Abstract
Autophagy and glycolysis are highly conserved biological processes involved in both physiological and pathological cellular programs, but the interplay between these processes is poorly understood. Here, we show that the glycolytic enzyme lactate dehydrogenase A (LDHA) is activated upon UNC-51–like kinase 1 (ULK1) activation under nutrient deprivation. Specifically, ULK1 directly interacts with LDHA, phosphorylates serine-196 when nutrients are scarce and promotes lactate production. Lactate connects autophagy and glycolysis through Vps34 lactylation (at lysine-356 and lysine-781), which is mediated by the acyltransferase KAT5/TIP60. Vps34 lactylation enhances the association of Vps34 with Beclin1, Atg14L, and UVRAG, and then increases Vps34 lipid kinase activity. Vps34 lactylation promotes autophagic flux and endolysosomal trafficking. Vps34 lactylation in skeletal muscle during intense exercise maintains muscle cell homeostasis and correlates with cancer progress by inducing cell autophagy. Together, our findings describe autophagy regulation mechanism and then integrate cell autophagy and glycolysis.
Vps34 lactylation plays an important role in the physiological and pathological processes by integrating autophagy and glycolysis.
INTRODUCTION
Macroautophagy (hereafter called autophagy) is a conserved lysosomal degradation process that contributes to cellular homeostasis and adaptation to stress by engulfing cytoplasmic components (including damaged organelles) in a double-membraned organelle (an autophagosome) and then delivers these components to lysosome for degradation (1, 2). Autophagy plays an important role in physiological and pathophysiological processes, including adaptation to metabolic stress (3, 4), removal of protein aggregates and damaged organelles, embryo preimplantation development (5), and human diseases (aging, neurodegenerative diseases, intracellular pathogen elimination, and tumorigenesis) (6).
Autophagy processes are regulated by core molecular machinery, including the UNC-51–like kinase 1 (ULK1) complex and the class III phosphatidylinositol 3-kinase (PI3K) complex, autophagy-related 12 (Atg12) conjugation system, Atg8 conjugation system, homotypic fusion and vacuole protein sorting tethering complex, and Stx17-SNAP29-Vamp8 complex (7). In addition, posttranslational modifications (PTMs), including phosphorylation, glycosylation, ubiquitination, lipidation, and acetylation, fine-tune the structure, activity, and function of the core autophagy machinery. The mechanistic target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK) regulates the early and late stages of autophagy by phosphorylating several core autophagy proteins, including ULK1 (8), WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) (9), vacuolar protein sorting 34 (Vps34) (10), UV radiation resistance-associated gene protein (UVRAG) (11), and Pacer (12). In addition to phosphorylation, autophagy core proteins are also regulated by ubiquitination (9), acetylation (12, 13), and glycosylation (14). Recently, several PTMs, including succinylation, crotonylation, SUMOylation, and lactylation, have been shown to play key roles in the regulation of protein function and gene transcription (15–18). However, whether these PTMs also regulate core autophagy machinery function remains unclear.
As a major source of cellular energy, glucose is metabolized via glycolysis into pyruvate, which can be catalyzed by lactate dehydrogenase (LDH) to generate lactate when oxygen is not available. Lactate as a metabolic intermediate not only involves central carbon metabolism but also plays important roles in regulating tumor immunity, antiviral responses, and endoplasmic reticulum (ER)–mitochondrial Mg2+ dynamics (19–23). Emerging evidence has shown that lactate dynamically drives the lysine lactylation of histones and nonhistone proteins to regulate gene expression and protein activity in macrophages, somatic cells, cancer cells, and brain cells in a glycolysis-dependent manner (18, 24–28). However, it is unclear whether lactate-mediated lactylation regulates the function of core autophagy machinery.
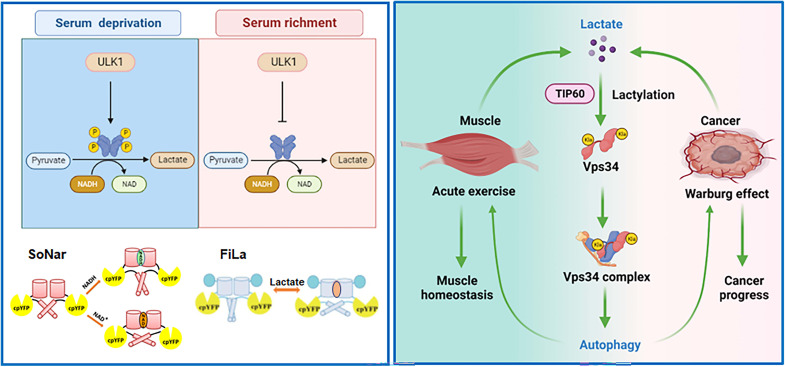
We hypothesized that lactate-mediated lactylation of the core autophagy machinery regulates autophagy activity. An anti–pan-lactylation antibody screening experiment showed that several core autophagy proteins were lactylated and that lactylation played a critical role in autophagy regulation. Vps34 is a catalytic subunit of the class III PI3K complex, which phosphorylates phosphatidylinositol (PI) to produce phosphatidylinositol 3-phosphate [PtdIns(3)P], promoting autophagy and endolysosomal trafficking. In this study, we found that ULK1 activated lactate dehydrogenase A (LDHA) by phosphorylation to promote lactate production, then lactate-mediated Vps34 lactylation. Lactylated Vps34 increased lipid kinase activity to enhance autophagy and endolysosomal degradation. In physiological and pathological processes, lactate-mediated Vps34 lactylation played a critical role in muscle homeostasis and tumor progression by promoting cell autophagy.
RESULTS
ULK1 induces LDHA Ser196 phosphorylation
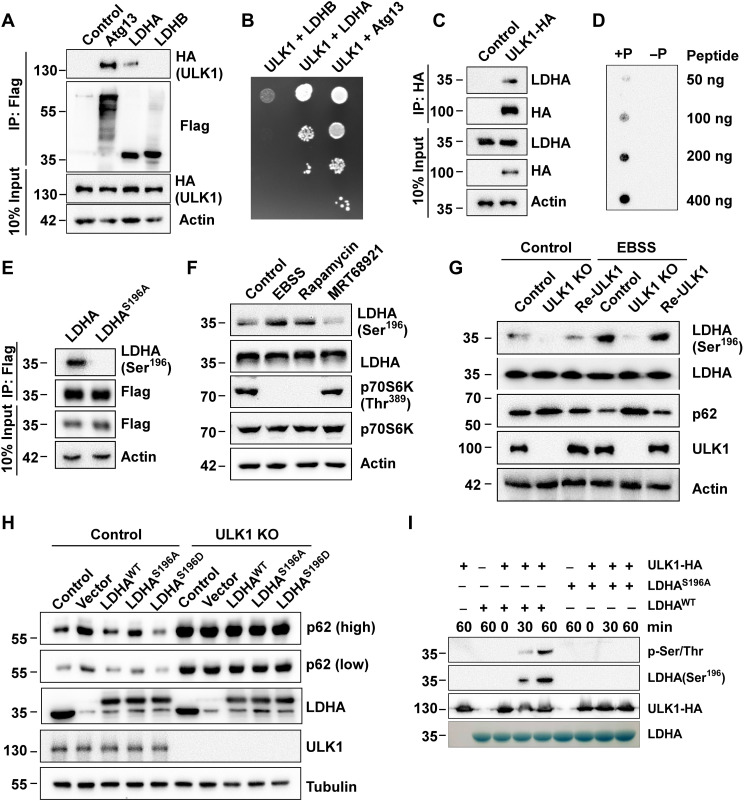
ULK1 is the most critical protein kinase involved in the early to late stages of autophagy. To comprehensively identify ULK1-interacting proteins, we performed tandem affinity purification. Mass spectrometry assays revealed the identities of several proteins, including RB1 inducible coiled-coil 1 (FIP200), Atg13, calcium binding and coiled-coil domain 2 (NDP52), hexokinase (HK), and phosphofructokinase 1 (PFK1), which had been previously reported to be associated with ULK1 (4, 29, 30), and another key glycolytic enzyme, LDHA, which had not been previously reported to bind ULK1. In this study, immunoprecipitation and yeast two-hybrid assays confirmed that ULK1 interacted with LDHA (Fig. 1, A to C, and fig. S1A). Because ULK1 mediates the phosphorylation of the glycolytic enzyme HK and PFK1, we hypothesized that ULK1 might be an LDHA kinase. To test this hypothesis, we performed an in vitro kinase assay and mass spectrometry analysis. We found that LDHA was phosphorylated at S196 by ULK1 (fig. S1B). LDHA S196 is a conserved residue across species ranging from zebrafish to humans (fig. S1C), and in the basis of the LDHA spatial structure, this residue was found in the loop domain close to the histidine site in the nicotinamide adenine dinucleotide (NADH) proton acceptor (fig. S1, D to F). Based on the above results, we speculated that phosphorylation of LDHA at serine-196 caused a conformational change in the loop structure, which was conducive for the interaction between the histidine residue and NADH.
Fig. 1. ULK1 induces LDHA Ser196 phosphorylation.
(A) ULK1 interacts with LDHA. Flag-tagged Atg13, LDHA, or LDHB were coexpressed with HA-ULK1 individually, and immunoprecipitation was performed using anti-Flag beads. Samples were analyzed by anti-HA Western blot. (B) Interaction assay of ULK1 and LDHA by yeast two-hybrid assay. pGBKT7-Atg13, pGBKT7-LDHA, or pGBKT7-LDHB and pGADT7-ULK1 were cotransformed into the yeast strain MJ109 and grown on the agar plate without leucine and tryptophan (YPD/−Leu/−Trp). Atg13 as positive control. (C) Interaction assay of ULK1 and endogenous LDHA. ULK1-HA was expressed in HEK293T cells. Immunoprecipitation was performed using anti-HA beads and analyzed by Western blot with antibody to LDHA. (D) Antibody specificity was analyzed by dot blot assay with a LDHA Ser196 phosphorylation antibody. +P peptide represent phosphorylation modification of LDHA Ser196. −P peptide represent nonphosphorylation modification of LDHA Ser196. (E) Antibody specificity test. LDHAWT-Flag or LDHAS196A-Flag was expressed in HEK293T cells. Immunoprecipitation was performed by anti-Flag beads, and LDHA Ser196 phosphorylation levels were analyzed by Western blot with phospho-LDHA Ser196 antibody. (F) LDHA Ser196 phosphorylation assay. HEK293T cells were treated with EBSS medium, mTOR inhibitor rapamycin, and ULK1 inhibitor MRT68921 for 4 hours. LDHA Ser196 phosphorylation was analyzed by phospho-LDHA Ser196 antibody. (G) LDHA Ser196 phosphorylation assay in ULK1-KO MEF cells. MEF cells and ULK1-KO MEF cells were transfected with vector or ULK1-HA and were cultured in complete medium or EBSS medium for 3 hours. LDHA Ser196 phosphorylation was analyzed by phospho-LDHA Ser196 antibody. (H) Autophagy assay in LDHA knockdown control cells and ULK1-KO cell (followed by expression of either LDHAWT, LDHAS196A, or LDHAS196D). (I) LDHA Ser196 phosphorylation assay in vitro. ULK1 was purified by immunoprecipitation from HEK293T cells, and its kinase activity in vitro was analyzed using purified LDHAWT-Flag and LDHAS196A-Flag from E. coli as substrate. LDHA Ser196 phosphorylation was analyzed by anti–phospho-LDHA Ser196 and anti–phospho-Ser/Thr antibody.
To verify the phosphorylation of LDHA S196, polyclonal antibodies that specifically recognize S196-phosphorylated LDHA were produced (Fig. 1D), and replacing the serine at position 196 with alanine (S196A) completely abolished LDHA phosphorylation (Fig. 1E). Moreover, ULK1 activation by Earle’s balanced salt solution (EBSS)–induced starvation or the mechanistic target of rapamycin (mTOR) inhibitor rapamycin enhanced LDHA S196 phosphorylation, and ULK1 inhibition by the ULK1 inhibitor MRT68921 reduced LDHA S196 phosphorylation (Fig. 1F). Consistently, LDHA S196 phosphorylation in ULK1-knockout (KO) mouse embryonic fibroblasts (MEFs) was reduced compared to that in control cells, and LDHA S196 phosphorylation was reestablished upon the ULK1 reexpression in both normal and EBSS medium (Fig. 1G). LDHA knockdown inhibited p62 degradation, and transfection of LDHAWT and LDHAS196D completely reversed the effects of LDHA knockdown, while LDHAS196A partially rescued these effects (Fig. 1H). In vitro kinase assay followed by Western blot further confirmed that Ser196 of LDHAWT was phosphorylated by ULK1, but mutant of LDHA (LDHAS96A) was not phosphorylated by ULK1 in vitro (Fig. 1I). Therefore, these results suggest that ULK1 as upstream regulator mediates LDHA S196 phosphorylation.
LDHA phosphorylation enhances its enzyme activity
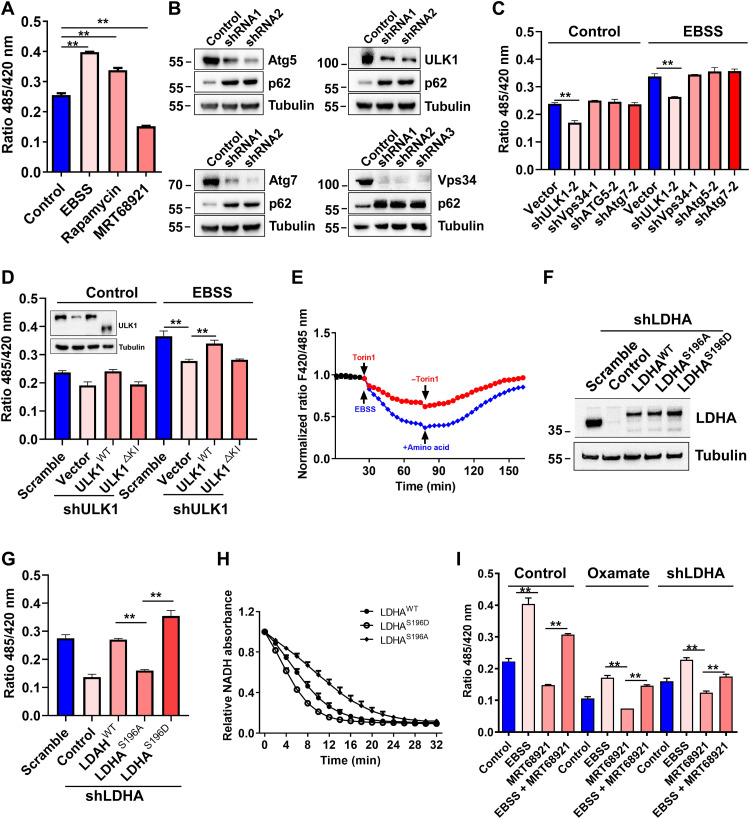
To test the biological effect of phosphorylation on LDHA enzyme activity, cytosolic lactate levels and NADH/NAD+ ratios were analyzed with the genetically encoded fluorescent sensors FiLa and SoNar, and FiLa-C and iNapc sensors are control senors of FiLa and SoNar (31, 32). ULK1 activation by EBSS medium or mTOR inhibitor rapamycin increased cytosolic lactate level, while ULK1 inhibitor MRT68921 decreased cytosolic lactate level (Fig. 2A). Consistently, knocking down ULK1 expression by short hairpin RNA (shRNA) also reduced cytosolic lactate levels in normal medium or EBSS medium, but silencing expression of other autophagic genes (namely, Vps34, Atg5, and Atg7) had no effect on cytosolic lactate level (Fig. 2, B and C). Moreover, recovery of ULK1WT expression in ULK1-knockdown H1299 cells reestablished cytosolic lactate level, while the expression of the ULK1ΔKI mutant (with a lost kinase domain) was not recovered (Fig. 2D). LDHA facilitates glycolytic flux by converting pyruvate to lactate and NADH to NAD+. We found that the cytosolic NADH/NAD+ ratio was reduced upon amino acid depletion (EBSS medium) or rapamycin treatment (fig. S2, A to C). Amino acid starvation and mTOR inhibitor Torin1 treatment reduced cytosolic NADH/NAD+ ratio, and then amino acid supplementation and excluding Torin1 restored the NADH/NAD+ ratio (Fig. 2E). Furthermore, LDHA silencing reduced cytosolic lactate level, and the recovery of LDHAWT and LDHAS196D expression restored cytosolic lactate level, while LDHAS196A expression failed to rescue LDHA enzyme activity (Fig. 2, F and G). These phenotypes were confirmed by measuring enzymatic activity in vitro and performing flow cytometry in vivo (Fig. 2H and fig. S2, D and E). In addition, inhibiting LDHA enzyme activity by oxamate or shRNA and ULK1 activity by MRT68921 reduced cytosolic lactate levels in cells cultured in EBSS medium (Fig. 2I). These results demonstrated that ULK1 mediated LDHA phosphorylation and enhanced its enzyme activity.
Fig. 2. LDHA phosphorylation enhances its enzyme activity.
(A) Cytosolic lactate level in EBSS, rapamycin (mTOR inhibitor), and ULK1 inhibitor MRT68921 by genetically encoded lactate sensor FiLa. FiLa-C was control senor of lactate FiLa. FiLa fluorescence ratio was corrected by FiLa-C. Data are shown as means ± SD; **P < 0.01, n = 3. (B) Silence efficiency of autophagy gene ULK1, Vps34, Atg5, or Atg7 knockdown H1299 cells by Western blot. (C) Cytosolic lactate level in autophagy gene ULK1, Vps34, Atg5, or Atg7 knockdown H1299 cells harboring genetically encoded lactate sensor FiLa that were treated with normal medium and EBSS medium for 3 hours. The shRNA of highest knockdown efficiency was chosen. Data are shown as means ± SD; **P < 0.01, n = 3. (D) Effect of ULK1 on cytosolic lactate level in H1299 cell line harboring genetically encoded lactate sensor FiLa treated with normal medium and EBSS medium for 3 hours. Data are shown as means ± SD; **P < 0.01, n = 3. (E) Dynamics of NADH/NAD+ ratio in H1299 cell line harboring genetically encoded lactate sensor SoNar in EBSS medium or EBSS medium supplementing amino acid and complete medium containing Torin1 or complete medium excluding Torin1. Data are shown as means ± SD; **P < 0.01, n = 3. (F) LDHA and its mutant expression in LDHA KD H1299 cells that was rescued by vector, LDHAWT, LDHAS196A, and LDHAS196D. (G) Cytosolic lactate level in LDHA KD H1299 cells that was rescued by vector, LDHAWT, LDHAS196A, and LDHAS196D. (H) LDHA enzyme assay in vitro. LDHAWT-Flag, LDHAS196A-Flag, and LDHAS196D-Flag were purified from HEK293T by anti-Flag beads. NADH absorbance were analyzed in microplate assay at 340 nm. Data are shown as means ± SD; **P < 0.01, n = 3. (I) Cytosolic lactate level in ULK1 activation and inhibition. Data are shown as means ± SD; **P < 0.01, n = 3.
Vps34 is lactylated at K356 and K781 by the acetyltransferase TIP60
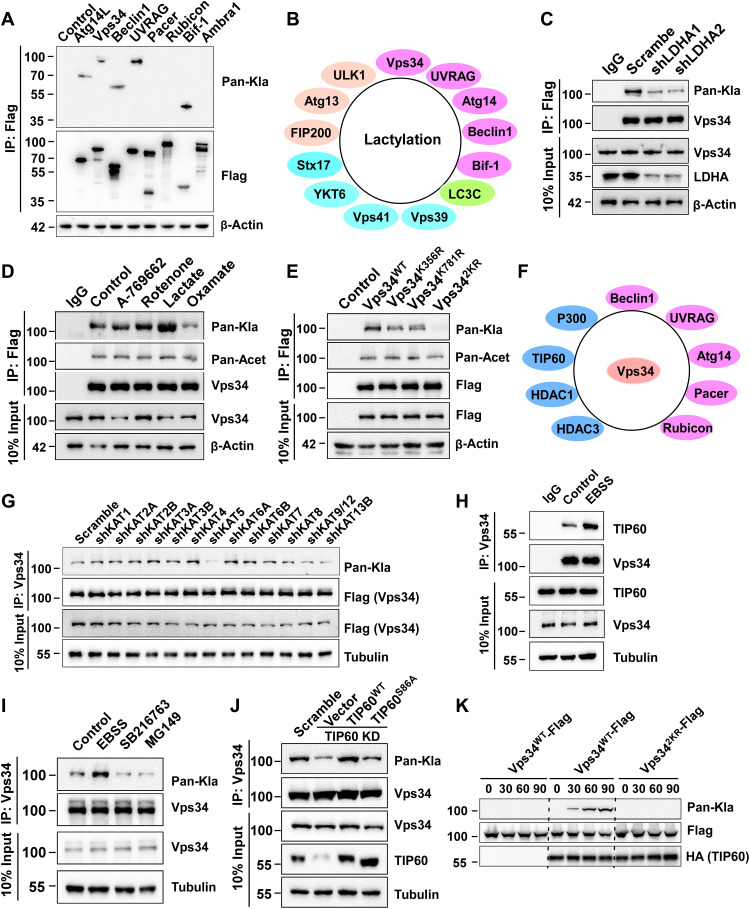
Lactylation, as a major posttranslational protein modification, regulates histone and HMGB1 biological activity in tumorigenesis and immune responses (18, 28). We also found that LDHA knockdown reduced lactate level and inhibited p62 degradation; we speculate whether lactate regulated autophagy through lactylation of autophagy core proteins. Therefore, we measured the lactylation of mammalian autophagy core proteins by coimmunoprecipitation and Western blot with an anti–pan-lactylation antibody. Intriguingly, many lactylated autophagy core proteins in different autophagy stages (Vps34, ULK1, UVRAG, etc.) were thus identified, indicating that lactylation played an important role in autophagy regulation (Fig. 3, A and B, and fig. S3, A to D). Class III PI3K Vps34 is a critical regulator of autophagy and endocytic sorting; therefore, Vps34 has been frequently selected as a research object (33, 34). We found that the Vps34 lactylation level reduced in LDHA-knockdown cells (Fig. 3C) and LDHA inhibitor oxamate-treated cells but increased in lactate, A-7669662 (AMPK activator), or rotenone-treated cells (Fig. 3D). To identify Vps34 lactylation sites, Vps34-Flag was purified after immunoprecipitation from human embryonic kidney (HEK) 293T cells treated with 10 mM lactate for 24 hours and analyzed by mass spectrometry, and two lysine residues (K356 and K781) were identified (fig. S3, E and F). K356 and K781 are conserved in higher mammals, indicating an evolutionarily conserved regulatory mechanism (fig. S3G). Lactylation of K356 and K781 shown in fig. S3H could cause the change of Vps34 conformation (fig. S3H). High lactate and EBSS starvation treatment promoted lactylation level of Vps34 K356 and K781 by mass spectrometry (fig. S3I). To verify these two lactylation sites, the two lysine residues were changed to arginine residues by site-directed mutagenesis, and these mutants were transfected into HEK293T cells. The lactylation levels of Vps34K356R and Vps34K781R were markedly reduced, and the lactylation level of the double-mutant Vps342KR was abolished (Fig. 3E).
Fig. 3. Vps34 at K356 and K781 is lactylated by acetyltransferase TIP60.
(A) Lactylation screening of PI3KC3 complex subunits. Flag-tagged Atg14L, Vps34, Beclin1, UVRAG, Pacer, Rubicon, Bif-1, and Ambra1 proteins was expressed individually in HEK293T cells and purified by immunoprecipitation; lactylation was analyzed by Western blot with Pan-Kla antibody. (B) Lactylation summary of autophagy associated proteins. (C) Endogenous Vps34 lactylation in LDHA knockdown H1299 cells. (D) Endogenous Vps34 lactylation in H1299 cells that were treated with A-769662 (100 nM), rotenone (10 nM), lactate (1 mM), and oxamate (10 mM) for 24 hours. Immunoprecipitation was performed and lactylation was analyzed by Western blot with Pan-Kla antibody. (E) Vps34 lactylation assay of Vps34 lactylation sites mutants in HEK293T cells. Vps34WT-Flag, Vps34K356R-Flag, Vps34K781R-Flag, and Vps342KR-Flag were transfected into HEK293T cells. Immunoprecipitation was performed and lactylation was analyzed by Western blot with Pan-Kla antibody. (F) Summary of interaction proteins with Vps34 by mass spectrometry. (G) Screening of Vps34 lactylation writer. Vps34WT-Flag was transfected into acyltransferase knockdown HEK293T cells (KAT1, KAT2A, KAT2B, KAT3A, KAT3B, KAT4, KAT5, KAT6A, KAT7, KAT8, KAT9, KAT12, and KAT13B). Vps34-Flag was purified by immunoprecipitation and lactylation was analyzed by Western blot with Pan-Kla antibody. (H) Vps34 interacts with TIP60. Endogenous Vps34 and TIP60 were immunoprecipitated from HEK293T cells, and interaction was analyzed by Western blot. (I) Endogenous Vps34 lactylation assay in HEK293T cells that were treated with EBSS medium, GSK3 inhibitor SB216763 (10 μM), and TIP60 inhibitor MG149 (20 μM). (J) Endogenous Vps34 lactylation assay in HEK293T cells (control shRNA, TIP60 KD/rescued by vector, TIP60WT, and TIP60S86A). (K) Vps34 lactylation assay in vitro. Vps34WT and Vps342KR were purified from E. coli, and TIP60-HA was purified from HEK293T cells. Vps34 lactylation was analyzed by Pan-Kla antibody.
Many studies have shown that acetyltransferases and deacetyltransferases not only mediate protein acetylation and deacetylation, respectively, but also catalyze other acylation modifications. To identify the Vps34 lactylation writer, an acetyltransferase shRNA library was constructed, and the Vps34 lactylation level was determined by coimmunoprecipitation and Western blotting with an anti–pan-lactylation antibody. Knocking down KAT5/TIP60 expression diminished Vps34 lactylation, but no other acetyltransferase exerted this effect (Fig. 3F and fig. S3, J and K). To identify the proteins that interact with Vps34, Vps34-Flag–overexpressing cells were treated with a dithiobis solution (succinimidyl propionate) (DSP) to induce cross-linking. The Vps34-Flag–interacting proteins were pulled down by coimmunoprecipitation and then were identified by mass spectrometry. In addition to the PI3KC3 subunits Beclin1, UVRAG, Atg14, Pacer, and Rubicon, Vps34 interacted with TIP60 and p300 (Fig. 3G). Endogenous and exogenous immunoprecipitation experiments confirmed that Vps34 interacted with TIP60 and EBSS starvation promoted interaction of Vps34 and TIP60 (Fig. 3H and fig. S3L). Knockdown of TIP60 impaired cell autophagy in cells deprived of serum (12, 35). Inhibiting of TIP60 activity by the TIP60 inhibitor MG149 or the TIP60 kinase GSK3 inhibitor SB216763 (35) reduced Vps34 lactylation, and enhancing TIP60 activity by amino acid or serum deprivation increased Vps34 lactylation level (Fig. 3I). Consistently, treatment with wild-type TIP60 reestablished the down-regulation of Vps34 lactylation in TIP60-knockdown HEK293T cells, but the loss-of-function mutant TIP60S86A failed to rescue Vps34 lactylation that had been reduced (Fig. 3J). In vitro lactylation assay showed that TIP60 lactylated Vps34 K356 and K781 by pan-Kla antibody and by mass spectrometry (Fig. 3K). These results showed that TIP60 mediated Vps34 lactylation.
Vps34 lactylation positively regulates its interaction with UVRAG and Beclin1 and lipid kinase activity
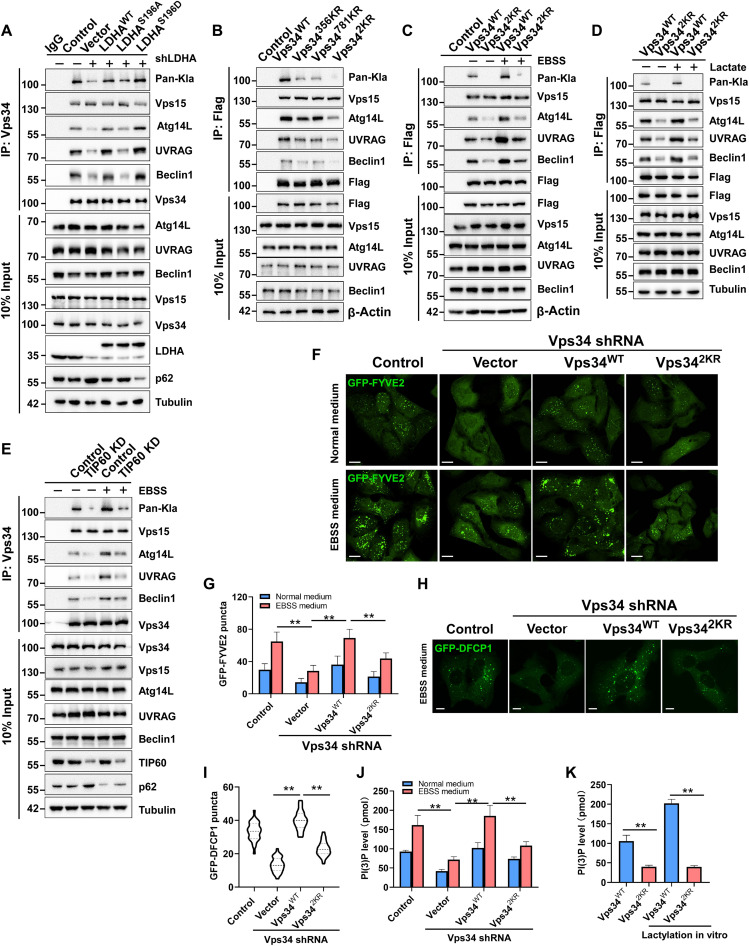
Vps34 forms two core complexes known as complex I and II. Complex I is composed of Vps34, Vps15 and ATG14L, whereas complex II includes UVRAG but not ATG14L. To investigate the biological function of Vps34 lactylation, the interaction of Vps34 with Vps15, Beclin1, ATG14L, and UVRAG was analyzed by immunoprecipitation and Western blot. Reducing Vps34 lactylation by shRNA-mediated LDHA knockdown impaired the association of Vps34 with Beclin1, ATG14L, and UVRAG, and the loss-of-function mutant LDHAS196A failed to rescue the association of Vps34 with these subunits in LDHA-knockdown cells, in contrast to the effect of wild-type LDHA treatment (Fig. 4A and fig.S4A). The Vps34K356R and Vps34K781R mutants destabilized the Vps34-Beclin1-ATG14L complex (I) and Vps34-Beclin1-UVRAG complex (II), respectively, whereas the Vps342KR mutant, with substitutions at both lactylation sites, reduced the association of Vps34 with Beclin1, ATG14L, and UVRAG in normal medium and EBSS medium (Fig. 4, B and C, and fig.S4, B and C). Moreover, high-lactate treatment promoted the association of Vps34WT with Beclin1, ATG14L, and UVRAG, rather than Vps342KR (Fig. 4D and fig. S4D). Consistently, enhancing Vps34 lactylation by lactate, A-7669662, and rotenone treatment facilitated the interaction of Vps34 with these subunits, while suppressing Vps34 lactylation by oxamate hampered the interaction of Vps34 with Beclin1 and UVRAG (fig. S4E). Furthermore, compared with that in the control cells, the association of Vps34 with Beclin1, ATG14L, and UVRAG was reduced when the Vps34 lactylation writer KAT5/TIP60 was knocked down in cells cultured in normal or EBSS medium (Fig. 4E and fig. S4F). The results indicated that Vps34 lactylation promoted the association of Vps34 with Beclin1, ATG14L, and UVRAG.
Fig. 4. Vps34 lactylation positively regulates its interaction with UVRAG and Beclin1 and lipid kinase activity.
(A) Interaction assay of Vps34 with Vps15, Beclin1, UVRAG, and Atg14L in LDHA knockdown HEK293T cells that rescued empty vector, LDHAS196A-Flag, and LDHAS196D-Flag. (B) Interaction assay of Vps34 lactylation sites mutants with Vps15, Beclin1, UVRAG, and Atg14L in HEK293T cells. Vps34WT-Flag, Vps34K356R-Flag, Vps34K781R-Flag, and Vps342KR-Flag were transfected into HEK293T cells. (C) Interaction assay of Vps34 lactylation sites mutants with Vps15, Beclin1, UVRAG, and Atg14L in HEK293T cells in normal medium or EBSS medium for 3 hours. Interaction assay as in (B). (D) Interaction assay of Vps34 with Vps15, Beclin1, UVRAG, and Atg14L in HEK293T cells that were cultured in normal medium or normal medium containing lactate (1 mM) for 24 hours. Interaction assay as in (A). (E) Interaction assay of Vps34 with Vps15, Beclin1, UVRAG and Atg14L in TIP60 knockdown HEK293T cells that were cultured normal medium or EBSS medium for 3 hours. Interaction assay as in (A). (F) Vps34 kinase activity assay by GFP-FYVE2. GFP-FYVE2 was expressed in U2OS cells (control shRNA, Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) and analyzed by confocal microscopy. Scale bar, 10 μm. (G) Quantification of the relative GFP-FYVE2 puncta. Data are shown as means ± SD; **P < 0.01, n = 30. (H) GFP-DFCP1 puncta assay in U2OS cells (control shRNA, Vps34 KD, and Vps34 KD/rescued by Vps34WT and Vps34r2KR) under EBSS medium for 2 hours. Scale bar, 10 μm. (I) Quantification of the relative GFP-DFCP1 puncta. Data are shown as means ± SD; **P < 0.01, n = 30. (J) PI(3)P level determined by PI(3)P ELISA kit in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR). **P < 0.01, n = 5. (K) Kinase activity of Vps34WT and Vps342KR in vitro by PI(3)P ELISA kit. Vps34WT and Vps342KR were purified from insect cells sf9, and TIP60-HA was purified from HEK293T cells. Vps34 lactylation was analyzed by Pan-Kla antibody. Data are shown as means ± SD; **P < 0.01, n = 3.
To determine whether Vps34 lactylation affects PI3KC3 kinase activity, a green fluorescent protein (GFP)–FYVE2 construct was used as a PtdIns(3)P [PI(3)P] sensor to monitor PtdIns(3)P biogenesis in cells. Lactate treatment increased cellular PtdIns(3)P levels, as indicated by an increase in the number of GFP-FYVE2–labeled puncta. In contrast, oxamate reduced the number of GFP-FYVE2–labeled puncta, similar to treatment with 3-methyladenine, a PI3KC3 kinase inhibitor (fig. S4, G and H). Furthermore, compared to those in the control cells cultured in normal medium or EBSS medium, the number of GFP-FYVE2–labeled puncta was reduced in Vps34-silenced cells. Moreover, transfection of Vps34WT in Vps34-silenced cells completely restored the effects of Vps34 knockdown, while transfection of Vps342KR only partially reversed these effects (Fig. 4, F and G). We further examined PtdIns(3)P levels by determining the localization of GFP-DFCP1 (a double FYVE domain–containing protein 1), which localizes to omegasomes in a PtdIns(3)P-dependent manner upon autophagy initiation (36, 37). Consistently, under starvation conditions, Vps34 silencing reduced the number of GFP-DFCP1 puncta compared to that in the control cells, recovery Vps34WT expression in Vps34-silenced cells completely stored the number of GFP-DFCP1 puncta, and Vps342KR expression only partially reversed these effects (Fig. 4, H and I). We determined PtdIns(3)P levels in vivo and in vitro by PtdIns(3)P mass enzyme-linked immunosorbent assay (ELISA) kit. The in vivo and in vitro ELISA results showed that Vps34 delactylation partially impaired Vps34 lipid kinase activity (Fig. 4, J and K and fig. S4I). Together, these results revealed that lactylation promoted Vps34 lipid kinase activity by enhancing the association of Vps34 with Beclin1, ATG14L, and UVRAG.
Vps34 lactylation facilitates autophagosome formation and maturation
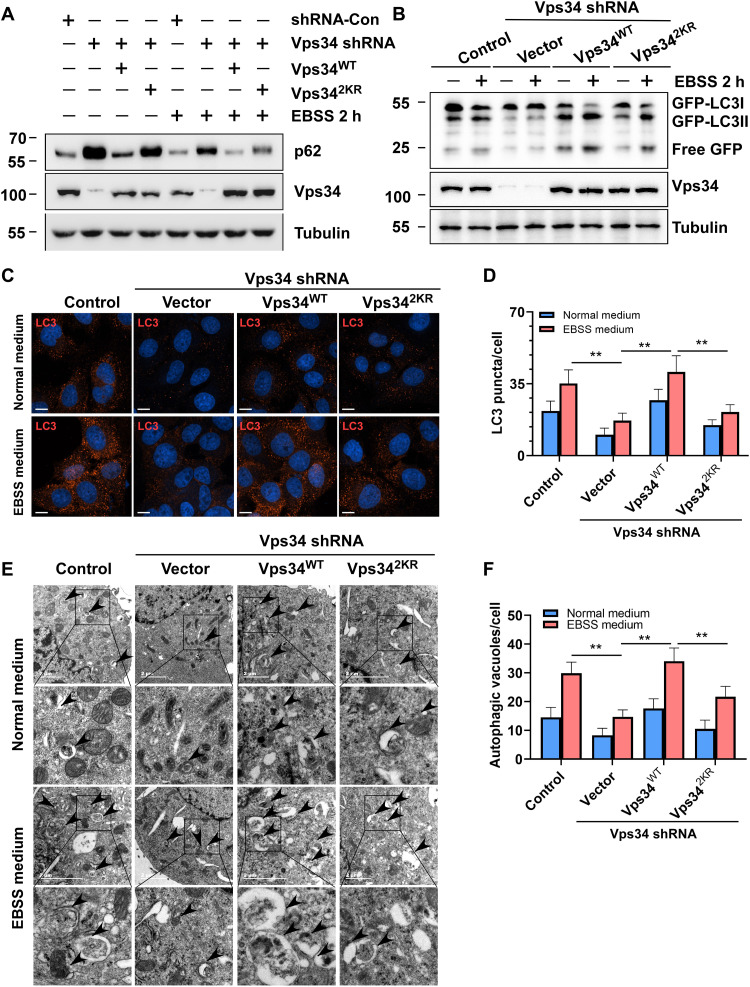
Vps34 plays an important role in autophagosome formation and maturation. To analyze the effect of Vps34 lactylation on autophagic flux, p62 degradation was analyzed by Western blot. Vps342KR partially inhibited p62 degradation, in contrast to the effect of Vps34WT at the cellular level in normal medium and under EBSS starvation conditions (Fig. 5A). The biological function of lactate and Vps34 lactylation on autophagic flux was validated with a GFP-LC3 cleavage assay. Notably, lactate treatment promoted GFP-LC3 cleavage, but oxamate treatment blocked GFP-LC3 cleavage (Fig.S5, A and B). In addition, GFP-LC3 cleavage in Vps34-knockdown HEK293T cells was inhibited in normal medium and EBSS medium, and recovery of Vps34WT expression promoted GFP-LC3 cleavage, and reexpression of Vps342KR only partially reversed these effects (Fig. 5B). These results were further confirmed by an endogenous LC3 puncta assay (Fig. 5, C and D). Moreover, transmission electron microscope assay showed that the number of autophagic vesicles was reduced in Vps34-knockdown H1299 cells compared to control cells cultured in normal medium and under EBSS-induced starvation conditions, Vps34WT expression reversed the reduction in the number of autophagic vesicles, while Vps342KR partially restored the number of vesicles (Fig. 5, E and F). Lactate treatment promotes LC3 turnover, but LDHA inhibitor oxamate treatment inhibited LC3 turnover (fig. S5, D and E). In addition, our previous studies have shown that in the late stage of autophagy, Pacer recruited the Vps34-Vps15-Beclin1-UVRAG complex to autophagosomes to promote autophagosome maturation by generating PtdIns(3)P (12, 38). To analyze whether Vps34 lactylation regulates autophagosome maturation, we overexpressed Vps34 mutants in Vps34 knockdown cells and performed tandem mCherry-GFP-LC3 reporter assay. Vps34 silencing largely blocked autophagosome maturation, as measured by the number of mCherry+GFP− puncta in U2OS cells cultured in either normal medium or EBSS starvation medium. Recovery of Vps34WT expression rescued autophagosome maturation in U2OS cells, while Vps342KR partially reversed this effect (fig. S5, F to H). These results demonstrated that Vps34 lactylation was required for autophagosome formation and maturation.
Fig. 5. Vps34 lactylation facilitates autophagosome formation and maturation.
(A) p62 degradation assay. HEK293T cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT and Vps342KR) were treated with normal medium or EBSS medium for 2 hours. p62 was analyzed by Western blot. (B) GFP-LC3 cleavage assay. HEK293T cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) were treated with normal medium or EBSS medium for 2 hours and analyzed by Western blot with GFP antibody. (C) LC3 puncta assay. U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) were treated with normal medium or EBSS medium for 2 hours. LC3 puncta was analyzed by confocal microscopy. Scale bar, 10 μm. (D) Quantification of LC3 puncta. Data are shown as means ± SD; **P < 0.01, n = 30. (E) Autophagic vacuole assay. U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) were treated with normal medium or EBSS medium for 2 hours. Autophagic vacuole was analyzed by transmission electron microscopy. (F) Quantification of autophagic vacuole. Data are shown as means ± SD; **P < 0.01, n = 20.
Vps34 lactylation facilitates endosome-lysosomal degradation
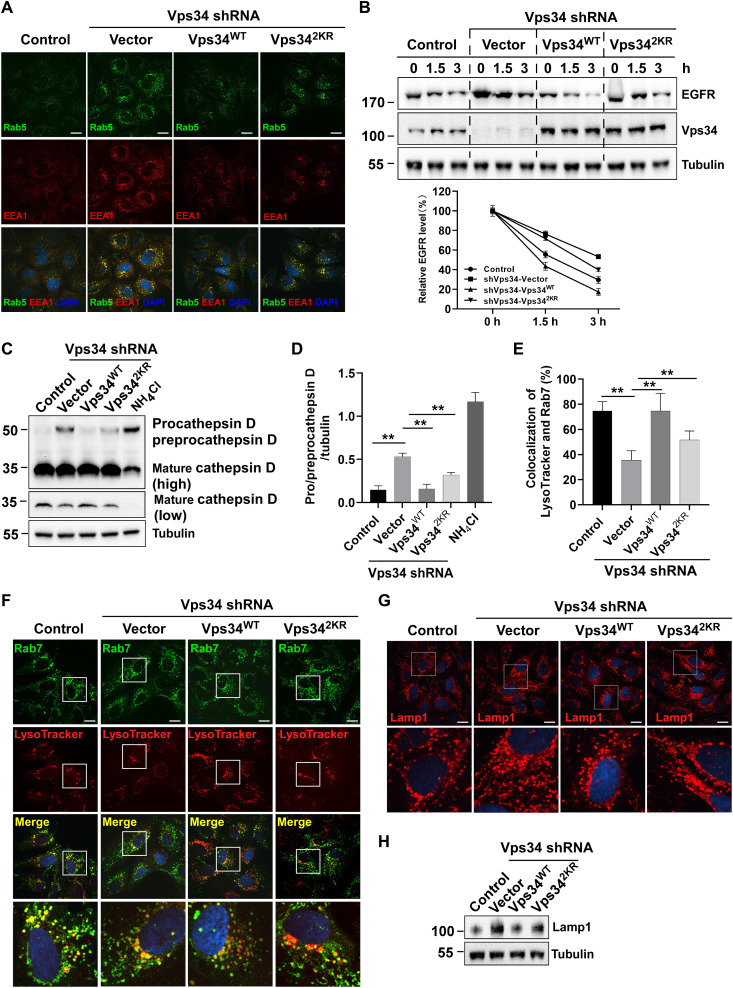
Vps34 produces PtdIns(3)P to recruit the early endosomal effectors, EEA1 and Rab5 on early endosome membranes and regulate Rab7 by recruitment of the guanosine triphosphatase–activating protein on late early endosome membranes (39, 40). Suppression of Vps34 expression showed accumulation of Rab5 and EEA1 enlarged endosomes. To determine whether Vps34 lactylation regulates this phenotype, we expressed Vps34WT and Vps342KR in Vps34-knockdown U2OS. Immunofluorescence microscopy revealed that reconstitution with Vps34WT corrected phenotype of enlarged endosomes accumulation, and Vps342KR partially rescued endosomes accumulation (Fig. 6A).
Fig. 6. Vps34 lactylation promotes endosome-lysosomal degradation.
(A) The Rab5 and EEA1 localization in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR). Rab5 and EEA1 colocalized on enlarged endosomes in Vps34 delactylation cells. Scale bar, 10 μm. (B) EGFR degradation assay. HEK293T cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) were treated with EGF (100 ng/ml) for 0,1.5, and 3 hours. EGFR were analyzed by Western blot. EGFR levels were quantified by ImageJ software and normalized by percentage of 0-hour EGFR level. (C) Cathepsin D maturation assay in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) was analyzed by Western blot with Cathepsin D to show procathepsin D, preprocathepsin, and mature cathepsin D. (D) The levels of procathepsin D and preprocathepsin D were quantified by ImageJ software. Data are shown as means ± SD; **P < 0.01, n = 3. (E) Percentage of Rab7 and LysoTracker colocalization. Data are shown as means ± SD; **P < 0.01, n = 30. (F) Rab7 and LysoTracker colocalization in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) was analyzed by confocal microscopy. Scale bar, 10 μm. (G) Lysosome staining assay by confocal microscopy with Lamp1 antibody in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR). Scale bar, 10 μm. (H). Lamp1 expression in U2OS cells (control shRNA and Vps34 KD/rescued by vector, Vps34WT, and Vps342KR) was analyzed by Western blot.
Although Vps34 knockdown had little effect on early endocytic trafficking, Vps34 played a critical role in late endosome activities such as epidermal growth factor receptor (EGFR) degradation and proteolytic cleavage of cathepsin D (41). EGFR turnover is a commonly used method to investigate endocytosis, endolysosomal trafficking, and degradation. We and others found that suppression of Vps34 expression by shRNA knockdown does not impede internalization of the EGFR by Immunofluorescence (41) (fig. S6, A and B). To analyze whether lactate-mediated Vps34 lactylation regulates EGFR degradation, endogenous EGFR degradation was analyzed by Western blot. Compared with that in control cells, EGFR degradation was increased in lactate-pretreated cells but blocked in oxamate-pretreated H1299 cells (fig. S6C). Moreover, Vps34 silencing inhibited EGFR degradation in Vps34-knockdown H1299 cells compared with control cells after EGF stimulation. Transfection of Vps34WT in Vps34-knockdown cells entirely promoted EGFR degradation, while Vps342KR transfection only partially restored EGFR degradation (Fig. 6B). We also assessed late endosome activity by Cathepsin D maturation. The proteolytic cleavage of Cathepsin D was severely compromised in Vps34-knockdown U2OS, recovery of Vps34WT expression rescued Cathepsin D maturation, and reexpression of Vps342KR only partially reversed this effect (Fig. 6). The proteolytic cleavage of the propre-cathepsin D intermediate became slowly but did not completely inhibit in Vps34-knockdown U2OS. This phenotype indicated that enlarged endosome-derived vacuoles was able to acquire lysosomal characteristics by limited fusion with preexisting lysosomes. Colocalization of late endosome marker Rab7 and LysoTracker red showed that Vps34-knockdown blocked fusion of late endosome and preexisting lysosomes, reexpression of Vps34WT rescued this phenotype but not Vps342KR (Fig. 6, E and F, and fig. S6, D and E). Because of insufficient endolysosomal fusion, we and other found that Lamp1 expression increased by immunofluorescence staining and Western blot in Vps34-knockdown U2OS (42), but Vps34WT recovered this phenotype, and Vps34 delactylation mutant lost this function (Fig. 6, G and H). These results showed that mutant of Vps34 lactylation sites impaired its biological function and led to insufficient lysosomal degradation.
Vps34 lactylation plays essential roles in skeletal muscle homeostasis
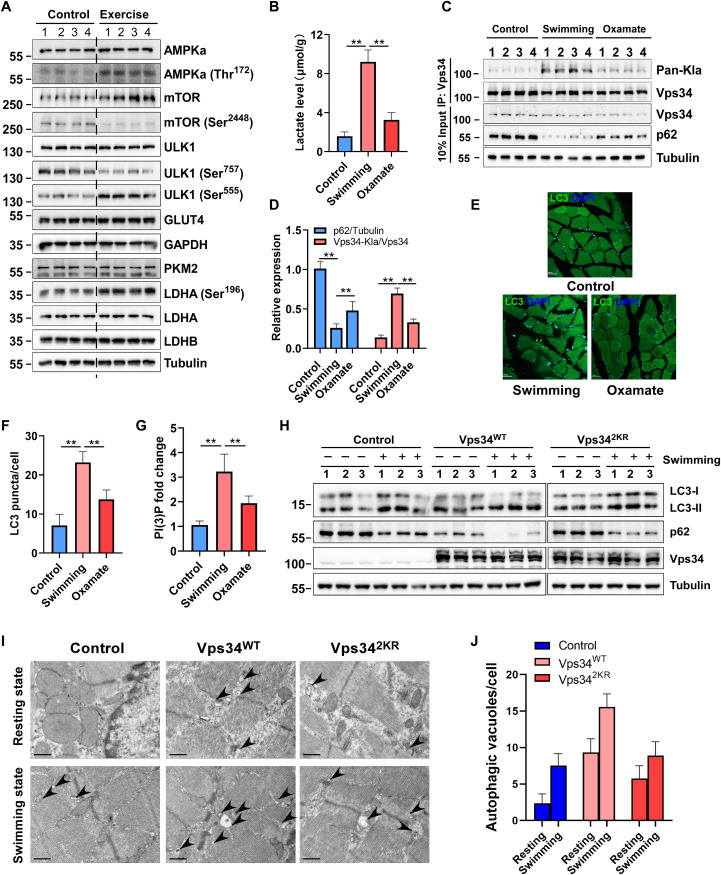
Autophagy plays a critical role in maintaining skeletal muscle homeostasis by eliminating abnormal mitochondria and inclusions (43). To assess the physiological function of Vps34 lactylation, we generated a forced swimming mouse model. Swimming for 3 hours did not affect expression level of total AMPKα, total ULK1, glucose transporter 4 (GLUT4) and glycolytic enzyme [glyceraldehyde-3-phosphate (GAPDH), PKM2, LDHA, and LDHB], but swimming induced up-regulation of AMPKα(Thr172), total mTOR, ULK1 (Ser555), LDHA (Ser196) and down-regulation of mTOR (Ser2448), and ULK1 (Ser757) (Fig. 7A and fig.S7A). Furthermore, exercise induced lactate production in the mouse muscle tissues, and this increase was partially reduced with oxamate treatment (Fig 7B). An evaluation of skeletal muscle biochemistry showed that swimming induced cell autophagy, as determined by measures of p62 degradation and LC3 puncta assay, and notably, the acquisition of these phenotypes was partially inhibited by oxamate treatment (Fig. 7, C to F). Swimming promoted Vps34 lactylation and increased the PtdIns(3)P level, and these phenotypes was partially inhibited by oxamate treatment (Fig. 7, C, D, and G). These results demonstrated that lactate-mediated Vps34 lactylation by ULK1-LDHA pathway was required for skeletal muscle homeostasis.
Fig. 7. Vps34 lactylation plays essential roles in skeletal muscle homeostasis.
(A) Signaling pathway assay in mouse skeletal muscle of control group and swimming group by Western blot. (B) Lactate level in mouse skeletal muscle of each group (control group, swimming group, and oxamate group). Data are shown as means ± SD; **P < 0.01, n = 4. (C) Vps34 lactylation level and autophagy flux in mouse skeletal muscle of each group (control group, swimming group, and oxamate group). Vps34 was purified by immunoprecipitation. Pan-Kla of Vps34, Vps34, and p62 proteins level were analyzed by Western blot. (D) Quantification of Western blot in (C). Data are shown as means ± SD; **P < 0.01, n = 4. (E) Representative images of LC3 puncta (autophagosomes) in skeletal muscle of each group (control group, swimming group, and oxamate group). (F) Quantification of LC3 puncta in (E). Data are shown as means ± SD; **P < 0.01, n = 4. (G) The PtdIns(3)P fold change in mouse skeletal muscle of each group (control group, swimming group, and oxamate group). Data are shown as means ± SD; **P < 0.01, n = 4. Vps34 was obtained by immunocoprecipitation from mouse skeletal muscle of each group (control group, swimming group, and oxamate group). Kinase activity was analyzed by PI(3)P ELISA kit. Data are shown as means ± SD; **P < 0.01, n = 4. (H) Autophagy flux assay in mouse muscle. Mice were intramusculary injected with control virus, rAAV-Vps34WT, or rAAV-Vps342KR (2 × 1011 CFU). After 4 weeks, the half of mice in each group were performed acute swimming exercise for 3 hours. Muscle autophagy levels were analyzed by p62 degradation and LC3 turnover by Western blot. (I) Autophagic vacuole assay in mouse muscle tissues by transmission electron microscopy. Scale bar, 1 μm. (J) Quantification of autophagic vacuole. Data are shown as means ± SD; **P < 0.01, n = 10.
To gain further evidence that Vps34 lactylation is involved in autophagy and skeletal muscle homeostasis in vivo, mice were treated with control virus, rAAV-Vps34WT, or rAAV-Vps342KR (1 × 1011 CFU) by intramuscular injection. Four weeks after viral injection, half of the mice in each group performed acute swimming exercise for 3 hours. The results showed that administration of Vps34WT in muscles enhanced autophagy level at resting state and swimming state than that of control, but administration of Vps342KR partially impaired muscle autophagy level than Vps34WT by p62 degradation and LC3 turnover (Fig. 7H and fig. S7B). Consistently, transmission electron microscopy analysis showed that Vps34WT overexpression promoted the formation of autophagic vacuoles at resting state and swimming state in mice muscle cells, but Vps342KR reduced its effect (Fig. 7, I and J). Morphological analyses did not reveal any abnormalities, such as necrosis or inflammation at resting state and swimming state by hematoxylin (HE) staining (fig. S7C), but swimming increased plasma membrane GLUT4 localization in the skeletal muscle of control rAAV, rAAV-Vps34WT, or rAAV-Vps342KR overexpression than that of resting state (fig. S7D). Vps34WT overexpression in skeletal muscle during resting and swimming states promoted the consumption of glycogen, but Vps342KR overexpression failed to exhibit increased consumption of glycogen (fig. S7, E and F). Mitochondrial number of skeletal muscles from control, Vps34WT, and Vps342KR overexpression mice at resting state and swimming state revealed no difference, but Vps34WT overexpression mice displayed increasing of mitophagy under both conditions than that of Vps342KR (as measured by the percentage of mitochondria encapsulated by autophagosome) (fig. S7, G and H). These results showed that Vps34 lactylation is required for skeletal muscle cell autophagy and cell homeostasis.
Vps34 lactylation correlates with cancer progress
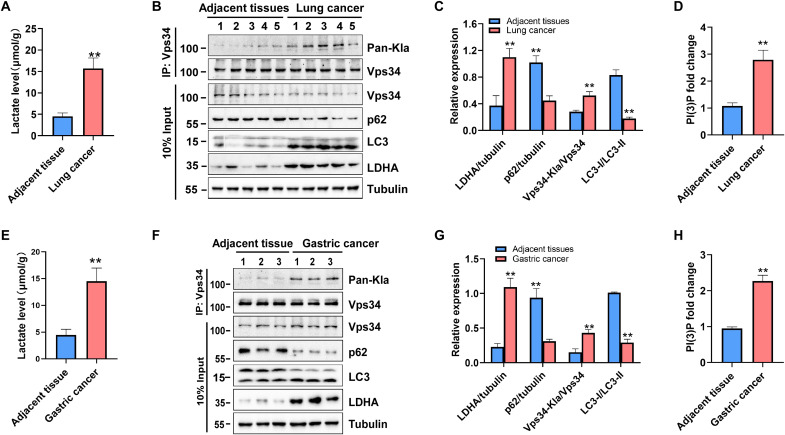
To obtain pathological function of Vps34 lactylation, we analyzed the relationship between lactate and Vps34 lactylation levels in human lung cancer and gastric cancer. Compared with that in adjacent tissues, the lactate level was increased in human lung and gastric cancers (Fig. 8, A and E). In addition, the degree of Vps34 lactylation was increased in human lung and gastric cancer tissues compared with that in adjacent tissues, and this lactylation increase was accompanied by up-regulation of LDHA expression and increased autophagic flux (Fig. 8, B, C, F, and G). Furthermore, the Vps34 kinase activity in human lung and gastric cancer tissues was enhanced compared with that in adjacent tissues, as determined with an in vitro PtdIns(3)P level assay (Fig. 8, D and H). The results indicated that lactate regulates autophagic activity in tumor tissues through Vps34 lactylation and correlates with cancer progress.
Fig. 8. Vps34 lactylation correlates with cancer progress.
(A) Lactate level in lung cancer and adjacent tissues. Data are shown as means ± SD; **P < 0.01, n = 5. (B) Vps34 lactylation level and autophagy flux in lung cancer and adjacent tissues. Tissues were lysed by TAP buffer, Vps34 was purified by immunoprecipitation, and protein level was analyzed by Western blot. (C) Quantification of Western blot in (B). Data are shown as means ± SD; **P < 0.01, n = 5. (D) The PI(3)P fold change in in lung cancer and adjacent tissues. Vps34 was purified by immunoprecipitation, in vitro Vps34 kinase assay was performed, and PI(3)P level was determined by the PI(3)P ELISA kit. Data are shown as means ± SD; **P < 0.01, n = 5. (E) Lactate level in gastric cancer and adjacent tissues. Data are shown as means ± SD; **P < 0.01, n = 3. (F) Vps34 lactylation level and autophagy flux in gastric cancer and adjacent tissues. Methods as K. (G) Quantification of Western blot in (F). Data are shown as means ± SD; **P < 0.01, n = 3. (H) The PI(3)P fold change in gastric cancer and adjacent tissues. Data are shown as means ± SD; **P < 0.01, n = 3.
DISCUSSION
ULK1 is the main serine/threonine kinase involved in the initiation of autophagy in response to nutrient deprivation (glucose and amino acid deprivation). As a serine/threonine kinase, ULK1 not only exhibits canonical functions in autophagy by phosphorylating autophagy machinery proteins such as Vps15, Beclin1, Atg14, Atg16L, and SQSTM1 (44–47) but also exhibits noncanonical functions in multiple pathways by phosphorylating substrates such as STING and glycolytic enzymes (hexokinase, PFK1, and enolase 1) and a gluconeogenic enzyme (fructose-1,6-bisphosphatase) (30, 48). In our study, we found that ULK1 interacts with the key glycolytic enzyme LDHA, phosphorylating LDHA at S196 to promote LDHA enzyme activity. Notably, LDHA S196 is conserved across species ranging from zebrafish to humans and located in the loop domain, as indicated by its spatial structure, which indicates that S196 is close to the histidine site in the NADH proton acceptor. LDHA glutamate 192, S196, and S319 maintain the spatial conformation of LDHA by hydrogen bonding to facilitate LDHA histidine-193 (H193) binding NADH. We speculated that LDHA S196 phosphorylation could change the LDHA loop molecular conformation to enhance the binding of LDHA H193 and NADH. Therefore, LDHA S196 phosphorylation promotes lactate production under ULK1 activation conditions induced by nutrient deprivation (Fig. 9).
Fig. 9. Model of ULK1-LDHA pathway and TIP60-Vps34 pathway in skeletal muscle homeostasis and cancer progress.
ULK1 phosphorylates LDHA serine-196 in serum deprivation, increases LDHA enzyme activity, and promotes lactate production. Lactate-mediated Vps34 lactylation further activates its lipid kinase activity and promotes cell autophagy in muscle cells and cancer cells. SoNar is NADH/NAD+ sensor, and Fila is lactate sensor.
Lactate has been previously considered a waste product of glucose metabolism, but an increasing number of studies have shown that lactate plays a critical role in regulating multiple biological processes, such as macrophage polarization, T helper cell differentiation, tumor immune surveillance, and Mg2+ handling movement between the ER and mitochondria (21, 49–51). Recent studies have shown that lactate-mediated lactylation, a PTM, plays an important role in regulating gene expression and protein function (18, 28). We found that multiple autophagy core machinery proteins are lactylated. These results indicated that lactate-mediated lactylation regulates the autophagy process. Vps34 is a catalytic subunit of the class III PI3K complex, which phosphorylates PI to produce PtdIns(3)P, which promotes autophagy and endolysosomal trafficking. Through mass spectrometry, we found that Vps34 K356 and K781 undergoes lactylation and that manipulating lactate levels modulates Vps34 lactylation in this study. Specifically, Vps34 K356 and K781 lactylation promotes the interaction of Vps34 with PI3KC3 complex I and II subunits to enhance Vps34 kinase activity and facilitate autophagy and endolysosomal trafficking. Although we still do not know how Vps34 K356 and K781 lactylation affects protein conformation, lactylation at these Vps34 sites enhances Vps34 enzyme activity.
In addition, Vps34 lactylation plays an essential role in both physiological and pathological processes. Intense exercise can cause muscle protein denaturation and mitochondrial damage through physiological processes (52). Therefore, to facilitate cellular adaptation to intense exercise, autophagy maintains muscle cell homeostasis by eliminating damaged organelles and proteins and restores metabolic balance (53). A preliminary study showed that autophagic flux is induced after acute unilateral high-resistance contractions by regulating mVps34 kinase activity and Bcl-2–mediated autophagy (54, 55). Our study also revealed that intense swimming increased Vps34 kinase activity in mice skeletal muscle. However, the molecular mechanism underlying this increase remains unclear. Notably, intense exercise is known to increase muscle lactate production. We found that the degree of Vps34 lactylation and the PtdIns(3)P level in mouse skeletal muscle during exercise are obviously enhanced compared with those in control mice and that these increases are accompanied by lactate production in skeletal muscle. Permanent delactylated mutant Vps342KR impaired Vps34 kinase activity and reduced autophagy level and removal of abnormal mitochondria caused by exercise. These results indicated that higher levels of muscle lactate, a signaling molecule, induces muscle cell autophagy by Vps34 lactylation (Fig. 9).
In cancer cells, aerobic glycolysis (the Warburg effect) is activated by altered growth factor signaling, hypoxic activation of hypoxia-inducible factor-1α transcription, and oncogene activation, which enhance cancer cell growth and malignant progression. Lactate levels in the tumor microenvironment are increased by the Warburg effect. Cell autophagy is a double-edged sword in tumorigenesis and cancer progression. In general, autophagy in healthy cells is thought to operate as a tumor suppressor mechanism to counteract the effects of pro-oncogenic stimuli (56), but autophagy can promote cancer progression by the removal damaged organelles and proteins and thereby increase the metabolic fitness of malignant cells to help them survive in established tumor lesions (57–59). We found that Vps34 lactylation increased in human lung cancer and gastric cancer tissues compared with that in normal adjacent tissues. These results suggested that Vps34 hyperlactylation in tumor tissues with high lactate levels could promote cancer cell autophagy to facilitate cancer progression (Fig. 9).
In conclusion, we provide evidence showing that the serine/threonine kinase ULK1 regulates the glycolysis pathway by phosphorylating LDHA at S196 to enhance LDHA activity and promotelactate production. Lactate-mediated Vps34 lactylation promotes its lipid kinase activity to facilitate cell autophagy and endolysosomal degradation. Vps34 lactylation plays a critical role in skeletal muscle homeostasis during exercise and tumor progression by regulating cell autophagy. This study describes an autophagy regulation mechanism and integrates two highly conserved life processes (glycolysis and autophagy).
MATERIALS AND METHODS
Cell lines, cell culture, and transfection
H1299 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and penicillin-streptomycin (100 U/ml). HEK293T, Hela, and U2OS cells were cultured in Dulbecco's minimum essential medium (DMEM) supplemented with 10% FBS, 2 mM l-glutamine, and penicillin-streptomycin (100 U/ml) in a humidified incubator at 37°C with 5% CO2. Genetically encoded lactate sensor and control cell lines H1299–FiLa-C and H1299-FiLa, NADH/NAD+ sensor and control sensor cell lines Hela-SoNar and Hela-iNapc, and U2OS GFP-FYVE2 cell line were obtained by lentivirus infection. Plasmids were transfected using Lipofectamine 2000. FiLa-C and iNapc sensors are control sensors of lactate FiLa and NADH/NAD+ SoNar.
Antibodies and reagents
Primary antibodies were obtained from Cell Signaling Technology [LDHA (#3582), PI3 Kinase Class III (#3358), ULK1 (Ser757) (#14202), P70S6K (#9202), P70S6K (Thr389) (#9234), EGFR (#54359), Atg14L (#5504), ULK1 (Ser555) (#5869), mTOR (Ser2448) (#5536), mTOR (#2983), AMPKα (#5831), AMPKα (Thr172) (#50081), GLUT4 (#5831), acetylated-lysine (#9441), Rab7 (#9367), Rab5 (#46449), Lamp1 (#15665), and Cathepsin D(#2284)], Proteintech [GAPDH (#60004-1-Ig), LDHB (#14824-1-AP), ULK1 (#20986-1-AP), P62/SQSTM1 (#18420-1-AP), Beclin 1 (#11306-1-AP), Beta-Actin (#66009-1-Ig), and KAT5/TIP60 (#10827-1-AP)], Roche [GFP (#G1546)], Merck Millipore [phospho-Ser/Thr-Pro (#05-368)], PTM BIO [Kla (#PTM-1401)], ABclonal [HA-Tag-HRP (#AE025)], MBL [Flag-Tag-HRP (#M185-7), UVRAG (#M160-3), and LC3 (#M152–3)], HUABIO [Beta-Tubulin (#M1305–2) and LDHA (Ser196) (Homemade)].
Rotenone (#R8875), sodium l-lactate (#71718), and sodium oxamate (#O2751) were obtained from Sigma-Aldrich. Selleck Chemicals was the source for torin1 (#S2827), rapamycin (#S1039), Trichostain A (TSA) (#S1045), and SB216763 (#S1075). MRT68921 (#HY-100006) was obtained from MedChemEpress. MG149 (#T6584), phosphatase inhibitor cocktail (100X) (#C0004), and protease inhibitor cocktail (100X) (#C0001) were obtained from TargetMol. L-Lactyl-CoA (CAS: 4625-32-5) was obtained from Shanghai Naford Biological Technology. Hieff TransTM Liposomal Transfection Reagent was obtained from Shanghai YEASEN (#40802ES03). Lipofectamine 2000 (#11668019) and EBSS (#24010043) were obtained from Thermo Fisher Scientific. DAPI (4′,6-diamidino-2-phenylindole) was obtained from Beyotime (#C1002). TRIzol reagent was obtained from Invitrogen (#15596026). LDHA recombinant protein was purified from Escherichia coli. Mouse EGF protein was obtained from Sino Biological Inc. (#50482-MNCH). Flag peptide was obtained from GenScript Biotech Corp. LDHA peptide (LGEHGDSSVPVWSGC) was synthesized by Hangzhou Huabio.
Human cancer and adjacent tissues
Human lung cancer (n = 5) and adjacent tissues (n = 5) were collected from Shanghai Chest Hospital. All lung cancer tissues were adenocarcinoma. Human gastric cancer (n = 3) and adjacent tissues (n = 3) were collected from the Second Affiliated Hospital of Zhejiang University and Shanghai Chest Hospital. All gastric cancer tissues were adenocarcinoma. The standard procedures were approved by the Ethics Committee for Medical Research (Institutional Review Board) at Shanghai Jiao Tong University Affiliated Shanghai Chest Hospital and the Second Affiliated Hospital of Zhejiang University. The informed consents were obtained from all patients.
Acute swimming exercise
C57BL/6 were purchased from the Shanghai SLAC Laboratory Animal Co. Ltd. and bred at normal room temperatures, on a regular 12-hour light and 12-hour dark cycle. Animal experiments were maintained according to the Guideline for Animal Care at East China University of Science and Technology. Fifteen animals were randomly divided into control group (n = 5), swimming group (n = 5), and oxamate group (n = 5) by random number table. Mice of the swimming group and oxamate group were acclimated to swimming for 20 min per day for 5 days. Before acute swimming exercise, mice in control group and swimming group were treated with normal saline (0.9% NaCl) and mice in oxamate group were administrated with 500 mg/kg of body weight of oxamate by intraperitoneal injection. After 30 min, mice in swimming group and oxamate group subsequently swam together in a glass water tank at a water temperature of 35° ± 1°C for 3 hours. Mice in the three groups were anesthetized, euthanized by CO2, and muscle tissues were obtained immediately after completion of swimming. The experiment was repeated three times.
Mouse models of Vps34 or mutant overexpression in muscle
For investigating the role of Vps34 lactylation in regulating autophagy in vivo, recombinant adeno-associated virus packing Vps34WT and Vps342KR were produced in HEK293T cells (Vigenebio, China). Mice were treated with the control virus, rAAV-Vps34WT, or rAAV-Vps342KR (1 × 1011 CFU) by intramuscular injection. Four weeks after viral injection, the half of mice in each group performed acute swimming exercise for 3 hours. The mice were sacrificed for the analysis of autophagy level and another indexs. The experiment was repeated three times.
Immunoprecipitation assay
Cells harvested by centrifugation were homogenized in lysis buffer [20 mM tris-HCl, (pH 7.4, 150 mM and NaCl, 0.5% NP-40 1 mM EDTA, supplemented with protease and phosphatase inhibitors) for 30 min at 4°C. For lactylation immunoprecipitation, 2 μM trichostatin A (TSA) was added Into TAP buffer. For weak protein-protein interaction assay, cells were treated with chemically reducible cross-linker DSP. Cell supernatants were incubated with antibody binding beads at 4°C for 6 hours. The beads were washed with lysis buffer three times, and then incubated in SDS loading buffer at 100°C for 5 min to SDS–polyacrylamide gel electrophoresis (PAGE).
Immunoblotting analysis
Cell lysate, tissue extracts, and IP sample were suspended in SDS sample buffer (100 mM tris-HCl, 2% SDS, 20 mM β-mercaptoethanol, and 10% glycerol (pH 7.5) and incubated at 100°C for 5 min. Samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membrane (PVDF). PVDF membrane was blocked by 5% bovine serum albumin (BSA) and incubated with the corresponding primary antibody. After washing with phosphate buffered saline with 0.05% Tween-20 (PBST) buffer three times, membranes were incubated with horseradish peroxidase (HRP)-conjugated second antibody at room temperature for 2 hours. Bands were scanned using enhanced chemiluminescence detection system. β-Actin and tubulin was used as an internal control.
Immunofluorescence microscopy
For GFP-FYVE2 puncta assay, U2OS GFP-FYVE2 cell line was cultured on coverslip and fixed by 4% paraformaldehyde for 20 min at room temperature for imaging. For endogenous Rab5, EEA1, EGFR, Lamp1, and Rab7, cells were fixed by 4% paraformaldehyde and blocked by 2.5% BSA and then incubated with appropriate primary antibodies for 12 hours at 4°C. Cells were then incubated with Alexa Fluor–conjugated secondary antibodies. For LC3 of muscle tissue, skeletal muscle was isolated from C57BL/6 J mice and then sectioned with a Leica VT1200 S Fully automated vibrating blade microtome. Muscle sections were maintained in phosphate-buffered saline (PBS) and then stained with LC3 primary antibodies and Alexa Fluor-conjugated secondary antibody for imaging. All fluorescence was detected using a laser scanning confocal microscope (Nikon Eclipse Ti-E and Zeiss LSM 510).
Mass spectrometry analysis
LDHA band and Vps34 band was reduced with dithiothreitol (DTT) and alkylated with iodoacetamide followed by digestion using 10 ng/μl Trypsin (Sigma-Aldrich, USA) and incubation overnight at 37°C. The digested samples were dried and desalted by C18 tip and reconstituted, loaded onto a capillary reverse-phase C18 column by an HPLC system (Thermo Fisher Scientific Easy-nLC 1000). Peptides were detected by an in-line mass spectrometer (Thermo Fisher Scientific LTQ Obitrap ETD). MS spectra were collected, and the top 20 abundant ions were sequentially isolated for MS/MS analysis.
Transmission electron microscopy
Cells transfected with Vps34 mutants and muscle tissues were fixed in 2.5% glutaraldehyde in PBS overnight at 4°C and washed three times for 15 min with 0.1 M phosphate buffer. Cells were fixed by 1% osmium tetroxide at pH 7.2 for 1 hour at room temperature and were then dyed with 2% uranyl acetate for 30 min. Then, samples were dehydrated through graded alcohols (50 to 100%) and embedded in EPON 812 resin and cured for 24 hours at 37°, 45°, and 60°C, respectively. Electron microscopy images of the samples were taken using Tecnai G2 Spirit transmission electron microscope (FEI Company).
Autophagy analysis
For assessment of autophagy of cell lines, cells were cultured in normal or starvation (EBSS) medium for 4 hours. After treatment, cells were lysed and we performed Western blot of total proteins for analysis of cell extracts with antibodies against LC3 and p62. For assessment of autophagy in vivo following exercise, skeletal muscle in control group, swimming group, and oxamate group were isolated and homogenized in lysis buffer, total tissues extracts were analyzed by Western blot with antibodies against LC3 and p62. Autophagy of skeletal muscle was also assessed by Immunofluorescence confocal microscopy with antibodies against LC3 to analyzed LC3 puncta.
Yeast two-hybrid assay
Atg13, LDHA, and LDHB complementary DNA (cDNA) were cloned into pGBKT7 expression vectors and ULK1 cDNA was cloned into pGADT7 expression vectors. The pGADT7-prey and pGBKT7-bait vectors were cotransformed into the yeast strain MJ109 and were selected in the yeast extract, peptone, and dextrose (YPD) medium lacking Leucine and tryptophan (YPD/−Leu/−Trp) for 3 days at 30°C. Restreak the single cloning and transferred into 3 ml of YPD medium (−Leu/−Trp) and continued to culture for 4 hours until the optical density at 600 nm (OD600) reached approximately 0.5. The yeast cells were diluted in a fivefold gradient and incubate the plates for 2 days at 30°C.
LDHA enzyme assay
LDHAWT-Flag, LDHAS196A-Flag, and LDHAS196D-Flag were transfected into HEK293T cells and immunoprecipitated by Flag beads. Proteins were eluted by eluting buffer [20 mM Hepes, 150 mM NaCl, 1 mM EDTA, 3*Flag peptide (200 μg/ml) (pH 7.5)]. Purified LDHA proteins (1 μg) was added to reaction buffer (0.2 M tris-HCl, 30 mM pyruvate, and 2 mM NADH (pH 7.4)]. The absorbance (340 nm) was measured by Synergy Neo2 Hybrid Multi-Mode Reader (BioTeck).
In vitro UKL1 kinase assay
In vitro kinase assay was performed as described previously (60). Briefly, LDHA protein were expressed and purified from BL-21(DE3). ULK1-HA kinase was expressed from HEK293T cells and purified by anti-HA beads. Recombinant LDHA-Flag was incubated with ULK1-HA in kinase buffer (10 mM tris, 15 mM NaCl, 10 mM MgCl2, and 0.5 mM DTT (pH 7.4) in a in 50 μl of reaction mixture at 37°C for 15 min and then stopped with sample buffer. The samples were subjected to SDS-PAGE and immunoblotting with specificity LDHA S196 phosphorylation antibody.
In vitro lactylation assay
Bacterially expressed Vps34WT-Flag and Vps342KR-Flag were purified on glutathione S-transferase beads. TIP60-HA was expressed from HEK293T and purified by anti-HA beads. Lactylation assays contained Vps34WT-Flag (500 ng) or Vps342KR-Flag (500 ng) and TIP60-HA (500 ng) in reaction buffer (50 mM Hepes, 30 mM KCl, 0.25 mM EDTA, 5.0 mM MgCl2, 5.0 mM sodium butyrate, 20 μM L-lactyl-CoA, (pH 7.8)]. Reactions were incubated at 30°C for 30 min. Samples were resolved by 10% SDS-PAGE and analyzed by mass spectrometry analysis (AIMS scientific Co., Ltd., Shanghai, China), immunoblotting or kinase assay.
Vps34 kinase assay
For in vitro Vps34 kinase activity, HEK293T cells were transfected by Vps34WT-Flag and Vps342KR-Flag vectors. Vps34 and Vps342KR proteins were expressed from HEK293T cells and purified by Anti-Flag beads. For endogenous Vps34 kinase activity in muscle tissues of three groups, endogenous Vps34 was immunoprecipitated for 8 hours at 4°C with 1 μg of rabbit anti-Vps34 antibodies using 5 mg of total lysate. The immune complexes were washed three times by lysis buffer. Kinase reaction buffer (50 mM tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 500 mM PI, and 1.25 mM ATP [ pH 7.4] were added to immune complexes (Vps34 or Vps342KR or endogenous Vps34) and incubated at 30°C for 1 hour. The reaction was terminated by adding 100 mM EDTA. PtdIns(3)P were extracted and determined by PtdIns(3)P Mass ELISA kit.
PtdIns(3)P ELISA assay
PtdIns(3)P were extracted according to PtdIns(3)P Mass ELISA kit following the instructions of the manufacturer. Briefly, cells were removed from the medium, 5 ml of ice-cold 0.5 M TCA was added, and we incubated cells on ice for 5 min and scraped the cells from dish with additional 0.5 M TCA. Cells were centrifuged at 3000 rpm for 7 min and discarded the supernatant. The pellet was resuspended by 3 ml 5% TCA/1 mM EDTA and centrifuged at 3000 rpm for 5 min. Acidic lipids were extracted by MeOH: CHCl3:12 N HCl (80:40:1) and PtdIns(3)P component were obtained from organic phase. Cell extraction samples were added to incubation plate. Add PtdIns(3)P 60 μl /well detector to sample wells. Seal the plate with a plate sealer and incubate on a plate shaker at room temperature for 1 hour. Wash the detection plate three times with PBST. Add 100 μl of diluted secondary detector to each well and incubate on a plate shaker at room temperature for 1 hour. Wash the detection plate three times with PBST. Add 100 μl TMB solution to each and develop for 30 min. Stop color development by 50 μL H2SO4. Read absorbance at 450 m on a plate reader. PI3P quantities can be estimated by the standard curve.
Lactate and NADH/NAD+ in live cell by microplate reader
Genetically encoded lactate sensor and control cell lines H1299-FiLa and H1299-FiLa, NADH/NAD+ sensor and control sensor cell lines Hela-SoNar and Hela-cpYFP were grown in 96-well black bottom plate. After 12 hours, the cells were treated with different compounds and incubated in HBSS buffer (pH 7.4) at 37°C. The ratios were determined by a Synergy Neo 2 Multi-Mode Microplate Reader (BioTek) with excitation filters 420 BP 10 nm and 485 BP 20 nm, and emission filter 528 BP 20 nm. Fluorescence values were background-corrected by subtracting the intensity of the cell samples not expressing FiLa sensors.
Lactate and NADH/NAD+ in live cell by flow cytometer
Genetically encoded lactate sensor and control cell lines H1299-FiLa and H1299-FiLa, NADH/NAD+ sensor and control sensor cell lines Hela-SoNar and Hela-cpYFP were grown in 96-well black bottom plate. After 12 hours, the cells were treated with different compounds and harvested by trypsinization. Cells were analyzed by flow cytometry within 10 min on a CytoFLEX flow cytometer (Beckman Coulter). Sensor was excited using laser lines at 405 nm and 488 nm. Emission filters were 525 BP 40 nm for both excitation wavelengths.
Quantitative real-time PCR analysis
Toal RNA was extracted from H1299 and HEK293T by TRIzol (Invitrogen) and cDNA was synthesized by HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd). Real-time PCR was performed by Taq Pro Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd) on the LightCycler 480 II system (Roche). Relative expression level of mRNA was normalized to endogenous control β-actin.
HE staining and periodic acid Schiff staining
Skeletal muscle was isolated from C57BL/6 J mice and then sectioned with a Leica VT1200 microtome. Sections were mounted on microscope slides, deparaffinized in three changes of xylene, and rehydrated through graded concentrations of ethanol. HE staining was performed to analyze histopathologic changes of muscle tissue according to standard protocols. For periodic acid Schiff staining, sections were treated with 1% periodic acid for 5 min, rinsed in distilled water, stained with Schiff’s reagent for 20 min, then counterstained with hematoxylin for 8 min, dehydrated in alcohol, cleared in xylene, and sealed by neutral gum.
Statistical analysis
To quantify the number of GFP-FYVE, LC3 puncta, LysoTracker positive vesicles and autophagosome vesicles, a total of 30 cells were recorded and analyzed using NIS-Elements AR Analysis program. These measurements were done on randomly selected fields of view. Two-tailed unpaired Student’s t test was performed for statistical analysis using GraphPad Prism software. All data are presented as means ± SD (**P < 0.01, *P < 0.05).
Acknowledgments
We are grateful to Q. Sun for reagents. We thank the Imaging Center of Zhejiang University School of Medicine for assistance with electron microscopy.
Funding: This study was supported by the National Key Research and Development Program of China (2021YFA0804900 and 2021YFC2101100 to X.C. and 2019YFA0904800 to Y.Z.); the National Nature Science Foundation of China (32170764 to X.C.; 32150030, 32030065, and 92049304 to Y.Z.; 32121005, 21937004, 32150028, and 91857202 to Y.Y.); Research Unit of New Techniques for Live-cell Metabolic Imaging (Chinese Academy of Medical Sciences, 2019RU01, 2019-I2M-5-013); Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism; Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone Stem Cell Strategic Biobank and Stem Cell Clinical Technology Transformation Platform (ZJ2018-ZD-004); Innovative research team of high-level local universities in Shanghai; the Nurture projects for basic research of Shanghai Chest Hospital (2021YNJCM09); the State Key Laboratory of Bioreactor Engineering; the Fundamental Research Funds for the Central Universities.
Author contributions: X.C. and YZ.Z. designed the experiments. X.C., M.J., X.Y., W.S., Q.Z., L.Z., Y.L., Y.Q., K.M., and R.Z. performed the experiments. X.L., Y.S., A.W., YJ.Z., Y.L., L.H.,., and W.C. contributed reagents. C.C., W.G., and J.F. provided clinical samples. X.C., Y.Y., and YZ.Z. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
REFERENCES AND NOTES
- 1.N. Mizushima, M. Komatsu, Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Y. Feng, D. He, Z. Yao, D. J. Klionsky, The machinery of macroautophagy. Cell Res. 24, 24–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. Kroemer, G. Marino, B. Levine, Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P. M. Wong, C. Puente, I. G. Ganley, X. Jiang, The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy 9, 124–137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Tsukamoto, A. Kuma, M. Murakami, C. Kishi, A. Yamamoto, N. Mizushima, Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008). [DOI] [PubMed] [Google Scholar]
- 6.N. Mizushima, A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527 (2018). [DOI] [PubMed] [Google Scholar]
- 7.M. Antonioli, M. Di Rienzo, M. Piacentini, G. M. Fimia, Emerging mechanisms in initiating and terminating autophagy. Trends Biochem. Sci. 42, 28–41 (2017). [DOI] [PubMed] [Google Scholar]
- 8.J. Kim, M. Kundu, B. Viollet, K. L. Guan, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.W. Wan, Z. You, L. Zhou, Y. Xu, C. Peng, T. Zhou, C. Yi, Y. Shi, W. Liu, mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol. Cell 72, 303–315 (2018). [DOI] [PubMed] [Google Scholar]
- 10.J. Kim, Y. C. Kim, C. Fang, R. C. Russell, J. H. Kim, W. Fan, R. Liu, Q. Zhong, K. L. Guan, Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Y. M. Kim, C. H. Jung, M. Seo, E. K. Kim, J. M. Park, S. S. Bae, D. H. Kim, mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 57, 207–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.X. Cheng, X. Ma, Q. Zhu, D. Song, X. Ding, L. Li, X. Jiang, X. Wang, R. Tian, H. Su, Z. Shen, S. Chen, T. Liu, W. Gong, W. Liu, Q. Sun, Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol. Cell 73, 788–802 (2019). [DOI] [PubMed] [Google Scholar]
- 13.H. Su, F. Yang, Q. Wang, Q. Shen, J. Huang, C. Peng, Y. Zhang, W. Wan, C. C. L. Wong, Q. Sun, F. Wang, T. Zhou, W. Liu, VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell 67, 907–921 (2017). [DOI] [PubMed] [Google Scholar]
- 14.K. E. Pyo, C. R. Kim, M. Lee, J. S. Kim, K. I. Kim, S. H. Baek, ULK1 O-glcnacylation is crucial for activating VPS34 via ATG14L during autophagy initiation. Cell Rep. 25, 2878–2890 (2018). [DOI] [PubMed] [Google Scholar]
- 15.G. Wang, J. G. Meyer, W. Cai, S. Softic, M. E. Li, E. Verdin, C. Newgard, B. Schilling, C. R. Kahn, Regulation of UCP1 and mitochondrial metabolism in brown adipose tissue by reversible succinylation. Mol. Cell 74, 844–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H. Yu, C. Bu, Y. Liu, T. Gong, X. Liu, S. Liu, X. Peng, W. Zhang, Y. Peng, J. Yang, L. He, Y. Zhang, X. Yi, X. Yang, L. Sun, Y. Shang, Z. Cheng, J. Liang, Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair. Sci. Adv. 6, eaay4697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y. Yang, W. Fiskus, B. Yong, P. Atadja, Y. Takahashi, T. K. Pandita, H. G. Wang, K. N. Bhalla, Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 6841–6846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D. Zhang, Z. Tang, H. Huang, G. Zhou, C. Cui, Y. Weng, W. Liu, S. Kim, S. Lee, M. Perez-Neut, J. Ding, D. Czyz, R. Hu, Z. Ye, M. He, Y. G. Zheng, H. A. Shuman, L. Dai, B. Ren, R. G. Roeder, L. Becker, Y. Zhao, Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.B. Faubert, K. Y. Li, L. Cai, C. T. Hensley, J. Kim, L. G. Zacharias, C. Yang, Q. N. Do, S. Doucette, D. Burguete, H. Li, G. Huet, Q. Yuan, T. Wigal, Y. Butt, M. Ni, J. Torrealba, D. Oliver, R. E. Lenkinski, C. R. Malloy, J. W. Wachsmann, J. D. Young, K. Kernstine, R. J. DeBerardinis, Lactate metabolism in human lung tumors. Cell 171, 358–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.W. Zhang, G. Wang, Z. G. Xu, H. Tu, F. Hu, J. Dai, Y. Chang, Y. Chen, Y. Lu, H. Zeng, Z. Cai, F. Han, C. Xu, G. Jin, L. Sun, B. S. Pan, S. W. Lai, C. C. Hsu, J. Xu, Z. Z. Chen, H. Y. Li, P. Seth, J. Hu, X. Zhang, H. Li, H. K. Lin, Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell 178, 176–189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. C. Daw, K. Ramachandran, B. T. Enslow, S. Maity, B. Bursic, M. J. Novello, C. S. Rubannelsonkumar, A. H. Mashal, J. Ravichandran, T. M. Bakewell, W. Wang, K. Li, T. R. Madaris, C. E. Shannon, L. Norton, S. Kandala, J. Caplan, S. Srikantan, P. B. Stathopulos, W. B. Reeves, M. Madesh, Lactate elicits ER-mitochondrial Mg2+ dynamics to integrate cellular metabolism. Cell 183, 474–489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.V. Pucino, M. Certo, V. Bulusu, D. Cucchi, K. Goldmann, E. Pontarini, R. Haas, J. Smith, S. E. Headland, K. Blighe, M. Ruscica, F. Humby, M. J. Lewis, J. J. Kamphorst, M. Bombardieri, C. Pitzalis, C. Mauro, Lactate buildup at the site of chronic inflammation promotes disease by inducing CD4+ T cell metabolic rewiring. Cell Metab. 30, 1055–1074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. Zhang, J. Muri, G. Fitzgerald, T. Gorski, R. Gianni-Barrera, E. Masschelein, G. D'Hulst, P. Gilardoni, G. Turiel, Z. Fan, T. Wang, M. Planque, P. Carmeliet, L. Pellerin, C. Wolfrum, S. M. Fendt, A. Banfi, C. Stockmann, I. Soro-Arnaiz, M. Kopf, K. De Bock, Endothelial lactate controls muscle regeneration from Ischemia by inducing M2-like macrophage polarization. Cell Metab. 31, 1136–1153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L. Li, K. Chen, T. Wang, Y. Wu, G. Xing, M. Chen, Z. Hao, C. Zhang, J. Zhang, B. Ma, Z. Liu, H. Yuan, Z. Liu, Q. Long, Y. Zhou, J. Qi, D. Zhao, M. Gao, D. Pei, J. Nie, D. Ye, G. Pan, X. Liu, Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat. Metab. 2, 882–892 (2020). [DOI] [PubMed] [Google Scholar]
- 25.H. Hagihara, H. Shoji, H. Otabi, A. Toyoda, K. Katoh, M. Namihira, T. Miyakawa, Protein lactylation induced by neural excitation. Cell Rep. 37, 109820 (2021). [DOI] [PubMed] [Google Scholar]
- 26.R. A. Irizarry-Caro, M. M. McDaniel, G. R. Overcast, V. G. Jain, T. D. Troutman, C. Pasare, TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proc. Natl. Acad. Sci. U.S.A. 117, 30628–30638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Yu, P. Chai, M. Xie, S. Ge, J. Ruan, X. Fan, R. Jia, Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 22, 85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.K. Yang, M. Fan, X. Wang, J. Xu, Y. Wang, F. Tu, P. S. Gill, T. Ha, L. Liu, D. L. Williams, C. Li, Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 29, 133–146 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N. Hosokawa, T. Hara, T. Kaizuka, C. Kishi, A. Takamura, Y. Miura, S. Iemura, T. Natsume, K. Takehana, N. Yamada, J. L. Guan, N. Oshiro, N. Mizushima, Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. Y. Li, Y. Sun, Y. Liang, Q. Liu, Y. Shi, C. S. Zhang, C. Zhang, L. Song, P. Zhang, X. Zhang, X. Li, T. Chen, H. Y. Huang, X. He, Y. Wang, Y. Q. Wu, S. Chen, M. Jiang, C. Chen, C. Xie, J. Y. Yang, Y. Lin, S. Zhao, Z. Ye, S. Y. Lin, D. T. Chiu, S. C. Lin, ULK1/2 constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol. Cell 62, 359–370 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Y. Zhao, Q. Hu, F. Cheng, N. Su, A. Wang, Y. Zou, H. Hu, X. Chen, H. M. Zhou, X. Huang, K. Yang, Q. Zhu, X. Wang, J. Yi, L. Zhu, X. Qian, L. Chen, Y. Tang, J. Loscalzo, Y. Yang, SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 21, 777–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.X. Li, Y. Zhang, L. Xu, A. Wang, Y. Zou, T. Li, L. Huang, W. Chen, S. Liu, K. Jiang, X. Zhang, D. Wang, L. Zhang, Z. Zhang, Z. Zhang, X. Chen, W. Jia, A. Zhao, X. Yan, H. Zhou, L. Zhu, X. Ma, Z. Ju, W. Jia, C. Wang, J. Loscalzo, Y. Yang, Y. Zhao, Ultrasensitive sensors reveal the spatiotemporal landscape of lactate metabolism in physiology and disease. Cell Metab. 35, 200–211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. Stjepanovic, S. Baskaran, M. G. Lin, J. H. Hurley, Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol. Cell 67, 528–534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Baskaran, L. A. Carlson, G. Stjepanovic, L. N. Young, D. J. Kim, P. Grob, R. E. Stanley, E. Nogales, J. H. Hurley, Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife 3, e05115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S. Y. Lin, T. Y. Li, Q. Liu, C. Zhang, X. Li, Y. Chen, S. M. Zhang, G. Lian, Q. Liu, K. Ruan, Z. Wang, C. S. Zhang, K. Y. Chien, J. Wu, Q. Li, J. Han, S. C. Lin, GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481 (2012). [DOI] [PubMed] [Google Scholar]
- 36.E. L. Axe, S. A. Walker, M. Manifava, P. Chandra, H. L. Roderick, A. Habermann, G. Griffiths, N. T. Ktistakis, Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.N. Jaber, Z. Dou, J. S. Chen, J. Catanzaro, Y. P. Jiang, L. M. Ballou, E. Selinger, X. Ouyang, R. Z. Lin, J. Zhang, W. X. Zong, Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. U.S.A. 109, 2003–2008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.X. Cheng, X. Ma, X. Ding, L. Li, X. Jiang, Z. Shen, S. Chen, W. Liu, W. Gong, Q. Sun, Pacer mediates the function of class III PI3K and HOPS complexes in autophagosome maturation by engaging Stx17. Mol. Cell 65, 1029–1043 (2017). [DOI] [PubMed] [Google Scholar]
- 39.N. Jaber, N. Mohd-Naim, Z. Wang, J. L. DeLeon, S. Kim, H. Zhong, N. Sheshadri, Z. Dou, A. L. Edinger, G. Du, V. M. Braga, W.-X. Zong, Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J. Cell Sci. 129, 4424–4435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.G. Juhasz, J. H. Hill, Y. Yan, M. Sass, E. H. Baehrecke, J. M. Backer, T. P. Neufeld, The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181, 655–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.E. E. Johnson, J. H. Overmeyer, W. T. Gunning, W. A. Maltese, Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J. Cell Sci. 119, 1219–1232 (2006). [DOI] [PubMed] [Google Scholar]
- 42.W. Bechtel, M. Helmstadter, J. Balica, B. Hartleben, B. Kiefer, F. Hrnjic, C. Schell, O. Kretz, S. Liu, F. Geist, D. Kerjaschki, G. Walz, T. B. Huber, Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J. Am. Soc. Nephrol. 24, 727–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E. Masiero, L. Agatea, C. Mammucari, B. Blaauw, E. Loro, M. Komatsu, D. Metzger, C. Reggiani, S. Schiaffino, M. Sandri, Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515 (2009). [DOI] [PubMed] [Google Scholar]
- 44.R. C. Russell, Y. Tian, H. Yuan, H. W. Park, Y. Y. Chang, J. Kim, H. Kim, T. P. Neufeld, A. Dillin, K. L. Guan, ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. F. Egan, M. G. Chun, M. Vamos, H. Zou, J. Rong, C. J. Miller, H. J. Lou, D. Raveendra-Panickar, C. C. Yang, D. J. Sheffler, P. Teriete, J. M. Asara, B. E. Turk, N. D. Cosford, R. J. Shaw, Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.T. J. Mercer, Y. Ohashi, S. Boeing, H. B. J. Jefferies, S. De Tito, H. Flynn, S. Tremel, W. Zhang, M. Wirth, D. Frith, A. P. Snijders, R. L. Williams, S. A. Tooze, Phosphoproteomic identification of ULK substrates reveals VPS15-dependent ULK/VPS34 interplay in the regulation of autophagy. EMBO J. 40, e105985 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. M. Park, M. Seo, C. H. Jung, D. Grunwald, M. Stone, N. M. Otto, E. Toso, Y. Ahn, M. Kyba, T. J. Griffin, L. Higgins, D. H. Kim, ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H. Konno, K. Konno, G. N. Barber, Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Peng, N. Yin, S. Chhangawala, K. Xu, C. S. Leslie, M. O. Li, Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O. R. Colegio, N. Q. Chu, A. L. Szabo, T. Chu, A. M. Rhebergen, V. Jairam, N. Cyrus, C. E. Brokowski, S. C. Eisenbarth, G. M. Phillips, G. W. Cline, A. J. Phillips, R. Medzhitov, Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. Brand, K. Singer, G. E. Koehl, M. Kolitzus, G. Schoenhammer, A. Thiel, C. Matos, C. Bruss, S. Klobuch, K. Peter, M. Kastenberger, C. Bogdan, U. Schleicher, A. Mackensen, E. Ullrich, S. Fichtner-Feigl, R. Kesselring, M. Mack, U. Ritter, M. Schmid, C. Blank, K. Dettmer, P. J. Oefner, P. Hoffmann, S. Walenta, E. K. Geissler, J. Pouyssegur, A. Villunger, A. Steven, B. Seliger, S. Schreml, S. Haferkamp, E. Kohl, S. Karrer, M. Berneburg, W. Herr, W. Mueller-Klieser, K. Renner, M. Kreutz, LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 24, 657–671 (2016). [DOI] [PubMed] [Google Scholar]
- 52.R. C. Laker, J. C. Drake, R. J. Wilson, V. A. Lira, B. M. Lewellen, K. A. Ryall, C. C. Fisher, M. Zhang, J. J. Saucerman, L. J. Goodyear, M. Kundu, Z. Yan, Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat. Commun. 8, 548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. Sebastian, A. Zorzano, Self-eating for muscle fitness: Autophagy in the control of energy metabolism. Dev. Cell 54, 268–281 (2020). [DOI] [PubMed] [Google Scholar]
- 54.C. He, M. C. Bassik, V. Moresi, K. Sun, Y. Wei, Z. Zou, Z. An, J. Loh, J. Fisher, Q. Sun, S. Korsmeyer, M. Packer, H. I. May, J. A. Hill, H. W. Virgin, C. Gilpin, G. Xiao, R. Bassel-Duby, P. E. Scherer, B. Levine, Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. G. MacKenzie, D. L. Hamilton, J. T. Murray, P. M. Taylor, K. Baar, mVps34 is activated following high-resistance contractions. J. Physiol. 587, 253–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D. J. Klionsky, G. Petroni, R. K. Amaravadi, E. H. Baehrecke, A. Ballabio, P. Boya, J. M. B.-S. Pedro, K. Cadwell, F. Cecconi, A. M. K. Choi, M. E. Choi, C. T. Chu, P. Codogno, M. I. Colombo, A. M. Cuervo, V. Deretic, I. Dikic, Z. Elazar, E. L. Eskelinen, G. M. Fimia, D. A. Gewirtz, D. R. Green, M. Hansen, M. Jaattela, T. Johansen, G. Juhasz, V. Karantza, C. Kraft, G. Kroemer, N. T. Ktistakis, S. Kumar, C. Lopez-Otin, K. F. Macleod, F. Madeo, J. Martinez, A. Melendez, N. Mizushima, C. Munz, J. M. Penninger, R. M. Perera, M. Piacentini, F. Reggiori, D. C. Rubinsztein, K. M. Ryan, J. Sadoshima, L. Santambrogio, L. Scorrano, H. U. Simon, A. K. Simon, A. Simonsen, A. Stolz, N. Tavernarakis, S. A. Tooze, T. Yoshimori, J. Yuan, Z. Yue, Q. Zhong, L. Galluzzi, F. Pietrocola, Autophagy in major human diseases. EMBO J 40, e108863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D. R. Miller, A. Thorburn, Autophagy and organelle homeostasis in cancer. Dev. Cell 56, 906–918 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Yang, G. Herter-Sprie, H. Zhang, E. Y. Lin, D. Biancur, X. Wang, J. Deng, J. Hai, S. Yang, K. K. Wong, A. C. Kimmelman, Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8, 276–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. C. Kimmelman, E. White, Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.B. Wang, B. A. Maxwell, J. H. Joo, Y. Gwon, J. Messing, A. Mishra, T. I. Shaw, A. L. Ward, H. Quan, S. M. Sakurada, S. M. Pruett-Miller, T. Bertorini, P. Vogel, H. J. Kim, J. Peng, J. P. Taylor, M. Kundu, ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol. Cell 74, 742–757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1