Abstract
DNA interstrand crosslinks (ICLs) pose a major obstacle for DNA replication and transcription if left unrepaired. The cellular response to ICLs requires the coordination of various DNA repair mechanisms. Homologous recombination (HR) intermediates generated in response to ICLs, require efficient and timely conversion by structure-selective endonucleases. Our knowledge on the precise coordination of this process remains incomplete. Here, we designed complementary genetic screens to map the machinery involved in the response to ICLs and identified FIRRM/C1orf112 as an indispensable factor in maintaining genome stability. FIRRM deficiency leads to hypersensitivity to ICL-inducing compounds, accumulation of DNA damage during S-G2 phase of the cell cycle, and chromosomal aberrations, and elicits a unique mutational signature previously observed in HR-deficient tumors. In addition, FIRRM is recruited to ICLs, controls MUS81 chromatin loading, and thereby affects resolution of HR intermediates. FIRRM deficiency in mice causes early embryonic lethality and accelerates tumor formation. Thus, FIRRM plays a critical role in the response to ICLs encountered during DNA replication.
Functional genetic screens identify FIRRM/C1orf112 as a crucial factor for genome stability in the interstrand crosslink response.
INTRODUCTION
The repair of interstrand crosslinks (ICLs) is crucial for cellular survival and involves the coordinated action of multiple DNA repair pathways including the Fanconi anemia pathway (FA), translesion synthesis (TLS), homologous recombination (HR), and nucleotide excision repair (NER) (1, 2). The pathway that specifically responds to ICLs is coupled to DNA replication and involves a network of 22 proteins, defective in a severe heritable syndrome known as FA (1–3). Upon stalling of replication forks at an ICL, the FA core complex is recruited to the site of DNA damage and ensures mono-ubiquitination of the FANCD2-FANCI heterodimer (1, 4). This event activates several downstream effectors, including specialized nucleases that perform dual nucleolytic incisions (5), which enables unhooking of the ICL lesion and subsequent HR (1–3, 6). Lesions that are left unrepaired can cause replication fork collapse and double strand breaks (DSBs) formation. Repair of these breaks involves HR, where a resected DSB end invades a homologous donor locus to enable strand exchange between the damaged chromatid and its unbroken sister by the strand exchange protein RAD51. This leads to the formation of an intermediate structure known as a D-loop, which primes DNA repair synthesis (7, 8). In some cases, covalent linkage between the two recombining DNA molecules creates a four-stranded DNA intermediate known as a Holliday junction (HJ) (9). These structures must be removed timely via structure-selective enzymes to maintain genome stability and ensure faithful chromosomal segregation (10–15).
Here, we used complementary genetic loss-of-function screens in haploid cells to map critical factors involved in maintaining genome stability upon ICL exposure. In addition to known factors (such as FA proteins), we identified C1orf112 [renamed FIRRM and also known as FLIP in Arabidopsis thaliana (16), MEICA1 in rice (17), and Apolo1 (18)] as an indispensable scaffold protein for maintaining genome stability. FIRRM affects the regulation of HR intermediates during ICL response and modulates the recruitment and the retention of MUS81 at sites of DNA damage. Embryonic lethality and accelerated tumorigenesis upon FIRRM loss emphasize the importance of this protein in maintaining genome integrity.
RESULTS
Haploid genetic screens identify FIRRM as an important modulator of the cellular response to ICLs
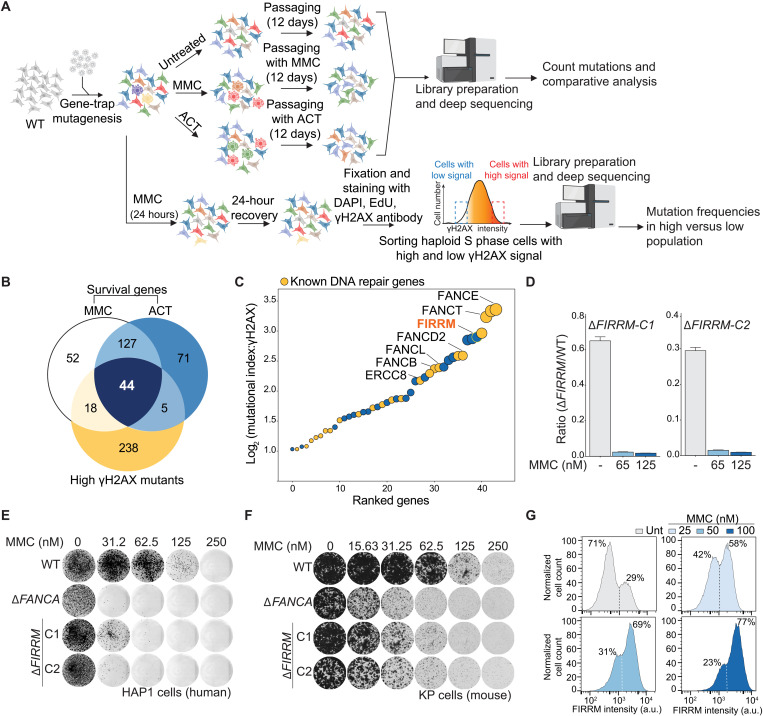
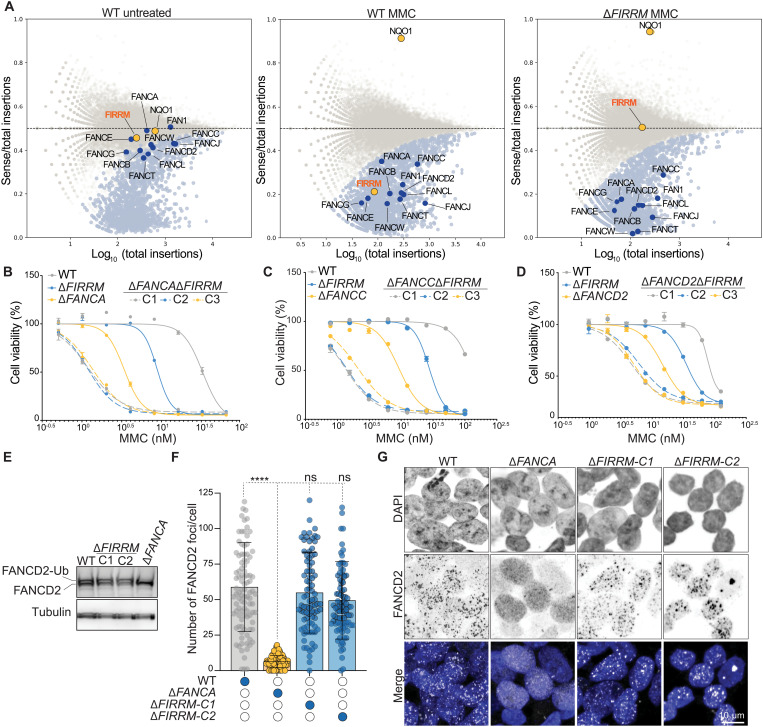
To systematically identify the core components involved in the ICL response, we performed genome-wide insertional mutagenesis in human haploid cells (HAP1) using gene-trap inactivation (19). The mutagenized cells were exposed to two classes of ICL-inducing compounds: mitomycin C (MMC), a chemotherapeutic agent (1), and acetaldehyde (ACT), a metabolite that is produced upon alcohol consumption (Fig. 1A) (20). Gene-trap insertions were identified in the treated cell populations and compared to the untreated controls, resulting in the identification of genes that displayed an underrepresentation of disruptive mutations specifically in the treated condition, which is indicative of impaired fitness. A high concordance between biological replicates and treatments was observed, and 171 significant genes were identified that caused sensitization to both MMC and ACT upon their inactivation (Fig. 1B; fig. S1, A to D; and table S1). Reassuringly, Gene Ontology enrichment analysis highlighted biological processes associated with DNA repair, ICL repair, DSB repair, and cellular responses to DNA damage (fig. S1E). Various DNA repair factors known to be important for proper functioning of FA, HR, and NER were identified (fig. S1F). In addition, genes encoding structure-selective endonucleases, such as MUS81-EME1 and GEN1 as well as BLM, helicase were found (fig. S1F), which are involved in the resolution and the dissolution of HJs (10, 11, 13, 21).
Fig. 1. Haploid genetics identify FIRRM as an important factor for genome stability in response to ICLs.
(A) Schematic overview of the haploid genetic screens performed using survival or γH2AX staining as readouts. (B) Venn diagram of the genes identified by the three genetic screens. (C) Ranking plot depicting the 44 genes commonly identified between the performed genetic screens. Genes are ranked on the basis of the mutational index of the γH2AX screen. Orange dots indicate known DNA repair genes, and blue dots indicate genes with an unknown function in ICL repair. (D) Competition growth assay with wild-type (GFP) and ΔFIRRM (mCherry) HAP1 cells treated with MMC (65 or 125 nM) for 8 days. Data are represented as mean ratio (mCherry/GFP) ± SEM. (E and F) Clonogenic survival in response to different concentrations of MMC in HAP1 cells (E) and mouse mammary tumor cells (K14Cre;Trp53−/−; KP) (F). WT, wild type. (G) HAP1 cells carrying an endogenous V5-tag at the C terminus of FIRRM were transduced with a 1:1 mix of nontargeting sgRNA and a sgRNA targeting FIRRM. After puromycin selection, the pool of cells was treated with different concentrations of MMC for 7 days. Samples were then fixed, stained with a V5 antibody, and analyzed by flow cytometry. All experiments were performed in triplicates except the genetic screens. For the survival-based screens, two biological replicates were performed, and for the γH2AX screen, one replicate was done. a.u., arbitrary unit.
In parallel, we performed a fluorescence-activated cell sorting (FACS)–based haploid genetic screen (22) using γH2AX as a readout for DNA damage. Cells were exposed to MMC for 24 hours and allowed to recover for an additional 24 hours before fixation, staining, and FACS sorting for 5-ethynyl-2′-deoxyuridine (EdU)–positive cells (S phase) with the 5% highest and lowest γH2AX signal (Fig. 1A). Subsequently, genes enriched for disruptive mutations in either cell population were identified. As expected, histone variant H2AX and the DNA damage scaffold protein MDC1 scored as positive modulators (23), whereas several ICL repair factors were identified as negative regulators (fig. S2, A and B). To select key genes involved in the cellular response to ICLs, we intersected ICL survival genes (MMC and ACT) with the γH2AX-negative regulators, which yielded a set of 44 factors (Fig. 1B), many of which have been previously associated with ICL repair (Fig. 1C, orange dots). The top scoring gene among the identified factors with an unknown function in ICL repair was FIRRM/C1orf112 (Fig. 1C; figs. S1, C, D, and F, S2, A and B, and S3, A and B; and table S2). FIRRM showed a similar phenotypic effect as the FA components such as FANCA and FANCD2 and did not affect 16 phenotypes unrelated to DNA damage response signaling previously studied using genome-wide mutagenesis in HAP1 cells (fig. S3C).
FIRRM maintains genome stability in response to ICLs
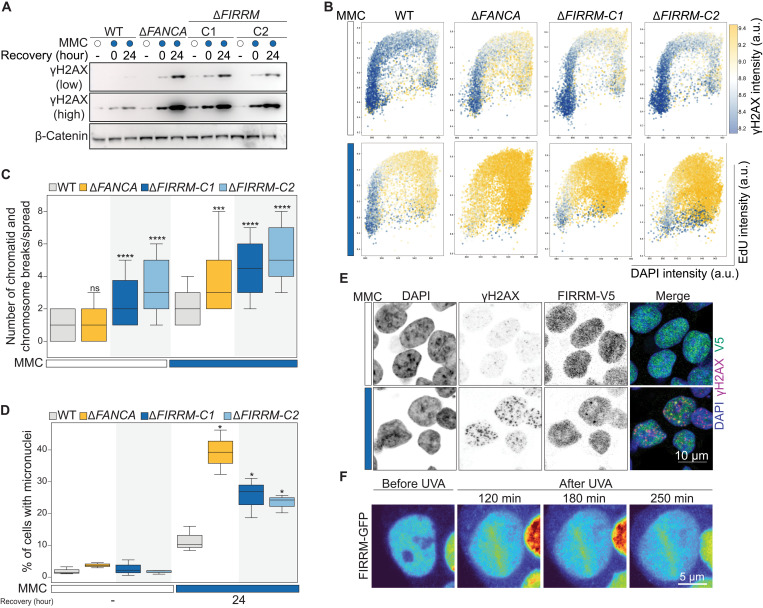
To validate our findings, we generated HAP1 cell lines in which FIRRM is mutated (ΔFIRRM), or the highly conserved HEAT repeat–containing domain of unknown function (DUF4487) is deleted using CRISPR-Cas9 (figs. S4A and S5, A to D) and measured their sensitivity to a panel of DNA damaging agents. Loss of FIRRM or the DUF4487 domain caused hypersensitivity to ICL-inducing compounds such as MMC, cisplatin and ACT in competition growth assays and in clonogenic survival assays (Fig. 1, D and E, and figs. S4, B to D and S5, E to F). No growth defect was observed in unchallenged conditions (Fig. 1E). Furthermore, FIRRM knockout cells showed impaired survival upon exposure to poly (ADP–ribose) polymerase (PARP) inhibitors or topoisomerase I inhibitors, but not to DSB-inducing agents or to replication stress generated by hydroxyurea (fig. S4, E to K). Similar observations were made for FIRRM knockout lines generated in a mouse mammary tumor cell line (K14cre;Trp53−/−, KP) (24) (Fig. 1F and fig. S5G). To exclude any clonal effects, we endogenously tagged FIRRM with a V5-tag at the C terminus in HAP1 cells and infected them with a lentiviral vector containing either a nontargeting single guide RNA (sgRNA) or sgRNAs targeting FIRRM. While the nontargeting sgRNA created a homogeneous cell population, the sgRNAs targeting FIRRM generated a heterogeneous pool of cells with different levels of FIRRM protein. (Fig. 1G and fig. S5, H to K). MMC treatment specifically decreased the cell population with low protein levels of FIRRM in a dose-dependent manner (Fig. 1G). As suggested by the genetic screens, FIRRM deficiency resulted in a strong accumulation of DNA damage marked by γH2AX during the S-G2 phase of the cell cycle in response to MMC. This was comparable to γH2AX levels detected in FA core complex–deficient cells (ΔFANCA) (Fig. 2, A to B, and fig. S6, A to C). In agreement with this observation, FIRRM-deficient cells exhibited elevated levels of chromosomal instability, highlighted by an accumulation of chromosomal breaks measured by metaphase spreads (Fig. 2C and fig. S6D). This was accompanied by a strong increase in micronuclei formation (Fig. 2D and fig. S6E), structures formed from lagging or broken chromosomes and directly associated with extensive chromosomal rearrangement and chromothripsis (25, 26). FIRRM protein levels were increased during S-G2 phase of the cell cycle and excluded from chromosomal regions during mitosis (fig. S7, A to F). Furthermore, we observed the formation of nuclear foci, which localized with a subset of γH2AX foci upon MMC exposure (Fig. 2E). Moreover, green fluorescent protein (GFP)–tagged FIRRM was enriched at psoralen-induced ICLs after ultraviolet A (UVA) laser microirradiation (Fig. 2F and fig. S7G). These data indicate that FIRRM is enriched at sites of DNA damage and plays a critical role in maintaining genome integrity in response to ICL-inducing agents.
Fig. 2. FIRRM loss causes increased levels of DNA damage and genome instability.
(A) Wild-type, FANCA, or FIRRM knockout HAP1 cells were treated with 50 nM MMC for 24 hours and left to recover for the indicated time points. Whole-cell extracts were subjected to immunoblot analysis using γH2AX antibodies. β-Catenin was used as a loading control. (B) Wild-type, ΔFANCA, or ΔFIRRM HAP1 cells were left untreated or exposed to 50 nM MMC for 24 hours, allowed to recover for 24 hours; stained with γH2AX, EdU, and (DAPI); and subjected to flow cytometry analysis. n = 3. (C) Quantification of chromosome and chromatid breaks per metaphase spread (means ± SEM); 60 metaphases of KP cells left untreated or treated with 50 nM MMC for 24 hours were analyzed for each condition and pooled from three independent experiments. (D) Quantification of micronuclei in indicated genotypes, unchallenged or upon 50 nM MMC treatment and 24 hours of recovery. n > 300 cells per condition. n = 3. (C and D) Significance was calculated by a Mann-Whitney U test. ns, not significant. *P < 0.05, ***P < 0.001, and ****P < 0.0001. (E) Immunofluorescence staining for γH2AX and FIRRM-V5 in HAP1 cells upon MMC treatment (50 nM, 24 hours) or left untreated. To visualize chromatin bound protein, cells were pre-extracted before fixation. n = 3. (F) Recruitment of GFP-FIRRM to laser UVA microirradiation sites after psoralen treatment in HAP1 cells. n = 2.
FIRRM loss induces a unique mutational signature
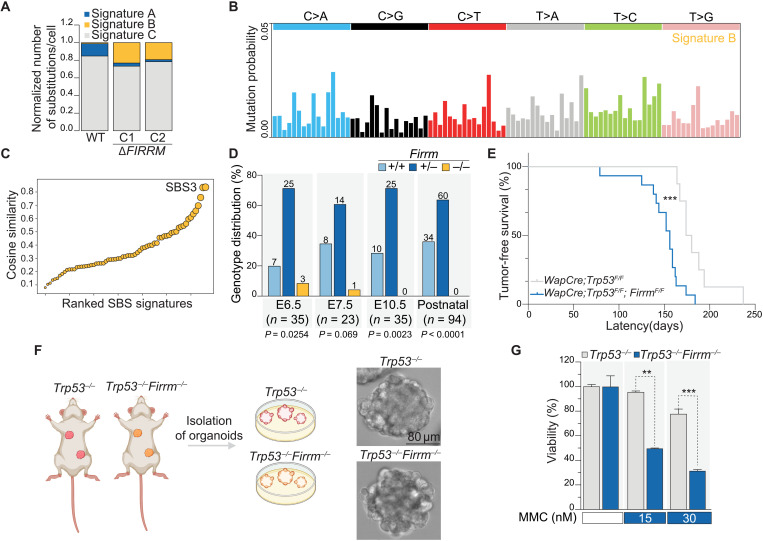
Given the fact that FIRRM deficiency induced genome instability, we asked whether long-term loss of FIRRM would trigger a specific mutational imprint across the genome. We determined the mutational landscape in wild-type and FIRRM-deficient HAP1 cells upon MMC treatment (EC20) or in unchallenged conditions using nanorate sequencing (NanoSeq) (27), a duplex sequencing protocol with a low error rate, which can be applied in heterogeneous cell populations (fig. S8A). Although MMC-treated samples presented with a twofold increase in mutational burden (fig. S8B), FIRRM deficiency only modestly affected the frequency and the pattern of point mutations (fig. S8, C to D). However, signature decomposition of the NanoSeq data highlighted distinct signatures (Fig. 3A and fig. S8E): Signature A was mainly detected upon MMC treatment (fig. S8F) and showed an increase in C>A, C>G, and C>T mutations. This pattern resembled the previously reported SBS4 signature (cosine similarity of 0.91) (fig. S8G), representing an imprint of bulky DNA adducts generated by polycyclic hydrocarbons found in tobacco smoke (similar to tobacco carcinogens, MMC also forms adducts on guanines) (28, 29). Signature B was uniquely observed in FIRRM knockout cells (Fig. 3, A to B) and matched with SBS3 (cosine similarity of 0.87) (Fig. 3C), which has been previously observed in BRCA1 and BRCA2 mutant breast cancer genomes and is thought to result from HR deficiency (30–34). However, small indels reported in the absence of BRCA1 and BRCA2 (31) were not detected in FIRRM-depleted cells (fig. S8, H to I).
Fig. 3. FIRRM loss causes early embryonic lethality and accelerates tumorigenesis.
(A) Contribution of signatures A, B, and C in untreated wild-type and ΔFIRRM HAP1 cells. (B) Signature decomposition of wild-type and ΔFIRRM HAP1 cells in untreated conditions. (C) Cosine similarity comparing signature B to existing single base substitution (SBS) signatures. (D) Recovery of Firrm+/+, Firrm+/−, and Firrm−/− embryos at different time points after performing timed matings. Numbers of analyzed embryos are indicated, and a Chi-square test was used to determine significance. (E) Kaplan-Meier survival curve depicting the mammary tumor-free survival of WapCre;Trp53F/F (n = 8) and WapCre;Trp53F/F;FirrmF/F (n = 15) female mice. Significance was calculated by a Mantel-Cox test. ***P < 0.001, median survival of 177 days (WapCre;Trp53F/F) versus 156 days (WapCre;Trp53F/F;FirrmF/F). (F) Schematic depiction of organoid isolation from mammary tumor–bearing animals and example images of the derived organoids. (G) Viability of mammary tumor organoids after exposure to MMC for 7 days. Values were normalized to untreated conditions. n = 2. P values were calculated using a two-tailed t test. **P < 0.01 and ***P < 0.001.
FIRRM loss causes embryonic lethality and accelerates tumorigenesis
To examine the physiological role of FIRRM in vivo, we generated Firrm knockout mice. While heterozygous mice (Firrm+/−) were born without any apparent defects, we did not observe any Firrm−/− offspring generated from F1 heterozygous crosses. Firrm−/− embryos were detected at early stages (E6.5 to E7.5) during embryonic development at sub-Mendelian ratios (Fig. 3D). Thus, FIRRM deficiency causes early embryonic lethality, as has been observed for several HR genes, such as BRCA1/2 and RAD51 (35, 36). Because FIRRM loss triggered an HR-deficiency-like mutational signature, we tested whether FIRRM deficiency, specifically in the mammary epithelium, would also alter tumor incidence. Mice carrying Cre-conditional FirrmF alleles were generated and crossed with our previously established Wap-Cre;Trp53F/F mammary tumor model (37). We found that homozygous loss of Firrm and Trp53 significantly accelerated mammary tumor formation compared to Trp53 loss alone (Fig. 3E). In addition, mammary tumor organoids derived from Trp53−/− or Trp53−/−;Firrm−/− tumors displayed hypersensitivity to MMC compared to Trp53−/− controls (Fig. 3, F to G), recapitulating our previously observed phenotype in cultured cells.
FIRRM acts independently of FIGNL1 and the FA pathway
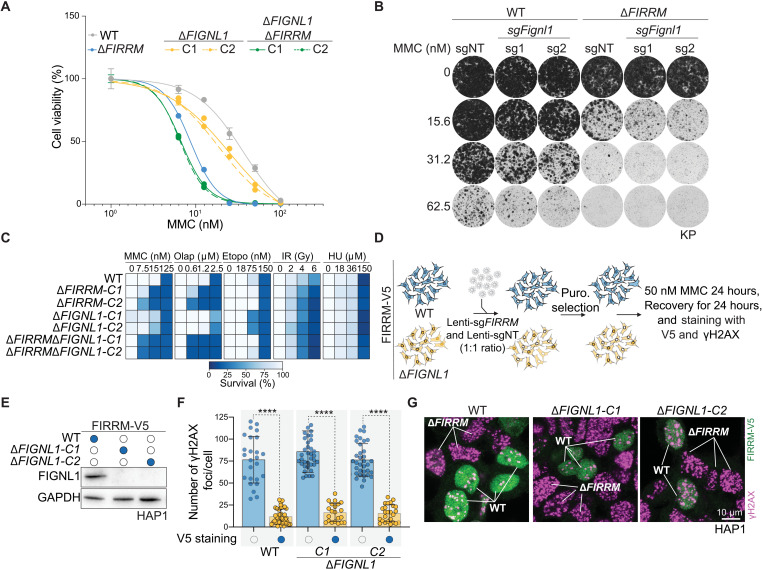
FIRRM has been found to interact with FIGL1 (FIGNL1 in humans) to limit meiotic crossovers in A. thaliana by modulating the DMC1/RAD51 activity in response to programmed DSBs generated by SPO11 (16). However, the effect of FIRRM was weaker than the impact observed for FIGNL1, suggesting that FIRRM could only be partially required for FIGNL1 activity. On the basis of these observations, and because FIGNL1 was not identified as a hit in our genetic screens, we hypothesized that FIRRM may have an additional function in ICL response independently of FIGNL1. To test this hypothesis, we knocked out FIGNL1 in a wild-type or FIRRM-deficient background and measured their sensitivity to MMC. As expected, FIGNL1 deficiency only modestly affected cellular survival after MMC exposure, while FIRRM single and FIRRM/FIGNL1 double mutant cells displayed hypersensitivity to MMC in different cell types (Fig. 4, A to B; fig. S9, A to D; and table S6). In line with this, FIRRM-deficient cells displayed distinct sensitivity profiles to different DNA damaging agents compared to FIGNL1-deficient cells (Fig. 4C). To exclude any clonal effects, we challenged a heterogeneous pool of cells with different levels of FIRRM protein with various concentrations of MMC in a wild-type or a FIGNL1-deficient background (fig. S9E). MMC treatment specifically depleted the cell population with low protein levels of FIRRM in a dose-dependent manner, regardless of whether the cells were wild type or FIGNL1 knockout (fig. S9, F to H). In addition, we measured γH2AX levels in the heterogeneous pool of cells with different levels of FIRRM protein either in a wild-type or FIGNL1-deficient background using our V5-tagged FIRRM cells (Fig. 4D). In line with the data above, loss of FIRRM increased γH2AX foci formation in both wild-type and FIGNL1 knockout cells (Fig. 4, E to G). The same results were obtained when assessing γH2AX levels in our clonal HAP1 knockout cell lines (fig. S10, A and B). Together, these data indicate that FIRRM has an additional function in the ICL response independent of FIGNL1, which we set out to further explore.
Fig. 4. FIGNL1-independent role of FIRRM in response to ICLs.
(A) Quantification of a clonogenic survival assay in response to different concentrations of MMC for wild-type, FIRRM, FIGNL1, single, or double knockouts in HAP1 cells. Data are shown as means ± SEM, normalized to untreated. n = 3. (B) Colony formation assay in KP wild-type or FIRRM knockout cells transduced with nontargeting (NT) or sgRNAs targeting Fignl1 and exposed to different concentrations of MMC for 8 days. n = 2. (C) HAP1 cells of indicated genotypes were exposed to the indicated DNA damaging agents for 8 days, and viability was quantified. Olap, olaparib; Etopo, etoposide; HU, hydroxyurea. Data are depicted as a heatmap, and values were normalized to untreated conditions. n = 2. (D) Schematic illustration of the experiment depicted in (E) to (G). In summary, FIRRM-V5 HAP1 wild-type or FIGNL1 knockout cells were transduced with a 1:1 mix of a nontargeting sgRNA and a sgRNA targeting FIRRM. After antibiotic selection, cells were exposed to 50 nM MMC for 24 hours, recovered for an additional 24 hours, and analyzed for FIRRM-V5 and γH2AX levels. (E) Western blot depicting the FIGNL1 knockout cells generated in the FIRRM-V5 cell line. GAPDH was used as a loading control. (F and G) Quantification (F) and representative images (G) of the experiment described in (D). Data are depicted as means ± SD. significance was calculated by a two-tailed t test. ****P < 0.0001. n = 2.
To investigate the role of FIRRM in the ICL response and examine the relation to other DNA repair factors, we performed a genetic screen in FIRRM knockout cells upon MMC treatment. Comparative analysis of this screen with the related screen carried out in wild-type cells revealed that dropout hits from both screens showed a high overlap of DNA repair factors, including many members of the FA pathway (Fig. 5A and fig. S11, A to B). Loss of FA factors showed additive sensitivity to MMC upon FIRRM depletion compared to the wild-type screens (Fig. 5A). This was further validated by survival assays, where double-mutant cells of FIRRM and different subunits of the FA pathway such as FANCA, FANCC, FANCD2, and FANCI exhibited stronger sensitivity to MMC compared to single mutants (Fig. 5, B to D, and fig. S11, C to K). Furthermore, FIRRM-deficient cells neither exhibited altered FANCD2 monoubiquitylation nor changed FANCD2 foci formation upon MMC treatment, in contrast to FANCA-deficient cells. (Fig. 5, E to G). In summary, loss of FIRRM or FA pathway factors leads to ICL hypersensitivity phenotypes; however, the activation and functionality of the FA pathway is retained in the absence of FIRRM.
Fig. 5. FIRRM acts independently of the FA pathway.
(A) Fishtail plots depicting the fitness genes in untreated wild-type HAP1 cells or MMC-treated wild-type and FIRRM knockout HAP1 cells. Hits with a false discovery rate (FDR)–corrected P value ≤ 0.05 are highlighted in light blue, and NQO1 and FIRRM are highlighted in orange. FA pathway members are marked in dark blue. (B to D) Quantification of clonogenic survival assays in HAP1 cells of indicated genotypes. Data are displayed as means ± SEM., normalized to untreated. (E) Whole-cell extracts of wild-type, ΔFANCA, or ΔFIRRM HAP1 cells treated with 50 nM MMC were immunoblotted for FANCD2. (F and G) Quantification (F) and representative images (G) of an immunofluorescence staining for FANCD2 in wild-type, ΔFANCA, or ΔFIRRM HAP1 cells exposed to 50 nM MMC. All experiments were performed in triplicates except the genetic screens, which were done in duplicates. ****P < 0.0001.
FIRRM mediates resolution of HR intermediates
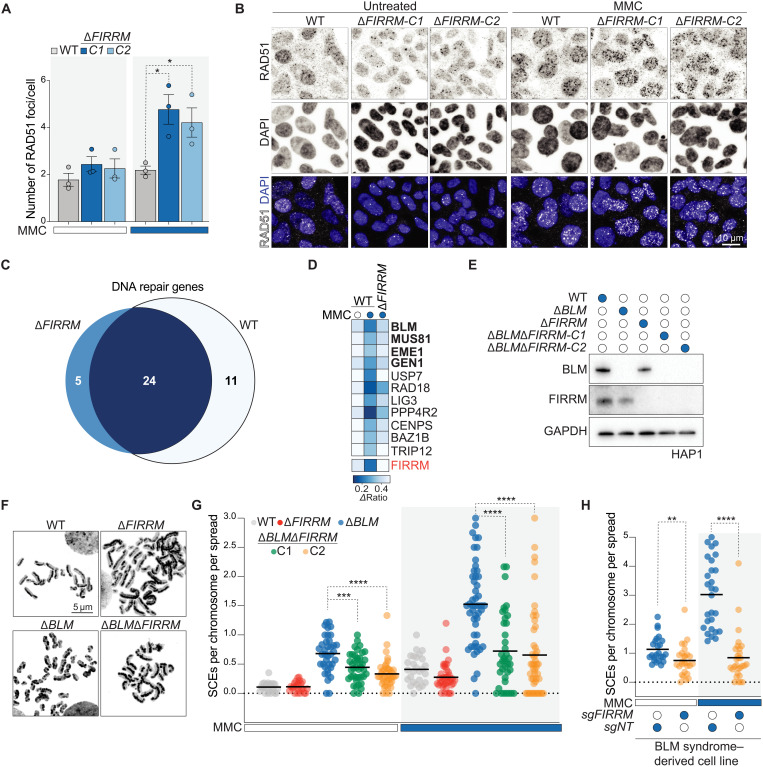
As demonstrated above, FIRRM is essential for embryonic survival in mice, as has also been observed for other HR genes, such as BRCA1/2 and RAD51. In addition, FIRRM loss triggers a specific mutational signature that has been previously observed in BRCA1 and BRCA2 mutant breast cancer genomes and is linked to HR deficiency. These data suggest a role for FIRRM in HR, which is further supported by the observation that FIRRM knockout cells displayed enhanced sensitivity to PARP inhibitors (PARPi) in wild-type and BRCA1/53BP1 double knockout cells (figs. S4, I to K and S12A). Next, we assessed the accumulation of both RPA and RAD51 in DNA repair foci. RPA readily formed MMC-induced foci in FIRRM-deficient cells at comparable levels to wild-type cells (fig. S12, B to C). However, FIRRM-depleted cells showed increased levels of RAD51 foci, which persisted on chromatin (Fig. 6, A to B, and fig. S12D), suggesting that the defect in FIRRM knockout cells occurs at a step downstream of DNA end resection and RAD51 recruitment to damaged replication forks. Although FIRRM depletion caused a mild defect in HR efficiency, as measured by the direct repeat–GFP (DR-GFP) assay (fig. S12, E to G), FIRRM-deficient cells displayed no sensitivity to ionizing radiation and etoposide (fig. S4, E and H). These data suggest that FIRRM functions specifically after the initiation of HR at damaged replication forks upon ICL exposure.
Fig. 6. FIRRM genetically interacts with factors resolving HR intermediates and promotes SCEs.
(A and B) HAP1 cells of indicated genotypes were left untreated or exposed to 50 nM MMC for 24 hours, pre-extracted, and stained for RAD51 and DAPI. Quantification of three independent biological replicates is shown in (A), and representative images are depicted in (B). P value was calculated by a two-tailed t test. At least 300 cells were analyzed per experiment, and data are depicted as means ± SEM. (C) Venn diagram of the DNA repair genes identified in the genetic screens in wild-type and FIRRM knockout cells after 12 days of MMC treatment. DNA repair genes that scored commonly in wild-type and FIRRM knockout cells (dark blue) and specifically in FIRRM knockout cells (light blue) are listed in fig. S11B. (D) Heatmap showing the DNA repair genes that were specifically identified in the wild-type but not in the FIRRM knockout screen. (E) Immunoblot of FIRRM and BLM single or double knockout HAP1 cell lines. (F) Representative images of metaphase spreads prepared from wild-type, ΔFIRRM, ΔBLM, or double knockout HAP1 cells treated with 50 nM MMC. (G) Quantification of SCE frequency. Every data point shows a metaphase that was scored blind for SCEs per chromosome per spread (>30 cells were analyzed for each condition). P value was calculated by the Mann-Whitney U test. (H) Quantification of SCE frequency in BS-derived patient cells (GM08505) transduced with a nontargeting sgRNA (sgNT) or sgFIRRM and exposed to 50 nM MMC or left untreated. P value was calculated by the Mann-Whitney U test. (B and E to H) Representative experiments of three independent experiments are displayed. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Comparative analysis of the wild-type and ΔFIRRM dropout screens highlighted a subset of genes, encoding factors that process HR intermediate structures, such as BLM, MUS81-EME1, and GEN1, which scored exclusively in the wild-type but not in FIRRM knockout cells upon MMC treatment (Fig. 6, C to D). This indicates that these proteins might function in the same pathway or act at the same level as FIRRM during ICL repair. Because these factors are involved in the dissolution and the resolution of HR intermediates in particular HJs (10, 12, 13, 21), we measured the end result of these structures after conversion by performing a sister chromatid exchange (SCE) assay on metaphase spreads of FIRRM and BLM single and double mutant cells. As expected, BLM-deficient cells displayed high levels of SCEs compared to wild-type cells (Fig. 6G) (11). In contrast, ΔFIRRM cells showed a mild decrease in SCEs after MMC treatment (Fig. 6, E to G). FIRRM deficiency significantly suppressed SCEs and reduced the number of harlequin chromosomes (chromosomes with more than five SCEs) in a BLM-deficient background (Fig. 6G and fig. S13A). The same observation was also made in a Bloom’s syndrome (BS)–derived cell line (GM08505) (Fig. 6H and fig. S13, B to C).
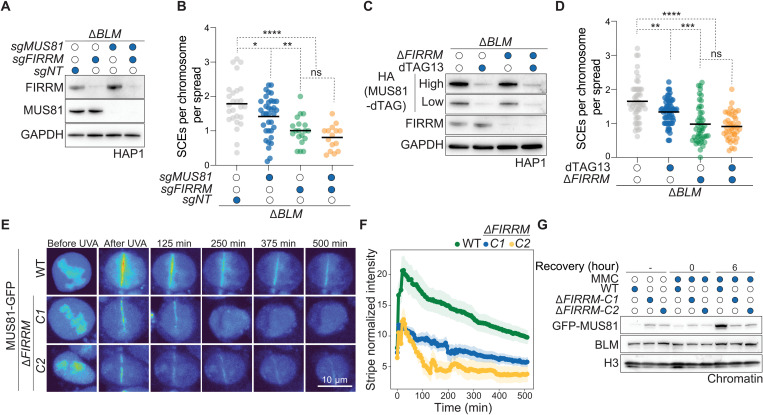
As previously reported, the high frequency of SCEs observed in BLM-deficient cells is caused by the activity of the resolution pathways, predominantly by the MUS81-EME1, SLX1-SLX4, and GEN1 nucleases (11, 12). MUS81 and EME1 were identified in our screens (but not SLX1 and SLX4), and GEN1 has been reported to modestly suppress SCE formation in BLM-deficient cells (11). Hence, we examined whether FIRRM affects SCE resolution by regulating either the activity or the recruitment of MUS81 to ICL repair sites. As observed above, FIRRM depletion led to a strong reduction of SCEs in a BLM-deficient background. MUS81 depletion caused a decrease in SCE frequency as previously reported (11). However, this effect was absent in FIRRM/MUS81 double-deficient cells (Fig. 7, A to B), suggesting that MUS81 functionality is hampered in the absence of FIRRM. Similar results were obtained by endogenous tagging of the MUS81 allele using the degradation tag (dTAG) technology (38) either in ΔBLM or ΔBLMΔFIRRM cells (Fig. 7, C to D). The predicted structure of FIRRM, using Alphafold2 (39), indicated a highly conserved region rich in HEAT repeats (characterized by helices linked by short loops), which are located in the annotated domain of unknown function (DUF4487) and are thought to mediate protein-protein interactions (fig. S5B). Thus, we hypothesized that FIRRM may mediate MUS81 recruitment to the ICL site. We found that GFP-tagged MUS81 was inefficiently recruited and poorly retained at sites of damage in FIRRM-deficient cells compared to wild-type controls upon psoralen/UVA treatment (Fig. 7, E to F). In agreement with this, MUS81 was not recruited to the chromatin after MMC treatment in FIRRM knockout cells, while the total MUS81 protein abundance was not affected. (Fig. 7G and fig. S13D). Moreover, FIRRM physically interacted with MUS81 in a BLM-independent manner (fig. S13, E to F). Collectively, our data show that FIRRM plays an important role during SCE resolution via the recruitment and the retention of MUS81 at sites of DNA damage.
Fig. 7. FIRRM mediates HR intermediate resolution via MUS81.
(A) Immunoblot of BLM-deficient HAP1 cells transduced with nontargeting, FIRRM, or MUS81 targeting sgRNAs. (B) Quantification of SCE frequency in BLM-deficient HAP1 cells treated with the indicated sgRNAs and challenged with 50 nM MMC. n = 2. (C) Immunoblot of HAP1 cells carrying a dTAG at the N terminus of MUS81 either in BLM single knockout (ΔBLM) or BLM and FIRRM double knockout (ΔBLMΔFIRRM) cells. Cells were pretreated with 250 nM dTAG13 for 24 hours, followed by treatment with 50 nM MMC and 250 nM dTAG13. (D) Quantification of SCEs of the conditions depicted in (C). (B and D) Every data point represents a metaphase that was scored blind for SCEs per chromosome per spread. P value was calculated by the Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (E and F) Recruitment of GFP-MUS81 to sites of laser UVA microirradiation after psoralen treatment in HAP1 cells (E). The graph represents the mean of stripe normalized intensity ± SEM for each time point (15 cells). n = 2 (F). (G) Chromatin fractions of wild-type or FIRRM knockout HAP1 cells untreated or treated with MMC at different recovery time points, which were immunoblotted with the indicated antibodies. n = 3.
DISCUSSION
In this study, we used complementary genetic loss-of-function screens in haploid cells (19, 22) to create a comprehensive dataset of the genes required for maintaining genome stability upon ICL exposure. In addition to known factors (such as FA proteins), we uncovered FIRRM, a poorly studied scaffold protein in human cells. Although FIRRM phenocopies several hallmarks of FA deficiency, including hypersensitivity to ICL-inducing agents, accumulation of DNA damage during S-G2 phase of the cell cycle, and chromosomal aberrations, the activation and functionality of the FA pathway are retained in the absence of FIRRM. Loss of FIRRM renders the cells exquisitely sensitive to ICL-inducing agents and PARPi but not to replication stress induced by hydroxyurea or to agents that directly generate DSBs. Our data suggest that FIRRM may affect DNA repair intermediates generated by agents that cause roadblocks during DNA replication. Moreover, we found that FIRRM is recruited to chromatin and to psoralen-induced ICLs during the later stages of the repair process, which is in line with previous work showing that FIRRM is recruited to psoralen–cross-linked chromatin later than FA factors using chromatin mass spectrometry in Xenopus egg extracts (40).
FIRRM has been previously linked to meiotic recombination in A. thaliana (16), where it physically interacts with FIGNL1 to limit meiotic crossovers by modulating DMC1/RAD51 activity. However, FIRRM depletion had less impact on DMC1/RAD51 foci increase and crossover frequencies than FIGNL1 loss, indicating that FIRRM activity is only partially required for FIGNL1 activity in plants. In mammalian cells, we find that, although FIRRM and FIGNL1 stabilize each other, they display distinct phenotypes. While FIRRM-deficient cells show hypersensitivity to ICL-inducing agents and accumulation of DNA damage, FIGNL1 knockout cells only show a very mild phenotype in these assays. Because FIGNL1 loss also affects FIRRM stability, it is tempting to suggest that the weak phenotype observed in FIGNL1-deficient cells might be caused by the lower FIRRM protein abundance. In addition, using coculture experiments, we found that FIRRM loss sensitizes cells to ICL-inducing agents and induces DNA damage in the absence of FIGNL1. Consistently, our data suggest that FIRRM has a function independent of FIGNL1 upon ICL exposure in mammalian cells.
By performing a genetic screen in FIRRM knockout cells, we uncovered a genetic interaction between FIRRM and structure-selective endonucleases (MUS81-EME1 and GEN1), as well as the BLM helicase. In human cells, BLM helicase acts together with topoisomerase IIIa, RMI1, and RMI2 (BTR complex) to dissolve HR intermediates such as double HJs (11, 13). The importance of this process is highlighted by the fact that mutations in BLM lead to a severe human genetic disorder known as BS, characterized by predisposition to a broad spectrum of early-onset cancers caused by genomic instability (1, 13). In addition, BS-derived cell lines show an increase in SCE frequency, which might be caused by alternative mechanisms for double HJ resolution (SLX-MUS and GEN1 nucleases) (10, 11).
Mechanistically, we show that FIRRM is recruited to process ICLs downstream of RAD51 to permit HR intermediate resolution via the recruitment and the retention of MUS81 at sites of DNA damage. In line with this, FIRRM loss potently suppresses the elevated levels of SCEs observed in BS cells and triggers genome instability, suggesting that FIRRM promotes SCEs in BLM-deficient cells. In plants, a FIRRM homolog has also been found to play an important role in meiotic recombination by acting as an anticrossover factor (16). Moreover, the rice homolog of FIRRM has been suggested to prevent the formation of abnormal heteroduplex intermediates and to regulate crossover formation during meiosis, where HR takes place between homologous chromosomes (17). In response to ICLs, these two functions seem to be conserved for mammalian FIRRM. First, it may regulate the formation of HR intermediates by dismantling RAD51 with or independently of FIGNL1. Second, FIRRM ensures the recruitment of structure selective endonucleases, such as MUS81, to act on unusual DNA structures that may be formed, for example, in the absence of the BTR complex. This is supported by an interaction of FIRRM and MUS81 during G2-M phase of the cell cycle; however, it remains to be elucidated how FIRRM influences the activity or recruitment of the BTR complex (fig. S14). It is conceivable that FIRRM may also recruit other proteins than MUS81, because MUS81 deficiency has a milder effect than FIRRM loss on SCE formation. However, it should be noted that MUS81 is not completely depleted in our setting due to its essentiality. Our genetic screens also highlighted EME1, a catalytically inactive regulatory subunit, as an important ICL response factor in a FIRRM-dependent manner. MUS81 interacts with EME1, which is required for the stability of the MUS81-EME1 complex (21). However, it remains unclear how EME1 modulates the MUS81-FIRRM interaction and whether this interaction is direct.
Furthermore, FIRRM deficiency is early embryonic lethal in mice, similar to what has been observed for several HR genes, such as Brca1/2 and Rad51, although direct comparison is complicated by differences in genetic background (35, 36, 41). We hypothesize that rapidly proliferating cells in FIRRM-deficient embryos may be exquisitely sensitive to endogenous DNA damage, such as aldehydes generated by metabolic processes, which may ultimately cause embryonic lethality. To study the role of FIRRM in tumorigenesis, we created a conditional mouse model where FIRRM is specifically deleted in the mammary epithelium. We found that homozygous loss of FIRRM accelerated mammary tumorigenesis induced by p53 loss. FIRRM might play a role not only in tumorigenesis but also in aging, because deleterious variants in FIRRM have been associated with increased ovarian aging and premature menopause (42).
Last, FIRRM loss triggers a specific mutational signature that has been previously observed in BRCA1 and BRCA2 mutant breast cancer genomes and is thought to result from HR deficiency (32–34). This is consistent with the observation that FIRRM deficiency sensitizes cancer cells to PARPi, even in PARPi-resistant genotypes such as BRCA1/53BP1 double-deficient cell lines. Whereas perturbation of FIRRM affects HR intermediate resolution and accelerates tumorigenesis, its loss may create a cancer cell vulnerability when combined with PARPi or ICL-inducing chemotherapeutics.
MATERIALS AND METHODS
Plasmids
Lentiviral sgRNA vectors were generated on the basis of lentiCRISPRv2 (Addgene 52961) or lentiGuide-Puro (Addgene 52963) combined with the Edit-R inducible Cas9 vector (Horizon). Guide RNA sequences used in this study are listed in table S3. To obtain the MUS81 overexpression construct, the cDNA sequence was obtained from pHAGE-P-CMVt-N-HA-GAW-MUS81 (Addgene 100157). The FIRRM sequence was codon-optimized and purchased from Twist Biosciences. Fragments were inserted into a modified version of pCW57.1 (Addgene 41393) containing an N-terminal GFP tag.
Reagents
The following compounds were used during this study: MMC (Tocris), ACT (Sigma-Aldrich), hydroxyurea (Sigma-Aldrich), etoposide (Selleckchem), cisplatin and actinomycin D (Thermo Fisher Scientific), camptothecin (Sigma-Aldrich), olaparib (Syncom), talazoparib (BMN-673, Selleckchem), veliparib (ABT-888, Selleckchem), doxycycline (Sigma-Aldrich), and dTAG-13 (Tocris).
Cell culture and generation of CRISPR-Cas9 edited cell lines
HAP1 cells (43) were cultured in Iscove’s modified Dulbecco’s medium (IMDM, Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich), 1% penicillin-streptomycin (Thermo Fisher Scientific), and 1% l-glutamine (Thermo Fisher Scientific). KP cells (KP3.33) (24) were grown in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 (DMEM/F-12) supplemented with 10% FCS, 1% penicillin-streptomycin, insulin (5 μg/ml; Sigma-Aldrich), and murine epidermal growth factor (5 ng/ml; Sigma-Aldrich). GM08505 (Coriell Biorepository) were maintained in minimum essential medium (MEM, Gibco) with 10% FCS, 1% penicillin-streptomycin, and 1% nonessential amino acids (Thermo Fisher Scientific). Human embryonic kidney 293T (HEK293T) [American Type Culture Collection (ATCC)] and U2OS DR-GFP cells (provided by J. Stark) were maintained in DMEM (Gibco) supplemented with 1% penicillin-streptomycin and 10% FCS. All cells were grown at 37°C and 5% CO2 and regularly tested for mycoplasma contamination.
For gene-editing experiments, cells at 80% confluency were transfected with lentiCRISPRv2 plasmids using Xfect (Takara) according to the manufacturer’s instructions. Twenty-four hours later, cells were refreshed with medium containing puromycin (HAP1, 1 μg/ml; KP, 2 μg/ml) and selected for an additional 48 hours. Single-cell clones were derived by FACS sorting (HAP1) or limiting dilution (KP). Successful editing of selected clones was verified by Sanger sequencing of the polymerase chain reaction (PCR) products, generated using primers listed in table S4 and TIDE analysis (44), or loss of the targeted protein was assessed by immunoblotting. Gene-editing outcomes are listed in tables S6 and S7.
Endogenous tagging of FIRRM and MUS81
A total of 106 HAP1 cells were seeded in a six-well plate. The following day, a synthesized guide RNA (IDT) and the corresponding trans-activating CRISPR (tracr) RNA (IDT) were mixed in equimolar concentrations to create a final duplex of 1 μM in nuclease-free duplex buffer. The complex was heated at 95°C for 5 min and allowed cooling down to room temperature (20° to 25°C). The sgRNA-tracrRNA complex (20 pmol) was then mixed with Cas9 nuclease (20 pmol; IDT) and a double-stranded DNA fragment containing a V5 and a Neogreen tag separated by a P2A site (V5-tag–P2A–Neogreen–stop codon) for C-terminal tagging of FIRRM and Neogreen–P2A–HA–dTAG for N-terminal tagging of MUS81. The mixture was then transfected in HAP1 cells using Lipofectamine CRISPRMAX Cas9 Transfection Reagent (CMAX00001, Thermo Fisher Scientific) according to the manufacturer’s instructions. After 3 days, Neogreen-positive cells were sorted in 96-well plates and the clones were tested for correct fragment integration using PCR and subsequent Sanger sequencing.
Lentiviral transductions
HEK293T cells (2.5 × 106) were plated in a 10-cm dish the day before they were transfected with packaging plasmids: 7.5 μg of VSV-G (Addgene 8454), 3.75 μg of psPAX-2 (Addgene 12260), and 10 μg of the target vectors using calcium phosphate. Viral supernatant was collected 48 hours after transfection, filtered, and used for transduction in the presence of polybrene (8 μg/ml; Sigma-Aldrich). For pCW57.1-GFP-MUS81 and pCW57.1-GFP-FIRRM constructs, viral supernatant was harvested, concentrated by ultracentrifugation at 20,000 rpm for 2 hours, and resuspended in 400 μl of phosphate-buffered saline (PBS; Gibco). To select for transduced cells, cells were treated with puromycin (KP, GM08505: 2 μg/ml; HAP1: 1 μg/ml) or blasticidin (15 μg/ml) for 48 hours.
Haploid genetic screens
ICL survival screens
The screens were performed as described previously (19). Briefly, gene-trap retrovirus used for the mutagenesis of HAP1 cells was produced using HEK293T cells transfected with a gene-trap vector containing blue fluorescent protein (BFP) to assess the efficiency of the mutagenesis (19) and Gag-pol, VSVg, and pAdv as packaging plasmids. The supernatant was harvested 48 and 72 hours after transfection, filtered, concentrated using Amicon filters, and stored at 4°C. Retrovirus of both harvests were combined, supplemented with protamine sulfate (8 μg/ml), and used to infect 40 × 106 wild-type or FIRRM knockout HAP1 cells. After expansion, the mutagenized cells were treated either with MMC (screen in wild-type cells: 65 nM or FIRRM knockout cells: 5 nM) or 1.25 mM ACT and cultured for 12 days, with splitting the cells every 3 days and refreshing the medium with MMC or ACT. Cells were then harvested with trypsin, resuspended in medium, pelleted, and washed once with PBS. Next, they were fixed with BD fix buffer I for 10 min at 37°C, washed twice with PBS, and permeabilized on ice with BD Perm Buffer III for 30 min. Cells were then stained for 1 hour at room temperature with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) to visualize G1 cells. A total of 32 × 106 G1 cells were then sorted using a BD FACSAria Fusion Cell Sorter. Genomic DNA was isolated, and insertion sites were amplified using a linear amplification PCR. The data analysis and the insertions were mapped as described in Blomen et al. (19) with some modifications. Briefly, the unique reads were aligned to the hg38 human genome with zero or one mismatch using Bowtie. The reads were then assigned to protein-coding genes taking the longest open reading frame transcript in consideration, excluding overlapping regions that cannot be attributed to a single gene. Unique alignments were counted in the intronic regions between the transcription initiation site and the stop codon. Enrichment of genes in the sense or antisense orientation of the gene-trap insertions were identified applying a false discovery rate–corrected binomial test (FDR-corrected P value cutoff of 0.05), and the genes were tested for significance after the chemical (MMC or ACT) or genetic (ΔFIRRM) perturbations compared to wild-type control cells using a bidirectional Fisher’s exact test with four independent control datasets (FDR-corrected P value cutoff of 0.05).
FACS-based phenotypic screen for γH2AX after MMC treatment
Wild-type HAP1 cells were mutagenized as described for the ICL survival screens. Cells were then expanded to 3 × 109, treated with 65 nM MMC for 24 hours, and left to recover in MMC-free medium for 24 hours after one PBS wash. Cells were incubated with 20 μM EdU (Invitrogen) for 1 hour to label cells undergoing DNA replication and then harvested with trypsin, resuspended, pelleted and washed once with PBS. Subsequently, the cells were fixed with 2% paraformaldehyde (PFA) for 10 min at RT, washed twice with PBS containing 10% FCS (FACS buffer), and permeabilized on ice with BD Perm Buffer III (BD) for 30 min. Next, cells were washed once with FACS buffer and stained for 1 hour at room temperature with anti-γH2AX antibody. After two washes with PBS, the secondary antibody (Alexa 488, Invitrogen) was added for 1 hour at room temperature. EdU labeling was performed using the Click-iT Plus EdU Flow Cytometry Kit (Alexa 647, Invitrogen) following the manufacturer’s instructions, and samples were further stained with DAPI. Last, cells were filtered through a 40-μm strainer (BD Falcon), and EdU-positive cells with ~5% of the lowest and highest of γH2AX were sorted using a BD FACSAria Fusion Cell Sorter. Genomic DNA was isolated using a DNA mini kit (QIAGEN), and library preparation for deep sequencing and data analysis were carried out as described by Brockmann et al. (22) with alignment to the hg38 human genome.
Colony formation assay
Five thousand cells were seeded in six-well plates, and the day after, DNA damaging agents were added at indicated concentrations. Cells were maintained for 8 days until visible colonies formed. Cellular viability was determined using the CellTiter-Blue reagent (Promega). After the measurement, plates were fixed with 3.7% formaldehyde and stained with a 0.1% crystal violet solution.
Two-color competition growth assay
A total of 2 × 105 wild-type HAP1 cells were stably labeled with enhanced GFP (eGFP) using viral transduction mixed with 2 × 105 ΔFIRRM stably expressing mCherry in 10-cm dishes. The following day, cells were treated with various DNA damaging agents at the indicated concentrations. After 8 days, the cells were analyzed by flow cytometry and the mCherry/eGFP ratios were calculated.
Flow cytometry analysis
A total of 3 × 106 HAP1 cells were seeded in 10-cm dishes and treated as indicated. To label S phase cells, samples were pulsed with 20 μM EdU (Invitrogen) 1 hour before collection. Cells were trypsinized, washed once with PBS, and fixed with BD Phosflow Fix Buffer I for 10 min at 37°C. Next, cells were washed once with PBS and permeabilized using BD Perm Buffer III for 30 min on ice. Samples were then washed once with FACS buffer (10% FCS in PBS) and incubated with primary antibodies (see table S5) in FACS buffer for 1 hour at room temperature. After two washes with FACS buffer, secondary antibodies (Alexa 647/488, Invitrogen) were added for 1 hour at room temperature and samples were washed again twice. EdU labeling was performed by incubating samples with 100 mM Tris-HCl (pH 8.5), 4 mM CuSO4, 10 mM ascorbic acid, and 10 μM Fluor-Azide 488 (1:1000; ATTO) for 30 min. Last, cells were washed once, stained with DAPI (2.5 μg/ml), and analyzed on a BD LSRFortessa Cell Analyzer.
Protein extracts and immunoblotting
Cell pellets were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with phosphatase and protease inhibitors (Roche). After sonication, protein concentrations were determined using the bicinchoninic acid assay (BCA; Pierce, Thermo Fisher Scientific). Equal amounts of protein were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Bio-Rad). Proteins were detected by primary antibodies mentioned in table S5 following standard procedures.
Chromatin fractionation
Cell pellets were resuspended in hypotonic buffer [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and 1 mM dithiothreitol (DTT)] supplemented with phosphatase and protease inhibitors (Roche) for 10 min. After centrifugation, cytosol fraction was removed and nuclei were washed in hypotonic buffer and resuspended in hypertonic buffer [20 mM Hepes (pH 7.9), 200 mM NaCl, 10% glycerol, 1 mM DTT, and 0.5% Triton X-100) with phosphatase and protease inhibitors for 20 min. Nuclear extracts were removed after centrifugation. To obtain chromatin fraction, pellets were resuspended in hypertonic buffer, sonicated (5′, 30%), and centrifuged.
Immunoprecipitation
HAP1 cells (15 × 106) stably expressing GFP-tagged FIRRM or MUS81 were seeded in 15-cm dishes, and protein expression was induced with doxycycline (2 μg/ml) for 24 hours. Next, cells were treated with 50 nM MMC or vehicle for an additional 24 hours in the presence of doxycycline. Cells were then washed with ice-cold PBS, scraped, and lysed with 0.5 ml of lysis buffer [20 mM Hepes (pH 8), 200 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 mM DTT, and 1× EDTA-free protease inhibitor cocktail (Roche)] on ice for 30 min. Subsequently, extracts were treated with Benzonase (Merck) for 30 min on ice and subsequently centrifuged at 20,000g for 15 min at 4°C and quantified using a BCA assay kit (Pierce, Thermo Fisher Scientific). One milligram of the extracts was incubated with 20 μl of GFP-Trap magnetic beads (ChromoTek) in immunoprecipitation (IP) buffer [50 mM Hepes (pH 8), 200 mM NaCl, 0.5% NP-40, and 1 × EDTA-free protease inhibitor cocktail (Roche, Basel)] overnight at 4°C, washed five times with the IP buffer, and eluted in sample buffer by boiling samples for 5 min at 95°C. Pull-downs and whole-cell extracts were separated by SDS-PAGE gels, followed by immunoblotting with indicated antibodies.
NanoSeq and analysis of mutational signatures
To assess the mutational landscape in wild-type cells (i.e., a pool of four different wild-type subclones) and FIRRM knockout cells, HAP1 cells were passaged in the presence of EC20 (effective concentration) of MMC (wild type: 50 nM; FIRRM-KO: 15 nM) for eight consecutive days. Subsequently, genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN). To determine the mutational impact of different knockouts with and without MMC treatment, we used the NanoSeq sequencing method (27). NanoSeq is a duplex-sequencing–based method, which avoids traditional end repair during library preparation to obtain extremely low error rates of <5 errors per billion base calls. To avoid end repair, we used the blunt-end HpyCH4V restriction enzyme. Samples were sequenced on Illumina NovaSeq S4 flow cells to a depth equivalent to 15× haploid human genome equivalents, allowing the calling of an average of 1.5 × 109 duplex bases per sample (~0.5× human haploid genomes; minimum of 9.1 × 108, maximum of 2.7 × 109 duplex bases). Sequencing data were preprocessed as in (27) (https://github.com/cancerit/NanoSeq/blob/master/README.md; last accessed 20 January 2023) and mapped to the GRCh38 human genome assembly. NanoSeq sequencing data were submitted to the European Nucleotide Archive (ENA) project under accession number EGAD00001010298. To filter out germline single-nucleotide polymorphisms, we used whole-genome sequencing data from the human HAP1 parental cell line, which was generated by paired-end sequencing on the Illumina HiSeq X sequencing platform. Reads were mapped to the GRCh38 reference genome. Sequencing data were submitted to the ENA under the accession nos. PRJEB26750, ERP108763, and SAMEA4663622. Because the parental HAP1 cell line comes from a different project and is distantly related to the one used for the samples at hand, much shared background between our samples was not filtered with the matched normal. We applied an additional filter to remove the shared background, discarding mutation calls in a sample when they are seen in any of the other samples. More precisely, mutations called in a sample were removed if 10 or more reads supported that mutation across all the other samples or if one of the samples had three or more supporting reads. Reassuringly, this filter mainly removed mutations in the C>A channel, typical of oxidative damage during in vitro growth. Burdens (mutations per base pair) and substitution and indel profiles were normalized as explained in (27), (https://github.com/cancerit/NanoSeq/blob/master/README.md; last accessed 20 January 2023). Signature extraction was done with R package Sigfit (45). We run 1000 iterations with two to five signatures to find out the optimal number of signatures according to goodness of fit. Both the cosine and L2 metrics agreed on three signatures as best explaining the data. We then extracted these three signatures using 10,000 iterations and fitted the signatures to each sample using 2000 iterations and discarding the first 1000 as warm up. Similarities with COSMIC signatures (46) were calculated using the lsa package cosine function (https://cran.r-project.org/web/packages/lsa/; last accessed 9 May 2022).
Generation of Firrm (BC055324) mouse models
All animal experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute and were executed according to national and European guidelines. Firrm knockout and Firrm conditional knockout animals were generated on an FVB/NRj background using pronuclear microinjection in mouse zygotes. The injection mixture consisted of water with Cas9 protein (200 ng/μl; IDT), two sgRNAs (25 ng/μl each) targeting the intronic sequence surrounding exon 7 of Firrm (5′-GTATGTCGTTCTCACCACGCAGG-3′; 5′-ATACTCATGGCTCCGCTGCGTGG-3′), and a long single-stranded DNA oligo (15 ng/μl) containing exon 7 of Firrm flanked by two loxP recombination sites and homology arms. Founder mice were screened for the conditional and the knockout allele by PCR, and modifications were verified by Sanger sequencing. FirrmF/F animals were crossed with Wap-Cre;Trp53F/F (37) to specifically abolish Firrm expression in the mammary epithelium.
Embryo isolations
Matings were started, and upon the presence of a copulation plug, females were considered to be E0.5. Females were euthanized at indicated times of embryonic development, and uteri were isolated. Subsequently, embryos were collected and lysed in DirectPCR lysis reagent (Viagen Biotech) for genotyping. Firrm locus was genotyped using the following primers: 5′-TGTGCACGTACGAAGTTCCTT-3′, 5′-GCCAAGATTTGGTGTGCTGG-3′, and 5′-AGGGTTGAACAACTCAAGTGC-3′.
Monitoring of tumor development
Animals were monitored for tumor growth once per week, starting from 3 weeks of age. Upon occurrence of a tumor, animals were checked twice per week. Mice were euthanized by CO2 once the tumor volume reached 1500 mm3, and tumor pieces were collected.
Organoid generation
Firrm−/−;Trp53−/− and Trp53−/− organoids were derived from cryopreserved mammary tumor pieces and processed as previously described by Duarte et al. (47). Briefly, tumor material was dissected and digested using collagenase A (2 mg/ml; Gibco) in advanced DMEM/F12, washed in growth medium (advanced DMEM/F12, 10 mM Hepes, GlutaMAX, and penicillin-streptomycin) and filtered through a cell strainer. Organoids were then cultured in growth medium further supplemented with B27 (Gibco), 125 μM N-acetyl-l-cysteine (Sigma-Aldrich), and murine epidermal growth factor (50 ng/ml; Sigma-Aldrich). Cells were embedded in 1:1 Culturex Reduced Growth Factor Basement Membrane Extract (BME) Type 2 (Trevigen) mixed with medium and maintained at 37°C and 5% CO2. To establish organoid cultures, cells were supplemented with 5 μM nutlin (Sigma-Aldrich) for the initial 3 weeks of culture.
Viability assay in murine tumor organoids
Organoids were collected in advanced DMEM/F12 and dissociated into a single-cell suspension using TrypLE (Gibco). Next, samples were counted, and 1 × 105 cells were plated in 24-well plates in 40-μl drops composed of 1:1 BME and organoid growth medium, and drops were left to solidify for 30 min at 37°C. Medium containing indicated concentrations of MMC was added to the wells, and the viability of the formed organoids was determined after 7 days using CellTiter-Blue reagent (Promega) following the manufacturer’s instructions.
Immunofluorescence
Cells were seeded in eight-well chambers (Thermo Fisher Scientific) and treated as indicated in the figure legends. To stain for FIRRM-V5, RAD51, or RPA foci in cells, slides were pre-extracted with 0.2% Triton X-100 in PBS for 2 min on ice to remove nonchromatin bound protein and washed with PBS twice. Cells were fixed with 2% PFA or ice-cold 100% methanol (for RPA staining in KP) for 10 min. After permeabilization for 1 hour in immunofluorescence (IF) buffer (0.2% Triton X-100 and 10% FCS in PBS), slides were incubated with primary antibodies in IF buffer for 1 hour at room temperature. Cells were then washed three times and incubated with secondary antibodies (Invitrogen) and DAPI (2.5 μg/ml; Sigma-Aldrich) for 1 hour. Then, slides were washed again and mounted in Aqua-Poly/Mount mounting medium (Polysciences). Images were taken on a Leica SP5 confocal microscope (Leica Microsystems) or ZEISS LSM 980 Airyscan. Quantification of DNA damage–induced foci was performed in Fiji using CLIJ2/x libraries (48) with a custom ImageJ macro (v1.3; https://github.com/BioImaging-NKI/Foci-analyzer; last accessed on 10 February 2023). Briefly, nuclei are recognized in the DAPI channel. DNA damage foci are detected by applying marker-controlled watershed (49) on local maxima in the foci channel, after which foci numbers and intensities are quantified per cell.
Laser microirradiation and live cell imaging
HAP1 cells expressing the indicated GFP-tagged proteins were grown on Nunc Lab Tek II chambered cover glasses (Thermo Fisher Scientific). Protein expression was induced using doxycycline (2 μg/ml) for 24 hours. Next, cells were pretreated with 10 μM 8-methoxypsoralen (Sigma-Aldrich) for 1 hour before UVA microirradiation using a 355-nm laser (CNI, AO-S-355-40 mW, power: 10 μW, power density: 50 to 100 W/cm2) focused through a 63× oil objective with numerical aperture of 1.4 to yield a stripe size of 20 × 0.5 μm. The imaging was performed on a Leica STELLARIS 8 confocal microscope, and a white light laser at 488 nm was used (10% power). Stripes were imaged for 10 hours after UVA microirradiation.
Chromosomal aberrations
To assess chromosomal aberrations, cells were either left untreated or incubated with 50 nM MMC for 24 hours. Afterward, cells were incubated in 80 nM calyculin A (Biomol) and collected in serum-free medium. Suspensions were incubated in 0.075 M KCl solution for 7 min at 37°C. Cells were pelleted and fixed dropwise with methanol:acetic acid (4:1), and then centrifugation and fixative replacement were repeated twice. Subsequently, cell suspension was dropped on microscope slides (Thermo Fisher Scientific) and left to air dry overnight. Telomere fluorescence in situ hybridization was performed by rehydrating spreads in PBS, fixing slides in 3.7% formaldehyde, and incubating slides in pepsin (1 mg/ml; Sigma-Aldrich) dissolved in 10 mM glycine (pH 2.0) for 10 min. Spreads were then washed in PBS and dehydrated in different ethanol dilutions (70, 95, and 100%) for 5 min each. To visualize telomeres, slides were stained with a PNA probe (Alexa Fluor 488 C-rich telomere probe, Eurogentec) in hybridization buffer [10 mM tris-HCl (pH 7.2), 70% formamide, and 0.5% blocking reagent (Merck)] and incubated for 8 min at 80°C. Slides were left to hybridize overnight and then washed twice with a wash buffer I [70% formamide and 10 mM tris-HCl (pH 7.0)] and thrice with wash buffer II [0.1 M tris-HCl (pH 7.0), 0.15 M NaCl, and 0.08% Tween 20]. Last, slides were dehydrated in an ethanol series as described above. Slides were then mounted in VECTASHIELD Antifade Mounting Medium (Vector Laboratories) and imaged using an Axio Imager Z2 microscope coupled to the Metafer 4 software (MetaSystems). Images were quantified manually, in a blinded fashion.
SCE assay
To assess SCEs, cells were treated with 10 μM 5-bromo-2′-deoxyuridine (Thermo Fisher Scientific) for 48 hours. Next, either fresh medium or medium containing 50 nM MMC was added. 18 hours later, KaryoMAX colcemid (0.3 μg/ml; Thermo Fisher Scientific) was added to the plates for 2.5 hours. After trypsinization, cell suspensions were centrifuged, resuspended in 0.075 M KCl, and incubated at 37°C for 7 min. Next, cells were fixed by dropwise addition in ice-cold 3:1 methanol:acetic acid and stored at 4°C overnight. The next day, cells were pelleted, resuspended in 1 ml of fixative, and dropped on microscope slides (Thermo Fisher Scientific). The day after, slides were rehydrated for 5 min in PBS and stained with Hoechst 33342 (2 μg/ml; Thermo Fisher Scientific) in 2× SSC (pH 7.0) for 15 min. Subsequently, slides were covered with a thin layer of 2× SSC and irradiated with UVC (5400 J/m2; UV Crosslinker CX-2000, VWR). Afterward, slides were dehydrated in a series of ethanol dilutions (70, 95, and 100%) 5 min each and left to air-dry. Slides were imaged and were quantified manually, in a blinded fashion.
DR-GFP assay
A total of 2.5 × 105 U2OS DR-GFP cells (provided by J. Stark) were transduced with lentiCRISPRv2 constructs containing a nontargeting sgRNA or sgRNAs targeting FIRRM or RAD51, respectively. Three days after transduction, 2.5 × 105 cells were plated in six-well plates and transfected with 7.5 μg of I-SceI–expressing plasmid coupled to mCherry using Xfect (Takara). Medium was refreshed the next day, and 48 hours after transfection, cells were collected, and before measuring samples on an LSR Fortessa flow cytometer (BD), DAPI (2.5 μg/ml) was added to label dead cells. Samples were gated for live cells (DAPI negative), and doublets were excluded on the basis of forward-side scatter. Subsequently, mCherry–I-SceI–positive cells were selected, and proportion of GFP-positive cells was determined and normalized to the wild-type control.
Statistical analysis
Statistical analysis used in this manuscript is indicated in the corresponding figure legends or Materials and Methods.
Acknowledgments
We would like to thank the T.R.B. and the J.J. laboratories for helpful discussions; members of Sixma laboratory, particularly S. Hsiao Lee; A. Murachelli and Patrick Celie from the NKI protein facility for the helpful discussion; and P. Knipscheer for reading the manuscript and helpful discussion. We also thank C. Lutz for help with the generation of lentiviruses, D. Zimmerli and H. van der Gulden for help with animal experiments, and the NKI Animal facility, Flow cytometry facility, and Genomics Core facility for technical support. Schematic illustrations were adapted from BioRender.com.
Funding: Research at the Netherlands Cancer institute is supported by institutional rants of the Dutch Cancer Society and the Dutch Ministry of Health, Welfare and Sport. The T.R.B. laboratory was supported by the NWO Vici grant 016.Vici.170.033 and the Oncode Institute. A.M. is supported by an EMBO Long Term Fellowship (ALTF 158-2018), and S.C.M. received a Boehringer Ingelheim Fonds PhD Fellowship. D.J.A. and M.D.C.V-H. were funded by the Wellcome Trust and Cancer Research UK. F.A. is supported by the Cancer Research UK (C57387/A21777). Work in J.J.’s laboratory is funded by the Oncode Institute, which is partly financed by the Dutch Cancer Society.
Author contributions: Conceptualization: A.M., S.C.M., J.J., and T.R.B. Methodology: A.M., S.C.M., F.A., M.D.C.V-H., I.v.d.H., D.JA., .B.v.d.B., K.J., M.H., A.P.D., E.v.d.B., and L.J.K. Investigation: A.M. and S.C.M. Visualization: A.M. and S.C.M. Funding acquisition: A.M., S.C.M., J.J., and T.R.B. Project administration: A.M., J.J., and T.R.B. Supervision: A.M., J.J., and T.R.B. Writing—original draft: A.M., S.C.M., J.J., and T.R.B. Writing—review and editing: A.M., S.C.M., J.J., T.R.B., F.A., and D.J.A.
Competing interests: T.R.B. is a co-founder and scientific advisor of Scenic Biotech. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All the generated raw DNA datasets of the genetic screens were deposited in the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov/sra/PRJNA934905). Data of the γH2AX FACS-based haploid genetic screen are also available on an interactive visualization platform (https://phenosaurus.nki.nl/). NanoSeq sequencing data are submitted to the ENA project under accession number EGAD00001010298. The HAP1 cells can be provided by the Netherlands Cancer Institute pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to T.R.B.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S3 to S7
Legends for tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.A. J. Deans, S. C. West, DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11, 467–480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. C. Kottemann, A. Smogorzewska, Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Raschle, P. Knipscheer, M. Enoiu, T. Angelov, J. Sun, J. D. Griffith, T. E. Ellenberger, O. D. Schärer, J. C. Walter, Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Wang, S. Wang, A. Dhar, C. Peralta, N. P. Pavletich, DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature 580, 278–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. Klein Douwel, R. A. C. M. Boonen, D. T. Long, A. A. Szypowska, M. Räschle, J. C. Walter, P. Knipscheer, XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 54, 460–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. Knipscheer, M. Räschle, A. Smogorzewska, M. Enoiu, T. V. Ho, O. D. Schärer, S. J. Elledge, J. C. Walter, The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.F. Paques, J. E. Haber, Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. W. Szostak, T. L. Orr-Weaver, R. J. Rothstein, F. W. Stahl, The double-strand-break repair model for recombination. Cell 33, 25–35 (1983). [DOI] [PubMed] [Google Scholar]
- 9.M. Bzymek, N. H. Thayer, S. D. Oh, N. Kleckner, N. Hunter, Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. C. Y. Ip, U. Rass, M. G. Blanco, H. R. Flynn, J. M. Skehel, S. C. West, Identification of Holliday junction resolvases from humans and yeast. Nature 456, 357–361 (2008). [DOI] [PubMed] [Google Scholar]
- 11.T. Wechsler, S. Newman, S. C. West, Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471, 642–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.H. D. M. Wyatt, S. Sarbajna, J. Matos, S. C. West, Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell 52, 234–247 (2013). [DOI] [PubMed] [Google Scholar]
- 13.L. Wu, I. D. Hickson, The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003). [DOI] [PubMed] [Google Scholar]
- 14.S. Fekairi, S. Scaglione, C. Chahwan, E. R. Taylor, A. Tissier, S. Coulon, M. Q. Dong, C. Ruse, J. R. Yates III, P. Russell, R. P. Fuchs, C. H. McGowan, P. H. L. Gaillard, Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J. M. Svendsen, A. Smogorzewska, M. E. Sowa, B. C. O'Connell, S. P. Gygi, S. J. Elledge, J. W. Harper, Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. B. Fernandes, M. Duhamel, M. Seguéla-Arnaud, N. Froger, C. Girard, S. Choinard, V. Solier, N. de Winne, G. de Jaeger, K. Gevaert, P. Andrey, M. Grelon, R. Guerois, R. Kumar, R. Mercier, FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLOS Genet. 14, e1007317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Q. Hu, Y. Li, H. Wang, Y. Shen, C. Zhang, G. du, D. Tang, Z. Cheng, Meiotic chromosome association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29, 1697–1708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Xu, M. Ali, W. Duan, X. Yuan, F. Garba, M. Mullen, B. Sun, I. Poser, H. Duan, J. Lu, R. Tian, Y. Ge, L. Chu, W. Pan, D. Wang, A. Hyman, H. Green, L. Li, Z. Dou, D. Liu, X. Liu, X. Yao, Feedback control of PLK1 by Apolo1 ensures accurate chromosome segregation. Cell Rep. 36, 109343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.V. A. Blomen, P. Májek, L. T. Jae, J. W. Bigenzahn, J. Nieuwenhuis, J. Staring, R. Sacco, F. R. van Diemen, N. Olk, A. Stukalov, C. Marceau, H. Janssen, J. E. Carette, K. L. Bennett, J. Colinge, G. Superti-Furga, T. R. Brummelkamp, Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 20.P. J. Brooks, M.-A. Enoch, D. Goldman, T.-K. Li, A. Yokoyama, The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLOS Med. 6, e50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. Sarbajna, S. C. West, Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 39, 409–419 (2014). [DOI] [PubMed] [Google Scholar]
- 22.M. Brockmann, V. A. Blomen, J. Nieuwenhuis, E. Stickel, M. Raaben, O. B. Bleijerveld, A. F. M. Altelaar, L. T. Jae, T. R. Brummelkamp, Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature 546, 307–311 (2017). [DOI] [PubMed] [Google Scholar]
- 23.G. S. Stewart, B. Wang, C. R. Bignell, A. M. Taylor, S. J. Elledge, MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003). [DOI] [PubMed] [Google Scholar]
- 24.B. Evers, R. Drost, E. Schut, M. de Bruin, E. van der Burg, P. W. B. Derksen, H. Holstege, X. Liu, E. van Drunen, H. B. Beverloo, G. C. M. Smith, N. M. B. Martin, A. Lau, M. J. O'Connor, J. Jonkers, Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin. Cancer Res. 14, 3916–3925 (2008). [DOI] [PubMed] [Google Scholar]
- 25.C. Z. Zhang, A. Spektor, H. Cornils, J. M. Francis, E. K. Jackson, S. Liu, M. Meyerson, D. Pellman, Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. A. Knouse, A. Amon, Cell biology: The micronucleus gets its big break. Nature 522, 162–163 (2015). [DOI] [PubMed] [Google Scholar]
- 27.F. Abascal, L. M. R. Harvey, E. Mitchell, A. R. J. Lawson, S. V. Lensing, P. Ellis, A. J. C. Russell, R. E. Alcantara, A. Baez-Ortega, Y. Wang, E. J. Kwa, H. Lee-Six, A. Cagan, T. H. H. Coorens, M. S. Chapman, S. Olafsson, S. Leonard, D. Jones, H. E. Machado, M. Davies, N. F. Øbro, K. T. Mahubani, K. Allinson, M. Gerstung, K. Saeb-Parsy, D. G. Kent, E. Laurenti, M. R. Stratton, R. Rahbari, P. J. Campbell, R. J. Osborne, I. Martincorena, Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021). [DOI] [PubMed] [Google Scholar]
- 28.S. Nik-Zainal, J. E. Kucab, S. Morganella, D. Glodzik, L. B. Alexandrov, V. M. Arlt, A. Weninger, M. Hollstein, M. R. Stratton, D. H. Phillips, The genome as a record of environmental exposure. Mutagenesis 30, 763–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. E. Kucab, X. Zou, S. Morganella, M. Joel, A. S. Nanda, E. Nagy, C. Gomez, A. Degasperi, R. Harris, S. P. Jackson, V. M. Arlt, D. H. Phillips, S. Nik-Zainal, A compendium of mutational signatures of environmental agents. Cell 177, 821–836.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Nik-Zainal, L. B. Alexandrov, D. C. Wedge, P. Van Loo, C. D. Greenman, K. Raine, D. Jones, J. Hinton, J. Marshall, L. A. Stebbings, A. Menzies, S. Martin, K. Leung, L. Chen, C. Leroy, M. Ramakrishna, R. Rance, K. W. Lau, L. J. Mudie, I. Varela, D. McBride, G. R. Bignell, S. L. Cooke, A. Shlien, J. Gamble, I. Whitmore, M. Maddison, P. S. Tarpey, H. R. Davies, E. Papaemmanuil, P. J. Stephens, S. McLaren, A. P. Butler, J. W. Teague, G. Jönsson, J. E. Garber, D. Silver, P. Miron, A. Fatima, S. Boyault, A. Langerød, A. Tutt, J. W. M. Martens, S. A. J. R. Aparicio, Å. Borg, A. V. Salomon, G. Thomas, A.-L. Børresen-Dale, A. L. Richardson, M. S. Neuberger, P. A. Futreal, P. J. Campbell, M. R. Stratton; Breast Cancer Working Group of the International Cancer Genome Consortium , Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Zamborszky, B. Szikriszt, J. Z. Gervai, O. Pipek, Á. Póti, M. Krzystanek, D. Ribli, J. M. Szalai-Gindl, I. Csabai, Z. Szallasi, C. Swanton, A. L. Richardson, D. Szüts, Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene 36, 746–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Nik-Zainal, H. Davies, J. Staaf, M. Ramakrishna, D. Glodzik, X. Zou, I. Martincorena, L. B. Alexandrov, S. Martin, D. C. Wedge, P. Van Loo, Y. S. Ju, M. Smid, A. B. Brinkman, S. Morganella, M. R. Aure, O. C. Lingjærde, A. Langerød, M. Ringnér, S.-M. Ahn, S. Boyault, J. E. Brock, A. Broeks, A. Butler, C. Desmedt, L. Dirix, S. Dronov, A. Fatima, J. A. Foekens, M. Gerstung, G. K. J. Hooijer, S. J. Jang, D. R. Jones, H.-Y. Kim, T. A. King, S. Krishnamurthy, H. J. Lee, J.-Y. Lee, Y. Li, S. McLaren, A. Menzies, V. Mustonen, S. O’Meara, I. Pauporté, X. Pivot, C. A. Purdie, K. Raine, K. Ramakrishnan, F. G. Rodríguez-González, G. Romieu, A. M. Sieuwerts, P. T. Simpson, R. Shepherd, L. Stebbings, O. A. Stefansson, J. Teague, S. Tommasi, I. Treilleux, G. G. Van den Eynden, P. Vermeulen, A. Vincent-Salomon, L. Yates, C. Caldas, L. van't Veer, A. Tutt, S. Knappskog, B. K. T. Tan, J. Jonkers, Å. Borg, N. T. Ueno, C. Sotiriou, A. Viari, P. A. Futreal, P. J. Campbell, P. N. Span, S. van Laere, S. R. Lakhani, J. E. Eyfjord, A. M. Thompson, E. Birney, H. G. Stunnenberg, M. J. van de Vijver, J. W. M. Martens, A. L. Børresen-Dale, A. L. Richardson, G. Kong, G. Thomas, M. R. Stratton, Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. C. Gulhan, J. J.-K. Lee, G. E. M. Melloni, I. Cortes-Ciriano, P. J. Park, Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat. Genet. 51, 912–919 (2019). [DOI] [PubMed] [Google Scholar]
- 34.F. Batalini, D. C. Gulhan, V. Mao, A. Tran, M. Polak, N. Xiong, N. Tayob, N. M. Tung, E. P. Winer, E. L. Mayer, S. Knappskog, P. E. Lønning, U. A. Matulonis, P. A. Konstantinopoulos, D. B. Solit, H. Won, H. P. Eikesdal, P. J. Park, G. M. Wulf, Mutational signature 3 detected from clinical panel sequencing is associated with responses to olaparib in breast and ovarian cancers. Clin. Cancer Res. 28, 4714–4723 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D. S. Lim, P. Hasty, A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133–7143 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. Evers, J. Jonkers, Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene 25, 5885–5897 (2006). [DOI] [PubMed] [Google Scholar]
- 37.P. W. B. Derksen, T. M. Braumuller, E. van der Burg, M. Hornsveld, E. Mesman, J. Wesseling, P. Krimpenfort, J. Jonkers, Mammary-specific inactivation of E-cadherin and p53 impairs functional gland development and leads to pleomorphic invasive lobular carcinoma in mice. Dis. Model. Mech. 4, 347–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.B. Nabet, J. M. Roberts, D. L. Buckley, J. Paulk, S. Dastjerdi, A. Yang, A. L. Leggett, M. A. Erb, M. A. Lawlor, A. Souza, T. G. Scott, S. Vittori, J. A. Perry, J. Qi, G. E. Winter, K. K. Wong, N. S. Gray, J. E. Bradner, The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Raschle, G. Smeenk, R. K. Hansen, T. Temu, Y. Oka, M. Y. Hein, N. Nagaraj, D. T. Long, J. C. Walter, K. Hofmann, Z. Storchova, J. Cox, S. Bekker-Jensen, N. Mailand, M. Mann, Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. K. Sharan, M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, A. Bradley, Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810 (1997). [DOI] [PubMed] [Google Scholar]
- 42.K. S. Ruth, F. R. Day, J. Hussain, A. Martínez-Marchal, C. E. Aiken, A. Azad, D. J. Thompson, L. Knoblochova, H. Abe, J. L. Tarry-Adkins, J. M. Gonzalez, P. Fontanillas, A. Claringbould, O. B. Bakker, P. Sulem, R. G. Walters, C. Terao, S. Turon, M. Horikoshi, K. Lin, N. C. Onland-Moret, A. Sankar, E. P. T. Hertz, P. N. Timshel, V. Shukla, R. Borup, K. W. Olsen, P. Aguilera, M. Ferrer-Roda, Y. Huang, S. Stankovic, P. R. H. J. Timmers, T. U. Ahearn, B. Z. Alizadeh, E. Naderi, I. L. Andrulis, A. M. Arnold, K. J. Aronson, A. Augustinsson, S. Bandinelli, C. M. Barbieri, R. N. Beaumont, H. Becher, M. W. Beckmann, S. Benonisdottir, S. Bergmann, M. Bochud, E. Boerwinkle, S. E. Bojesen, M. K. Bolla, D. I. Boomsma, N. Bowker, J. A. Brody, L. Broer, J. E. Buring, A. Campbell, H. Campbell, J. E. Castelao, E. Catamo, S. J. Chanock, G. Chenevix-Trench, M. Ciullo, T. Corre, F. J. Couch, A. Cox, L. Crisponi, S. S. Cross, F. Cucca, K. Czene, G. D. Smith, E. J. C. N. de Geus, R. de Mutsert, I. de Vivo, E. W. Demerath, J. Dennis, A. M. Dunning, M. Dwek, M. Eriksson, T. Esko, P. A. Fasching, J. D. Faul, L. Ferrucci, N. Franceschini, T. M. Frayling, M. Gago-Dominguez, M. Mezzavilla, M. García-Closas, C. Gieger, G. G. Giles, H. Grallert, D. F. Gudbjartsson, V. Gudnason, P. Guénel, C. A. Haiman, N. Håkansson, P. Hall, C. Hayward, C. He, W. He, G. Heiss, M. K. Høffding, J. L. Hopper, J. J. Hottenga, F. Hu, D. Hunter, M. A. Ikram, R. D. Jackson, M. D. R. Joaquim, E. M. John, P. K. Joshi, D. Karasik, S. L. R. Kardia, C. Kartsonaki, R. Karlsson, C. M. Kitahara, I. Kolcic, C. Kooperberg, P. Kraft, A. W. Kurian, Z. Kutalik, M. la Bianca, G. LaChance, C. Langenberg, L. J. Launer, J. S. E. Laven, D. A. Lawlor, L. le Marchand, J. Li, A. Lindblom, S. Lindstrom, T. Lindstrom, M. Linet, Y. M. Liu, S. Liu, J.’. Luan, R. Mägi, P. K. E. Magnusson, M. Mangino, A. Mannermaa, B. Marco, J. Marten, N. G. Martin, H. Mbarek, B. McKnight, S. E. Medland, C. Meisinger, T. Meitinger, C. Menni, A. Metspalu, L. Milani, R. L. Milne, G. W. Montgomery, D. O. Mook-Kanamori, A. Mulas, A. M. Mulligan, A. Murray, M. A. Nalls, A. Newman, R. Noordam, T. Nutile, D. R. Nyholt, A. F. Olshan, H. Olsson, J. N. Painter, A. V. Patel, N. L. Pedersen, N. Perjakova, A. Peters, U. Peters, P. D. P. Pharoah, O. Polasek, E. Porcu, B. M. Psaty, I. Rahman, G. Rennert, H. S. Rennert, P. M. Ridker, S. M. Ring, A. Robino, L. M. Rose, F. R. Rosendaal, J. Rossouw, I. Rudan, R. Rueedi, D. Ruggiero, C. F. Sala, E. Saloustros, D. P. Sandler, S. Sanna, E. J. Sawyer, C. Sarnowski, D. Schlessinger, M. K. Schmidt, M. J. Schoemaker, K. E. Schraut, C. Scott, S. Shekari, A. Shrikhande, A. V. Smith, B. H. Smith, J. A. Smith, R. Sorice, M. C. Southey, T. D. Spector, J. J. Spinelli, M. Stampfer, D. Stöckl, J. B. J. van Meurs, K. Strauch, U. Styrkarsdottir, A. J. Swerdlow, T. Tanaka, L. R. Teras, A. Teumer, U. Þorsteinsdottir, N. J. Timpson, D. Toniolo, M. Traglia, M. A. Troester, T. Truong, J. Tyrrell, A. G. Uitterlinden, S. Ulivi, C. M. Vachon, V. Vitart, U. Völker, P. Vollenweider, H. Völzke, Q. Wang, N. J. Wareham, C. R. Weinberg, D. R. Weir, A. N. Wilcox, K. W. van Dijk, G. Willemsen, J. F. Wilson, B. H. R. Wolffenbuttel, A. Wolk, A. R. Wood, W. Zhao, M. Zygmunt; Biobank-based Integrative Omics Study (BIOS) Consortium; eQTLGen Consortium; Biobank Japan Project; China Kadoorie Biobank Collaborative Group; kConFab Investigators; LifeLines Cohort Study; InterAct consortium; 23andMe Research Team, Z. Chen, L. Li, L. Franke, S. Burgess, P. Deelen, T. H. Pers, M. L. Grøndahl, C. Y. Andersen, A. Pujol, A. J. Lopez-Contreras, J. A. Daniel, K. Stefansson, J. Chang-Claude, Y. T. van der Schouw, K. L. Lunetta, D. I. Chasman, D. F. Easton, J. A. Visser, S. E. Ozanne, S. H. Namekawa, P. Solc, J. M. Murabito, K. K. Ong, E. R. Hoffmann, A. Murray, I. Roig, J. R. B. Perry, Genetic insights into biological mechanisms governing human ovarian ageing. Nature 596, 393–397 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. E. Carette, J. Pruszak, M. Varadarajan, V. A. Blomen, S. Gokhale, F. D. Camargo, M. Wernig, R. Jaenisch, T. R. Brummelkamp, Generation of iPSCs from cultured human malignant cells. Blood 115, 4039–4042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. K. Brinkman, T. Chen, M. Amendola, B. van Steensel, Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K. B.-O. Gori, A, sigfit: Flexible Bayesian inference of mutational signatures. bioRxiv 372896 [Preprint]. 17 January 2020.
- 46.L. B. Alexandrov, J. Kim, N. J. Haradhvala, M. N. Huang, A. W. Tian Ng, Y. Wu, A. Boot, K. R. Covington, D. A. Gordenin, E. N. Bergstrom, S. M. A. Islam, N. Lopez-Bigas, L. J. Klimczak, J. R. McPherson, S. Morganella, R. Sabarinathan, D. A. Wheeler, V. Mustonen; PCAWG Mutational Signatures Working Group, G. Getz, S. G. Rozen, R. Sabarinathan; PCAWG Consortium , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. A. Duarte, E. Gogola, N. Sachs, M. Barazas, S. Annunziato, J. R de Ruiter, A. Velds, S. Blatter, J. M. Houthuijzen, M. van de Ven, H. Clevers, P. Borst, J. Jonkers, S. Rottenberg, BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat. Methods 15, 134–140 (2018). [DOI] [PubMed] [Google Scholar]
- 48.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. Legland, I. Arganda-Carreras, P. Andrey, MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S3 to S7
Legends for tables S1 and S2
Tables S1 and S2