. Movies were synchronized on the basis of observing twitching, which was set to 470 min after fertilization.
For fixed embryos, complete z-stacks were acquired at the confocal microscope and sorted manually according to morphology. Images were filtered by a Gaussian blur (kernel size = 0.75 pixel) and background-subtracted. Subsequently, images were loaded into the machine learning software Ilastik (59). In Ilastik, pixel prediction maps were generated by training on the basis of background and centriolar signal, pooling the training for SAS-6 and SAS-4. The pixel prediction maps were then used for final segmentation. Thereafter, on the basis of the shape of the focal signal, centrioles were classified into singlets and doublets. Subsequently, the automatically determined counts were verified in a small sample by manual counting, with good correlations between the two (see fig. S1, B to D).
Nuclei numbers were determined in Imaris by segmenting surfaces based on thresholding by absolute intensity, which was set based on visual inspection of the segmentation result and a very characteristic inflection point in the histogram generally splitting the signal from noise. Split touching objects (region growing) was enabled with a seed diameter of 1.08 μm. Nuclei were filtered on the basis of an empirical quality threshold that was determined on the basis of visual inspection. For assessing the quality of the image processing pipeline, nuclei number was determined by manual counting at four different stages in four different embryos. These numbers were compared to the numbers detected in Imaris. Whereas differences were small until ~270 min after fertilization (1 ± 20), they increased markedly after 300 min (−159 ± 71). Visual assessment of the raw and segmented images showed that nuclei at this stage cannot be always clearly distinguished because nuclei in neighboring cells are very close to one another. All worms progressed normally through morphogenesis.
Tracking of centriole fate in the embryo
We developed the following workflow to track GFP::SAS-7 foci in individual cells. Live embryos expressing the centriolar marker GFP::SAS-7 throughout the embryo, and the nuclear marker HIS-24::mCherry in a subset of cells, were acquired at the modified lattice light-sheet microscope using 3- and 1-min time intervals. Thereafter, in Imaris, the nuclear channel was Gaussian filtered (kernel size = 0.216 μm) and the centriole channel was median- filtered (kernel size = 3 × 3 × 1 pixel). Using spot segmentation, nuclei were segmented and tracked using the Brownian motion algorithm. The diameter of cells was estimated on the basis of measurements at the 1.5-fold stage. Because of the lower z-resolution, 0.5 μm was added to the z-diameter, giving the nuclei an ellipsoid shape that fitted well with the overall form observed in Imaris. Tracks were manually corrected. Subsequently, GFP::SAS-7 foci were segmented using spot segmentation with an estimated diameter of 0.5 μm in xy and 1 μm in z. Spots were filtered on the basis of a quality threshold.
Last, using a custom MATLAB program, GFP::SAS-7 foci were associated to the nucleus at every time point if the edge-to-edge distance between nucleus and GFP::SAS-7 was within 0.1 μm. This threshold was empirically determined comparing the manual annotation with the outcome of the analysis. Nuclei were identified manually on the basis of data available on the Expression Patterns in Caenorhabditis (EPIC) database (http://epic2.gs.washington.edu/Epic2/) (60, 61), which were fed into WormGUIDES Atlas (62) to create a model of time-resolved localization of the cell expressing a marker.
Because of the noise in the tracks, a sliding average with a window size of 4 was applied. The timing of GFP::SAS-7 foci disappearance was set when the average number detected fell below 0.5 and did not recover within the next five frames. Plots and pairwise log-rank tests were generated in Python. The results obtained with the 3-min time intervals were verified for nine cells with 1-min time interval recordings similarly acquired on the modified lattice light-sheet microscope and processed in Imaris. This led to essentially the same result and was therefore included in the dataset (see fig. S6 for numbers).
Identification of cells in the L1 larva
Confocal microscopy data were used to score for presence or absence of centriole foci. For cell identification, proliferating and intestinal cells were identified first on the basis of existing maps and nuclear morphology (11, 26). Afterwards, nonproliferating cells maintaining GFP::SAS-7 or GFP::SAS-4 foci were identified. Cells most reliably maintaining foci were verified using cell-specific markers (grl-2p::GFP for AMsoL/R, col-34p::HIS-24::mCherry for rectal epithelial cells).
Correlative light electron microscopy
For CLEM experiments, gravid worms were bleached with bleaching solution. The resulting eggs were washed three times with egg buffer [5 mM Hepes (pH 6.9), 110 mM NaCl, 4 mM KCl, 5 mM MgCl2, and 5 mM CaCl2] (46). After staging eggs to the 1.5-fold stage, worms were transferred with a mouth pipette into 100 μl of chitinase (2 mg/ml) and incubated for 30 min at 24°C. With the mouth pipette, eggs were transferred in egg buffer with Hoechst 33342 (1 μg/ml) on a poly-l-lysine–coated MatTek dish with a glass bottom and a grid to identify the embryos. Subsequently, embryos were imaged on a Leica SP8 inverted microscope with hybrid photon counting detectors (HyD). The embryos were then fixed overnight using 1% glutaraldehyde and 0.9% paraformaldehyde in cacodylate buffer [0.05 M (pH 7.4), 0.09 M sucrose, and 0.9 mM MgCl2] at 4°C. Afterwards, embryos were washed three times for 5 min in 0.05 M cacodylate with 0.09 M sucrose. Thereafter, embryos were cryoprotected in 0.05 M cacodylate buffer (pH 7.0) with 2% glycerol and 20% dimethyl sulfoxide. After two freeze-thaw cycles, embryos were washed three times for 5 min in cacodylate buffer (0.05 M) with 0.09 M sucrose. Embryos were postfixed in 1% osmium tetroxide with 0.8% potassium ferrocyanide in cacodylate buffer (0.1 M, pH 7.2) for 40 min before washing three times for 5 min in 0.05 M cacodylate buffer (pH 7.0). After 15-min treatment with 0.2% tannic acid in 0.05 M cacodylate buffer, embryos were washed for 5 min in 0.05 M cacodylate buffer (pH 7.0), three times in ddH2O and 5 min in sodium acetate (pH 5.2). Thereafter, embryos were stained in 1% uranyl acetate in sodium acetate (pH 5.2) for 1 hour and subsequently washed three times for 5 min in sodium acetate (pH 5.2), and then washed three times for 5 min in ddH2O. Thereafter, embryos were dehydrated in a graded alcohol series (1 × 50%, 1 × 70%, 2 × 96%, 2 × 100%) for 10 min after each change. Lastly, the specimens were embedded in hard EPON on coated glass slides and placed in a 65°C oven overnight. Slices were cut at 50-nm intervals and imaged using a TecnaiSpirit (FEI Company) operated at 80 kV and equipped with an Eagle CCD camera (FEI Company). Using the relative position of nuclei and other landmarks in the embryo, we first identified cells of interest, and then focused on searching the centriole at the position predicted based on the fluorescent signal, using initially at 1.24 nm/pixel, which was generally followed by taking images at 1 nm/pixel.
Ethanol fixation
Adult worms were ethanol (EtOH)-fixed as previously described (14); this led to less cytoplasmic signal and a clearer signal for centriolar localization throughout the z-stack in adult worms. Adult worms were washed off the plate into a reaction tube using PBS-T and subsequently washed once using PBS-T. After removing most of the PBS-T, 1 ml of 100% EtOH was added and the tube was incubated for 3 min at room temperature. After removing all EtOH, 50 μl of 1:1 dilution M9 + VECTASHIELD with Hoechst 33258 (1 μg/ml) was added. After rehydration for 1 min, worms were mounted on a slide with a coverslip.
Overexpression of ELT-7
The strains sas-7(is1[rfp::sas-7 + loxP])III, wIs125[hsp-16-2::elt-7 hsp-16-41::elt-7]; rrIs1 [elt-2::GFP + unc-119(+)], glo-1(zu931)X, and sas-7(is1[rfp::sas-7 + loxP])III, rrIs1 [elt-2::GFP + unc-119(+)]; glo-1(zu931)X for control were used in ELT-7 overexpression experiments. For global overexpression during embryogenesis, gravid adults were bleached and the obtained embryos were incubated for 3 to 6 hours before being heat-shocked at 34°C for 30 min. Embryos were allowed to develop overnight.
For localized heat shocks, embryos before the bean stage were exposed in the anterior region to an infrared laser of λ = 1550 nm at an intensity of 5.1 to 6.2 mW for 45 min using an inverted Zeiss Axiovert 200 microscope equipped with a Roper CoolSNAP HQ camera and a 63× C-Achromat water objective (NA = 1.2) with a correction ring. The embryos were then recovered and left to hatch overnight. In the control condition, 72% (n = 32) of worms hatched, whereas 37% (n = 52) of hsp16-2/41p::ELT-7–expressing worms hatched.
L1 larvae were anesthetized using 100 mM NaN3 in M9, and then imaged on a 2% agarose pad using a Zeiss ObserverD.1 inverted microscope. Control worms not carrying hsp16-2/41p::ELT-7 were heat-shocked in the same way.
Scoring of RFP::SAS-7 foci after blocking transdifferentiation by sem-4(n1971)
For transdifferentiation experiments, sem-4(n1971) worms were grown to the desired stage, anesthetized using NaN3, and subsequently imaged on an agarose pad using the SP8 confocal microscope. Originally, L3 (n = 17) and L4 (n = 24) larvae were scored for sem-4(n1971) phenotypes. Thirty percent of worms at each stage did not have the characteristic nuclear configuration shown in Fig. 4H; these were therefore excluded from the analysis since they did not exhibit the sem-4 phenotype, as previously reported to occur in this background (33).
Acknowledgments
Lattice light-sheet microscopy was performed at the Advanced Imaging Center (AIC) at Janelia Research Campus, a facility jointly supported by the Gordon and Betty Moore Foundation and Howard Hughes Medical Institute. We are grateful to the AIC, in particular to C. Hobson, S. Khuon, and R. Lee, for help in acquiring and analyzing data on the modified lattice light-sheet microscope. We are thankful to K. O’Connell, P. Askjaer, S. Jarriault, and J. Rothman for worm strains and insightful advice. We thank N. Bhatla for providing a digital version of the hermaphrodite lineage tree. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank G. Knott and M.-C. Croisier-Coeytaux (BioEM platform of the School of Life Sciences, EPFL) for CLEM work, as well as O. Burri (BIOP platform of the School of Life Sciences, EPFL) for the MATLAB script associating centrioles and nuclei and for suggestions about image analysis. O. Dudin, G. G. Rodriguez, and A. Woglar are acknowledged for critical reading of the manuscript.
Funding: This work was supported by Swiss National Science Foundation grant 310030_197749 to P.G.
Author contributions: Conceptualization: N.K. and P.G. Methodology: N.K. and P.G. Investigation: N.K. and P.G. Visualization: N.K. and P.G. Funding acquisition: P.G. Project administration: P.G. Supervision: P.G. Writing—original draft: N.K. and P.G. Writing—review and editing: N.K. and P.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The relevant data and code (if not provided in the main text or the Supplementary Materials) were deposited in Zenodo and can be accessed through https://zenodo.org/record/7774077 or DOI 10.5281/zenodo.7774077.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Legend for movie S1
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.P. Gönczy, G. N. Hatzopoulos, Centriole assembly at a glance. J. Cell Sci. 132, jcs228833 (2019). [DOI] [PubMed] [Google Scholar]
- 2.L. Pintard, B. Bowerman, Mitotic Cell Division in Caenorhabditis elegans. Genetics 211, 35–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Z. Carvalho-Santos, J. Azimzadeh, J. B. Pereira-Leal, M. Bettencourt-Dias, Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. G. Riparbelli, V. Persico, M. Gottardo, G. Callaini The developing Drosophila eye: An oncoming model to study centriole reduction. J. Cell Sci. 131 (2018). [DOI] [PubMed]
- 5.A. M. Tassin, B. Maro, M. Bornens, Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100, 35–46 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Bugnard, K. J. M. Zaal, E. Ralston, Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskeleton 60, 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- 7.A. P. Mahowald, J. M. Strassheim, Intercellular migration of centrioles in the germarium of Drosophila melanogaster. J. Cell Biol. 45, 306–320 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. P. Mahowald, J. H. Caulton, M. K. Edwards, A. D. Floyd, Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster. Exp. Cell Res. 118, 404–410 (1979). [DOI] [PubMed] [Google Scholar]
- 9.K. P. Schoenfelder, R. A. Montague, S. V. Paramore, A. L. Lennox, A. P. Mahowald, D. T. Fox, Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development 141, 3551–3560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Y. Zheng, R. A. Buchwalter, C. Zheng, E. M. Wight, J. V. Chen, T. L. Megraw, A perinuclear microtubule-organizing centre controls nuclear positioning and basement membrane secretion. Nat. Cell Biol. 22, 297–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. E. Sulston, H. R. Horvitz, Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977). [DOI] [PubMed] [Google Scholar]
- 12.J. E. Sulston, E. Schierenberg, J. G. White, J. N. Thomson, The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 13.M. Ohta, A. Desai, K. Oegema, How centrioles acquire the ability to reproduce. eLife 6, e25358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Woglar, M. Pierron, F. Z. Schneider, K. Jha, C. Busso, P. Gönczy, Molecular architecture of the C. elegans centriole. PLOS Biol. 20, e3001784 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.F. R. Balestra, L. Von Tobel, P. Gönczy, Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res. 25, 642–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Kirkham, T. Müller-Reichert, K. Oegema, S. Grill, A. A. Hyman, SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 (2003). [DOI] [PubMed] [Google Scholar]
- 17.S. Leidel, P. Gönczy, SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439 (2003). [DOI] [PubMed] [Google Scholar]
- 18.S. Leidel, M. Delattre, L. Cerutti, K. Baumer, P. Gönczy, SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125 (2005). [DOI] [PubMed] [Google Scholar]
- 19.A. Dammermann, T. Müller-Reichert, L. Pelletier, B. Habermann, A. Desai, K. Oegema, Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829 (2004). [DOI] [PubMed] [Google Scholar]
- 20.P. N. Inglis, G. Ou, M. R. Leroux, J. M. Scholey, The sensory cilia of Caenorhabditis elegans. WormBook 10.1895/wormbook.1.126.2, 1–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. Serwas, T. Y. Su, M. Roessler, S. Wang, A. Dammermann, Centrioles initiate cilia assembly but are dispensable for maturation and maintenance in C. elegans. J. Cell Biol. 216, 1659–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.W. Li, P. Yi, Z. Zhu, X. Zhang, W. Li, G. Ou, Centriole translocation and degeneration during ciliogenesis in Caenorhabditis elegans neurons. EMBO J. 36, 2553–2566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. Lu, R. Roy, Centrosome/cell cycle uncoupling and elimination in the endoreduplicating intestinal cells of C. elegans. PLOS ONE 9, e110958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. X. Huang, M.-K. Wong, V. W. S. Ho, L.-Y. Chan, D. Xie, R. Li, X. Ren, G. Guan, Y. Ma, B. Hu, H. Yan, Z. Zhao, Initial characterization of gap phase introduction in every cell cycle of C. elegans embryogenesis. Front. Cell Dev. Biol. 10 (2022). [DOI] [PMC free article] [PubMed]
- 25.B.-C. Chen, W. R. Legant, K. Wang, L. Shao, D. E. Milkie, M. W. Davidson, C. Janetopoulos, X. S. Wu, J. A. Hammer 3rd, Z. Liu, B. P. English, Y. Mimori-Kiyosue, D. P. Romero, A. T. Ritter, J. Lippincott-Schwartz, L. Fritz-Laylin, R. D. Mullins, D. M. Mitchell, J. N. Bembenek, A.-C. Reymann, R. Böhme, S. W. Grill, J. T. Wang, G. Seydoux, U. S. Tulu, D. P. Kiehart, E. Betzig, Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.F. Long, H. Peng, X. Liu, S. K. Kim, E. Myers, A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat. Methods 6, 667–672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.K. F. O’Connell, C. Caron, K. R. Kopish, D. D. Hurd, K. J. Kemphues, Y. Li, J. G. White, The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558 (2001). [DOI] [PubMed] [Google Scholar]
- 28.B. A. Edgar, N. Zielke, C. Gutierrez, Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15, 197–210 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Z. F. Altun, D. H. Hall, WormAtlas Hermaphrodite Handbook –Alimentary System– Intestine (WormAtlas, 2009).
- 30.M. Sammut, S. J. Cook, K. C. Q. Nguyen, T. Felton, D. H. Hall, S. W. Emmons, R. J. Poole, A. Barrios, Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526, 385–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. R. Riddle, A. Weintraub, K. C. Q. Nguyen, D. H. Hall, J. H. Rothman, Transdifferentiation and remodeling of post-embryonic C. elegans cells by a single transcription factor. Development 140, 4844–4849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. G. White, E. Southgate, J. N. Thomson, S. Brenner, The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 33.S. Jarriault, Y. Schwab, I. Greenwald, A Caenorhabditis elegans model for epithelial–neuronal transdifferentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 3790–3795 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. Manandhar, H. Schatten, P. Sutovsky, Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 72, 2–13 (2005). [DOI] [PubMed] [Google Scholar]
- 35.T. Boveri, Zellen-Studien: Ueber die Natur der Centrosomen (G. Fischer, 1900).
- 36.G. Schatten, The centrosome and its mode of inheritance: The reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 165, 299–335 (1994). [DOI] [PubMed] [Google Scholar]
- 37.M. Delattre, P. Gönczy, The arithmetic of centrosome biogenesis. J. Cell Sci. 117, 1619–1630 (2004). [DOI] [PubMed] [Google Scholar]
- 38.A. H. Sathananthan, K. Selvaraj, M. L. Girijashankar, V. Ganesh, P. Selvaraj, A. O. Trounson, From oogonia to mature oocytes: Inactivation of the maternal centrosome in humans. Microsc. Res. Tech. 69, 396–407 (2006). [DOI] [PubMed] [Google Scholar]
- 39.A. Pimenta-Marques, I. Bento, C. A. M. Lopes, P. Duarte, S. C. Jana, M. Bettencourt-Dias, A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 353, aaf4866 (2016). [DOI] [PubMed] [Google Scholar]
- 40.M. Pierron, N. Kalbfuss, J. Borrego-pinto, P. Lénárt, P. Gönczy, Centriole foci persist in starfish oocytes despite Polo-like kinase 1 inactivation or loss of microtubule nucleation activity. Mol. Biol. Cell 31, 873–880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Magescas, S. Eskinazi, M. V. Tran, J. L. Feldman, Centriole-less pericentriolar material serves as a microtubule organizing center at the base of C. elegans sensory cilia. Curr. Biol. 31, 2410–2417.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Garbrecht, T. Laos, E. Holzer, M. Dillinger, A. Dammermann, An acentriolar centrosome at the C. elegans ciliary base. Curr. Biol. 31, 2418–2428.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 43.M. Bornens, The centrosome in cells and organisms. Science 335, 422–426 (2012). [DOI] [PubMed] [Google Scholar]
- 44.E. T. O’Toole, T. H. Giddings, J. R. McIntosh, S. K. Dutcher, Three-dimensional organization of basal bodies from wild-type and δ-tubulin deletion strains of Chlamydomonas reinhardtii. Mol. Biol. Cell 14, 2999–3012 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.L. Pelletier, E. O’Toole, A. Schwager, A. A. Hyman, T. Müller-Reichert, Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 (2006). [DOI] [PubMed] [Google Scholar]
- 46.K. Sugioka, D. R. Hamill, J. B. Lowry, M. E. McNeely, M. Enrick, A. C. Richter, L. E. Kiebler, J. R. Priess, B. Bowerman, Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. eLife 6, e20353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. T. Wang, D. Kong, C. R. Hoerner, J. Loncarek, T. Stearns, Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. eLife 6, e29061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Bezler, P. Gönczy, Mutual antagonism between the anaphase promoting complex and the spindle assembly checkpoint contributes to mitotic timing in Caenorhabditis elegans. Genetics 186, 1271–1283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Y. L. Wong, J. V. Anzola, R. L. Davis, M. Yoon, A. Motamedi, A. Kroll, C. P. Seo, J. E. Hsia, S. K. Kim, J. W. Mitchell, B. J. Mitchell, A. Desai, T. C. Gahman, A. K. Shiau, K. Oegema, Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Khodjakov, C. L. Rieder, Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. Basto, J. Lau, T. Vinogradova, A. Gardiol, C. G. Woods, A. Khodjakov, J. W. Raff, Flies without Centrioles. Cell 125, 1375–1386 (2006). [DOI] [PubMed] [Google Scholar]
- 52.S. Elmore, Apoptosis: A review of programmed cell death. Toxicol. Pathol. 35, 495–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. Brenner, The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T.-L. Liu, S. Upadhyayula, D. E. Milkie, V. Singh, K. Wang, I. A. Swinburne, K. R. Mosaliganti, Z. M. Collins, T. W. Hiscock, J. Shea, A. Q. Kohrman, T. N. Medwig, D. Dambournet, R. Forster, B. Cunniff, Y. Ruan, H. Yashiro, S. Scholpp, E. M. Meyerowitz, D. Hockemeyer, D. G. Drubin, B. L. Martin, D. Q. Matus, M. Koyama, S. G. Megason, T. Kirchhausen, E. Betzig, Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. D. Edelstein, M. A. Tsuchida, N. Amodaj, H. Pinkard, R. D. Vale, N. Stuurman, Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R. V. Aroian, C. Field, G. Pruliere, C. Kenyon, B. M. Alberts, Isolation of actin-associated proteins from Caenorhabditis elegans oocytes and their localization in the early embryo. EMBO J. 16, 1541–1549 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Delattre, S. Leidel, K. Wani, K. Baumer, J. Bamat, H. Schnabel, R. Feichtinger, R. Schnabel, P. Gönczy, Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656–664 (2004). [DOI] [PubMed] [Google Scholar]
- 58.M. Arzt, J. Deschamps, C. Schmied, T. Pietzsch, D. Schmidt, P. Tomancak, R. Haase, F. Jug, LABKIT: Labeling and segmentation toolkit for big image data. Front. Comput. Sci. 4, 10.3389/fcomp.2022.777728, (2022). [Google Scholar]
- 59.S. Berg, D. Kutra, T. Kroeger, C. N. Straehle, B. X. Kausler, C. Haubold, M. Schiegg, J. Ales, T. Beier, M. Rudy, K. Eren, J. I. Cervantes, B. Xu, F. Beuttenmueller, A. Wolny, C. Zhang, U. Koethe, F. A. Hamprecht, A. Kreshuk, ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 60.D. L. Mace, P. Weisdepp, L. Gevirtzman, T. Boyle, R. H. Waterston, A high-fidelity cell lineage tracing method for obtaining systematic spatiotemporal gene expression patterns in Caenorhabditis elegans. G3: Genes Genomes Genet. 3, 851–863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.J. I. Murray, T. J. Boyle, E. Preston, D. Vafeados, B. Mericle, P. Weisdepp, Z. Zhao, Z. Bao, M. Boeck, R. H. Waterston, Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 22, 1282–1294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. Santella, R. Catena, I. Kovacevic, P. Shah, Z. Yu, J. Marquina-solis, A. Kumar, Y. Wu, J. Schaff, D. Colón-ramos, H. Shroff, W. A. Mohler, Z. Bao, WormGUIDES: An interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC Bioinformatics 16, 189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. J. Beanan, S. Strome, Characterization of a germ-line proliferation mutation in C. elegans. Development 116, 755–766 (1992). [DOI] [PubMed] [Google Scholar]
- 64.G. Gómez-Saldivar, A. Fernandez, Y. Hirano, M. Mauro, A. Lai, C. Ayuso, T. Haraguchi, Y. Hiraoka, F. Piano, P. Askjaer, Identification of conserved MEL-28/ELYS domains with essential roles in nuclear assembly and chromosome segregation. PLOS Genet. 12, e1006131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.K. Koh, J. H. Rothman, ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128, 2867–2880 (2001). [DOI] [PubMed] [Google Scholar]
- 66.K. Klinkert, N. Levernier, P. Gross, C. Gentili, L. von Tobel, M. Pierron, C. Busso, S. Herrman, S. W. Grill, K. Kruse, P. Gönczy, Aurora A depletion reveals centrosome-independent polarization mechanism in Caenorhabditis elegans. eLife 8, e44552 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.S. J. McKay, R. Johnsen, J. Khattra, J. Asano, D. L. Baillie, S. Chan, N. Dube, L. Fang, B. Goszczynski, E. Ha, E. Halfnight, R. Hollebakken, P. Huang, K. Hung, V. Jensen, S. J. M. Jones, H. Kai, D. Li, A. Mah, M. Marra, J. McGhee, R. Newbury, A. Pouzyrev, D. L. Riddle, E. Sonnhammer, H. Tian, D. Tu, J. R. Tyson, G. Vatcher, A. Warner, K. Wong, Z. Zhao, D. G. Moerman, Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68, 159–170 (2003). [DOI] [PubMed] [Google Scholar]
- 68.A. C. Erpf, L. Stenzel, N. Memar, M. Antoniolli, M. Osepashvili, R. Schnabel, B. Conradt, T. Mikeladze-Dvali, PCMD-1 organizes centrosome matrix assembly in C. elegans. Curr. Biol. 29, 1324–1336.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 69.E. M. Sommermann, K. R. Strohmaier, M. F. Maduro, J. H. Rothman, Endoderm development in Caenorhabditis elegans: The synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Dev. Biol. 347, 154–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.K. Kagias, A. Ahier, N. Fischer, S. Jarriault, Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, 6596–6601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R. E. Palmer, T. Inoue, D. R. Sherwood, L. I. Jiang, P. W. Sternberg, Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 252, 202–213 (2002). [DOI] [PubMed] [Google Scholar]
- 72.M. Basson, H. R. Horvitz, The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes Dev. 10, 1953–1965 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Legend for movie S1
References
Movie S1