Abstract
For most paramyxoviruses, syncytium formation requires the expression of both surface glycoproteins (HN and F) in the same cell, and evidence suggests that fusion involves a specific interaction between the HN and F proteins (X. Hu et al., J. Virol. 66:1528–1534, 1992). The stalk region of the Newcastle disease virus (NDV) HN protein has been implicated in both fusion promotion and virus specificity of that activity. The NDV F protein contains two heptad repeat motifs which have been shown by site-directed mutagenesis to be critical for fusion (R. Buckland et al., J. Gen. Virol. 73:1703–1707, 1992; T. Sergel-Germano et al., J. Virol. 68:7654–7658, 1994; J. Reitter et al., J. Virol. 69:5995–6004, 1995). Heptad repeat motifs mediate protein-protein interactions by enabling the formation of coiled coils. Upon analysis of the stalk region of the NDV HN protein, we identified two heptad repeats. Secondary structure analysis of these repeats suggested the potential for these regions to form alpha helices. To investigate the importance of this sequence motif for fusion promotion, we mutated the hydrophobic a-position amino acids of each heptad repeat to alanine or methionine. In addition, hydrophobic amino acids in other positions were also changed to alanine. Every mutant protein retained levels of attachment activity that was greater than or equal to the wild-type protein activity and bound to conformation-specific monoclonal as well as polyclonal antisera. Neuraminidase activity was variably affected. Every mutation, however, showed a dramatic decrease in fusion promotion activity. The phenotypes of these mutant proteins indicate that individual amino acids within the heptad repeat region of the stalk domain of the HN protein are important for the fusion promotion activity of the protein. These data are consistent with the idea that the HN protein associates with the F protein via specific interactions between the heptad repeat regions of both proteins.
Newcastle disease virus (NDV) is one of many paramyxoviruses that requires two surface glycoproteins in order fuse with uninfected cells. In paramyxovirus-mediated fusion, the fusion (F) protein is thought to directly mediate the fusion event, and with the exception of simian virus 5 (SV5), the viral attachment protein is also necessary (9). Thus, the hemagglutinin-neuraminidase (HN) protein, which serves as the attachment protein for NDV, has three functions: attachment, neuraminidase (NA) activity, and an undefined role in fusion termed fusion promotion.
The requirement for the HN protein in fusion is virus specific, and recent work from several laboratories suggest that the presumed stalk domain of various HN proteins confers this specificity. Deng et al. constructed chimeric HN proteins containing regions from human parainfluenza virus type 3 (hPIV3) and NDV (5). Their results suggest that both the presumed transmembrane domain as well as a portion of the presumed stalk region of the HN protein confer F protein specificity for fusion. In a similar approach using parainfluenza virus 2 (PIV2) and simian virus 41 (SV41) chimeras, Tsurudome et al. also found that the presumed stalk region of the HN protein defines F protein specificity (25). Additionally, they reported that the globular head was necessary for maximal fusion promotion. However, they found that PIV2 and SV41 chimeras did not require a transmembrane sequence specific to either PIV2 or SV41 for fusion promotion. Tanabayashi and Compans also created chimeric HN proteins combining Sendai virus (SeV) and hPIV3 and found that only the stalk region of the HN protein was important for fusion specificity (24). Thus, while there is disagreement about the role of the transmembrane region and the globular head domain in virus specificity, it is clear that the stalk regions of HN proteins from various paramyxoviruses are crucial for F protein specificity. We have previously expressed HN proteins containing mutations in the stalk domain (22). These mutant proteins separated fusion promotion activity from attachment activity and led us to conclude that the stalk region of the NDV HN protein is critical for fusion promotion.
Virus specificity of the HN protein argues for an interaction between the HN and F proteins required for fusion (6), and as described above, studies of chimeric HN proteins as well as point mutations suggest that it is the stalk domain that interacts with the F protein. While no clear studies of F protein chimeras have shown which domains of the F protein are important for an interaction with the HN protein, mutational analysis of the F protein has shown that several domains, including the fusion peptide as well as the heptad repeat regions HR1 and HR2 are important in fusion (2, 8, 9, 20, 23).
Heptad repeat regions are often involved in protein-protein interactions. Given the importance of heptad repeat domains in the F protein, the transmembrane-proximal location of one of them, as well as the apparent role of the transmembrane-proximal presumed stalk region of the HN protein in fusion promotion, we explored the potential for the presence of heptad repeats in this region of the HN protein. We found heptad repeat domains in all paramyxovirus and rubulavirus attachment proteins. Furthermore, use of secondary structure prediction software revealed that the heptad repeats from all the viruses analyzed showed a high probability of forming alpha helices.
We explored the importance of individual amino acids within these potential helices by mutation. The hydrophobic a-position amino acids were the first residues chosen for mutagenesis because the a positions of heptad repeats are often important for mediating protein-protein interactions. Thus, we hypothesized that such mutations would have the potential to cause a more deleterious effect on fusion promotion than mutations in other positions of the helices. Indeed, we found that all proteins altered in the a positions negatively affected fusion. However, mutations in other positions of the helix also negatively affected fusion. All mutant proteins had wild-type levels of hemagglutination (HA) and variable NA activity. These results argue that a specific amino acid sequence within the stalk is important for the fusion promotion activity of the HN protein, a result that would be expected if the region is involved in a specific interaction with the F protein.
MATERIALS AND METHODS
Cells.
Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with nonessential amino acids, vitamins, glutamine, penicillin-streptomycin, and 10% fetal calf serum.
Antibodies.
Anti-NDV was raised against UV-inactivated NDV (strain AV) virions (22). Monoclonal antibodies specific for NDV HN protein were a generous gift of Ron Iorio. Antibodies used were anti-2b, anti-3a, anti-4a, anti-1,4c, and anti-2,3c (7).
Site-directed mutagenesis.
Positive-sense oligonucleotides were synthesized by DNA International, Operon, or Life Technologies. The oligonucleotides used for mutagenesis (written 5′ to 3′; bases that were altered are underlined) were L74A (GGAAAGATTACATCTGCAGCCGGCTCCAATCAGGATGTAG), V81A (GGTTCCAATCAGGATGTCGCGGATAGGATATACAAGC), V88A (GGATATACAAGCAGGCAGCTCTTGAATCTCCG), L96A (GGCAGCGCTAAACACCG), I103A (GAATCTATAGCAATGAATGC), L110A (CAATAACATCCGCCTCTTATC), L74M (GGAAAGATTACATCTGCAATGGGTTCCAATCAGGATGTAG), L96M (CTTGAATCTCCGTTGGCAATGCTAAACACCGAATCTATA), L90A (GGATATACAAGCAGGTGGCCGCGGAATCTCCGTTGGC), L97A (GAATCTCCGTTGGCATTGGCCAACACCGAATCTATAATT), and I102A (CTAAACACCGAATCCGCGATTATGAATGCAATAACATCC). Double mutants were made by sequential mutagenesis. Oligonucleotide-directed mutagenesis of pSVL (Pharmacia) containing the HN gene (17) was accomplished by using a Morph site-specific plasmid DNA mutagenesis kit (5 Prime→3 Prime, Inc.) or a Chameleon double-stranded, site-directed mutagenesis kit (Stratagene), using the methods and reagents supplied with each kit. Mutant pSVL-HN cDNAs were identified by sequencing or by the introduction of a novel restriction site into the mutant gene. Each HN mutant gene was then fully sequenced to ensure that no extraneous mutations were generated in other parts of the gene.
Transient gene expression.
Two methods were used to express HN cDNAs in Cos-7 cells. DEAE-dextran transfection was performed by a modification of the method of Levesque et al. as described previously (12, 20). Lipofectin (Gibco) transfections were done essentially as suggested by the manufacturer and were described previously (13) except that cells were incubated with the Lipofectin-OptiMem-DNA mixture at 37°C for 20 to 24 h.
Radiolabeling, lysis, and immunoprecipitation of protein.
At 48 h posttransfection, cells were radiolabeled for 2 h at 37°C in DMEM containing 70% of the cysteine of standard medium and lacking methionine. The labeling medium contained 0.15 mCi of [35S]methionine-[35S]cysteine (EXPRE35S35S; New England Nuclear) per ml. The cells were chased in nonradioactive medium for 2 h (4 h for cell surface assays).
At the end of the chase period, cells were washed once in phosphate-buffered saline (PBS) and lysed in reticulocyte standard buffer buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl) containing 0.5% sodium deoxycholate, 2.5 mg of N-ethylmaleimide per ml, 2 mg of iodoacetamide per ml, and 1% Triton X-100. Lysates were homogenized by passage through a 21-gauge needle four times. After lysis, the nuclei were removed by centrifugation.
Cell lysates were incubated with antisera for 1 h at room temperature. Fixed, killed Staphylococcus aureus cells (Boehringer Mannheim) resuspended in PBS–0.5% polyoxyethylenesorbitan monolaurate–1 mg of bovine serum albumin per ml were added to the lysate in the presence of 0.4% sodium dodecyl sulfate (SDS) and incubated at room temperature with agitation for 30 min. The S. aureus cells were pelleted, and the supernatants were removed. The pellets were washed three times with PBS–1% Triton X-100–0.5% sodium deoxycholate–0.1% SDS. The S. aureus cells were then resuspended in sample buffer and stored at −20°C until analysis by SDS-polyacrylamide gel electrophoresis (PAGE). Samples were incubated at 100°C for 5 min prior to loading on SDS–8% polyacrylamide gels.
Fusion assay.
After a 48-h incubation in DMEM, 20 of the largest fusion areas were counted for each mutant and averaged as described previously (22). Values obtained for fusion activities were taken from three separate experiments and averaged.
Cell surface assay.
Transfected cells were radiolabeled as described above and chased for 4 h in nonradioactive medium. Analysis of protein at the cell surface was done as described previously (22). Proteins at the cell surface were quantitated from autoradiographs by densitometry, and values obtained were taken from at least three separate experiments and averaged.
Attachment assay.
At 48-h posttransfection, attachment was assayed as described previously (16).
NA assay.
At 48 h posttransfection, NA was assayed as described previously (16). Values obtained for NA activities were taken from three separate experiments and averaged.
RESULTS
Mutagenesis of HN protein stalk domain.
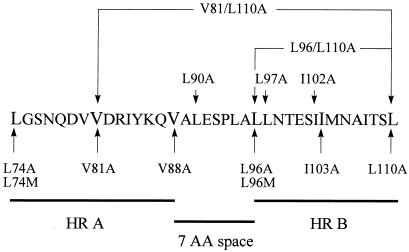
The stalk region of the NDV HN protein has been defined as amino acids 49 to 146 (4, 11). Visual inspection of this sequence showed that it contains two heptad repeats of hydrophobic amino acids (leucine, valine, isoleucine) separated by a space of seven amino acids (Fig. 1). The heptad repeat motif is found in many proteins and is thought to impart an alpha-helical secondary structure (14). Secondary structure prediction software (1) predicts that the heptad repeats in the stalk region of the HN protein do indeed have the potential to form two alpha helices with an intervening region of seven amino acids (Fig. 2).
FIG. 1.
Amino acid sequence for the wild-type HN protein showing residues 74 to 110. The heptad repeat a positions are shown in a larger font, heptad repeat regions A (HR A) and B (HR B) are denoted by a line below the sequence, and the seven-amino-acid (7 AA) sequence between the heptad repeats is denoted by a line below the sequence. Mutations of the wild-type protein are indicated with arrows.
FIG. 2.
Secondary structure prediction of paramyxoviruses and rubulaviruses. Secondary structure prediction of NDV amino acids 74 to 110, using Baylor College of Medicine SSP (segment-oriented prediction) and NNSSP (nearest-neighbor prediction) programs (1). The corresponding regions of other rubulaviruses and paramyxoviruses were similarly analyzed. + denotes area predicted to be alpha-helical; a line above the sequence denotes a space between the two heptad repeats. para, parainfluenza virus.
The stalk region of other paramyxovirus HN proteins were similarly analyzed for secondary structure. The attachment proteins of the paramyxoviruses and rubulaviruses SV41, mumps virus (MuV), SV5, hPIV3, SeV, and avian parainfluenza virus 4 also contain heptad repeats of hydrophobic amino acids with the potential to form alpha helices, although morbilliviruses and pneumoviruses lacked predicted helical structure in this region. Furthermore, most sequences were predicted to form two alpha helices with an intervening space as observed in the NDV HN protein.
To investigate the importance of each of these NDV HN protein heptad repeat regions for fusion as well as determine the relative importance of specific amino acids and helical structure, conservative mutations of hydrophobic residues were made, changing heptad repeat a-position residues to alanine. Alanine was chosen because it has a short side chain and therefore should not disrupt a potential helical structure (1a). Mutations were made to generate the mutants L74A, V81A, V88A, L96A, I103A, and L110A (Fig. 1).
Expression of mutant proteins.
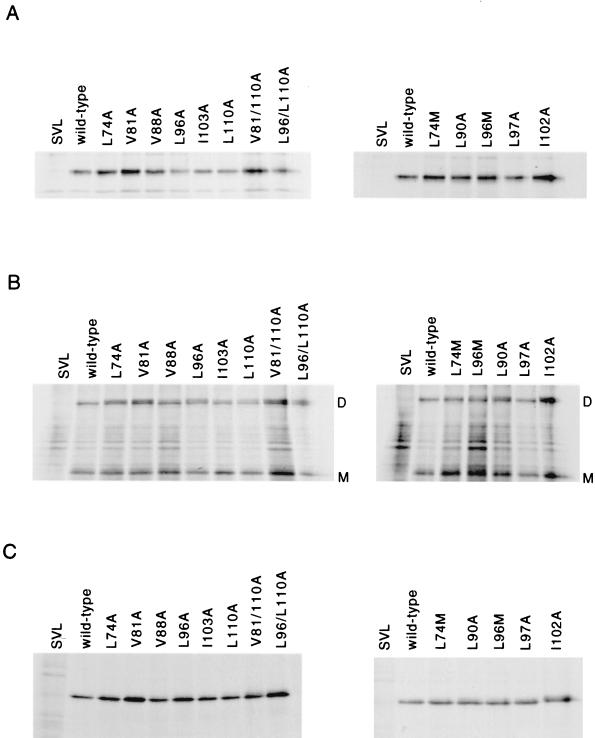
To characterize the expression of these mutant proteins, Cos cells were transfected with either wild-type or mutant HN cDNA as described in Materials and Methods. HN proteins were immunoprecipitated with anti-NDV antisera and analyzed by SDS-PAGE under reducing (Fig. 3A) or nonreducing (Fig. 3B) conditions. Mutant HN proteins were immunoprecipitated in various amounts, although the amounts precipitated were typically at least as high as the wild-type protein (Fig. 3A). Disulfide-linked dimers were observed for each of the mutant proteins, suggesting that the conformation necessary for dimer formation was present (Fig. 3B). Cytoplasmic extracts containing each of the mutant proteins were immunoprecipitated with five different conformation-specific monoclonal antibodies (Fig. 3C; Materials and Methods). In every case, the mutant proteins were precipitated at levels as least as high as observed for the wild-type protein, supporting the idea that the proteins were folded correctly. Clearly, all the mutant HN proteins were expressed and stable within the cell for at least a 2-h chase period.
FIG. 3.
Expression of mutant HN proteins. At 48 h posttransfection, cells transfected with wild-type or mutant cDNAs were radiolabeled for 2 h and chased for 2 h in nonradioactive medium. Cells were lysed; postnuclear supernatants were immunoprecipitated with polyclonal antisera and analyzed by SDS-PAGE on 8% gels in the presence (A) or absence (B) of reducing agent. (C) Proteins in postnuclear supernatants were immunoprecipitated with the conformation-specific monoclonal antibody anti-2b, which is representative of results obtained with four other monoclonal antibodies, as described in Materials and Methods. SVL, vector; wild-type, wild-type HN protein; D, disulfide-linked HN protein dimer; M, monomeric HN protein. A Molecular Dynamics densitometer was used to image the autoradiograph, and the figure was generated from Adobe Photoshop without enhancement. The images presented accurately represent the original autoradiographs.
Cell surface proteins were analyzed by SDS-PAGE under reducing or nonreducing conditions (not shown), and amounts detected were quantitated by densitometry. All mutant proteins were detected at the cell surface, although at different levels (Table 1). Disulfide-linked dimers were observed for each mutant protein at the cell surface, although the apparent size of the dimers observed varied slightly from that of the wild-type protein in some experiments.
TABLE 1.
Biological activities of mutant proteins
| HN | % of wild-type activity
|
|||
|---|---|---|---|---|
| Cell surface (mean ± SD) | NA (mean ± SD) | NA(CS)a | HAb | |
| L74A | 102 ± 29 | 124 ± 20 | 122 | ++ |
| V81A | 128 ± 10 | 201 ± 59 | 157 | +++ |
| V88A | 81 ± 13 | 21 ± 6 | 26 | + |
| L96A | 77 ± 20 | 70 ± 13 | 91 | ++ |
| I103A | 59 ± 26 | 10 ± 3 | 17 | ++ |
| L110A | 76 ± 21 | 11 ± 7 | 14 | ++ |
| V81/L110A | 112 ± 20 | 43 ± 15 | 38 | +++ |
| L96/L110A | 55 ± 22 | 12 ± 1 | 22 | +++ |
| L74M | 102 ± 7 | 54 ± 0 | 53 | +/− |
| L96M | 72 ± 21 | 69 ± 7 | 96 | + |
| L90A | 67 ± 8 | 59 ± 5 | 88 | + |
| L97A | 79 ± 14 | 79 ± 7 | 100 | + |
| I102A | 111 ± 19 | 136 ± 20 | 123 | +++ |
NA(CS), NA value normalized to CS expression.
+/−, slightly lower than wild-type level of binding; +, wild-type level of binding; ++, greater than wild-type level of binding; +++, much greater than wild-type level of binding.
Biological activities of mutant proteins.
The effect of each mutation on the three activities (attachment, NA, and fusion promotion) was determined. We analyzed attachment activity by assaying erythrocyte binding or HA. Chicken erythrocytes were bound to the surface of Cos cells expressing either wild-type or mutant proteins (Table 1). In cells expressing vector alone, virtually no binding was observed, while bound erythrocytes were seen in cells expressing the wild-type HN protein. The mutant V88A bound erythrocytes at least at wild-type levels, while the other mutant HN proteins bound at greater than wild-type levels. Clearly, all mutant proteins retained attachment activity.
NA activity of each mutant protein was determined by quantitating the ability of each mutant protein to cleave the substrate neuraminlactose (Table 1). L74A and V81A had greater NA activity than the wild-type protein. The other mutant proteins had decreased NA activity ranging from 14 to 91%. Interestingly, two mutant proteins (I103A and L110A) had little NA activity and greater than wild-type levels of attachment activity. These data suggest that NA and HA activities of the NDV HN protein can be genetically separated as has been previously reported (21).
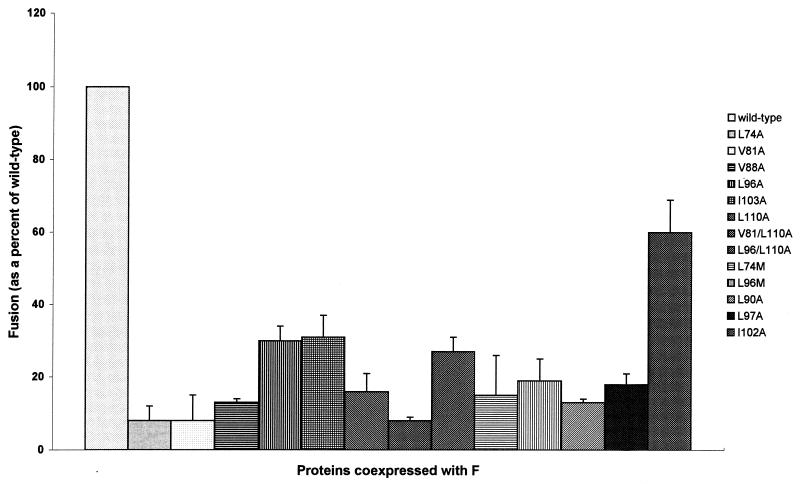
Fusion promotion was determined by analyzing syncytium formation after coexpression of each mutant protein with the NDV F protein in Cos cells (Fig. 4). Every mutation negatively affected fusion, although to various degrees. Mutations in the first heptad repeat decreased activity to 8 to 13% of the wild-type protein. Mutations in the second heptad repeat had slightly less effect, as fusion activity decreased to 16 to 31% of that of the wild-type protein. Thus, a conservative substitution of any one of these hydrophobic amino acid residues produced an HN protein with greatly decreased fusion activity.
FIG. 4.
Syncytium formation promoted by mutant HN proteins. At 48 h posttransfection, 20 of the largest syncytia were counted. The background fusion from cells expressing vector alone was subtracted, and values were normalized to cell surface expression.
Expression of proteins with double substitutions.
After determining that alanine substitutions for a-position amino acids negatively affected fusion, we asked if mutant proteins with double substitutions would further inhibit fusion. Similar double mutations in the F protein had been previously shown to decrease fusion to a much greater degree than single mutants (20). Thus, the double mutants V81/L110A and L96/L110A were generated. These mutants were expressed at the cell surface and appeared to be folded correctly (Fig. 3; Table 1). Both mutants bound erythrocytes at higher levels than the wild-type protein, as was observed for each of the corresponding single mutant proteins (Table 1). NA activities for both proteins were much lower than observed for the wild-type protein but not as low as was observed for the single mutant L110A (Table 1). Surprisingly, the fusion activity of each mutant was not decreased further than the activities of the single substitution mutants (Fig. 4).
Mutant proteins containing methionine substitutions.
We next asked if we could further disrupt fusion promotion with the substitution of a bulky amino acid for leucine in the a- position of each heptad repeat. Methionine residues have a longer side chain than leucine and could therefore more efficiently inhibit the HN protein heptad repeats from interacting with the F protein by steric hindrance. Alternatively, the methionine residue may substitute for leucine and restore activity. The mutant proteins L74M and L96M were generated to explore these possibilities (Fig. 1).
Immunoprecipitated mutant proteins, analyzed under reducing conditions (Fig. 3A), showed that each of these mutants was as stable as the wild-type protein and, like the wild-type HN protein, was recognized by both NDV antisera and conformation-specific monoclonal antibodies (Fig. 3C). Disulfide-linked dimers were observed for each of the mutant proteins (Fig. 3B). Mutant HN proteins expressed at the cell surface were immunoprecipitated and analyzed by SDS-PAGE under reducing and nonreducing conditions (not shown). As observed for the a-position alanine mutants, these proteins were expressed at the cell surface (Table 1) and formed disulfide-linked dimers.
Biological activities of methionine mutant proteins.
The NA activity of L74M dropped from 122% (observed for L74A) to 53% (Table 1), and there was a decrease in HA levels from above to slightly below the wild-type level (Table 1). Little change in NA activity was observed for L96M (compared to L96A); however, HA activity decreased from above to equal to the wild-type level. Fusion promotion activity of L74M was slightly higher than that observed for L74A (8% versus 15%), but a decrease was observed for L96M (19% versus 30% for L96A) (Fig. 4). While this decrease was substantial, it was not lower than levels observed for some of the other heptad repeat mutants.
Substitutions in other positions of the predicted helices.
Coiled-coil interactions are mediated by hydrophobic or neutral amino acids in the a and d positions of a heptad repeat. To address whether alanine substitutions in heptad repeat positions other than a influenced the fusion promotion activity of the HN protein, the following mutants were generated. Leucine 97 (b position) and isoleucine 102 (g position), which are positioned in the second heptad repeat region of the stalk domain, were mutated, resulting in L97A and I102A, respectively. A residue between the two repeats, Leu 90, was also mutated (L90A) (Fig. 1).
Biological activities of alanine mutant proteins in other positions of the predicted helices.
Alanine substitutions for hydrophobic residues in a b position (L97A), in a g position (I102A), or between the heptad repeats (L90A) generated proteins with wild-type epitopes, stability, and expression levels (Fig. 3). These mutations had little or no effect on the NA activities of the proteins (Table 1). L90A and L97A had wild-type HA activities, while HA activity was increased to a very high level for I102A (Table 1). Fusion promotion activity was decreased to 13% of the wild-type level for L90A, a level observed for the other substitutions in heptad repeat 1 (Fig. 4). L97A also showed a decrease in fusion (18% of the wild-type level) comparable to levels observed for other substitutions in heptad repeat 2. I102A had less effect on fusion promotion than any of the other mutants, with fusion promotion at 60% of the wild-type level.
DISCUSSION
Heptad repeat motifs are important for fusion activity and are found in the fusion proteins of a variety of viruses, including retroviruses (envelope protein), coronaviruses (peplomer protein), paramyxoviruses (F protein), and influenza viruses (HA protein) (3). Heptad repeats in many of these proteins have been shown both by site-directed mutagenesis and peptide inhibition studies to be critical for fusion (2, 10, 19, 26–28). Indeed, for the NDV F protein it has been shown that mutations of heptad repeat 1, which is adjacent to the fusion peptide (23), as well as the transmembrane adjacent heptad repeat 2 (the leucine zipper) domains abrogate fusion (2, 20). Furthermore, peptides with sequences from two heptad repeats inhibit fusion (29, 30). The membrane-spanning region of the NDV HN protein also contains a heptad repeat of leucine residues which, when mutated, destabilized the tetrameric structure of the mature protein and altered the biological activities of the protein, including fusion promotion (15).
A heptad repeat motif is that in which a hydrophobic amino acid is repeated every seven (heptad) residues; such motifs are designated a through g (14). Heptad repeats which contain hydrophobic or neutral residues in the a and d positions of the repeat can form alpha helices and are able to interact with other heptad repeats by forming coiled coils (3, 14). Proteins which interact in this matter are diverse and include c-Fos–c-Jun heterodimer, the catabolite gene activator protein in Escherichia coli, GCN4 in yeast, and the influenza virus HA protein (14). Clearly, the coiled coils are involved in protein-protein interactions important in many diverse systems.
Chimeric studies of paramyxovirus HN proteins have shown that the presumed stalk domains of various HN proteins confer F protein specificity in fusion promotion (5, 24, 25). One interpretation of these data is that the stalk domain of the HN protein is important for interactions with the F protein. Because of the importance of paramyxovirus F protein heptad repeat motifs, we wanted to investigate a region containing two heptad repeats in the presumed stalk domain of the NDV HN protein (amino acids 74 to 110). This region of the HN protein was analyzed for secondary structure by using prediction software from the Baylor College of Medicine (Fig. 2). Two alpha-helical heptad repeat regions separated by a nonhelical region of seven amino acids were predicted. The structure of proteins with amino acid substitutions presented here were similarly analyzed (Fig. 5). Importantly, none of the substitutions resulted in a decrease of predicted alpha-helical structure. Furthermore, the seven-amino-acid region between the two helices was predicted to gain helical structure in the mutants V88A, L96A, I103A, and L96/L110A.
FIG. 5.
Secondary structure prediction of mutant HN proteins. Secondary structure prediction (NNSSP program) of amino acids 74 to 110 for the wild-type HN protein and mutant HN proteins. + denotes area predicted to have alpha-helical structure.
The paramyxoviruses SV41, MuV, SV5, hPIV3, avian parainfluenza virus 4, and SeV HN proteins were similarly analyzed for secondary structure and found to contain heptad repeat regions predicted to form alpha helices (Fig. 2). All but one of the viruses analyzed (SeV) were predicted to contain a nonhelical space between the two helices. Disruptions of alpha-helical regions such as these are known as discontinuities and may introduce fixed bends or flexible regions or provide boundaries between coiled coils (18). Discontinuities are quite common in coiled coils and consist of several groups (14, 18). Non-helical regions do not have the structure of an alpha helix (these are present in all but one of the paramyxoviruses analyzed), skip residues are the addition of extra amino acids in the heptad repeat (observed for NDV, SV41, and SeV), and stutters occur when three residues are dropped from the heptad repeat (MuV and SV5 have one stutter; hPIV3 has two stutters in a row). Clearly such discontinuities potentially impart many different structures to alpha-helical regions of proteins. Conservation of heptad repeats, presumed alpha-helical regions, as well as discontinuities in the presumed stalk of paramyxovirus HN proteins suggest that these structural determinants may be important to the structure and function of the protein.
We generated four sets of mutants to begin to elucidate the mechanism by which the heptad repeat domain of the HN protein may contribute to fusion. The first set of mutants changed the hydrophobic a-position residues of the first (more amino terminal) heptad repeat to the hydrophobic residue alanine generating L74A, V81A, and V88A. Similarly, a second set of mutants L96A, I103A, and L110A were generated in the a-position residues of the second heptad repeat. A double mutation containing an a-position substitution in each repeat (V81/L110A) and a double mutation with two changes in the second heptad repeat (L96/L110A) were generated as well. A third set of mutants introduced a methionine residue in place of leucine in heptad repeat a-positions, generating L74M and L96M. Because methionine is bulkier than leucine, these substitutions could potentially sterically prevent a possible HN-F protein interaction and thus more efficiently inhibit fusion. Alternatively, the methionine could substitute for leucine, restoring activity. A final set of mutations introduced alanines into heptad repeat positions other than the a-position, creating L97A and I102A, and into the seven-amino-acid space between the two heptad repeats generating L90A.
All of the mutant HN proteins appeared to fold correctly and to retain wild-type epitopes as determined by immunoprecipitation with polyclonal as well as conformationally sensitive monoclonal antisera against the HN protein. Furthermore, that the oligomeric structure was not disrupted was shown by the formation of disulfide-linked dimers. In addition, sucrose density gradients showed no shifts in sedimentation from the tetramer position to a monomer or dimer position, which would indicate a loosely associated or absent tetramer (not shown).
All of the mutant proteins were able to bind erythrocytes. Indeed, most of these mutations resulted in mutant proteins with increased, in some cases substantially greater, abilities to bind erythrocytes. These results argue that specific amino acids in the stalk domain of the HN protein are not critical for attachment activity because the domain is highly tolerant of amino acid substitutions which appear for the most part to increase its activity.
NA activities of the mutant proteins varied greatly with V81A, having 157% of wild-type NA activity and L110A having 14% of wild-type NA activity. Approximately half of the mutant proteins had wild-type or slightly lower NA activities, while the other half showed decreased NA activities. There was not an obvious pattern of preferred amino acids in specific heptad repeat positions for NA activity. The presence of mutants with less than wild-type activity, however, would argue that individual amino acids in this region appear to be important for NA activity. While these results suggest that the overall conformations of the mutant proteins may be abnormal, the presence of epitopes similar to the wild-type protein as well as wild-type levels of oligomerization suggest that any conformational alterations are subtle. Levels of NA activity do not correlate in any obvious way with the fusion activities of the mutants.
All of the mutant proteins negatively affected fusion. Defects in fusion promotion were not due to defects in HA activity, as all of the mutant proteins were able to bind erythrocytes. These results reinforce the previous conclusion that fusion promotion and attachment (as well as NA activity) can be genetically separated (22). Furthermore and most importantly, these mutant proteins (L74A, V81A, L96A, L97A, I102A, L96M, and L90A) illustrate a requirement in the HN protein for specific amino acids in this region of the protein for fusion promotion. Single, extremely conservative changes (L74A and V81A) virtually eliminated fusion promotion activity. Additionally, these mutations illustrate that the a-positions of the helices were not more critical than other positions for fusion promotion. These results suggest that the presumed alpha-helical structure of the heptad repeats is not sufficient for fusion, although one cannot rule out the possibility that a helical structure is necessary for fusion promotion, as none of the mutants generated were predicted to lessen the probability of forming an alpha helix.
As mentioned previously, paramyxovirus F proteins contain two heptad repeat regions which are conserved and have been shown to be critical for fusion. We propose that it is possible for the conserved heptad repeat region of the paramyxovirus HN protein to interact with the heptad repeats of the F protein, since the helical nature of these regions in both proteins presents the possibility of coiled-coil interactions between the proteins. The importance of specific residues for fusion promotion may indicate specific interactions between the proteins. One intriguing possibility is that the HN heptad repeats may bind to the heptad repeats HR1 and HR2 of the F protein, serving to keep these two regions apart. The discontinuity between the helices would give the HN protein the flexibility to participate in such an interaction. Upon binding of the HN protein to its receptor, a conformational change may occur in both proteins, disrupting the HN-F interaction and resulting in the release of the fusion peptide into the target membrane. Individual amino acids would create a level of specificity which agrees with the observations that HN and F proteins from different paramyxoviruses do not complement each other to promote fusion.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant AI30572.
REFERENCES
- 1.Baylor College of Medicine. February 1998, revision date. SSP and NNSSP programs. [Online]. http://dot.imgen.bcm.tmc.edu:9331/seq-search/struc-predict.html. [February 1999, last date accessed.]
- 1a.Branden C, Tooze J. Introduction to protein structure. New York, N.Y: Garland Publishing, Inc.; 1991. [Google Scholar]
- 2.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 3.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 4.Colman P M, Hoyne P A, Lawrence M C. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol. 1993;67:2972–2980. doi: 10.1128/jvi.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, R., Z., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457–469. [DOI] [PubMed]
- 6.Hu X, Ray R, Compans R W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio R M, Glickman R L, Riel A M, Sheehan J P, Bratt M A. Functional and neutralization profile of seven overlapping antigenic sites on the HN glycoprotein of Newcastle disease virus: monoclonal antibodies to some sites prevent attachment. Virus Res. 1989;13:245–262. doi: 10.1016/0168-1702(89)90019-1. [DOI] [PubMed] [Google Scholar]
- 8.Joshi S B, Dutch R E, Lamb R A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 9.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 10.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langedijk J P M, Daus F J, van Oirschot J T. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levesque J-P, Sanilvestri P, Hatzfeld A, Hatzfeld J. DNA transformation in Cos cells: a low cost serum free method compared to Lipofectin. BioTechniques. 1991;11:313–318. [PubMed] [Google Scholar]
- 13.Li Z, Sergel T, Razvi E, Morrison T. Effect of cleavage mutants on syncytium formation directed by the wild-type fusion protein of Newcastle disease virus. J Virol. 1998;72:3789–3795. doi: 10.1128/jvi.72.5.3789-3795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 15.McGinnes L, Sergel T, Morrison T. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology. 1996;196:101–110. doi: 10.1006/viro.1993.1458. [DOI] [PubMed] [Google Scholar]
- 16.Morrison T G, McGinnes L W. Avian cells expressing the Newcastle disease virus HN protein are resistant to NDV infection. Virology. 1989;171:10–17. doi: 10.1016/0042-6822(89)90505-9. [DOI] [PubMed] [Google Scholar]
- 17.Morrison T G, McQuain C, McGinnes L W. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J Virol. 1991;65:813–822. doi: 10.1128/jvi.65.2.813-822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oas T G, Endow S A. Springs and hinges: dynamic coiled coils and discontinuities. Trends Biochem Sci. 1994;19:52–54. doi: 10.1016/0968-0004(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 19.Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reitter J N, Sergel T, Morrison T G. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sergel T, McGinnes L W, Morrison T G. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology. 1993;196:831–834. doi: 10.1006/viro.1993.1541. [DOI] [PubMed] [Google Scholar]
- 22.Sergel T, McGinnes L W, Peeples M E, Morrison T G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 23.Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanabayashi K, Compans R W. Functional interaction of Paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsurudome M, Kawano M, Yuasa T, Nishio M, Hiroshi K, Ito Y. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology. 1995;213:190–203. doi: 10.1006/viro.1995.1559. [DOI] [PubMed] [Google Scholar]
- 26.Wild C, Dubay J W, Greenwell T, Baird J, Oas T G, McDanal C, Hunter E, Mathews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild T F, Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- 28.Yao Q, Compans R W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 29.Young J K, Hicks R P, Wright G E, Morrison T G. The role of leucine residues in the structure and function of a leucine zipper peptide inhibitor of paramyxovirus (NDV) fusion. Virology. 1998;243:21–31. doi: 10.1006/viro.1998.9044. [DOI] [PubMed] [Google Scholar]
- 30.Young, J., et al. Unpublished data.