Abstract
Introduction
This study aimed to determine whether miR-146a and miR-499 polymorphisms are associated with susceptibility to systemic lupus erythematosus (SLE).
Methods
We searched the MEDLINE, EMBASE, and Cochrane databases. We performed a meta-analysis on the association of miR-146a rs2910164, rs2431697, rs57095329, and miR-499 rs3746444 polymorphisms with susceptibility to SLE.
Results
Twenty-one studies from 17 reports, with 18,910 patients and 29,622 controls, were included in the meta-analysis. Meta-analysis revealed no association between SLE and the rs2910164 C allele (odds ratio [OR] = 0.999, 95% confidence interval [CI] = 0.816–1.222, p = 0.990). Stratification by ethnicity indicated no association between the miR-146a C allele and SLE in Arab or Latin American populations. The meta-analysis revealed an association between SLE and the miR-499 rs374644 CC + CT genotype in the overall group (OR = 1.313, 95% CI = 1.015–1.698, p = 0.038). Furthermore, meta-analysis revealed a significant association between SLE and the miR-146a rs2431697 C allele in the overall group (OR = 0.746, 95% CI = 0.697–0.798, p = 0.038). The miR-146a rs2431697 C allele is protective against SLE risk. Stratification by ethnicity indicated an association between the miR-146a rs2431697 C allele and SLE in Asian and European, but not Arab populations. The meta-analysis showed an association between the miR-146a rs57095329 G allele and SLE in Asian, but not Arab populations.
Discussion/Conclusion
This meta-analysis suggests that the miR-146a rs2431697 polymorphism is a protective factor against the risk of SLE, and that the miR-146a rs57095329 and miR-499 rs3746444 polymorphisms are associated with susceptibility to SLE. However, miR-146a rs2910164 was not associated with susceptibility to SLE.
Keywords: Systemic lupus erythematosus, MicroRNA, Polymorphism, Meta-analysis
Introduction
Systemic lupus erythematosus (SLE) is a typical autoimmune disease characterized by organ damage, immune-complex deposition, high levels of autoantibody production, and B-cell hyperactivity. Although the exact cause of SLE is unknown, genetic factors are recognized as significant contributing components [1, 2].
MicroRNAs (miRNAs) are noncoding RNA molecules of approximately 22 nucleotides that are involved in the control of transcription and translation [3]. They function by binding to the 3’ untranslated region of the target mRNA, resulting in degradation of the mRNA or inhibition of translation [4]. Thus, miRNAs are involved in cell division, proliferation, and apoptosis [5]. Single-nucleotide polymorphisms (SNPs) in miRNA sequences can alter the expression of these RNAs and altered expression of miR-146a has been linked to autoimmune diseases. Both miR-146a and miR-499 are implicated in the production of inflammatory cytokines [6, 7].
Some studies have shown an association between the miR-146a rs2910164, rs2431697, rs57095329, and miR-499 rs3746444 polymorphisms and SLE, whereas others have found no such associations [8–23]. These disparities are likely due to the small sample sizes, low statistical power, and/or clinical heterogeneity. To overcome the limitations of individual studies, resolve discrepancies, and reduce the likelihood of random errors being responsible for false-positive or false-negative associations, we performed a meta-analysis to evaluate the association between SLE susceptibility and miR-146a and miR-499 polymorphisms.
Methods
Identification of Eligible Studies and Data Extraction
We searched for published studies that examined the association between miR-146a and miR-499 polymorphisms and SLE. The MEDLINE, EMBASE, and Cochrane databases were used to identify published articles in which miR-146a and/or miR-499 polymorphisms were analyzed in patients with inflammatory arthritis. Combinations of keywords, such as “microRNA,” “miR-146a,” “miR-499,” “polymorphism,” and “lupus,” were entered as Medical Subject Headings (MeSH) and text words. We also manually searched the reference list in the shortlisted studies to identify additional studies that were not indexed in the MEDLINE, EMBASE, and Cochrane databases. No restrictions were placed on language, race, ethnicity, or geographical area. Studies were included if: (1) they were published prior to August 2022; (2) they were case-control studies on the association between the miR-146a rs2910164, rs2431697, rs57095329, and miR-499 rs3746444 polymorphisms and SLE; and (3) provided sufficient genotypic data to calculate the odds ratio (OR). Exclusion criteria were as follows: (1) studies containing overlapping data, (2) studies in which the numbers of null and wild genotypes/alleles could not be ascertained, and (3) studies in which family members had been studied (e.g., a transmission disequilibrium test) because those analyses were based on linkage considerations. Data relevant to the meta-analysis were extracted from the original studies by two independent reviewers. Discrepancies between reviewers were resolved through consensus or by a third reviewer when necessary. The following information was obtained for each study: author, year of publication, ethnicity of the study population, demographics, and the number of cases and controls. Allele frequencies were calculated from genotype distributions.
Evaluations of Statistical Associations
A χ2 test was used to determine whether the observed genotype frequencies conformed to Hardy-Weinberg equilibrium. Meta-analyses were performed using (1) allelic contrast, (2) homozygote contrast, (3) recessive, and (4) dominant models. Subgroup analyses were performed according to ethnicity to evaluate ethnicity-specific effects. Point estimates of OR and 95% confidence intervals (CIs) were calculated for each study. Cochran’s Q-statistic was used to assess within- and between-study variation and heterogeneity. This heterogeneity test assessed the null hypothesis that all the studies evaluated the same effect. I2 values were used to quantify the effect of heterogeneity, and values ranged between 0% and 100%, representing the proportion of between-study variability attributable to heterogeneity rather than chance [24]. I2 values of 25%, 50%, and 75% were defined as low, moderate, and high estimates, respectively. The fixed-effects model assumes that a genetic factor has the same effect on disease susceptibility across all studies and that observed variations between studies are caused by chance alone. The random-effects model assumes that different studies show substantial diversity and assesses both within-study sampling error and between-study variance. When study groups are homogeneous, the two models are similar; if not, the random-effects model usually provides wider CIs than the fixed-effects model. Furthermore, a random-effects model was used in the presence of significant between-study heterogeneity [25]. Statistical manipulations were performed using the Comprehensive Meta-Analysis software (Biostat Inc., Englewood, NJ, USA).
Evaluation of Publication Bias
Funnel plots are often used to detect publication bias. However, owing to the limitations of funnel plotting, which requires a range of studies of varying sizes involving subjective judgments, publication bias was evaluated using Egger’s linear regression test [26], which measures funnel plot asymmetry using a natural logarithm scale of the OR.
Results
Studies Included in the Meta-Analysis
Three hundred and seven reports were identified by electronic and manual searching, and 19 were selected for full-text review based on title and abstract details. Two reports were excluded because one had no data on SLE and the other was a review article; thus, 17 reports met the inclusion criteria [8–23]. Two of these reports contained data on three different groups [11, 16] and these were analyzed independently. Therefore, 21 separate studies were considered in the meta-analysis, which included 18,910 patients and 29,622 controls. Seven studies examined the rs2910164 (miR-146a) polymorphism, 12 examined the rs2431697 (miR-146a)polymorphism, seven examined the rs57095329 (miR-146a) polymorphism, and three examined the rs3746444 (miR-499) polymorphism. Thus, a meta-analysis was performed on the miR-146a rs2910164, rs2431697, rs57095329, and miR-499 s3746444 polymorphisms. Selected characteristics of these studies related to the association between the miR-146a and miR-499 polymorphisms and disease are summarized in Table 1.
Table 1.
Characteristics of the studies included in the meta-analysis
| Author (Ref) | Country | Ethnicity | Subjects | Polymorphisms studied | |
|---|---|---|---|---|---|
| case | control | ||||
| El-Akhras et al., 2022 [23] | Egypt | Arab | 113 | 104 | miR-146a: Rs2910164, rs2431697, rs57095329 |
| Mohammed et al., 2021 [22] | Egypt | Arab | 120 | 100 | miR-146a: rs57095329 |
| Ahmadi et al., 2020 [20] | Iran | Arab | 237 | 50 | miR-146a: Rs2910164, miR-499: rs3746444 |
| Fouda et al., 2020 [21] | Egypt | Arab | 65 | 40 | miR-146a: Rs2431697, rs57095329 |
| Labib et al., 2018 [19] | Egypt | Arab | 80 | 120 | miR-146a: Rs2910164 |
| Aleman-Avila et al., 2017 [18] | Mexico | LA | 486 | 407 | miR-146a: Rs2910164, miR-499: rs3746444 |
| Tang et al., 2015 [17] | China | Asian | 353 | 322 | miR-146a: Rs2431697 |
| Sheng-1 et al., 2015 [16] | China | Asian | 1,047 | 1,205 | miR-146a: Rs2431697 |
| Sheng-2 et al., 2015 [16] | China | Asian | 2202 | 2208 | miR-146a: Rs2431697 |
| Sheng-3 et al., 2015 [16] | China | Asian | 1,307 | 6,038 | miR-146a: Rs2431697 |
| Lee et al., 2015 [15] | Korea | Asian | 1,596 | 2540 | miR-146a: Rs2431697 |
| Jimenez-Morales et al., 2012 [35] | Mexico | LA | 531 | 367 | miR-146a: Rs2910164 |
| Lofgren et al., 2012 [14] | Spain | European | 1,324 | 1,453 | miR-146a: Rs2910164, rs2431697 |
| Leng et al., 2012 [13] | China | Asian | 858 | 967 | miR-146a: Rs57095329 |
| Luo-1 et al., 2011 [11] | China | Asian | 2,352 | 1,080 | miR-146a: Rs2910164, rs57095329 |
| Luo-2 et al., 2011 [11] | China | Asian | 1,152 | 1,080 | miR-146a: Rs57095329 |
| Luo-3 et al., 2011 [11] | China | Asian | 464 | 1,152 | miR-146a: Rs57095329 |
| Zhang et al., 2011 [12] | China | Asian | 213 | 209 | miR-146a: Rs2910164, miR-499: rs3746444 |
| Chung et al., 2011 [10] | USA | Mixed | 811 | 4,813 | miR-146a: Rs2431697 |
| Han et al., 2009 [9] | China | Asian | 1,047 | 1,205 | miR-146a: Rs2431697 |
| Harley et al., 2008 [8] | USA | Mixed | 2,552 | 4,162 | miR-146a: Rs2431697 |
Ref, reference; LA, Latin American.
Meta-Analysis of the Relationship between miR-146a rs2910164 and miR-499 rs374644 Polymorphisms and SLE
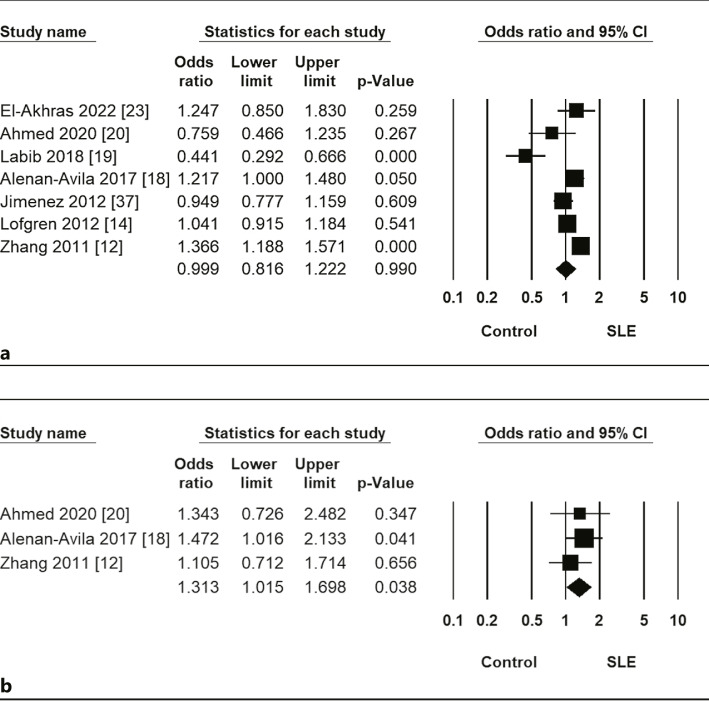
Meta-analysis revealed no association between SLE and the rs2910164 C allele (OR = 0.999, 95% CI = 0.816–1.222, p = 0.990) (Fig. 1; Table 2). Stratification by ethnicity indicated no association between the miR-146a C allele and SLE in Arab or Latin American populations (Table 2). Furthermore, no association was found using the homozygote contrast, recessive, and dominant models between the miR-146a rs2910164 polymorphism and SLE (Table 2). In contrast, the meta-analysis revealed a significant association between SLE and the miR-499 rs374644 CC + CT genotype in the overall group (OR = 1.313, 95% CI = 1.015–1.698, p = 0.038) (Fig. 1; Table 2).
Fig. 1.
OR and 95% CIs of individual studies and pooled data for the association between miR-146a rs2910164 C allele (a) and miR-499 rs3746444 CC + CT genotype (b) and SLE in all subjects.
Table 2.
Meta-analysis of the association between the miR-146a rs2910164 and miR-499 rs3746444 polymorphism and SLE
|
Polymorphism |
Population | Studies, N | Test of association | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p value | model | p value | I 2 | |||
| miR-146a rs2910164 C versus G |
Overall | 7 | 0.999 | 0.816–1.222 | 0.990 | R | <0.001 | 82.6 |
| Arab | 3 | 0.749 | 0.399–1.406 | 0.369 | R | 0.001 | 84.7 | |
| Latin American | 2 | 1.075 | 0.843–1.372 | 0.558 | R | 0.082 | 66.9 | |
| CC versus CG + GG (recessive) | Overall | 7 | 0.909 | 0.639–1.293 | 0.595 | R | 0.003 | 69.5 |
| Arab | 3 | 0.608 | 0.208–1.783 | 0.365 | R | 0.009 | 78.9 | |
| Latin American | 2 | 1.146 | 0.556–2.364 | 0.712 | R | 0.014 | 83.3 | |
| CC + CG versus GG (dominant) | Overall | 7 | 0.940 | 0.768–1.151 | 0.549 | R | 0.040 | 54.4 |
| Arab | 3 | 0.744 | 0.379–1.459 | 0.390 | R | 0.022 | 73.8 | |
| Latin American | 2 | 1.077 | 0.892–1.301 | 0.442 | F | 0.452 | 0 | |
| CC versus GG | Overall | 7 | 0.842 | 0.543–1.304 | 0.441 | R | 0.001 | 75.0 |
| Arab | 3 | 0.547 | 0.143–2.089 | 0.378 | R | 0.002 | 84.1 | |
| Latin American | 2 | 1.175 | 0.572–2.414 | 0.661 | R | 0.021 | 81.2 | |
| miR-499 rs3746444 C versus T | Overall | 3 | 1.214 | 0.962–1.533 | 0.103 | F | 0.569 | 0 |
| CC versus CT + TT (recessive) | Overall | 3 | 0.751 | 0.273–2.064 | 2.064 | F | 0.613 | 0 |
| CC + CT versus TT (dominant) | Overall | 3 | 1.313 | 1.015–1.698 | 0.038 | F | 0.618 | 0 |
| CC versus TT | Overall | 3 | 0.786 | 0.285–2.167 | 0.641 | F | 0.663 | 0 |
OR, odds ratio; CI, confidence interval; R, random-effects model; F, fixed-effects model; NA, not available.
Meta-Analysis of the Relationship between the miR-146a rs2431697 and rs57095329 Polymorphisms and SLE
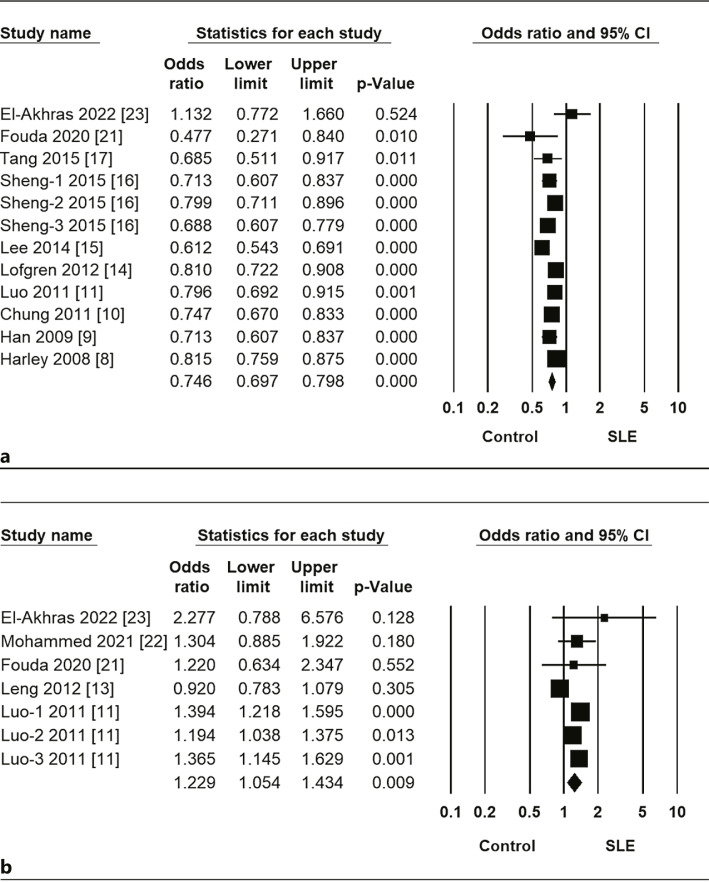
Meta-analysis revealed a significant association between SLE and the miR-146a rs2431697 C allele in the overall group (OR = 0.746, 95% CI = 0.697–0.798, p = 0.038) (Fig. 2, 3; Table 3). The miR-146a rs2431697 C allele was found to be protective against SLE. Stratification by ethnicity indicated an association between the miR-146a rs2431697 C allele and SLE in Asian and European but not Arab populations (Table 3). Meta-analysis showed a significant association between SLE and the miR-146a rs57095329 G allele in the overall group (OR = 1.229, 95% CI = 1.054–1.434, p = 0.009) (Fig. 2; Table 3). Stratification by ethnicity indicated an association between the miR-146a rs57095329 G allele and SLE in Asian but not Arab populations (Table 3).
Fig. 2.
OR and 95% CIs of individual studies and pooled data for the association between (a) miR-146a rs2431697 and (b) rs57095329 polymorphisms and SLE in all subjects.
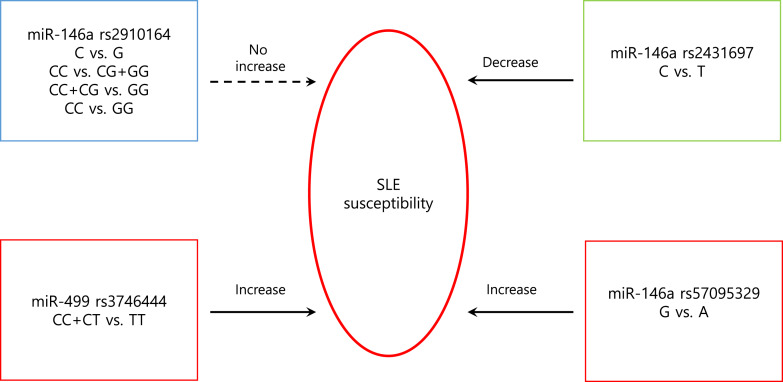
Fig. 3.
A schematic illustration to describe the sequence and polymorphisms of miR-146a and miR-499 for SLE susceptibility.
Table 3.
Meta-analysis of the association between the miR-146a rs2431697and miR-146a rs57095329 polymorphism and SLE
|
Polymorphism |
Population | Studies, N | Test of association | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p value | model | p value | I 2 | |||
| Rs2431697 C versus T |
Overall | 12 | 0.746 | 0.697–0.798 | <0.001 | R | 0.002 | 62.5 |
| Asian | 7 | 0.715 | 0.659–0.744 | <0.001 | R | 0.050 | 52.3 | |
| European | 3 | 0.797 | 0.757–0.841 | <0.001 | F | 0.405 | 0 | |
| Arab | 2 | 0.755 | 0.324–1.757 | 0.514 | R | 0.013 | 83.7 | |
| Rs57095329 G versus A |
Overall | 7 | 1.229 | 1.054–1.434 | 0.009 | R | 0.005 | 68.1 |
| Asian | 4 | 1.203 | 1.003–1.444 | 0.047 | R | 0.001 | 82.6 | |
| Arab | 3 | 1.350 | 0.982–1.855 | 0.065 | F | 0.590 | 0 | |
OR, odds ratio; CI, confidence interval; R, random-effects model; F, fixed-effects model; NA, not available; LA, Latin American.
Heterogeneity and Publication Bias
Between-study heterogeneity was found in the relationship between miR-146a polymorphisms and SLE (Tables 2, 3). However, there was no heterogeneity in the meta-analysis of the miR-499 rs3746444 polymorphism (Table 2). The distribution of the genotypes of the miR-146a and miR-499 polymorphisms in the control groups was consistent with Hardy-Weinberg equilibrium. It was difficult to correlate the funnel plot, which is typically used to detect publication bias, because the number of studies included in the analysis was relatively small. Egger’s regression test showed no evidence of publication bias (p > 0.1).
Discussion
In the current meta-analysis, we investigated studies on the relationship between the polymorphisms in miR-146a rs2910164, rs2431697, rs57095329, and miR-499 rs3746444 and susceptibility to SLE. We did not find any correlation between SLE and miR-146a rs2910164 polymorphism. However, under the dominant model, the meta-analysis revealed a strong association between SLE and the miR-499 rs374644 polymorphism. In addition, the meta-analysis revealed a correlation between SLE and the miR-146a rs2431697 polymorphism. The rs2431697 C allele of miR-146a is a protective factor against SLE risk. Furthermore, miR-146a rs57095329 was significantly associated with SLE.
miR-146a is one of the earliest recognized SLE-related miRNAs. It targets tumor necrosis factor receptor-associated factor 6, interleukin (IL)-1 receptor-associated kinase, interferon (IFN) regulator factor 5, and signal transducer and activator of transcription 1 to suppress the type 1 IFN pathway [27]. Increased type 1 IFN signaling is important for the development of SLE. The SNP rs2431697 is located in the high potential regulatory region of miR-146a. miR-146a expression is upregulated in peripheral blood mononuclear cells when the rs2431697 C allele is present [14]. Thus, the C allele of rs2431697 may influence the development of SLE by enhancing miR-146a expression. In SLE patients, increased miR146a expression results in decreased production of type I IFNs and activation of type I IFN signaling [28]. We discovered a relationship between miR-146a rs2431697 and a reduced incidence of SLE, which is consistent with the results of previous functional investigations [14, 29]. The genetic variant rs57095329 in the promoter region of the miR-146a gene may increase miR-146a production by altering the affinity of the Ets-1 33 ligand [11]. SNP-rs3746444 is found in the pre-miRNA region of hsa-mir-499 and may alter target mRNA binding to 3p mature miRNAs as well as the pre-miRNA maturation of 5p and 3p miRNAs [30]. IL-17Rb, IL-23a, IL-2R, IL-6, IL-2, and IL-18R are among the targets of hsa-mir-499, and they all play a role in the development and pathophysiology of SLE. The miR-146a rs2910164 polymorphism was not associated with increased risk of SLE in our meta-analysis. The miR-146a rs2910164 polymorphism may have an impact on target mRNA binding and mature miR-146a expression [31]. The GG genotype of the miR-146a polymorphism confers increased expression of mature miR-146a. However, our results do not agree with those of functional studies. Owing to the conflicting findings, the functional importance of rs2910164 remains unclear. Additionally, we cannot completely rule out the possibility that the lack of correlation may be due to the limited number of studies included, their weak statistical significance, or type II error.
The current study has some limitations that must be considered. First, the analysis may have been affected by heterogeneity and confounding variables. In particular, publication bias may have impacted our findings as research with contradictory findings may not have been published. Second, as most of the patients in our ethnicity-specific meta-analysis were of Asian and Arabic descent, our findings only apply to these two ethnic groups. Further research on various other ethnic communities is required. Third, owing to lack of data, we were unable to stratify and assess variables such as sex and clinical or environmental variables. Additionally, polymorphisms may be linked to clinical symptoms and disease risk. Fourth, the number of studies and subjects in the ethnic subgroup analysis were limited; as a result, our analysis may have been underpowered. Nonetheless, this meta-analysis enhances prior knowledge and provides some additional benefits. We improved the statistical power and resolution by merging the results of several independent analyses to provide data that are more accurate than those of individual studies [32–34]. Furthermore, this meta-analysis was performed meticulously and systematically based on the most recent literature.
In conclusion, this meta-analysis of published data suggests that the miR-146a rs2431697 polymorphism is protective against the development of SLE, whereas the miR-146a rs57095329 and miR-499 rs3746444 polymorphisms are linked to susceptibility to SLE. However, the miR-146a rs2910164 polymorphism was not associated with SLE susceptibility. Further research is needed to fully understand the involvement of miRNA genes in SLE pathogenesis in various ethnic groups.
Statement of Ethics
An ethics statement is not applicable because this study is based exclusively on published literature.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
There were no funding sources.
Author Contributions
Young Ho Lee was involved in conception and design of study, acquisition of data, analysis and/or interpretation of data, drafting the manuscript, and revising the manuscript critically for important intellectual content. Gwan Gyu Song was involved in conception and design of study, analysis and interpretation of data, and drafting the manuscript.
Funding Statement
There were no funding sources.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.
References
- 1. Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39(12):10627–35. 10.1007/s11033-012-1952-x. [DOI] [PubMed] [Google Scholar]
- 2. Shin JM, Kim D, Kwon YC, Ahn GY, Lee J, Park Y, et al. Clinical and genetic risk factors associated with the presence of lupus nephritis. J Rheum Dis. 2021;28(3):150–8. 10.4078/jrd.2021.28.3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001 Oct 26;294(5543):853–8. 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008 Feb;9(2):102–14. 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5. Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009 Mar;10(3):399–416. 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J, Bi J, Liu X, Li K, Di J, Wang B. Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Mol Biol Rep. 2012 Apr;39(4):4571–9. 10.1007/s11033-011-1247-7. [DOI] [PubMed] [Google Scholar]
- 7. Xu WD, Lu MM, Pan HF, Ye DQ. Association of microRNA-146a with autoimmune diseases. Inflammation. 2012 Aug;35(4):1525–9. 10.1007/s10753-012-9467-0. [DOI] [PubMed] [Google Scholar]
- 8. Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008 Feb;40(2):204–10. 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE). Hum Mol Genet. 2009 Mar 15;18(6):1171–80. 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011 Mar;7(3):e1001323. 10.1371/journal.pgen.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011 Jun;7(6):e1002128. 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J, Yang B, Ying B, Li D, Shi Y, Song X, et al. Association of pre-microRNAs genetic variants with susceptibility in systemic lupus erythematosus. Mol Biol Rep. 2011 Mar;38(3):1463–8. 10.1007/s11033-010-0252-6. [DOI] [PubMed] [Google Scholar]
- 13. Leng RX, Wang W, Cen H, Zhou M, Feng CC, Zhu Y, et al. Gene-gene and gene-sex epistatic interactions of MiR146a, IRF5, IKZF1, ETS1 and IL21 in systemic lupus erythematosus. PLoS One. 2012;7(12):e51090. 10.1371/journal.pone.0051090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lofgren SE, Frostegard J, Truedsson L, Pons-Estel BA, D’Alfonso S, Witte T, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012 Apr;13(3):268–74. 10.1038/gene.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee YH, Bae SC. The miR-146a polymorphism and susceptibility to systemic lupus erythematosus and rheumatoid arthritis: a meta-analysis. Z Rheumatol. 2015 Mar;74(2):153–6. 10.1007/s00393-014-1509-6. [DOI] [PubMed] [Google Scholar]
- 16. Sheng YJ, Xu JH, Wu YG, Zuo XB, Gao JP, Lin Y, et al. Association analyses confirm five susceptibility loci for systemic lupus erythematosus in the Han Chinese population. Arthritis Res Ther. 2015 Mar 28;17(1):85. 10.1186/s13075-015-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang ZM, Wang P, Chang PP, Hasahya T, Xing H, Wang JP, et al. Association between rs2431697 T allele on 5q33.3 and systemic lupus erythematosus: case-control study and meta-analysis. Clin Rheumatol. 2015 Nov;34(11):1893–902. 10.1007/s10067-015-3045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aleman-Avila I, Jimenez-Morales M, Beltran-Ramirez O, Barbosa-Cobos RE, Jimenez-Morales S, Sanchez-Munoz F, et al. Functional polymorphisms in pre-miR146a and pre-miR499 are associated with systemic lupus erythematosus but not with rheumatoid arthritis or Graves’ disease in Mexican patients. Oncotarget. 2017 Nov 3;8(54):91876–86. 10.18632/oncotarget.19621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labib DA, Shaker OG, El Refai RM, Ghoniem SA, Elmazny A. Association between miRNA-146a and polymorphisms of its target gene, IRAK1, regarding susceptibility to and clinical features of systemic lupus erythematous and multiple sclerosis. Lab Med. 2019;50(1):34–41. 10.1093/labmed/lmy033. [DOI] [PubMed] [Google Scholar]
- 20. Ahmadi K, Soleimani A, Soleimani Motlagh S, Baharvand Ahmadi S, Almasian M, Kiani AA. Polymorphisms of Pre-miR-499 rs3746444 T/C and Pre-miR-146a rs2910164 C/G in the autoimmune diseases of rheumatoid arthritis and systemic lupus erythematosus in the West of Iran. Iran J Public Health. 2020 Apr;49(4):782–90. [PMC free article] [PubMed] [Google Scholar]
- 21. Fouda ME, Nour El Din DM, Mahgoub MY, Elashkar AE, Abdel Halim WA. Genetic variants of microRNA-146a gene: an indicator of systemic lupus erythematosus susceptibility, lupus nephritis, and disease activity. Mol Biol Rep. 2020;47(10):7459–66. 10.1007/s11033-020-05802-y. [DOI] [PubMed] [Google Scholar]
- 22. Mohammed SR, Shaker OG, Mohammed AA, Fouad NA, Hussein HA, Ahmed NA, et al. Impact of miR-155 (rs767649 A>T) and miR-146a (rs57095329 A>G) polymorphisms in system lupus erythematosus susceptibility in an Egyptian cohort. Eur Rev Med Pharmacol Sci. 2021 Feb;25(3):1425–35. [DOI] [PubMed] [Google Scholar]
- 23. El-Akhras BA, Ali YBM, El-Masry SA, Bassyouni IH, El-Sayed IH, Talaat RM. mir-146a genetic polymorphisms in systemic lupus erythematosus patients: correlation with disease manifestations. Noncoding RNA Res. 2022 Sep;7(3):142–9. 10.1016/j.ncrna.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002 Jun 15;21(11):1539–58. 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986 Sep;7(3):177–88. 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26. Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997 Sep 13;315(7109):629–34. 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA‐146a contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–75. 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 29. Hou G, Harley ITW, Lu X, Zhou T, Xu N, Yao C, et al. SLE non-coding genetic risk variant determines the epigenetic dysfunction of an immune cell specific enhancer that controls disease-critical microRNA expression. Nat Commun. 2021;12(1):135–19. 10.1038/s41467-020-20460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008 May 20;105(20):7269–74. 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee YH, Choi SJ, Ji JD, Song GG. Association between toll-like receptor polymorphisms and systemic lupus erythematosus: a meta-analysis update. Lupus. 2016;25(6):593–601. 10.1177/0961203315622823. [DOI] [PubMed] [Google Scholar]
- 33. Lee YH, Song GG. Association between signal transducers and activators of transcription 4 rs7574865 polymorphism and systemic lupus erythematosus: a meta-analysis. J Rheum Dis. 2020;27(4):277–84. 10.4078/jrd.2020.27.4.277. [DOI] [Google Scholar]
- 34. Lee YH, Song GG. Mendelian randomization research on the relationship between rheumatoid arthritis and systemic lupus erythematosus and the risk of autistic spectrum disorder. J Rheum Dis. 2022;29(1):46–51. 10.4078/jrd.2022.29.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiménez-Morales S, Gamboa-Becerra R, Baca V, Del Río-Navarro BE, López-Ley DY, Velázquez-Cruz R, et al. miR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue antigens. 2012;80(4):317–21. 10.1111/j.1399-0039.2012.01929.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.