Abstract
Tracking and eradicating Staphylococcus aureus in the periprosthetic microenvironment are critical for preventing periprosthetic joint infection (PJI), yet effective strategies remain elusive. Here, we report an implant nanoparticle coating that locoregionally yields bactericidal super chimeric antigen receptor macrophages (CAR-MΦs) to prevent PJI. We demonstrate that the plasmid-laden nanoparticle from the coating can introduce S. aureus–targeted CAR genes and caspase-11 short hairpin RNA (CASP11 shRNA) into macrophage nuclei to generate super CAR-MΦs in mouse models. CASP11 shRNA allowed mitochondria to be recruited around phagosomes containing phagocytosed bacteria to deliver mitochondria-generated bactericidal reactive oxygen species. These super CAR-MΦs targeted and eradicated S. aureus and conferred robust bactericidal immunologic activity at the bone-implant interface. Furthermore, the coating biodegradability precisely matched the bone regeneration process, achieving satisfactory osteogenesis. Overall, our work establishes a locoregional treatment strategy for priming macrophage-specific bactericidal immunity with broad application in patients suffering from multidrug-resistant bacterial infection.
Priming macrophage-specific bactericidal immunity prevents periprosthetic joint infection and expedites osseointegration.
INTRODUCTION
Periprosthetic joint infection (PJI), one of the most devastating complications after artificial arthroplasty, represents a substantial clinical challenge (1, 2). Management of PJI requires complex treatment strategies, including multiple surgical revisions and long-term antimicrobial treatment (3, 4). However, little clinical progress has been made in the prevention and eradication of PJI. Although many organisms have been linked to PJI, Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), remains the most prevalent and destructive causative pathogen (5–7). Because implant-infecting S. aureus can elude both innate and adaptive host defenses as well as biocides and antibiotic chemotherapies (7, 8), tracking and eradicating S. aureus in the implant microenvironment are critical for preventing PJI. However, effective strategies remain elusive.
Macrophages (MΦs) are critical to the innate immune response to S. aureus and commence their antimicrobial activity after internalizing the pathogen (9, 10). However, this immune defense mechanism is not commonly activated in the presence of postoperative infections, which allows infections to persist (11). Furthermore, the implant itself triggers a local tissue response, leading to a niche of immune depression that suppresses the phagocytic and bactericidal activities of MΦs (12, 13). In addition, S. aureus has evolved immune escape mechanisms for thwarting phagolysosomal killing, thus permitting their intracellular survival and allowing for recurrent infections (14–16). Given the unique effector functions of MΦs and their chemotaxis toward bacteria (17, 18), we hypothesized that genetically engineering the periprosthetic MΦs to express S. aureus–specific chimeric antigen receptors (CARs) could redirect the phagocytic and bactericidal activity of these cells against S. aureus, thereby preventing bacterial immune evasion via a host-directed strategy and subsequently eliminating PJI.
S. aureus surface protein A (SasA), an essential virulence factor for the survival of S. aureus during invasive infections, is prevalent and highly conserved among clinical S. aureus strains (19, 20). SasA promotes adhesion to host cells and tissue, contributes to biofilm formation by stimulating attachment to the host or implants, helps bacteria evade host innate immune responses, and accelerates the progression of infection (21–23). SasA is considered a potential target for immunotherapy against S. aureus infections. During S. aureus infection, MΦs are not effective at eradicating internalized S. aureus (10, 24). This phenomenon is largely due to the hijacking of MΦ mitochondria via the caspase-11 (CASP11) activation triggered by intracellular S. aureus, which decouples the mitochondrion-derived reactive oxygen species (mROS) defense mechanism, thereby allowing S. aureus within MΦs to resist succumbing to mROS-mediated clearance (25, 26). Together, these features of S. aureus infection, i.e., phagocytosis interfere and thwarted phagolysosomal killing, present an opportunity for CAR-MΦ–mediated immunotherapy to be used in the treatment of PJI for overcoming bacterial immune evasion and eradicating the infection.
In situ CAR-MΦ engineering at the peri-implant represents a rational therapeutic approach as an alternative to the current clinical-scale manufacturing of CAR cells because it avoids the need for an assortment of elaborate protocols to isolate, genetically modify, and selectively expand the redirected cells before infusing them back into patients (27–30). Here, we investigated a nanoparticle coating for the locoregional in situ induction of super CAR-MΦ at the bone-implant interface, which simplified CAR-MΦ preparation and minimized the systemic adverse effects of MΦ reprogramming (Fig. 1). We simultaneously decreased the expression of CASP11 in CAR-MΦs by using short hairpin RNA (shRNA) and found that this treatment notably enhanced the phagolysosomal killing effect of SasA-specific CAR-MΦs. Together, our results suggest that this nanoparticle coating enabled peri-implant MΦs to target and eradicate S. aureus in a seek-and-destroy fashion while promoting rapid bone osseointegration at the bone-implant interfaces. These results present a potential approach for the prevention and treatment of PJI that warrants further assessment in clinical trials.
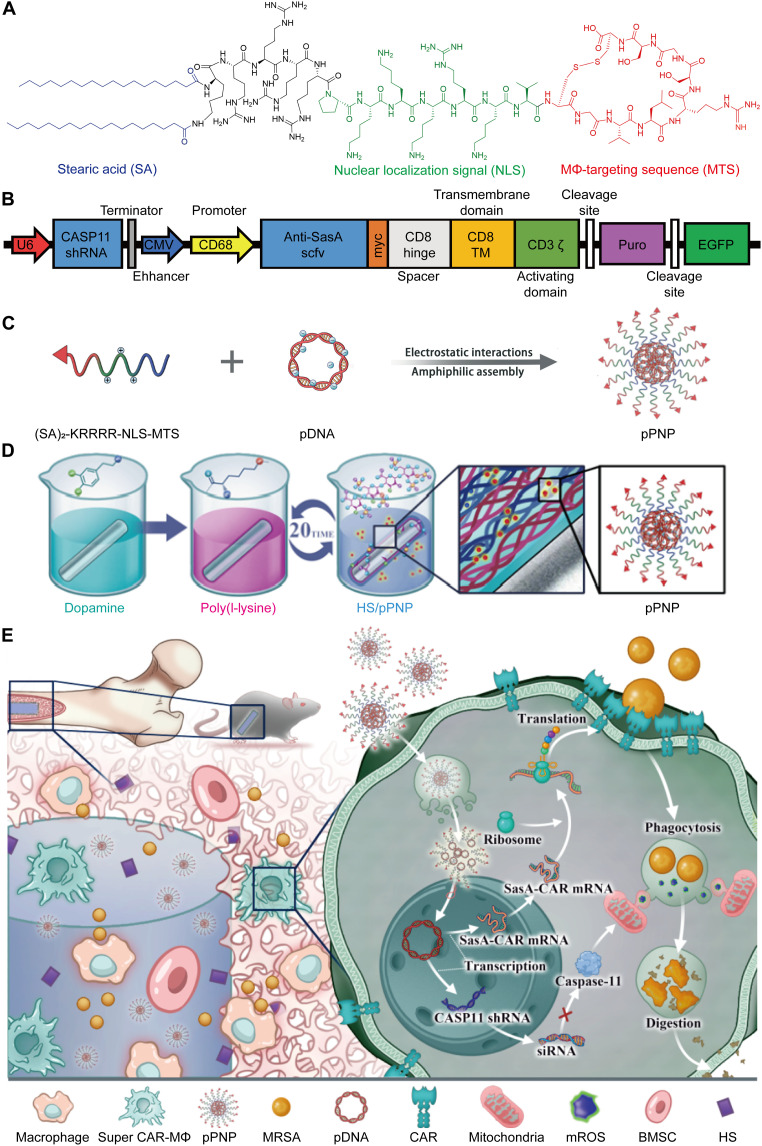
Fig. 1. Schematic illustration of the locoregional generation of S. aureus–specific super CAR-MΦs at the bone-implant interface for preventing PJI.
(A) Chemical structure of the peptide-SA monomer. (B) Diagram of anti-SasA CAR and CASP11 short hairpin RNA (shRNA) structure in plasmid DNA (pDNA). CMV, cytomegalovirus; EGFP, enhanced green fluorescent protein. (C) Schematic illustration of the preparation of pDNA-laden peptide nanoparticle (pPNP). (D) Schematic illustration of the pPNP coating on an implant (Ti-pPNP). HS, heparan sulfate. (E) pPNP coating generates S. aureus–specific super CAR-MΦs for tracking and eradicating S. aureus infection; this approach orchestrates periprosthetic anti-infection and osseointegration in an arthroplasty mouse model. BMSC, bone mesenchymal stem cell; siRNA, small interfering RNA.
RESULTS AND DISCUSSION
Synthesis and characterization of the nanoparticle coating
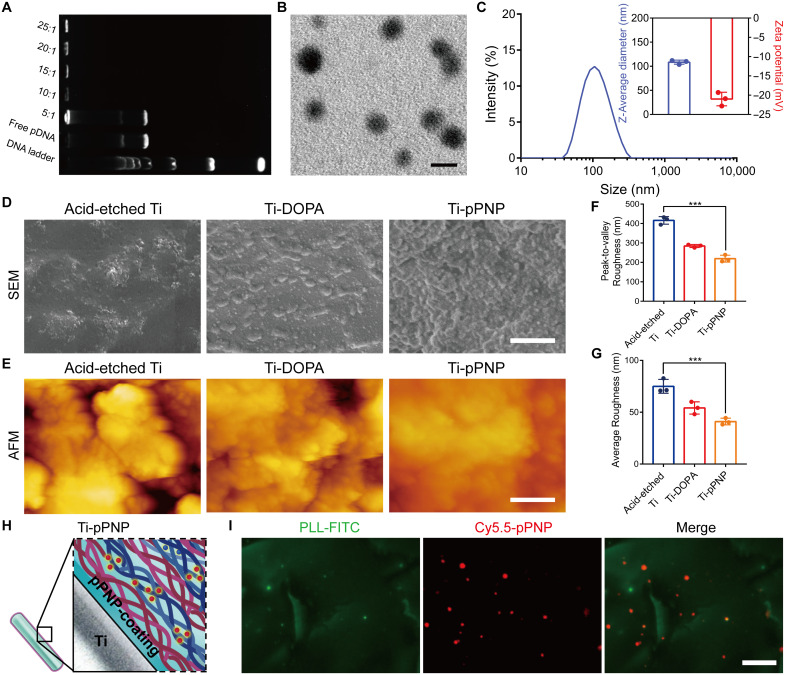
To achieve the goal of MΦ targeting and efficient nuclear-targeted gene delivery, the peptide nanoparticle (PNP) was designed and synthesized. Using peptides containing a MΦ-targeting sequence (MTS) (31) and a nuclear localization signal (NLS) (32) as the hydrophilic moiety and stearic acid (SA) as the hydrophobic domain, we first synthesized an amphiphilic peptide-SA monomer, (SA)2-KRRRR-NLS-MTS (Fig. 1A). The chemical structure was confirmed by mass spectra (fig. S1). The amphiphilic peptide-SA monomers could readily self-assemble and encapsulate plasmid DNA (pDNA) (Fig. 1B and fig. S2) to form core-shell–structured PNP (Fig. 1C) in an aqueous solution for sequential targeted delivery. As shown in Fig. 2A, pDNA was completely loaded by the PNP when the weight ratio of peptide-SA monomers to the pDNA was greater than 10:1. This result indicates that there is an increase in the interaction between pDNA and peptide-SA with an increased amount of peptide-SA monomers. The pDNA-laden PNP (pPNP) was revealed to have a well-defined spherical structure under transmission electron microscopy (TEM) (Fig. 2B), with an average diameter of 108.5 nm (Fig. 2C). The stability of pPNPs in deoxyribonuclease I (DNase I) or fetal bovine serum was analyzed by gel retardation assay, and the results show that the pDNA was stable in DNase I or fetal bovine serum after it was condensed with PNPs (fig. S3A). We further evaluated the stability of pDNA in a PNP-encapsulated manner on day 14. As shown in fig. S3B, pDNA in PNP was detectable by day 14, confirming that PNP could protect pDNA from degradation under physiological conditions.
Fig. 2. Design and characterization of the pPNP coating.
(A) Gel retardation assay of the pPNP. (B) Transmission electron microscopy (TEM) images of PNPs with a weight ratio of peptide-SA:pDNA of 10:1. Scale bar, 100 nm. (C) Zeta potential and size distributions of PNPs with a weight ratio of SA monomers:pDNA of 10:1. (D) Surface morphology and cross-sectional view of different titanium-based surfaces investigated by scanning electron microscopy (SEM). Scale bar, 1 μm. (E) Three-dimensional reconstructed surface roughness images of different titanium-based surfaces as investigated by atomic force microscopy (AFM). Scale bar, 1 μm. (F and G) Quantitative measurement of surface roughness parameters of different titanium-based surfaces. (H) Schematic illustration of the coating structure. (I) Representative fluorescent images of coating, in which poly-l-lysine (PLL) was labeled with fluorescein isothiocyanate (FITC) and pPNP with Cy5.5 (Cy5.5-labeled pPNP). Scale bar, 25 μm. Data are presented as means ± SD. n = 3 independent experiments per group, ***P < 0.001.
We next constructed a self-assembling coating on titanium (Ti) implants for locoregionally immobilizing therapeutics for periprosthetic infection at the bone-implant interface (Fig. 1D). In the first step, the surface of the Ti implants was roughened using an acid-etching method to improve the adhesion efficiency of dopamine (DOPA) on the Ti surface. The catechol and quinone groups in the DOPA coating can react with amine-containing polymers via a Michael-type addition or Schiff base reaction (33, 34). Therefore, in the second step, we used the cationic and amine-containing polymer poly-l-lysine (PLL) to initiate the layer-by-layer process. Heparan sulfate (HS)–containing pPNP was selected as a negatively charged polyelectrolyte with which to form nanolayers in the third step. Layers of positively charged PLL were alternated with layers of negatively charged HS containing pPNP to form the pPNP coating. As shown in Fig. 2D, we first used scanning electron microscopy (SEM) to show the surface morphology of the prepared pPNP coating. The SEM image confirmed that the pPNP coating was successfully self-assembled. Then, we used atomic force microscopy (AFM), revealing the change in the surface topography of different surfaces, from the relatively coarse surface of the acid-etched Ti implant (Ti) to the smooth ones after coating (Fig. 2, E to G). Moreover, our contact angle measurements indicate an increase in the hydrophilicity of the pPNP-coated Ti implant surface compared with the native Ti implant surface (fig. S4), which promotes biological interactions between the implant and bone tissue. X-ray photoelectron spectroscopy (XPS) showed area-specific changes in the N 1s, C 1s, and S 2p peaks, further demonstrating that the pPNP coating was successfully fabricated (fig. S5). As demonstrated by confocal imaging, the Cy5.5 signals from pPNP exhibited a uniform distribution pattern in the coating, further indicating the encapsulation of pPNP in the coating (Fig. 2, H and I). We next tested the release profile of HS and pDNA from the pPNP coating. As shown in fig. S6, HS and pPNP each exhibited an obvious sustained release from the coating over 14 days. Around 11.7% of pDNA was released from the pPNPs in the coating, and 75.6% of pDNA was released in a PNP-encapsulated manner. These results collectively indicate that the pPNP coating was successfully constructed and had the potential to mediate gene delivery.
pPNP-mediated SasA-CAR programming in MΦs
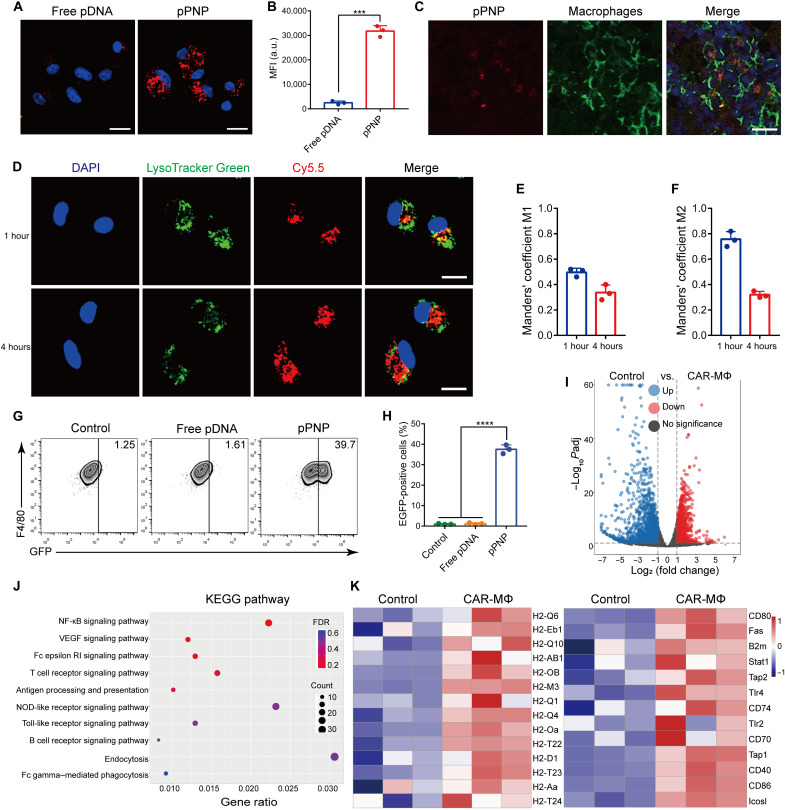
Multiple types of MΦ cells, including bone marrow–derived MΦs (BMDMs) and RAW 264.7 cells, were used to test the efficiency of pPNP-mediated gene delivery, which allows MΦs to express corresponding domains, including the CD8 stalk and transmembrane domains, the CD3ζ endodomain, and the anti-SasA scFv (single-chain fragment variable) domain (fig. S7). In both BMDMs and RAW 264.7 cells, substantial enhancement was achieved in the in vitro cellular internalization of pDNA after the pDNA was encapsulated in PNP (Fig. 3A and fig. S8A). These results were further confirmed with flow cytometry, indicating that the pPNP efficiently delivered the genes into MΦs (Fig. 3B and fig. S8B). Consistently, immunofluorescence staining of the bone tissues around a pPNP-coated bone implant depicted well-dispersed pPNP that also colocalized well with MΦs (Fig. 3C). Together, these data indicate that pPNPs enable MΦ-targeted delivery of pDNA at the bone-implant interface. When the incubation time was increased, the resulting wide distribution of Cy5.5-labeled pDNA in the cytoplasm of BMDMs indicates that the pPNP escaped the lysosome and entered the cytosol (Fig. 3D). These observations were confirmed by quantitative analyses of the colocalization of pPNP with endo/lysosomes in confocal fluorescence images using Manders’ coefficients M1 and M2 (35). Manders’ coefficient M1 denotes the fraction of Cy5.5-labeled pDNA overlapping with LysoTracker Green, and M2 denotes the fraction of LysoTracker Green overlapping with Cy5.5-labeled pDNA. These coefficients should each be close to 1 if the LysoTracker Green and Cy5.5-labeled pDNA are highly colocalized. As shown in Fig. 3 (E and F), M1 was much less than 1 at all time points (particularly 4 hours), and the M2 data reveal that few endo/lysosomes contained pDNA at 4 hours, indicating that the pPNP was able to accomplish endo/lysosomal escape, thus allowing for the cytosolic delivery of pDNA.
Fig. 3. pPNP-mediated SasA-CAR programming in MΦs.
(A) Confocal images of BMDMs treated with free pDNA or pPNP. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars, 20 μm. (B) Cellular uptake of free pDNA or pPNP by BMDMs as measured through flow cytometry analysis. MFI, mean fluorescence intensity; a.u., arbitrary units. (C) Immunofluorescence staining showing that pPNPs (red) colocalized with F4/80+ MΦs (green). Scale bar, 25 μm. (D) Typical confocal images of BMDMs incubated with pPNPs for 1 or 4 hours at 37°C. The cell nuclei were stained using DAPI (blue), the endo/lysosomes were stained using LysoTracker Green (green), and pDNAs were labeled with Cy5.5 (red). Scale bars, 20 μm. (E and F) Quantitative analysis of the colocalization of Cy5.5-labeled pDNA with endo/lysosomes labeled with LysoTracker Green. (G and H) Percentage of EGFP-positive BMDMs treated with free pDNA or pPNP. (I) Volcano plot of genes differentially expressed in BMDMs versus CAR-MΦs. (J) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the identified differentially expressed genes. NF-κB, nuclear factor κB; VEGF, vascular endothelial growth factor. (K) Heatmap of differentially expressed antigen-presentation genes and inflammatory genes between BMDMs and pPNP-transduced CAR-MΦs. Data are presented as means ± SD. n = 3 independent experiments per group, ***P < 0.001.
We next investigated whether pPNP-mediated gene delivery could induce SasA-CAR expression in MΦs. After incubation with the pPNP, enhanced green fluorescent protein (EGFP) abundance in MΦs was measured using confocal microscopy and flow cytometry analysis. As shown in fig. S9, among the two groups (the pPNP group and the free pDNA group), the pPNP group had the highest percentage of EGFP-positive cells. Similarly, in the pPNP group, the percentages of EGFP-positive BMDMs and RAW 264.7 cells were 37.53 ± 2.15% and 40.77 ± 2.94%, respectively, which are considerably higher than those of the free pDNA–treated cells (Fig. 3, G and H, and fig. S10). We further tested the specificity of the CD68 promoter that drives the specific expression of the CAR gene in MΦs. As shown in fig. S11, the CD68 promoter enabled the CAR gene to be transcribed in only MΦs, thereby avoiding off-target effects. These data provide direct evidence that the functional CAR was expressed on MΦs after their treatment with pPNP.
To uncover the immune profiles of CAR engineered-MΦs, we next performed a transcriptomics analysis of MΦs after pPNP-mediated treatment. The volcano plot of differentially expressed genes in MΦs treated with pPNPs shows an obvious separation of gene expression (Fig. 3I). A Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed the comprehensive immune reaction of BMDMs, reflecting the main activated signaling pathways, such as endocytosis, nuclear factor κB (NF-κB) signaling pathway, and antigen processing and presentation (Fig. 3J). Furthermore, the expression changes of antigen presentation and inflammatory factors between BMDMs and CAR-MΦs were singled out and replotted in Fig. 3K. From a heatmap analysis of the secreted factors, we found that many antigen presentation and inflammatory genes, including H2-Oa, H2-Q4, H2-M3, CD80, Tap2, and Tlr4, were up-regulated in the pPNP group. The H2 gene, which is located on the 17th chromosome of mice and is related to the major histocompatibility complex and antigen presentation of mice (36), was obviously up-regulated. In addition, the overexpressed CD80, Tap2, and Tlr4 were attributed to the pro-inflammatory signals. We further validated the induction of a pro-inflammatory M1 phenotype using immunofluorescence analysis and enzyme-linked immunosorbent assay (ELISA), which demonstrated similar results, such as increased secretion of the M1 markers CD80, interleukin-1β (IL-1β), and tumor necrosis factor–α (TNF-α) and reduced expression of the M2 markers arginase 1 (Arg-1) and IL-10 in pPNP-treated MΦs (fig. S12). These results indicated that pPNP-mediated CAR engineering in MΦs could enhance polarization into the pro-inflammatory M1 phenotype. In total, pPNP treatment was beneficial to reinvigorate MΦ activity and enhance the expression of pro-inflammatory, antibacterial, and antigen presentation genes.
Bactericidal activity of super CAR-MΦ in vitro
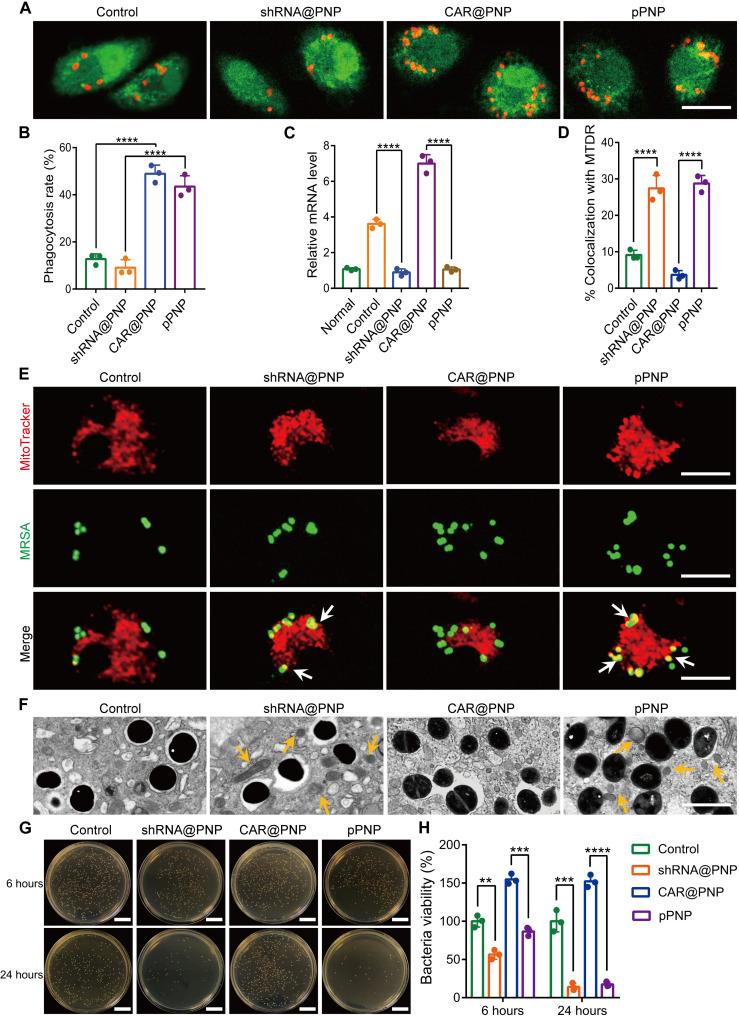
MΦs are crucial for host defense against MRSA, whereas MRSA frequently evades engulfment by these cells and also blocks their intracellular killing (14, 15). To test whether creating super CAR-MΦs would be beneficial for bacterial clearance, we evaluated the phagocytosis and phagolysosomal killing of MRSA by different treatment groups of BMDMs. We first evaluated the CAR-mediated phagocytosis of MΦs through confocal microscopy and flow cytometry. The CAR modification of MΦs did increase their robust phagocytosis of MRSA (Fig. 4A). The percentage of BMDMs that phagocytized MRSA in the pPNP treatment group was 5.2-fold higher than that in the control group (Fig. 4B and fig. S13). These data suggest that engineered CAR MΦs had elevated phagocytotic activity against MRSA.
Fig. 4. Bactericidal activity of super CAR-MΦs in vitro.
(A) Representative images of the phagocytosis of MRSA by MΦs treated with each formulation. Scale bar, 10 μm. (B) Rate of phagocytosis by MΦs after 4 hours of incubation with MRSA at 37°C. (C) Relative mRNA levels of CASP11 in different treatment groups as verified by quantitative reverse transcription polymerase chain reaction. (D) Quantification of MRSA colocalized with MitoTracker Deep Red. (E) Typical confocal images of MRSA-infected BMDMs in the indicated treatment groups at 4 hours after infection. White arrows indicate the colocalization of MRSA with MitoTracker. Scale bars, 10 μm. (F) Qualitative TEM images of MRSA-infected CAR-MΦs at 4 hours after infection. Yellow arrows indicate mitochondria. Scale bar, 10 μm. (G and H) Representative images (G) and quantitative analysis (H) of the intracellular survival of MRSA in BMDMs (multiplicity of infection = 5:1) in the indicated treatment groups. Scale bars, 2 cm. Data are presented as means ± SD. n = 3 independent experiments per group, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Intracellular MRSA-triggered CASP11 activation actively prevents the recruitment of mitochondria to the vicinity of the phagolysosomes MRSA resides in, allowing these bacteria to avoid being destroyed by intracellular killing (37). To determine whether CASP11 shRNA effectively reduced the CASP11 expression in MRSA-infected MΦs, we tested the mRNA expression level of CASP11. CASP11-specific shRNA led to a notable repression of the mRNA of its target gene (Fig. 4C). Subsequently, to investigate whether CASP11 deficiency allows an increased association of MRSA-containing vacuoles with mitochondria, we analyzed the colocalization events between MRSA and mitochondria stained with MitoTracker Deep Red in different treatment groups. As indicated in Fig. 4 (D and E), the CASP11 inhibition groups displayed a higher percentage of bacteria colocalized with MitoTracker compared with the corresponding groups without CASP11 inhibition. Furthermore, TEM revealed that more mitochondria were in proximity to bacteria inside the BMDMs in the CASP11-inhibited groups compared with those in the corresponding groups without CASP11 inhibition (Fig. 4F). Evidence suggests that the bactericidal activity of mROS directly contributes to killing phagocytosed bacteria (37, 38). Thus, we measured mitochondrial superoxide production using the MitoSOX Red reagent (fig. S14). Compared with noninfected control cells, the superoxide levels were suppressed in normal MΦs infected with MRSA. MΦs lacking CASP11 exhibited higher MitoSOX fluorescence in response to MRSA than did cells without CASP11 inhibition. These data indicate that in pPNP-engineered super CAR-MΦs, the mitochondria are recruited to phagosomes containing phagocytosed bacteria to actively deliver oxidative phosphorylation-generated bactericidal mROS.
Next, to evaluate the bactericidal activity of the super CAR-MΦs, we quantified the intracellular survival of MRSA in BMDMs treated by control, shRNA@PNP, CAR@PNP, or pPNP. Among these groups, the pPNP group showed the strongest bactericidal activity at all tested end time points, with a percentage of inhibition of 83.6% (Fig. 4, G and H). The bactericidal activity in the shRNA@PNP was markedly reduced (Fig. 4H), suggesting the importance of simultaneously decreasing the expression of CASP11 by shRNA. These results together demonstrate the potent bactericidal activity of super CAR-MΦs.
Anti-infection and immunomodulation effects of pPNP coating in vivo
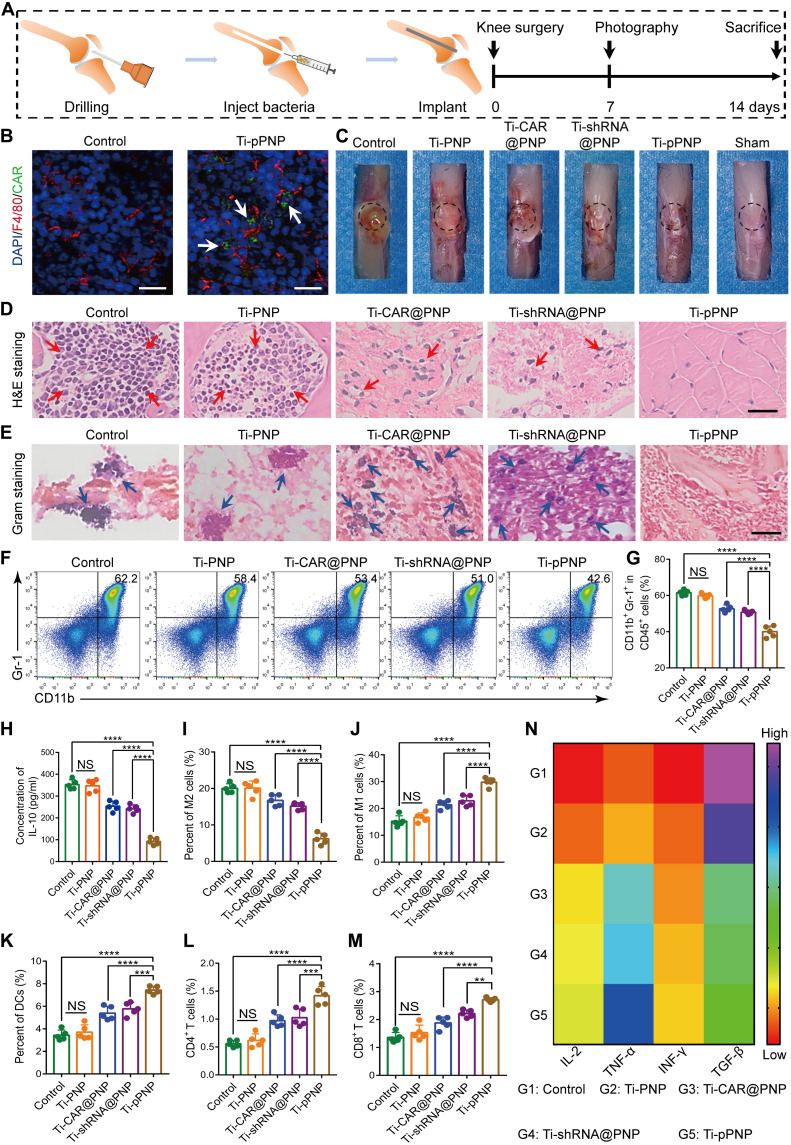
Given the observed potent in vitro bactericidal activity of super CAR-MΦs, we tested their bactericidal effects in a MRSA-infected orthopedic implant mouse model (Fig. 5A). The CAR expression in bone tissues at the bone-implant interface was determined with EGFP. As shown in Fig. 5B, a large number of MΦs were gathered in the infected bone, and some of them expressed EGFP. To evaluate the bactericidal activity of Ti-pPNP, the implant was harvested 7 days after surgery. We observed macroscopic inflammatory changes in infected murine joints, including edema that decreased with the lower MRSA bioburden in the Ti-pPNP group (Fig. 5C). The number of MRSA colonies in the Ti-pPNP group was significantly less than that in the control, Ti-PNP, Ti-shRNA@PNP, or Ti-CAR@PNP groups (fig. S15A). The corresponding antibacterial efficiency of Ti-pPNP is more than 90%; thus, it has excellent bactericidal activity in vivo (fig. S15B). In addition, hematoxylin and eosin (H&E) and Gram staining were used to investigate the inflammatory reaction and the residual bacteria, respectively, in the bone tissue surrounding the implant. As displayed in Fig. 5D (red arrows), typical infections of bone tissue were observed through H&E staining in the control, Ti-PNP, Ti-shRNA@PNP, Ti-CAR@PNP, and Ti-pPNP groups. In these groups, we observed large numbers of inflammatory cells infiltrated into the tissues, indicating PJI. Many scattered bacteria were found in the Gram-stained sections from the control and Ti-PNP groups (Fig. 5E, blue arrows). In contrast, lower numbers of inflammatory cells and residual bacteria were found in the Ti-pPNP–treated group. Collectively, these data indicate that CAR and CASP11 shRNA additively exert robust antibacterial effects.
Fig. 5. pPNP coating-induced anti-infection and immunomodulation effects in vivo.
(A) Schematic illustration of the experimental design. (B) EGFP-positive MΦs (white arrows) at the bone-implant interfaces of mice after their treatment with pPNP. Blue, DAPI-stained cell nuclei; red, F4/80+ MΦs. Scale bars, 25 μm. (C) Representative mouse knee joint images at 7 days after implantation. (D) Hematoxylin and eosin (H&E) staining of the bone tissues surrounding the implants. Red arrows indicate the infiltration of inflammatory cells. Scale bar, 100 μm. (E) Gram staining of the bone tissues surrounding the implants. Blue arrows indicate scattered bacteria. Scale bar, 50 μm. (F and G) Representative flow cytometry plots (F) and quantitative analysis (G) of CD11b+Gr1+ myeloid-derived suppressor cells in MRSA-infected bone tissues at day 14 after infection. (H) IL-10 levels in implant-associated tissue, as measured by ELISA. (I and J) Representative flow cytometry quantitative analysis of M2-like MΦs (F4/80+CD206+) (I) and M1-like MΦs (F4/80+CD80+) (J) in the implant-associated tissues of mice in the indicated treatment groups. (K) Mature dendritic cells (DCs) in the draining lymph nodes. (L and M) Representative flow cytometry quantitative analysis of the percentage of CD3+CD4+ (L) or CD3+CD8+ (M) T cells in the implant-associated tissues of mice in the indicated treatment groups. (N) Heatmap of the IL-2, TNF-α, IFN-γ, and TGF-β expression profiles in implant-associated tissues. Data are presented as means ± SD. n = 5 mice per group, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
S. aureus biofilm formation allows the bacteria to avoid immune recognition and thereby limit the establishment of effective antibacterial immunologic responses (39, 40). To clarify whether the Ti-pPNP treatment could reverse pathogen-induced immunosuppression, we first evaluated the levels of myeloid-derived suppressor cells (MDSCs) around the infected region. As shown in Fig. 5 (F and G), the frequency of MDSCs in implant-associated tissue was significantly lower for the Ti-pPNP group at 14 days after infection. This coincided with a lower bacterial burden and implies the establishment of antibacterial immunologic responses during pathogen clearance. IL-10 production by MDSCs is critical for the immune-suppressive properties of PJI (12). As expected, the levels of IL-10 secretion were reduced in animals after treatment with Ti-pPNP (Fig. 5H). In addition, we found that the Ti-pPNP–redirected MΦ phagocytic activity against S. aureus elicited a higher level of antibacterial immunologic responses, regulating the pro-inflammatory polarization of MΦs (Fig. 5, I and J, and fig. S16A), promoting the maturation of dendritic cells (Fig. 5K and fig. S16B), and activating CD4+/CD8+ T cells (Fig. 5, L and M, and fig. S16C) against the residual S. aureus. Moreover, as presented in Fig. 5N, compared with other groups, the levels of TNF-α, IL-2, and interferon-γ (IFN-γ) production were markedly higher in the Ti-pPNP group, whereas the expression level of transforming growth factor–β (TGF-β) was lower, demonstrating that the Ti-pPNP fostered an antibacterial immunologic microenvironment around the implant. These results collectively suggest that the Ti-pPNP treatment reversed the infection-induced immunosuppression milieu, thereby conferring robust bactericidal immunologic activity at the bone-implant interface in a host-directed strategy.
Promoted osteogenesis of pPNP coating
Implant surfaces are carefully designed for optimal biocompatibility and osseointegration. As shown in fig. S17A, the spread of bone mesenchymal stem cells (BMSCs) adhered on the surface in the Ti-pPNP group was comparable to those in the control and Ti-DOPA groups. Furthermore, the cell viability of BMSCs in these samples at day 7 after treatment showed minimal difference, which proves that the pPNP coating exhibited an excellent cytocompatibility with BMSCs after long-term culture (fig. S17B). We further tested the expression of the osteogenic-related genes alkaline phosphatase (ALP), runt-related transcription factor 2 (RUNX2), osteocalcin (OCN), and type I collagen (COL-I) after 14 days. From the quantitative reverse transcription polymerase chain reaction (qRT-PCR) results (fig. S18A), the expressions of ALP, RUNX2, OCN, and COL-I in the Ti-pPNP group maintained the highest levels among all the samples. The OCN and osteopontin (OPN) immunofluorescence staining results also confirm that the most efficient enhancement of osteogenic differentiation occurred in the Ti-pPNP–derived BMSCs (fig. S18B). We further determined the primary safety of pPNP by a cytotoxicity assay in vitro. As shown in fig. S19, no significant changes in cell viability were observed in pPNP-treated MΦs. Furthermore, we performed blood analyses and histological analyses of the treated mice to evaluate the potential systemic toxicity of degradation byproducts. Throughout the study period of up to 4 weeks, the blood (fig. S20A) and histological (fig. S20B) analyses of animals with the implanted Ti-pPNP yielded results comparable to those of healthy animals, lacking any marked signs of systemic toxicity. These results suggest that our pPNP coating had excellent biocompatibility.
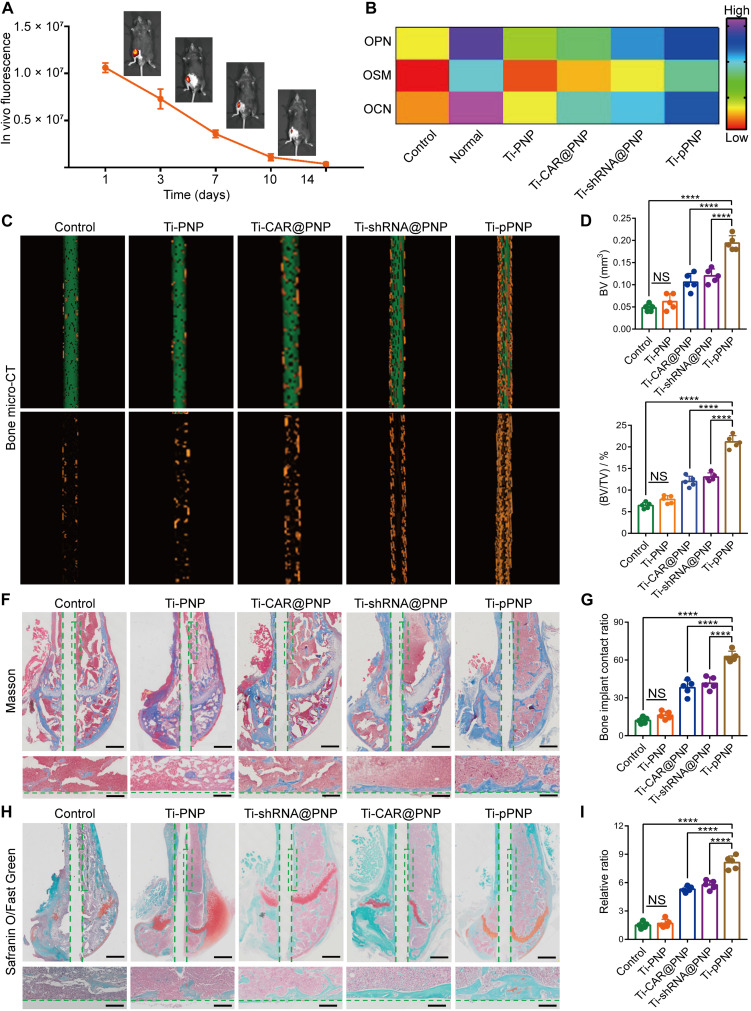
An ideal bone implant should precisely match the process of bone regeneration and achieve satisfactory osteogenesis at the bone-implant interfaces (41, 42). Thus, we investigated the in vivo degradability of the pPNP coating in a PJI mouse model. The in vivo biodegradability of the pPNP coating was assessed using in vivo imaging systems (IVIS) and labeling the pPNP coating with Cy5.5. After implantation, we found a steady decrease in the fluorescence signal over time, down to no detectable level at day 14 after implantation (Fig. 6A). The results indicate that the pPNP coating is largely resorbed after 2 weeks and will not compromise osseointegration. We next investigated the expression of osteogenic factors using ELISA analyses. The secretion of osteogenic-related cytokines, including OPN, oncostatin M (OSM), and OCN, was elevated after treatment with Ti-pPNP, demonstrating the strong potential of this approach to inducing osteogenesis (Fig. 6B). Next, a micro–computed tomography (micro-CT) analysis was used to determine the level of neonatal bone formation at the interface of bone and implant. Compared with other groups, the Ti-pPNP group was observed to have the highest amount of neonatal bone formation (Fig. 6C, yellow highlight). A quantitative analysis further confirmed this result. Tissues in the Ti-pPNP group had an elevated bone volume (BV) (Fig. 6D) and the highest percentage of BV to tissue volume (BV/TV) (Fig. 6E). The BV/TV value in the Ti-pPNP group (21.15% ± 1.47%) was 1.76- and 1.62-fold higher as compared with that in the Ti-CAR@PNP (12% ± 1.23%) and Ti-shRNA@PNP (13.04% ± 0.94%) groups, respectively. Moreover, quantitative analyses of bone mineral density (BMD) demonstrated that the mean BMD in pPNP-treated mice was markedly increased compared with that in mice from the other groups (fig. S21). The newly formed bone was further assessed by using Masson staining (Fig. 6, F and G) and Safranin O/Fast Green staining (Fig. 6, H and I), and similar results were obtained. Collectively, the above data demonstrate that the devised coating clearly improved bone-implant integration.
Fig. 6. Promoted osteogenesis of pPNP coating.
(A) In vivo biodegradation of pPNP coating labeled with Cy5.5, visualized using in vivo fluorescence imaging. The fluorescence scale shown was obtained directly from the in vivo imaging systems imaging scanner. (B) Heatmap of the OPN, OSM (oncostatin M), and OCN (osteocalcin) expression profiles in bone tissues. (C) Micro–computed tomography (micro-CT) images of implants in mouse femurs and three-dimensional reconstructed images showing the neonatal bone generation surrounding the implants (green, implant; yellow, bone tissue). (D and E) Quantitative statistics of the bone volume (BV) (D) and the percentage of BV to tissue volume (%, BV/TV) (E) calculated from the micro-CT data. (F) Representative Masson staining (blue, bone tissue) for the neonatal bone surrounding the implants. Scale bars, 200 and 50 μm. (G) Bone area ratios calculated from Masson-stained samples. The bone-implant contact ratio was calculated by comparing the accumulated length of the edge, where the implant contacts the bone tissue, to the implant circumference based on the images of Masson staining analysis. (H) Representative Safranin O/Fast Green staining (green, bone tissue) for the neonatal bone surrounding the implants. Scale bars, 200 and 50 μm. (I) Histomorphometric measurements of osteogenesis. The relative ratio was defined as the percentage of osteogenesis with a region extending 20 μm from the implant surface. Data are presented as means ± SD. n = 5 mice per group, ****P < 0.0001.
Effects of pPNP coating on anti-hematogenous infection
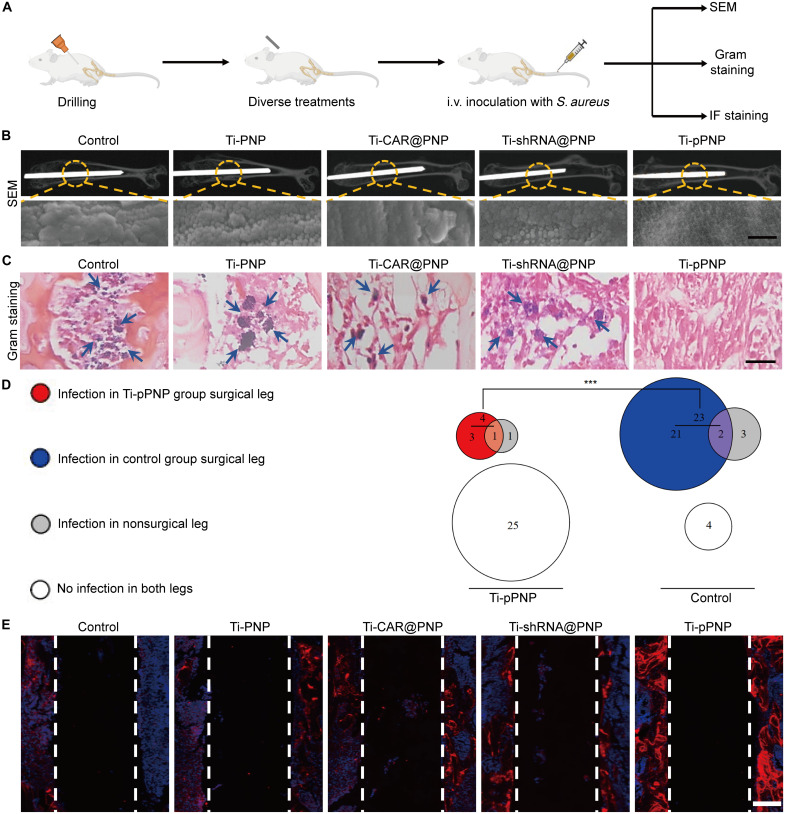
In orthopedic prosthetic infections, hematogenous implant-related infections are an important clinical problem because bacteria can spread from the bloodstream to a previously well-functioning implant and result in infectious complications and even orthopedic surgery failure (43, 44). To evaluate the broad applicability of the pPNP coating, we established a S. aureus hematogenous implant infection mouse model as previously reported (Fig. 7A) (45). Briefly, we first placed the designed implant into the right femurs of mice using an aseptic surgical technique. The mice were then intravenously infected with S. aureus at 1 week after surgery, and the infection was monitored on day 14 after inoculation. We performed SEM to evaluate biofilm formation on the implants at 2 weeks after injection. As seen in Fig. 7B, the implants from mice in the Ti-pPNP group had only infrequent coccoid bacteria and lacked appreciable biofilm formation, whereas the implants from the mice in the other groups had dense biofilm aggregates. In addition, Gram staining for the evaluation of infection revealed no active MRSA in the Ti-pPNP group (Fig. 7C). After removing the mouse femur containing the implant and homogenizing it, we found that the Ti-pPNP group had nearly 83% fewer mice with an infection in the surgical legs compared with the control group (4 versus 23). Moreover, the number of mice with no detectable colony-forming units (CFUs) of bacteria in both the surgical and nonsurgical legs in the Ti-pPNP group was sixfold higher than that in the control group (25 versus 4) (Fig. 7D), suggesting that the Ti-pPNP group had a lower overall propensity of infection. Because of the major concern of osseointegration on artificial implants, the details of the newly formed bone tissue at the bone-implant interface were further studied by OCN immunofluorescence staining (Fig. 7E). We clearly observed a large area of bone matrix (red stain) in the Ti-pPNP group. As shown in fig. S22, compared with other treatment groups, the levels of TNF-α, IL-2, and IFN-γ production were markedly higher in the Ti-pPNP group, whereas the expression level of TGF-β was lower, demonstrating that the Ti-pPNP fostered an antibacterial immunologic microenvironment around the implant. Moreover, the secretion of osteogenic-related cytokines, including OPN, OSM, and OCN, was elevated after treatment with Ti-pPNP, demonstrating the strong potential of this approach to inducing osteogenesis. Thus, the pPNP coating not only improved the osteogenic activities but also prevented the MRSA-associated infection. Together, these findings demonstrate that the pPNP coating has the potential to reduce the risk of hematogenous implant infection in vivo.
Fig. 7. Effects of pPNP coating on anti-hematogenous infection.
(A) The study protocol of a prophylactic regimen with a hematogenous S. aureus implant infection mouse model. (B) Representative SEM images of biofilm formation on the implants harvested on day 14 after inoculation; n = 5 mice per group. Scale bar, 200 nm. (C) Gram staining of the bone tissues surrounding the implants. Blue arrows indicate scattered bacteria; n = 5 mice per group. Scale bar, 50 μm. (D) Euler diagram depicting the number of mice with or without infection in the surgical or nonsurgical legs of mice in the Ti or Ti-pPNP groups (n = 30 mice per group). (E) Representative fluorescent images showing the expression of OCN at the bone-implant interface (n = 5 mice per group). Scale bar, 100 μm. ***P < 0.001.
In summary, our findings establish that periprosthetic MΦs can be genetically engineered to yield super CAR-MΦs with the proposed pPNP coating, thereby enabling them to eradicate S. aureus to prevent PJI and therapeutic resistance. Furthermore, the in vivo biodegradability of our proposed implant coating precisely matches the bone regeneration process and achieves satisfactory osteogenesis at the bone-implant interfaces. Therefore, our work may provide a more efficient host-directed immunotherapeutic strategy for patients suffering from PJI or multidrug-resistant bacterial infection, and this new approach warrants further clinical validation.
MATERIALS AND METHODS
Materials
(SA)2-KRRRR-NLS-MTS was custom-synthesized by Shanghai Apeptide Co. Ltd. All of the plasmids used in our work were customized by vectorbuilder.com. Titanium (Ti) plates (1-cm long by 1-cm wide by 1-mm thick; 99.5% purity) and Ti implants (1 mm in diameter, 10 mm in length, 99.5% purity) were obtained from BIORTHO VET (Jiangsu, China). HS, PLL (Molecular weight. 70,000 to 150,000), and DOPA hydrochloride (98%) were purchased from MCE. All other chemicals and solvents of an analytical nature were procured locally. The 4′,6-diamidino-2-phenylindole (DAPI) kit was obtained from Thermo Fisher Scientific. Macrophage colony-stimulating factor was obtained from Sino Biological. LysoTracker was provided by Dalian Meilun Biotech Co., Ltd. or GeneTex. PerCP/Cy5.5-labeled anti-CD11b, antigen-presenting cell (APC)/Cy7-labeled anti-CD3, APC-labeled anti-CD4, phycoerythrin (PE)/Cy7-labeled anti-CD8, fluorescein isothiocyanate (FITC)–labeled anti-F4/80, PerCP/Cy5.5-labeled anti-F4/80, PE/Cy7-labeled anti-Gr1, PE-labeled anti-CD206, PE/Dazzle594-labeled anti-CD80, and APC-labeled anti-CD86 antibodies were all purchased from BD Biosciences or BioLegend Inc. ELISA kits for TNF-α, IFN-γ, IL-10, TGF-β, IL-2, OCN, OPN, and OSM were procured from Abcam. The anti-OCN and anti-OPN antibodies for immunofluorescence analysis were obtained from Abcam or Cell Signaling Technology.
Animals
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Shandong University of Traditional Chinese Medicine and Shandong University, and experimental protocols were approved by the Animal Ethics Committee of Shandong University of Traditional Chinese Medicine and Shandong University. Schemas of the used mouse models are shown in detail in the first panel of each relevant figure. C57BL/6J mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and housed by the Shandong University of Traditional Chinese Medicine and Shandong University Medical School Animal Care Facility under pathogen-free conditions.
Cell isolation and cell culture
BMDMs were harvested from mouse femurs for producing mouse MΦs and cultured in Dulbecco’s minimum essential medium (containing 10% fetal bovine serum, penicillin/streptomycin, glutamine, and Hepes). RAW 264.7 cell line cells and BMSCs were purchased from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Science. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Bacterial culture and in vitro antibacterial assay
MRSA (ATCC43300) was purchased from American Type Culture Collection, and an individual colony was separated from the LB agar plate, stored in the 4°C refrigerator, added to fresh LB broth, and then shaken at 200 rpm overnight at 37°C. The overnight bacterial solution was diluted 1:200 and shaken for another 3 hours to make sure the bacteria were in the log-growth phase before their use in an experiment. The intracellular antimicrobial assay was conducted in accordance with a previously reported method (46). Briefly, after different groups of BMDMs were subjected to the appropriate treatment, the BMDMs were incubated with MRSA at a multiplicity of infection of 5 for 6 or 24 hours. After being washed with phosphate-buffered saline (PBS), cells were incubated for an additional 1 hour in medium containing gentamicin (100 μg ml−1) to clear the extracellular bacteria. Cells were then washed with PBS and lysed with 0.1% Triton X-100. The resulting lysates were cultured on LB agar to determine the number of bacterial CFUs.
pDNA construction
The pDNA used in this study was prepared with piggyBac transposon gene expression vectors. The CASP11 shRNA was expressed under the control of the U6 promoter, and the S. aureus–specific CAR gene was expressed under the control of the MΦ-specific CD68 promoter (47). The CAR gene was composed of a mouse CD8 signal peptide, SasA-specific scFv, myc-tag, mouse CD8 hinge sequence, CD8 transmembrane region, and mouse CD3ζ endodomains. To evaluate the gene-transfer efficiency and monitor in situ MΦ programming, constructs with reporter proteins were generated via fusing GFP or luciferase with the CAR domain, separated by a P2A sequence. To strengthen gene expression, a woodchuck hepatitis virus posttranscriptional regulatory element was inserted between the EGFP/luciferase and the rabbit beta-globin polyadenylation signal.
Synthesis and characterization of the pPNP
(SA)2-KRRRR-NLS-MTS (2 mg) was dissolved in 10 μl of dimethyl sulfoxide and diluted with 1 ml of diethylpyrocarbonate-treated water solution with 35 μl of pDNA (weight ratio of peptide-SA monomer:pDNA = 10:1). The mixture was vortexed to obtain pPNPs. The dynamic light scattering and zeta potential analysis of the resultant nanomicelle were conducted with a Zeta Sizer Nano ZS90 (Malvern Instruments Ltd.). The morphology of the pPNPs was characterized using TEM performed on a JEM-1200 EX II electron microscope after negative staining with 2% phosphotungstic acid.
Synthesis and characterization of pPNP coating
Commercially pure Ti plates (1-cm long by 1-cm wide × 1-mm thick; 99.5% purity) and Ti implants (1 mm in diameter, 10 mm in length, 99.5% purity) were prepared with their surfaces machined and acid-etched with 67% (w/w) sulfuric acid (H2SO4) at 120°C for 75 s. Acid-etched Ti plates and implants were incubated in DOPA solution (2 mg ml−1, pH 8.5) for 16 hours. The alternating layer-by-layer operation was then performed as follows. First, Ti plates and implants were immersed in PLL (0.5 mg ml−1, pH 7.4) solution for 10 min. After being rinsed with distilled water for 1 min, the samples were immersed in a solution of HS (0.5 mg ml−1, pH 7.4) containing nanoparticles (108 nanoparticles ml−1, pH 7.4) for 10 min. The above two-step treatment was defined as one coating cycle. The number of layer coating cycles for all the experiments was set to 20. Nanolayers were cross-linked with an aqueous solution of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (100 mg ml−1) and N-hydroxysuccinimide (20 mg ml−1). Surface-modified Ti plates and implants were characterized by SEM (FEI QUANTA FEG250 field emission scanning; accelerating voltage, 8 kV; working distance, 7.2 mm), AFM (E-SEEEP, elastic coefficient 5 to 37 N/m; resonance frequency, 96 to 175 kHz), XPS (Escalab 250Xi multifunctional surface analyzer; Al Kα x-ray source; working voltage, 14.6 kV; working current, 10 mA), and water contact angle (Theta Flex).
Agarose gel electrophoresis assay
Agarose gel electrophoresis was conducted to study the complete complexation of (SA)2-peptide with pDNA. Nanoparticles were prepared with (SA)2-peptide and pDNA at ratios of 5:1, 10:1, 15:1, 20:1, or 25:1. Ten microliters of solution containing 0.3 μg of pDNA was mixed with DNA loading buffer and loaded into parallel wells of a 1.2% (w/v) agarose gel containing 0.5 μl ml−1 NA-Green (Beyotime) in tris-acetate-EDTA buffer to evaluate the DNA-complexation effect of the (SA)2-peptide compared with that of naked pDNA. Electrophoresis was performed at 90 V for 60 min, and gel retardation was visualized by using an Amersham Imager 600 RGB.
Cellular uptake and nanoparticle-mediated gene transfection in vitro
MΦs were cultured with each formulation, and the cellular uptake of the pPNPs was assessed both qualitatively and quantitatively. The cells were first incubated with free pDNA or pPNP at a plasmid dose of 5 μg ml−1. Four hours later, the cells were washed three times with ice-cold PBS and examined using flow cytometry. For Confocal laser scanning microscope (CLSM) observations, the nuclei were stained with DAPI (blue), and then subjected to CLSM analysis (CLSM 780, Carl Zeiss Inc.).
For the in vitro gene transfection assay, RAW 264.7 cells, BMDMs, BMSCs, or T cells were incubated with saline and pPNP. After a 48-hour incubation, the percentage of EGFP-positive cells was analyzed by flow cytometry.
Transcriptome sequencing and data analysis
After being subjected to pPNP-mediated treatment, MΦs were treated with TRIzol reagent (Beyotime Biotechnology) and stored at −80°C before sequencing. The RNA sequencing was performed using an Illumina HiSeq X10 (Illumina, USA). The gene expression value was transformed as log10 [TPM (transcripts per million reads) +1]. The RNA sequencing data were normalized via the fragments per kilobase per million reads method.
Macrophage phagocytosis analysis
BMDMs that were pretreated with PBS, CAR@PNP, shRNA@PNP, or pPNP were cocultured with 5(6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) dye-stained (Dalian Meilun Biotechnology Co. Ltd.) bacteria. After being cocultured for 4 hours, the BMDMs were harvested, stained with anti-F4/80 labeled with PerCP/Cy5.5, and then analyzed with flow cytometry. For the qualitative analysis of phagocytosis, bacteria were prestained with rhodamine B 5-isothiocyanate (Dalian Meilun Biotechnology Co., Ltd.). Afterward, MΦs and bacteria were coincubated at a ratio of MΦs to bacteria of 1:5 for 4 hours at 37°C. The cells were lastly observed with a fluorescence microscope.
Analysis of vacuole-containing bacteria and mitochondria in infected MΦs
BMDMs and CFSE-stained bacteria pretreated with PBS, CAR@PNP, shRNA@PNP, or pPNP were coincubated at a ratio of MΦs to bacteria of 1:5 for 4 hour at 37°C. Afterward, the mitochondria of the infected MΦs were stained with MitoTracker Red CMXRos (Invitrogen). The association of vacuole-containing bacteria with mitochondria in the cells was observed with fluorescence microscope. In addition, infected MΦs were fixed with 2.5% glutaraldehyde (TedPella, 18426) in 0.1 M phosphate buffer, pH 7.4 containing 0.1 M sucrose, and the relationship between the bacteria and the mitochondria of the infected MΦs was further qualitatively analyzed with a JEM-1200EX II electron microscope.
Mitochondrial ROS assay
To evaluate mitochondrial superoxide production, BMDMs were incubated with 2 μM MitoSOX dye (Thermo Fisher Scientific, M36008) diluted in cell imaging solution [10 mM Hepes, bovine serum albumin (1 mg ml−1), glucose (1 mg ml−1), 1 mM MgCl2, and 1.8 mM CaCl2 in PBS) at 37°C in a humidified atmosphere containing 5% CO2. The cells were then washed with PBS and further incubated with cell imaging solution containing gentamicin (10 μg ml−1). The fluorescence was read using a SpectraMax i3x microplate reader at 510/580 nm, and the cell number in each well was determined using a SpectraMax MiniMax300 Imaging Cytometer. Fluorescence values were first normalized to the cell number in each well, and then all conditions for each group were normalized to the nontreated control group.
qRT-PCR assay
For real-time quantitative PCR analysis, total RNA were isolated using TRIzol (Invitrogen, Carlsbad, CA, USA), and the isolated RNA were then subjected to reverse transcription using a PrimeScript RT Reagent kit (Takara, Dalian, China). Real-time PCR analysis was conducted with a SYBR Premix Ex Taq II (Takara) using a CFX96 Real-Time System (Bio-Rad, USA). Relative expression was calculated with the 2−ΔΔCT method. The sequences of the primers used for these assays are listed in table S1.
Mouse model of implant-related infection
During the implantation surgery, mice were anesthetized using inhalational 1 to 5% isoflurane mixed with oxygen. The distal femur was accessed via medial parapatellar arthrotomy with lateral displacement of the quadriceps–patellar complex. An intramedullary femoral canal was manually reamed with a 25-gauge needle. To establish the PJI models, 10 μl of S. aureus (1 × 105 CFU ml−1) was injected into the medullary cavity carefully and slowly. To prevent leaking of the bacterial suspension from the injection track, the syringe was removed 30 s later. After that, control, Ti-PNP, Ti-shRNA@PNP, Ti-CAR@PNP, or Ti-pPNP implants (length, 10 mm; diameter, 1 mm) were surgically placed in a retrograde fashion with 1 mm protruding into the knee joint space. Last, the quadriceps–patellar complex was reduced to the midline, and the surgical site was closed with absorbable Vicryl 5-0 sutures. For investigating the antibacterial capability of different implants, mice were euthanized by CO2 inhalation at 7 days after implantation, and the implants were harvested and ultrasonicated for 5 min in 2 ml of PBS. The isolated bacteria were diluted, inoculated on LB medium, and cultured at 37°C for 24 hours for counting the bacterial CFUs. Osteogenesis was investigated with the mouse model of implant-related infection after 4 weeks.
To establish the hematogenous orthopedic infection models, 100 μl of S. aureus (1 × 107 CFU ml−1) was injected into the tail vein at 1 week after surgery. The antibacterial effect and osteogenesis of different implants were investigated after 14 days.
Flow cytometry assay in vivo
For analysis of the immune subsets present within the infection microenvironment around the implant, the bone marrow around infection foci and nearby lymph nodes were harvested and filtered to obtain single-cell suspensions. These isolated cells were treated with red blood cell lysis buffer (Beyotime) and washed with PBS. The cells were subsequently stained with different antibodies including anti-CD11b-PerCP/Cy5.5, anti-CD3-APC/Cy7, anti-CD4-APC, anti-CD8-PE/Cy7, anti-F4/80-FITC, anti-Gr1-PE/Cy7, anti-CD206-PE, anti-CD80-PE/Dazzle594, and anti-CD86-APC. After being stained with fluorescently labeled antibodies, the cells were washed in PBS with EDTA. Flow cytometry data were acquired by a BD FACSCanto II flow cytometer using the BD FACS Diva 8 software (BD Bioscience). Data were analyzed using FlowJo software.
In vivo evaluation of the biocompatibility and degradability of the nanoparticle coating and the persistence and biodistribution of the super CAR- MΦs
To investigate the nanoparticle coating biocompatibility, at 1, 2, and 4 weeks after implantation, 200 μl of blood was collected from the eye socket venous plexus for use in a blood analysis. Last, the animals were euthanized by CO2 inhalation. The heart, liver, spleen, lungs, and kidneys were excised and fixed in 10% formalin for 24 hours for histological analyses. To investigate the nanoparticle coating degradability, a nanoparticle coating labeled with Cy5.5 was surgically placed into the distal femur of mice. Mice were anesthetized via the inhalation of isoflurane (2%), and in vivo fluorescence imaging was performed by using the IVIS Lumina II in vivo imaging system (PerkinElmer, Hopkinton, MA, USA).
Micro-CT analysis
The bone tissues surrounding the implantation site were scanned at a 36-μm resolution on a SkyScan 1172 micro-CT system with a rotation step of 0.15°/180°. Image parameters were as follows: source voltage, 80 kV; exposure time, 0.37 s; 1-mm Al filter to optimize the contrast; and source current, 112 μA. Scans were reconstructed using SkyScan NRecon software and analyzed by CTAN software to generate three-dimensional images. During the micro-CT analysis, we did not pull out the implant from the bone tissue, and the selected region of interest was defined on the basis of the cylindrical implant. The newly formed bone tissues around the implants were further determined by micro-CT. The standardized threshold (>220 HU) was set as mineralized tissue. After reconstruction, the BMD, new BV, and percentage of BV to TV (%, BV/TV) were calculated by CT Analyser software (version 1.11; SkyScan).
Histopathological, immunohistochemical, and immunofluorescence analyses
To investigate the inflammatory reaction, the bone tissues surrounding the implants were harvested 2 weeks after implantation and then fixed with 4% paraformaldehyde, decalcified by EDTA, dehydrated in an ascending graded series of ethanol solutions, and embedded in paraffin. The sectioned tissues were subjected to H&E and Gram staining to observe the inflammation level and the remaining bacteria, respectively. To investigate the bone integration surrounding the implants, the bone tissues were harvested 4 weeks after implantation. The samples were stained with Masson staining or Safranin O/Fast Green staining. For immunofluorescence staining, paraffin-embedded bone tissue sections were stained overnight with primary antibodies against OCN or OPN (Abcam) at 4°C. The sections were then incubated with an appropriate secondary antibody labeled with Alexa Fluor (Life Technologies) for 1 hour at room temperature, followed by counter-staining with DAPI. To examine the colocalization of nanoparticles and MΦs, samples were stained with a primary antibody against F4/80 (Abcam) at 4°C overnight. The sections were then incubated with an appropriate secondary antibody labeled with Alexa Fluor (Life Technologies) for 1 hour at room temperature, followed by counter-staining with DAPI.
Determination of proteins by ELISA
The bone tissue homogenate was collected and frozen at −80°C before use. The levels of TNF-α, IL-2, IFN-γ, TGF-β, OSM, OPN, and OCN were measured using corresponding ELISA Quantitation Kits (Abcam) according to the manufacturer’s instructions. Results were normalized to the total amount of protein in each tissue sample.
Statistical analysis
Results are presented as the means ± SD. Error bars represent the SD of the mean from independent samples. Animals were randomly grouped before the initiation of the treatment. Data were compared using a one-way analysis of variance (ANOVA), two-tailed Student’s t test, or two-tailed Fisher’s exact test, as indicated in the figure legends. All statistical analyses were carried out with the Prism software (GraphPad). Statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
Acknowledgments
We thank Y. Yu, X.-M. Yu, J. Zhang, M.-L. Wu, and L.-M. Wang in Advanced Medical Research Institute/Translational Medicine Core Facility of Advanced Medical Research Institute, Shandong University for the technical support.
Funding: This work was supported by the National Natural Science Foundation of China (82173763), the ISF-NSFC Joint Scientific Research Program (52161145501), Funds for Youth Interdisciplinary and Innovation Research Groups of Shandong University (2020QNQT003), the Fundamental Research Funds of Shandong Province (ZR2022ZD18 and ZR2022ZD17), the Shandong Provincial Key Research and Development Program (Major Scientific and Technological Innovation Project) (2021CXGC010515 and 2019JZZY021013), and the Shandong Provincial Natural Science Foundation (ZR2020MH260).
Author contributions: Z.Li, S.Z., and X.J. conceived and designed the concept of the experiments. Z.Li, S.Z., Y.L., and C.T. conducted the material characterizations. Z.Liao, S.Z., Z.F., Z.M., C.S., Q.C., J.Z., X.Z., and H.X. conducted and analyzed most of the in vitro and in vivo experiments. C.C., Z.Y., M.H., Y.W., Z.Li, and G.Y. contributed to the histological analysis with specialized techniques. B.S. and K.Z. provided important experimental insights and participated in the bioinformatics analysis. Z.Li, S.Z., W.L., and X.J. cowrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Table S1
Legend for source data
Other Supplementary Material for this manuscript includes the following:
Source data
REFERENCES AND NOTES
- 1.B. H. Kapadia, R. A. Berg, J. A. Daley, J. Fritz, A. Bhave, M. A. Mont, Periprosthetic joint infection. Lancet 387, 386–394 (2016). [DOI] [PubMed] [Google Scholar]
- 2.J. Tande Aaron, R. Patel, Prosthetic joint infection. Clin. Microbiol. Rev. 27, 302–345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. R. Osmon, E. F. Berbari, A. R. Berendt, D. Lew, W. Zimmerli, J. M. Steckelberg, N. Rao, A. Hanssen, W. R. Wilson, Diagnosis and management of prosthetic joint infection: Clinical practice guidelines by the infectious diseases society of americaa. Clin. Infect. Dis. 56, e1–e25 (2013). [DOI] [PubMed] [Google Scholar]
- 4.P. Izakovicova, O. Borens, A. Trampuz, Periprosthetic joint infection: Current concepts and outlook. EFORT Open Rev. 4, 482–494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland, V. G. Fowler Jr., Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. Wang, M. Shi, F. Xu, Y. Qiu, P. Zhang, K. Shen, Q. Zhao, J. Yu, Y. Zhang, Graphdiyne-modified TiO2 nanofibers with osteoinductive and enhanced photocatalytic antibacterial activities to prevent implant infection. Nat. Commun. 11, 4465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C. R. Arciola, D. Campoccia, L. Montanaro, Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409 (2018). [DOI] [PubMed] [Google Scholar]
- 8.E. A. Masters, B. F. Ricciardi, K. L. D. M. Bentley, T. F. Moriarty, E. M. Schwarz, G. Muthukrishnan, Skeletal infections: Microbial pathogenesis, immunity and clinical management. Nat. Rev. Microbiol. 20, 385–400 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. S. Flannagan, V. Jaumouille, S. Grinstein, The cell biology of phagocytosis. Annu. Rev. Pathol. 7, 61–98 (2012). [DOI] [PubMed] [Google Scholar]
- 10.R. S. Flannagan, R. C. Kuiack, M. J. McGavin, D. E. Heinrichs, Staphylococcus aureus uses the GraXRS regulatory system to sense and adapt to the acidified phagolysosome in macrophages. MBio 9, e01143-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. D. Zoller, V. Hegde, Z. D. C. Burke, H. Y. Park, C. R. Ishmael, G. W. Blumstein, W. Sheppard, C. Hamad, A. H. Loftin, D. O. Johansen, R. A. Smith, M. M. Sprague, K. R. Hori, S. J. Clarkson, R. Borthwell, S. I. Simon, J. F. Miller, S. D. Nelson, N. M. Bernthal, Evading the host response: Staphylococcus “hiding” in cortical bone canalicular system causes increased bacterial burden. Bone Research 8, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. E. Heim, M. E. Bosch, K. J. Yamada, A. L. Aldrich, S. S. Chaudhari, D. Klinkebiel, C. M. Gries, A. A. Alqarzaee, Y. Li, V. C. Thomas, E. Seto, A. R. Karpf, T. Kielian, Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 5, 1271–1284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.C. E. Heim, K. J. Yamada, R. Fallet, J. Odvody, D. M. Schwarz, E. R. Lyden, M. J. Anderson, R. Alter, D. Vidlak, C. W. Hartman, B. S. Konigsberg, C. A. Cornett, K. L. Garvin, N. Mohamed, A. S. Anderson, T. Kielian, Orthopaedic surgery elicits a systemic anti-inflammatory signature. J. Clin. Med. 9, 2123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.V. Thammavongsa, H. K. Kim, D. Missiakas, O. Schneewind, Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. H. E. Kaufmann, A. Dorhoi, Molecular determinants in phagocyte-bacteria interactions. Immunity 44, 476–491 (2016). [DOI] [PubMed] [Google Scholar]
- 16.D. M. Sivaloganathan, M. P. Brynildsen, Phagosome–bacteria interactions from the bottom up. Annu. Rev. Chem. Biomol. Eng. 12, 309–331 (2021). [DOI] [PubMed] [Google Scholar]
- 17.F. Y. McWhorter, C. T. Davis, W. F. Liu, Physical and mechanical regulation of macrophage phenotype and function. Cell. Mol. Life Sci. 72, 1303–1316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Amin Yavari, S. M. Castenmiller, J. A. G. van Strijp, M. Croes, Combating implant infections: Shifting focus from bacteria to host. Adv. Mater. 32, 2002962 (2020). [DOI] [PubMed] [Google Scholar]
- 19.F. M. Roche, R. Massey, S. J. Peacock, N. P. J. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, T. J. Foster, Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149, 643–654 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Y. Yang, M. Qian, S. Yi, S. Liu, B. Li, R. Yu, Q. Guo, X. Zhang, C. Yu, J. Li, J. Xu, W. Chen, Monoclonal antibody targeting Staphylococcus aureus surface protein A (SasA) protect against Staphylococcus aureus sepsis and peritonitis in mice. PLOS ONE 11, e0149460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.T. J. Foster, J. A. Geoghegan, V. K. Ganesh, M. Hook, Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. M. Patti, B. L. Allen, M. J. McGavin, M. Hook, MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617 (1994). [DOI] [PubMed] [Google Scholar]
- 23.T. J. Foster, Surface proteins of Staphylococcus aureus. Microbiol. Spectr. 7, 7–4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.X. Hou, X. Zhang, W. Zhao, C. Zeng, B. Deng, D. W. McComb, S. Du, C. Zhang, W. Li, Y. Dong, Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 15, 41–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. Krause, K. Daily, S. Estfanous, K. Hamilton, A. Badr, A. Abu Khweek, R. Hegazi, M. N. Anne, B. Klamer, X. Zhang, M. A. Gavrilin, V. Pancholi, A. O. Amer, Caspase-11 counteracts mitochondrial ROS-mediated clearance of Staphylococcus aureus in macrophages. EMBO Rep. 20, e48109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. J. Foster, Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 (2005). [DOI] [PubMed] [Google Scholar]
- 27.A. D. Fesnak, C. H. June, B. L. Levine, Engineered T cells: The promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. H. June, M. Sadelain, Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.E. Liu, D. Marin, P. Banerjee, H. A. Macapinlac, P. Thompson, R. Basar, L. Nassif Kerbauy, B. Overman, P. Thall, M. Kaplan, V. Nandivada, I. Kaur, A. Nunez Cortes, K. Cao, M. Daher, C. Hosing, E. N. Cohen, P. Kebriaei, R. Mehta, S. Neelapu, Y. Nieto, M. Wang, W. Wierda, M. Keating, R. Champlin, E. J. Shpall, K. Rezvani, Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. T. Smith, S. B. Stephan, H. F. Moffett, L. E. McKnight, W. Ji, D. Reiman, E. Bonagofski, M. E. Wohlfahrt, S. P. S. Pillai, M. T. Stephan, In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 12, 813–820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. Kim, H.-B. Pang, J. Kang, J.-H. Park, E. Ruoslahti, M. J. Sailor, Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus. Nat. Commun. 9, 1969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. J. Brandén, A. J. Mohamed, C. I. E. Smith, A peptide nucleic acid–Nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 17, 784–787 (1999). [DOI] [PubMed] [Google Scholar]
- 33.N. Huang, S. Zhang, L. Yang, M. Liu, H. Li, Y. Zhang, S. Yao, Multifunctional electrochemical platforms based on the michael addition/Schiff base reaction of polydopamine modified reduced graphene oxide: Construction and application. ACS Appl. Mater. Interfaces 7, 17935–17946 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Z. Jia, M. Wen, Y. Cheng, Y. Zheng, Strategic advances in spatiotemporal control of bioinspired phenolic chemistries in materials science. Adv. Funct. Mater. 31, 2008821 (2021). [Google Scholar]
- 35.E. M. M. Manders, F. J. Verbeek, J. A. Aten, Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 36.J. Klein, The major histocompatibility complex of the mouse. Science 203, 516–521 (1979). [DOI] [PubMed] [Google Scholar]
- 37.C. J. Hall, R. H. Boyle, J. W. Astin, M. V. Flores, S. H. Oehlers, L. E. Sanderson, F. Ellett, G. J. Lieschke, K. E. Crosier, P. S. Crosier, Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ROS production. Cell Metab. 18, 265–278 (2013). [DOI] [PubMed] [Google Scholar]
- 38.B. H. Abuaita, T. L. Schultz, M. X. O’Riordan, Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O. Ciofu, C. Moser, P. Ø. Jensen, N. Høiby, Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 20, 621–635 (2022). [DOI] [PubMed] [Google Scholar]
- 40.K. Schilcher, R. Horswill Alexander, Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84, e00026-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G. L. Koons, M. Diba, A. G. Mikos, Materials design for bone-tissue engineering. Nat. Rev. Mater. 5, 584–603 (2020). [Google Scholar]
- 42.J. Wu, Z. Feng, L. Chen, Y. Li, H. Bian, J. Geng, Z.-H. Zheng, X. Fu, Z. Pei, Y. Qin, L. Yang, Y. Zhao, K. Wang, R. Chen, Q. He, G. Nan, X. Jiang, Z.-N. Chen, P. Zhu, TNF antagonist sensitizes synovial fibroblasts to ferroptotic cell death in collagen-induced arthritis mouse models. Nat. Commun. 13, 676 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.B. S. Konigsberg, C. J. Della Valle, N. T. Ting, F. Qiu, S. M. Sporer, Acute hematogenous infection following total hip and knee arthroplasty. J. Arthroplasty 29, 469–472 (2014). [DOI] [PubMed] [Google Scholar]
- 44.A. J. Tande, B. R. Palraj, D. R. Osmon, E. F. Berbari, L. M. Baddour, C. M. Lohse, J. M. Steckelberg, W. R. Wilson, M. R. Sohail, Clinical presentation, risk factors, and outcomes of hematogenous prosthetic joint infection in patients with Staphylococcus aureus bacteremia. Am. J. Med. 129, 221.e11–221.e20 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Y. Wang, L. I. Cheng, D. R. Helfer, A. G. Ashbaugh, R. J. Miller, A. J. Tzomides, J. M. Thompson, R. V. Ortines, A. S. Tsai, H. Liu, C. A. Dillen, N. K. Archer, T. S. Cohen, C. Tkaczyk, C. K. Stover, B. R. Sellman, L. S. Miller, Mouse model of hematogenous implant-related Staphylococcus aureus biofilm infection reveals therapeutic targets. Proc. Natl. Acad. Sci. U.S.A. 114, E5094–E5102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Garzoni, W. L. Kelley, Staphylococcus aureus: New evidence for intracellular persistence. Trends Microbiol. 17, 59–65 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Y. L. Luo, C. F. Xu, H. J. Li, Z. T. Cao, J. Liu, J. L. Wang, X. J. Du, X. Z. Yang, Z. Gu, J. Wang, Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano 12, 994–1005 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Table S1
Legend for source data
Source data