Abstract
Bovine viral diarrhea virus (BVDV), a member of the genus Pestivirus in the family Flaviviridae, has a positive-stranded RNA genome consisting of a single open reading frame and untranslated regions (UTRs) at the 5′ and 3′ ends. Computer modeling suggested the 3′ UTR comprised single-stranded regions as well as stem-loop structures—features that were suspected of being essentially implicated in the viral RNA replication pathway. Employing a subgenomic BVDV RNA (DI9c) that was shown to function as an autonomous RNA replicon (S.-E. Behrens, C. W. Grassmann, H. J. Thiel, G. Meyers, and N. Tautz, J. Virol. 72:2364–2372, 1998) the goal of this study was to determine the RNA secondary structure of the 3′ UTR by experimental means and to investigate the significance of defined RNA motifs for the RNA replication pathway. Enzymatic and chemical structure probing revealed mainly the conserved terminal part (termed 3′C) of the DI9c 3′ UTR containing distinctive RNA motifs, i.e., a stable stem-loop, SL I, near the RNA 3′ terminus and a considerably less stable stem-loop, SL II, that forms the 5′ portion of 3′C. SL I and SL II are separated by a long single-stranded intervening sequence, denoted SS. The 3′-terminal four C residues of the viral RNA were confirmed to be single stranded as well. Other intramolecular interactions, e.g., with upstream DI9c RNA sequences, were not detected under the experimental conditions used. Mutagenesis of the DI9c RNA demonstrated that the SL I and SS motifs do indeed play essential roles during RNA replication. Abolition of RNA stems, which ought to maintain the overall folding of SL I, as well as substitution of certain single-stranded nucleotides located in the SS region or SL I loop region, gave rise to DI9c derivatives unable to replicate. Conversely, SL I stems comprising compensatory base exchanges turned out to support replication, but mostly to a lower degree than the original structure. Surprisingly, replacement of a number of residues, although they were previously defined as constituents of a highly conserved stretch of sequence of the SS motif, had little effect on the replication ability of DI9c. In summary, these results indicate that RNA structure as well as sequence elements harbored within the 3′C region of the BVDV 3′ UTR create a common cis-acting element of the replication process. The data further point at possible interaction sites of host and/or viral proteins and thus provide valuable information for future experiments intended to identify and characterize these factors.
Bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and border disease virus (BDV) of sheep are the constituent members of the genus Pestivirus, and all represent important animal pathogens (reviewed in reference 67).
Pestiviruses are small enveloped viruses; the viral genome is a single-stranded positive-strand RNA molecule with a size of approximately 12.3 kb, which consists of a single, long open reading frame (ORF) and untranslated regions (UTRs) at the 5′ and 3′ ends. Translation of the ORF yields a polyprotein that is cleaved by cellular and virus-encoded proteases into at least 11 mature structural and nonstructural (NS) proteins (56, 67).
Along with flaviviruses and hepatitis C virus (HCV), pestiviruses are classified in the family Flaviviridae (25). The genomic organization of pestiviruses and the characteristics of gene expression, i.e., the mode of polyprotein translation and processing, however, resemble more those of HCV than those of flaviviruses (31, 56).
Infection studies (27) already suggested that BVDV follows a replication strategy similar to those of other positive-strand RNA viruses, such as poliovirus (72): concomitant with the translation of the viral genome, the nascent viral proteins presumably associate with the RNA genome and hypothetical host factors to form replication complexes. These catalyze the transcription of complementary negative-strand RNA molecules, which than act as templates for the synthesis of novel genomic RNA molecules. Detailed studies of the biochemical processes underlying pestiviral replication are significantly facilitated by the successful composition of genomic DNA copies capable of producing infectious RNA transcripts (cRNA) in vitro (43–45, 47, 57, 70). BVDV cRNA-based transfection experiments revealed that the replication process occurs exclusively in the host cell cytoplasm and proceeds in an asymmetric manner, i.e., with respect to the negative-strand RNA intermediate, an excess of progeny positive-strand RNA is synthesized. Further experiments led to the discovery that a subgenomic 7.8-kb BVDV RNA molecule (denoted DI9c), comprising only the 5′ and 3′ UTRs of the viral genome as well as the coding regions of the pestivirus autoprotease Npro and the mature nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B, functions as an autonomous RNA replicon: upon transfection into eukaryotic host cells, DI9c RNA supports both steps of the replication pathway even in the absence of helper virus (7).
The termini of viral RNA genomes are known to harbor cis-acting signals, usually originating from both conserved sequence and structural motifs, which function during modulation of translation and replication events via interaction with proteins or other parts of the RNA molecule (62, 63). A number of extensive stem-loop structures span the 5′ UTR of picornaviruses (51, 35) as well as the 5′ UTRs of HCV and pestiviruses (14, 30, 54, 68). They contribute to internal ribosomal entry sites which mediate cap-independent translation of the viral ORF (38). The immediate 5′ terminus of the poliovirus genome has been shown to fold into a “cloverleaf-like” structure; viral and host proteins interact with this motif at specific sites, thus constituting a ribonucleoprotein complex that exhibits catalytic activity during the second replication step (1, 2). The 3′ ends of the genomes of many positive-strand RNA plant viruses have been shown to form tRNA-like motifs (19), which often contain internal pseudoknot structures (53). Similar features were found in the 3′ end of the genome of bacteriophage Qβ (12). Such secondary or tertiary RNA structure motifs are therefore suspected to represent important functional elements of a “negative-strand promoter” that is involved in the formation of the initiation complex of viral replication (20).
Although the termini of the genomic RNAs of the different members of the family Flaviviridae are also conceivably implicated in RNA replication, knowledge about their functional significance is still preliminary. The 3′ ends of the genomes of many flaviviruses are predicted to consist of a conserved stem-loop motif with an internal pseudoknot structure (13, 59, 71). Progress in understanding the functional importance of a conserved 98-base structural element at the 3′ terminus of the HCV genome (11, 37, 64, 65) is unfortunately hindered by the current lack of a convenient animal model and in vitro culture system to follow HCV replication.
In this report, we examine the RNA secondary structure of a BVDV 3′ UTR by experimental means, demonstrating that the 3′-terminal part of this region contains characteristic RNA structure as well as exposed single-stranded sequence elements. The most striking of the these RNA motifs were subsequently determined by genetic approaches to represent essential cis-encoded signals of the RNA replication process.
MATERIALS AND METHODS
Cells.
BHK-21 (baby hamster kidney) cells were a gift from J. Cox (Federal Research Centre for Virus Diseases of Animals, Tübingen, Germany). The cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and nonessential amino acids.
Construction of recombinant plasmids.
Restriction and subcloning procedures were done following standard protocols. Restriction and modifying enzymes were purchased from New England Biolabs (NEB) (Schwalbach, Germany), Pharmacia (Freiburg, Germany), and Boehringer Mannheim (Mannheim, Germany). All primers used for mutagenesis as well as the 5′ IRD71-labeled oligonucleotides (see below) utilized for sequencing were obtained from MWG Biotech GmbH (Eberbach, Germany). Construction of the DI9c cDNA clone pA/BVDV/D9 was described previously (45). The diverse 3′ UTR mutations were introduced into pA/BVDV/D9 as follows. Initially, a PstI-SmaI fragment of pA/BVDV/D9 encoding the 3′ terminus of the RNA (corresponding to nucleotides 11967 to 12281 of the full-length BVDV CP7 genome [45, 66]; nomenclature according to Deng and Brock [21]) was ligated into the multiple cloning site (MCS) of pBluescript KS vector (Stratagene, La Jolla, Calif.) cut with the same enzymes. This construct (pPS) served as a template for primer-directed mutagenesis via PCR amplification with the M13 reverse primer as sense primer and oligonucleotides containing mutated 3′ UTR sequences as antisense primers (Table 1). PCR products were gel purified, digested with PstI-AatII (position 12241 of BVDV CP7) (primers 1 to 8 and 10) or PstI-SmaI (primers 9*, 11*, and 12 to 15), and cloned between the corresponding restriction sites of subclone pPS. Stem double-mutant subclones 9 and 11 (see Fig. 5) were composed of subclones 8 and 9*, and 10 and 11*, respectively, using AatII and XmnI. Substitution of the mutated inserts for the wild-type sequence was verified by dideoxy cycle sequencing (see below). By cutting with PstI-ScaI, the mutations were next inserted into a second cloning intermediate (pCAS) previously created by cloning the 1,220-bp ClaI-SmaI fragment (ClaI at position 11061 of BVDV CP7) of pA/BVDV/D9 into pBluescript KS. Finally, either the ClaI-AatII (mutations 1 to 8 and 10) or the ClaI-SmaI (mutations 9 and 11 to 15) fragment of the respective pCAS construct was introduced into the pA/BVDV/D9 plasmid cut with the corresponding enzymes. All mutations were confirmed by dideoxy sequencing.
TABLE 1.
Oligonucleotide primers used for mutagenesis of the DI9c 3′ UTR
| Primer | Sequencea |
|---|---|
| 1 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TAA AGT GCC ACG TGcC ATT GAG 3′ |
| 2 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TAA AGA CAT GTdG TGC ATT GAG 3′ |
| 3 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TAA AAT GCT GTG TGC 3′ |
| 4 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TAA GGT GCT GTG TGC 3′ |
| 5 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TAG AGT GCT GTG TGC 3′ |
| 6 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC TGA AGT GCT GTG TGC 3′ |
| 7 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TAC AGC CAA AGT GCT GTG TGC 3′ |
| 8 | 5′ CTA TGG ACG TCbG GGT GTA CCC TCA TTG TCGC TAA AGT GCT GTG TGC 3′ |
| 9* | 5′ ATC CCC GGGe GGC GAC ATA AAG GTC TTC CC 3′ |
| 10 | 5′ CTA TGG ACG TCbC CCT GTA CCC TCA 3′ |
| 11* | 5′ ATC CCC GGGe GGG CTG TTA AAG GTC TTG GGT AGT CCA ACT ATG 3′ |
| 12 | 5′ ATC CCC GGGe GGG CTG TTA AAG GTC TTC CCT ACA GGT ACT ATG GAC GTC 3′ |
| 13 | 5′ ATC CCC GGGe GGG CTG TTA AAG GTC TTC CCT ACA GGT ACT AAC CTG GTC 3′ |
| 14 | 5′ ATC CCC GGGe GGG CTG TTA AAG GTC TTC CCT AGT CCA ACC GTG GAC GTC GGG 3′ |
| 15 | 5′ ATC CCC GGGe GGG CTG TTA AAG GTC TTC CCT AGT CCA GTT ATG GAC GTC GGG 3′ |
Mutated bases are in boldface italics.
AatII site.
PmlI site.
AflIII site.
SmaI site.
FIG. 5.
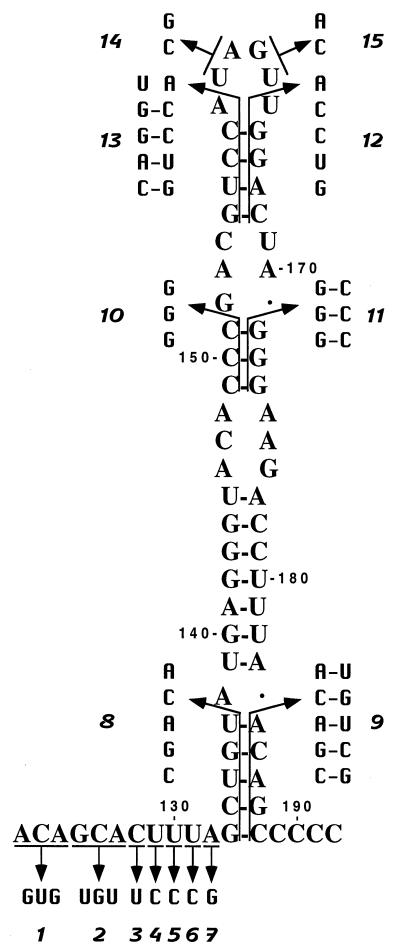
Mutagenesis of the 3′ end of the BVDV DI9c 3′ UTR. A schematic drawing of the diverse mutations which were introduced into the 3′ UTR of DI9c RNA is shown. The 3′ terminus of the RNA is depicted in its secondary structure suggested in Fig. 4; the numbering scheme is the same as in the previous figures. The substitutions are indicated by arrows, and the resulting DI9c RNA mutants are denoted with numbers 1 to 15.
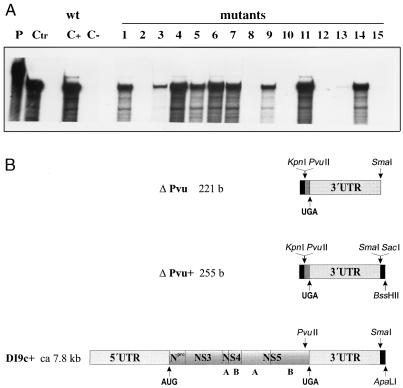
pΔPvu is a pBluescript KS derivative which was originated from pPS: pPS was cut with KpnI (pBluescript MCS) and PvuII (position 12070 of BVDV CP7) and both sites were blunted with T4 DNA polymerase (NEB) and religated. To generate in vitro transcripts for determination of the RNA secondary structure by enzymatic probing, pΔPvu was linearized with SmaI (Fig. 1) (see below). Removal of the SmaI-SacI fragment from the MCS of pΔPvu, blunting with T4 DNA polymerase, and religation led to plasmid pΔPvu+ (Fig. 1). In order to prepare transcripts that could be applied to both enzymatic and chemical probing of the RNA structure (see below), pΔPvu+ was linearized with BssHII located downstream within the pBluescript MCS (Fig. 1).
FIG. 1.
(A) Primary structure of the 3′ UTR of BVDV DI9c RNA. The numbers above the nucleotide sequence represent the coordinates utilized throughout this work to describe the structure-probing experiments. The polyprotein stop codon UGA is indicated by boldface italics. The boldface nucleotides represent the constituents of the conserved region (3′C) of the 3′ UTR as previously suggested by Deng and Brock (22). The underlined nucleotides are 100% conserved among different representative pestivirus strains (6). (B) Schematic representation of the RNA transcripts used in this study. RNA sequences corresponding to the UTRs are represented as dark shaded boxes. The positions of translation start and stop codons are indicated by arrows. Regions of the RNA encoding either partial or entire viral proteins are depicted as light shaded boxes. RNA portions originated by transcription of vector sequences are shown as solid boxes. Arrows also mark the location of restriction endonuclease cleavage sites within the cDNA constructs that were utilized during the cloning procedures or for runoff transcription (for details, see Materials and Methods and Results). ΔPvu RNA contains a sequence of 12 bases (b) at its 5′ extremity derived by transcription of vector sequences (from the start point of T3 promoter-driven transcription to the KpnI site within the pBluescript polylinker) and 17 bases encoding the carboxy terminus of the NS5B protein (PvuII [12070 of BVDV CP7] [66, 45] to the UGA stop at 12090 of BVDV CP7, corresponding to position 1 of the scheme in panel A) and the entire 3′ UTR (192 bases). ΔPvu+ RNA includes the entire ΔPvu sequence as well as an additional 3′-terminally located 34 bases, also derived by transcription of vector sequences (SacI to the BssHII site of the pBluescript polylinker) to allow hybridization of a T7 primer. DI9c+ RNA consists of the BVDV ORF encoding the viral proteins Npro, NS3, NS4A, NS4B, NS5A, and NS5B and the flanking 5′ and 3′ UTRs. In the lower diagram showing a schematic drawing of DI9c+ RNA, the UTRs are depicted greatly oversized with respect to the polyprotein coding region; the latter encompasses approximately 90% (7.2 of 7.8 kb) of the total length of the RNA. Compared to the DI9c RNA replicon, DI9c+ contains an additional 160 bases derived from vector sequences (spanning the region between the SmaI and ApaLI sites of pA/BVDV/D9) to facilitate annealing of a KS primer.
For in vitro transcription and replication experiments, all pA/BVDV/D9 derivatives were linearized with SmaI. DI9c RNA used for chemical probing was transcribed from the template that had been prior restricted at the ApaLI site, which is 3′ terminally located within the original pACYC 177 vector (NEB) of pA/BVDV/D9. Plasmid constructs applied to generate the radiolabeled RNA probes for the RNase protection assay were described previously (7).
DNA sequencing.
Dideoxy sequencing of double-stranded DNA was carried out by applying either a cycle-sequencing kit (Amersham) and a DNA sequencer 4000L (Li-Cor; MWG) or the T7 DNA polymerase dideoxy sequencing kit from Pharmacia. Computer-aided analysis of sequence data was performed with the Genetics Computer Group software.
Radioactive end labeling of RNA and DNA oligonucleotides.
RNAs were in vitro transcribed and purified as described previously (7). Prior to being 5′ end labeled, the RNA was dephosphorylated with alkaline phosphatase (Boehringer Mannheim) by a standard protocol. Initially, 10 μg of RNA was dephosphorylated with 3 U of alkaline phosphatase at 50°C for 1 h; subsequently, it was phenol-chloroform extracted and ethanol precipitated.
Oligonucleotides (MWG) or dephosphorylated RNAs were 5′ end labeled by applying T4 polynucleotide kinase (AGS GmbH, Heidelberg, Germany) and [γ-32P]ATP (5,000 Ci/mmol; Amersham) as follows: 10 pmol of RNA or oligonucleotide was incubated in a 10-μl reaction mixture containing 1 μl of 10× buffer (AGS), 20 U of RNaseOUT (Gibco-BRL, Bethesda, Md.), 10 U of T4 polynucleotide kinase, and 50 μCi of [γ-32P]ATP at 37°C for 30 min. RNA 3′ end labeling was carried out by ligation of [α-32P]pCp (3,000 Ci/mmol; Amersham) with T4 RNA ligase (Gibco-BRL) following a standard protocol (24).
The radiolabeled RNAs were purified by electrophoresis and subsequent elution from denaturing polyacrylamide gels. Radiolabeled oligonucleotides were separated from unincorporated nucleotides by gel filtration through a 1-ml Sephadex G-50 spin column. The radiolabeled nucleic acids were phenol-chloroform extracted and ethanol precipitated with tRNA as a carrier.
Enzymatic probing of RNA secondary structure.
Enzymatic probing of in vitro-transcribed and end-labeled RNA was performed at 0°C for 30 min, with RNases T1 (0.7 U), U2 (4 U), and PhyM (4 U) and Bacillus cereus RNase (1 U) (RNA sequencing enzyme kit from Pharmacia). 32P-labeled RNA molecules (5 × 104 cpm) were digested in a 5-μl reaction volume containing 30 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, and 4 μg of tRNA. To stop the reaction, an equal volume of formamide sample buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol) was added. In parallel, the corresponding RNA sequences were determined by digestion of end-labeled RNA with the four RNases (each at 1 U) at 55°C for 5 to 20 min in different urea-citrate buffers (T1 and PhyM, 20 mM sodium citrate [pH 5.0], 7 M urea, 1 mM EDTA; U2, 20 mM sodium citrate [pH 3.5], 7 M urea, 1 mM EDTA; and B. cereus, 20 mM sodium citrate [pH 5], 1 mM EDTA). Alkaline hydrolysis was performed at 94°C for 90 s in 50 mM NaOH to produce an RNA ladder. The locations of specific cleavage sites were determined electrophoretically on 7.5 or 10% Tris (100 mM)–boric acid (100 mM)–EDTA (2.5 mM) polyacrylamide gels containing 7 M urea (see figure legends).
Chemical probing of RNA secondary structure.
The modifying agents used were dimethyl sulfate (DMS; Fluka) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT; Sigma, Deisenhofen, Germany). DMS modification was performed with 1 μg of in vitro-transcribed RNA and 0.5 or 1 μl of DMS in a 200-μl reaction mixture containing 20 mM HEPES-KOH (pH 7.9), 60 mM KCl, and 12 mM MgCl2 at 30°C for 5 min. The reaction was stopped by addition of an equal volume of DMS stop buffer containing 0.6 M sodium acetate, 0.4 M β-mercaptoethanol, 0.4 M Tris-HCl (pH 7.5), and 10 mM EDTA. The CMCT modification was carried out with 1 μg of RNA and 210 or 420 μg of CMCT in a 25-μl reaction volume containing 80 mM sodium borate (pH 8.1), 60 mM KCl, and 12 mM MgCl2 for 10 min at 30°C. The reaction was stopped with 375 μl of CMCT stop buffer containing 0.3 M sodium acetate, 0.2 M PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid)–HCl (pH 6.4), and 5 mM EDTA. The modified RNA was then phenol-chloroform extracted and ethanol precipitated with 10 μg of tRNA as a carrier.
Primer extension analysis.
Chemically modified RNA molecules (0.5 μg) were mixed with 2 × 105 cpm of 5′-end-labeled DNA primer (T7, 5′-AAT ACG ACT CAC TAT AGG GC-3′; KS, 5′-AAC TAG TGG ATC CCC CGG-3′; Stratagene). Primer extension was performed at 37°C for 50 min with 100 U of Superscript II RNase H− reverse transcriptase (Gibco-BRL) at standard conditions as recommended by the manufacturer. The locations of modification sites were determined by polyacrylamide gel electrophoresis of the primer extension products (see above). In parallel, the corresponding DNA sequences were determined by dideoxynucleotide sequencing (T7 DNA sequencing kit; Pharmacia) with the same 5′-end-labeled primers.
Transfection of BHK-21 cells.
the transfection procedure for BHK-21 cells has been described previously (7). In brief, the cells were washed twice with phosphate-buffered saline (PBS; 20 mM Na-PO4 [pH 7.4], 130 mM NaCl) and transfected with 2 μg of RNA transcript by electroporation (2 pulses; 200 Ω; 25 μF; 1,500 V) applying a model II gene pulser (Bio-Rad, Munich, Germany). The quality of the transfected RNA transcripts and efficiency of RNA transfection were controlled by an immunofluorescence assay employing a monoclonal antibody directed against the BVDV NS3 protein (7).
RNase protection assay.
The method used to determine RNA replication was as described previously with slight modifications (7). Transfected BHK cells grown for 24 h in 100-mm-diameter plates were washed once with PBS, harvested, and lysed in 375 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 0.5% Nonidet P-40) at 0°C. The nuclei were removed by centrifugation for 2 min at 1,000 × g. The cytoplasmic supernatant was supplemented with 8 μl of 10% (wt/vol) sodium dodecyl sulfate, and proteins were removed by digestion with 50 μg of proteinase K (Boehringer Mannheim) at 37°C for 1 h. After phenol-chloroform extraction, the nucleic acids were precipitated with ethanol and the washed pellet was dissolved in 180 μl of hybridization buffer (80% [vol/vol] formamide, 40 mM PIPES-HCl [pH 6.4], 400 mM NaCl, 1 mM EDTA). Dissolved RNA (12 to 30 μl) was denatured at 85°C for 2.5 min and subjected to hybridization at 45°C overnight with radiolabeled DI9c sense or antisense probes (105 cpm; ca. 109 cpm/μg). Subsequently, 350 μl of RNase digestion buffer (10 mM Tris-HCl [pH 7.5], 1 M NaCl, 5 mM EDTA), as well as 25 U of RNase T1 and 3.5 μg of RNase A, was added, and digestion was performed for at least 1 h at 37°C. To obtain efficient negative-strand detection, the excess of positive-strand RNA was removed by performing a cycle of hybridization and RNase treatment before applying the radiolabeled probe in a second hybridization and protection procedure (7). After the addition of 20 μl of 10% sodium dodecyl sulfate, proteinase K digestion (50 μg), phenol-chloroform extraction, and ethanol precipitation, the protected fragments were analyzed electrophoretically on 5% denaturing polyacrylamide gels (see above).
Quantification of the protected RNA fragments was performed with a Fuji Bio imaging analyzer and the corresponding software, TINA version 2.09.
RT-PCR.
Reverse transcription (RT) to verify the stability of the diverse DI9c mutants posttransfection was performed essentially as described previously (7) with appropriate deoxyoligonucleotide primers.
RESULTS
Several factors recommended the RNA replicon BVDV DI9c as the most suitable system for a genetic study of cis-encoded RNA elements conceivably involved in the viral replication process, such as the 3′ UTR. (i) Because they are significantly shorter, DI9c cDNA constructs are easier to handle than cDNAs that encode the full-length viral genome. (ii) DI9c and derivative cRNA transcripts exhibit remarkable stability under the experimental conditions used (7) (see below); moreover, emerging revertants do not possess the capacity to spread. (iii) By far the most important factor is that the subgenomic RNA provides the opportunity to examine the replication process apart from RNA packaging and/or virion assembly. This considerably facilitates studies aimed at defining the essential requirements of the RNA replication process by applying site-directed mutagenesis (“reverse genetics”).
Primary structure of the BVDV DI9c 3′ UTR.
The 3′ UTR of DI9c RNA represents a functional chimera which, due to the cloning procedure of the cDNA construct, had been assembled from different BVDV sources (for details, see reference 45); most of it, however, was derived from the BVDV strain CP7 (66). Its sequence, comprising 192 nucleotides, was reconfirmed and is depicted in Fig. 1A. In order to prevent confusion during presentation of the structure-probing experiments described below, a numbering scheme was chosen for the 3′ UTR starting with the first residue of the UGA translation stop codon and ending with the C residue as originated by runoff transcription at the SmaI restriction site of the cDNA construct. Comparison of 3′ UTRs among different pestivirus strains suggested that it is apparently composed of a variable region (3′V) and a conserved region (3′C) (22) (see below). Application of this alignment to the DI9c 3′ UTR revealed the respective variable part, 3′V, to consist of 90 nucleotides at the 5′ end, whereas in analogy to all other BVDV and CSFV strains, the conserved 3′C region includes the 3′ terminus and encompasses 102 nucleotides (Fig. 1A).
Structure probing of the BVDV DI9c 3′ end.
In the first series of experiments we wanted to determine the RNA secondary structure of the DI9c 3′ UTR. For this purpose, two experimental procedures were employed that basically allowed the identification of nucleotides in single-stranded conformation within the native molecule (17, 61). On the one hand, structure probing was performed by partial RNase digestion of 5′- or 3′-end-labeled in vitro transcripts by applying a variety of single-strand-specific ribonucleases with different substrate specificities. On the other hand, unpaired nucleotides, even those that were sterically hindered from access by nucleases, were identified by chemical modification of the native RNA transcripts and a subsequent primer extension assay in which cDNA was synthesized by reverse transcriptase from a 5′-end-labeled downstream hybridized oligonucleotide (see Materials and Methods and below).
Most of the RNase structure-probing experiments were carried out on runoff RNA transcripts generated from the plasmid template pΔPvu, prior linearized at the SmaI site (Fig. 1B). The RNA molecules (denoted ΔPvu RNA) contained a short 3′-terminal part of the ORF as well as the entire 3′ UTR and terminated with five C residues, which corresponds to the authentic BVDV genomic 3′ end (Fig. 1A) (45). Structure probing with RNases as well as with chemicals was also carried out on transcripts derived from pΔPvu+ linearized at the BssHII site further downstream (Fig. 1B); hence, these RNA molecules contained additional vector sequences to allow binding of a complementary oligonucleotide primer (termed ΔPvu+ RNA) for RT. Furthermore, probing was performed on the entire DI9c RNA replicon with an additional primer binding site; it was generated by runoff transcription from the DI9c-encoding plasmid prior linearized at the downstream ApaLI site (denoted DI9c+ [Fig. 1B]).
During initial structure-probing attempts, the RNA transcripts were denatured and then slowly cooled prior to exposure to RNases and chemicals in order to receive a maximum amount of native molecules. However, omission of this procedure had no significant effect on the probing profiles obtained (shown below), which suggested that the RNA secondary structure of the DI9c 3′ UTR as determined throughout these experiments forms rapidly and reproducibly under the chosen conditions (data not shown).
Structure probing of the BVDV DI9c 3′ UTR by RNases.
Initially, 5′-end-labeled native ΔPvu RNA was tested for its susceptibility to RNase T1 (G specific), B. cereus RNase (U and C specific), RNase PhyM (U and A specific), and RNase U2 (A specific). Analysis of the cleavage products by electrophoresis on different denaturing polyacrylamide gel systems (see Materials and Methods) (Fig. 2) enabled detection of single-stranded nucleotides in the region encompassing residues 1 to approximately 180, almost the entire DI9c 3′ UTR. Identification of the individual RNase-sensitive nucleotides within the native RNA was achieved via side-by-side electrophoresis of sequence ladders generated in parallel by alkaline hydrolysis of ΔPvu RNA or digestion with the same set of RNases, but under denaturing conditions (Fig. 2).
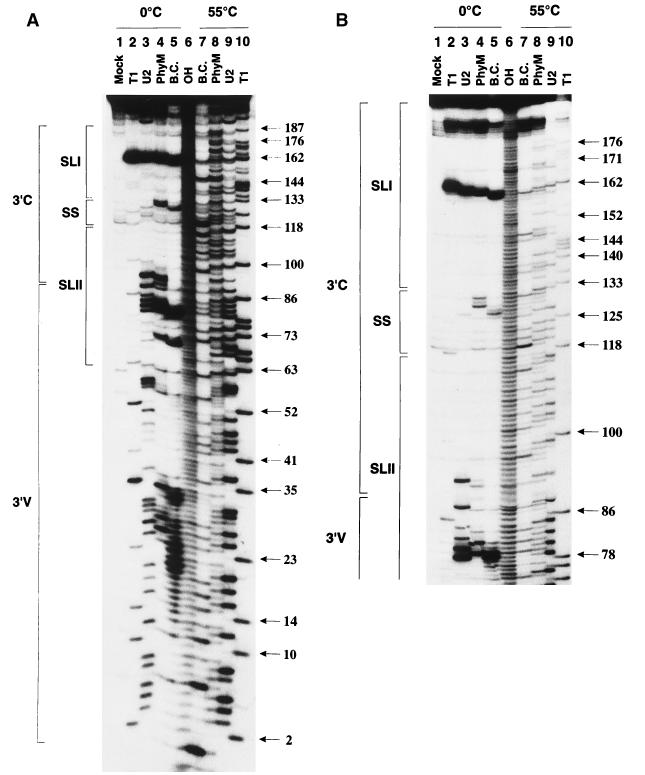
FIG. 2.
Enzymatic probing of the BVDV DI9c 3′ UTR RNA secondary structure. Cleavage products obtained by digestion of 5′-end-labeled ΔPvu RNA with single-strand-specific RNase T1 (G specific), U2 (A specific), PhyM (U and A specific), or B. cereus (B.C.) (U and C specific) (see Materials and Methods) were electrophoretically separated on a 10 (A) or 7.5% (B) denaturing polyacrylamide gel. Lanes 2 to 5, ΔPvu RNA digested under nondenaturing conditions at 0°C; lanes 6 to 10, ΔPvu RNA digested under denaturing conditions at 55°C, generating a sequence ladder to facilitate identification of the single-strand-specific cleavage sites within the native molecule that are monitored in lanes 2 to 5; lane 1, control incubation of ΔPvu RNA without RNases, performed under nondenaturing conditions (Mock); lane 6, RNA ladder obtained by alkaline hydrolysis (OH). The numbers on the right represent the positions of guanosine residues within the RNA sequence of the 3′ UTR as identified by digestion with RNase T1 under nondenaturing conditions (lane 10). The regions representing the variable 3′V and the conserved 3′C portions of the 3′ UTR (Fig. 1) are marked on the left, as well as the suggested SL I, SL II, and SS motifs of the RNA secondary structure (Fig. 4). Interestingly, B. cereus-directed cleavages (normally C and U specific) were also noticed in the case of A residues 69, 79, 126, and 159. These observations may reflect a common property of this enzyme under certain experimental conditions or may be the result of a contamination with a different RNase due to its mode of preparation. Similar data were obtained by other laboratories, e.g., during structure-probing experiments with B. cereus RNase on the HCV 3′ UTR (11).
Exposure of the natively folded ΔPvu RNA to partial digestion with each of the RNases yielded a characteristic and reproducible cleavage profile. As shown in Fig. 2A, almost the entire upstream portion of the 3′V region turned out to be accessible to the diverse single-strand-specific RNases. Conversely, the remainder of the DI9c 3′ UTR, encompassing the 3′ portion of the 3′V region and the entire 3′C region was observed to consist of highly susceptible as well as protected areas (Fig. 2A, residues ∼60 to 192). Interestingly, of the entire DI9c 3′ UTR, the region detected as most sensitive to hydrolysis by RNases resided within this latter part of the 3′ UTR, including nucleotides AUAG (159 to 162) (Fig. 2). Other accessible nucleotide stretches were detected further upstream of this motif, comprising residues ACUUUA (127 to 132), UA (92 and 93) and UAU (88 to 90), AUAUAUAG (79 to 86), and AUUA (69 to 72) (Fig. 2). RNase structure probing performed on 3′-end-labeled ΔPvu RNA yielded a congruent pattern of cleavage products as well as RNase digestion of DI9c+ RNA; the latter was analyzed by primer extension (data not shown). Indistinguishable results were obtained if 5′-end-labeled ΔPvu+ RNA was employed in the different RNase digestion procedures (not shown). From these results we concluded that the 3′-terminal part of DI9c 3′ UTR exhibited a defined RNA secondary structure. The data, moreover, imply that additional 5′-terminal and 3′-terminal sequences have negligible influence on the formation of this structure.
Thermodynamically favored RNA secondary structures of the 3′ UTR of the BVDV strains NADL, Osloss, and SD-1 were recently proposed by Deng and Brock (22), applying the FOLD program of Zuker and Stiegler (73). Since these proposals apparently reflected some of the above structure-probing data, a similar prediction was employed for the 3′-terminal 137 nucleotides of the DI9c 3′ UTR with the improved version of the program (34). The prediction proved to be of significant use not only in supporting the interpretation of our structure-probing experiments but also, after consideration of all data (see below), in the drawing up of a final suggestion as to the nature of the RNA secondary structure of this part of the DI9c 3′ UTR (see Fig. 4). As an initial approach, the nucleotide stretches that were determined to be exposed to the different RNases were classified as constituents of certain single-stranded regions within the RNA as follows. (i) Together with the UU (nucleotides 163 to 164) located further downstream (see below), the most efficiently cleaved AUAG (159 to 162) residues would represent components of a single-stranded hexaloop, which is part of a huge stem-loop structure (encompassing positions 133 to 188) near the 3′ terminus of the BVDV DI9c RNA (see Fig. 4). Such a distinctive structural motif (termed SL I) would also explain the remarkable RNase protection observed for almost all the residues ranging from positions 133 to 180 (Fig. 2B). (ii) ACUUUA (127 to 132) would belong to a single-stranded stretch of the RNA, consequently designated the SS region of the BVDV 3′ UTR. (iii) As outlined above, significant RNase cleavages were monitored for nucleotides suspected to compose a 12-nucleotide loop (88 to 99) as well as two bulged regions (comprising residues 69 to 72 and 111, and 79 to 84 and 103 and 104, respectively) of a second extensive stem-loop structure (SL II) formed by the downstream part of the 3′V region and the upstream part of the 3′C region (comprising positions 63 to 117 [see Fig. 4]). However, digestion was also detected within a region suggested to form a short stem structure, involving nucleotides 85 to 87 and 100 to 102 (Fig. 2A; also see Fig. 4). This latter region, in contrast, was observed to exhibit very little susceptibility to chemical modification (Fig. 3 and 4), supporting the idea of an RNA double strand made up of these residues. Since besides AU (85 and 102) and UG (87 and 100), only a single GC base pair (86 and 101) is supposed to be involved in the formation of such a stem, its susceptibility to RNases may be explained by its rather weak constitution. Alternatively, a huge SL II loop may be feasible, constituted of nucleotides 79 to 104.
FIG. 4.
Secondary-structure model of the 3′-terminal region of the BVDV DI9c 3′ UTR. The depicted model of the RNA secondary structure summarizes all experimental results superimposed on one of several computer predictions, which exhibited the lowest ΔGmax value (−36 kcal/mol). (A) Model of the RNA secondary structure with the numbering scheme that was initially introduced for the 3′ UTR primary structure (Fig. 1A). The asterisks indicate base pairing as suggested by the computer prediction (e.g., GC [positions 118 and 119] was predicted to anneal with GC [positions 125 and 126]), which during the different probing procedures turned out to be absolutely unlikely. The three different RNA secondary-structure motifs, SL I, SS, and SL II, suggested by the data comprise nucleotides 63 to 188 of the BVDV DI9c 3′ UTR. The model also includes short sequences at the 5′ and 3′ ends of this region (positions 56 to 62 and 189 to 192, respectively) which were found to be single-stranded. The 100% conserved nucleotides are indicated by underlining. (B) Summary of the experimental data shown in Fig. 2 and 3. Nucleotides within the native RNA that were determined to susceptible to RNases are labeled with sans-serif letters (U, RNase U2; T, RNase T1; M, RNase PhyM; B, B. cereus RNase). The cleavage efficiency is reflected by the size of the letter. Nucleotides within the native RNA that were determined to be susceptible to chemicals are denoted by italics (d, DMS, c, CMCT). Complementary bases that were found to be accessible to RNases as well as chemicals were concluded to exist mostly in single-stranded conformation. Accordingly, slightly different interpretations of the structure are conceivable.
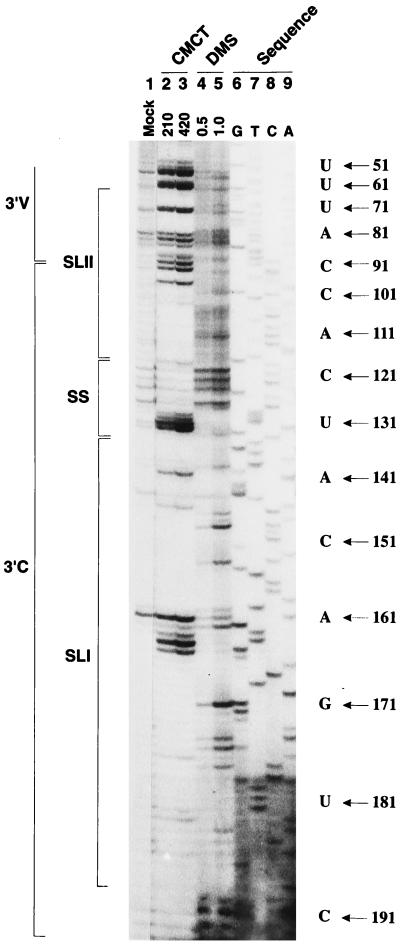
FIG. 3.
Chemical probing of the BVDV DI9c 3′ UTR RNA secondary structure. The single-strand-specific chemicals DMS (0.5 or 1 μl) and CMCT (210 or 420 μg) were used to modify 1 μg of transcribed ΔPvu+ RNA (see Materials and Methods). The products of the subsequent primer extension reaction, carried out with 5′-end-labeled T7 primer, were electrophoretically analyzed on a 7.5% denaturing polyacrylamide gel. Lane 1, control incubation of RNA performed in CMCT buffer (Mock; the visible primer extension products are most probably due to a minor contamination with RNase); lanes 2 and 3, CMCT modification (G and U specific); lanes 4 and 5, DMS modification (A and C specific); lanes 6 to 9, DNA sequence ladder obtained by dideoxy sequencing of the original ΔPvu plasmid with 5′-end-labeled T7 primer as well. The positions within the BVDV DI9c 3′ UTR are given on the right side. For comparison with the pattern obtained by primer extension, note that cDNA products synthesized from digested and modified RNA are usually displaced by 1 nucleotide relative to the DNA sequencing lanes, since the last nucleotide incorporated by reverse transcriptase is complementary to the one on the 3′ side of the modified or digested position in the RNA template. As in Fig. 2, borders of the variable (3′V) and conserved (3′C) regions as well as the SL I, SL II, and SS motifs of the RNA secondary-structure model (Fig. 4) are indicated on the left.
Determination of the BVDV DI9c 3′ UTR secondary structure by chemical modification.
We next wanted to further confirm and extend the data on the RNA secondary structure of the DI9c 3′ UTR by treating natively folded ΔPvu+ or DI9c+ RNA with the chemicals DMS and CMCT, respectively. DMS specifically modifies unpaired bases in the order of preference (most to least) G-N7, A-N1, and C-N3; CMCT modifies U-N3 and G-N2, in that order (8). The sites of chemical modification were subsequently determined by primer extension as described above; identification of the particular nucleotides was possible via side-by-side electrophoresis of a DNA sequencing reaction, performed on the ΔPvu+ plasmid with the same primer (Fig. 3). Since methylation at the N7 position of G does not induce termination of the reverse transcriptase, DMS was consequently considered as an AC-specific agent and CMCT was considered as a UG-specific agent.
As shown in Fig. 3, almost the entire native 3′ UTR, starting at approximately A (56) and ending at C (192) (the RNA 3′ terminus), could be investigated for the presence of single-stranded nucleotides. With the exception of the above-mentioned stem region, 85 to 87 and 100 to 102, within the SL II motif, structure probing by chemical treatment gave rise to a primer extension pattern that turned out to be in complete agreement with the RNase digestion experiments described above (Fig. 4); identical results were obtained by chemical probing of DI9c+ RNA (not shown). However, with regard to the RNase probing data, a number of additional unpaired nucleotides were detected by chemical modification (Fig. 3): (i) residues 118 to 132 were clearly defined as susceptible to chemical modification within the native RNA, thus confirming that the predicted SS region consisted of an uninterrupted stretch of 15 unpaired nucleotides; (ii) CCCC (189 to 192) was reproducibly modified by DMS, demonstrating the identity of the 3′ end of the genome as an RNA single strand; and (iii) nucleotides that compose three predicted bulges within the SL I motif, as well as UU (163 and 164), the “missing” part of the SL I hexaloop (see above), were shown to be accessible (Fig. 3 and 4). Chemical modification was also observed for certain cases of U or A residues located immediately adjacent to unpaired regions, which, according to the summarizing model shown in Fig. 4, should be base paired (U [117], UA [137 and 183], and UA [145 and 177]). This result could be interpreted in two ways: chemical modification may be the consequence of a real “looping out” of these bases, or it may just reflect “breathing” of the stem structure due to the low melting temperature of UA and GU base pairs. Since none of these residues has been observed to be available for RNase digestion, we favor the latter possibility (Fig. 4).
With the combination of data obtained by RNase and chemical probing, we were able to suggest a reasonable RNA secondary-structure model for that part of the BVDV DI9c 3′ UTR comprising the 3′ end of the 3′V region as well as the entire 3′C region (nucleotides 56 to 192 [Fig. 4]). Its RNA sequence apparently folds into a less stable stem-loop structure (SL II) at the 5′ end and a rather stable stem-loop structure (SL I) at the 3′ end (see Discussion). The two motifs are separated by a surprisingly long single-stranded intervening sequence (SS).
Functional significance of sequence and structural elements contained within the 3′ terminus of the DI9c RNA for the replication process.
The above-mentioned data demonstrated the presence of a pronounced and stable structural motif, namely, the SL I stem-loop within the 3′-terminal portion of the BVDV DI9c 3′ UTR, as well as a number of particularly exposed unpaired nucleotides, which constitute the SS region and the hexaloop of SL I (Fig. 4B). These features, which had been previously shown and/or suggested to be particularly conserved among pestiviruses (6, 22) (Fig. 1A and 4A), were suspected to represent important cis-acting signals of the RNA replication process.
As an initial approach to address this hypothesis, 15 different mutations were introduced into the DI9c RNA 3′ UTR via in vitro transcription of recombinant cDNA constructs, which affected either the SL I or the SS motif (see Materials and Methods; an overview of all mutations is given in Fig. 5). Eleven nucleotides (ACAGCACUUUA; positions 122 to 132) of the SS region were exchanged by performing nucleotide transition, i.e., purine residues were replaced by purines and pyrimidines by pyrimidines (mutations 1 to 7). Analogous interventions led to the exchange of four residues of the SL I hexaloop: UA (160 and 161) was replaced by CG, and GU (162 and 163) was replaced by AC (mutations 14 and 15). In addition, six mutations were introduced into SL I; mutations 8, 10, and 12 were aimed at completely inhibiting formation of three of the four stem structures, which ought to fold the backbone of the SL I structure. The other three mutations were performed in such a way that the overall structure of SL I remained intact; stems, however, were replaced by different double-stranded RNA regions of identical length. These were composed of complementary stretches of nucleotides, which exhibited reverse orientation with regard to the original DI9c sequence (mutations 9, 11, and 13). In other words, mutations 9, 11, and 13 represented compensatory base exchanges of mutations 8, 10, and 12, respectively (Fig. 5).
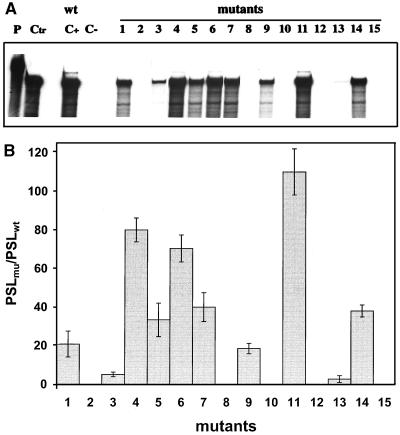
The replication abilities of the DI9c RNA derivatives were judged 24 h posttransfection into BHK cells either by monitoring synthesis of the viral nonstructural protein NS3 by immunofluorescence analysis (reference 7 and data not shown) or by measuring replication directly on the level of the newly synthesized RNA products by using RNase protection (7). In the course of the reported experiments, both assay systems were verified to detect neither input (transfected) cRNA nor protein expression of input RNA alone. Accordingly, viral genomes lacking the RNA polymerase-encoding region or containing other lethal mutations were shown to be definitely negative in the assays (Fig. 6). The possibility of cross-contamination by wild-type DI9c or emerging revertants was ruled out by RT-PCR (see Materials and Methods) carried out on total cytoplasmic RNA 24 h posttransfection and by subsequent sequencing of the PCR fragments (data not shown). Surprisingly, as shown in Fig. 6, DI9c RNA molecules carrying mutations 1 to 7 turned out to exhibit rather different abilities to support replication: substitution of UGU for GCA (125 to 127) or a U residue for C (128) gave rise to replicon derivatives that were either absolutely or nearly unable to replicate. In contrast, other transitions were found to have a much weaker effect (Fig. 6); in particular, substitutions within the UUUA (129 to 132) region yielded DI9c molecules that were moderately replication competent (approximately 40 to 80% of the wild-type level). Similar observations were made during replication experiments directed at examining the consequences of mutations within the loop of SL I. While replacement of GU (162 and 163) caused a complete inhibitory effect, exchange of UA (160 and 161) resulted in only a decrease of replication ability to 50% of that of the wild-type replicon (Fig. 6). Abolishment of the diverse stem regions of SL I totally destroyed the replication capability of the respective RNAs, whereas preservation of these stems—though with a different nucleotide sequence—allowed replication to occur at either a low level (mutants 9 and 13) or a high level (mutant 11).
FIG. 6.
Monitoring the RNA replication of mutant BVDV DI9c derivatives. (A) RNase protection assay performed as described in Materials and Methods at 24 h posttransfection into BHK-21 cells. The autoradiogram shows the protected RNA products of one representative experiment with a 32P-labeled antisense probe; replication ability was monitored for novel synthesized positive-strand RNA. Monitoring of a negative-strand RNA intermediate yielded identical tendencies (it should be noted that the assay performed does not allow any conclusion as to the ratio of negative-strand versus positive-strand RNA accumulation) (data not shown). Lane P, input antisense probe; lane Ctr, 400 ng of in vitro-transcribed DI9c RNA hybridized with antisense probe; lane C+, RNase protection of RNA derived from the cytoplasmic fraction of BHK cells that were previously transfected with DI9c wild-type (wt) replicon RNA; lane C−, identical experiment carried out on BHK cells previously transfected with DI9c RNA without the 3′ end (DI9cΔ3′; see reference 2) as a negative control; lanes 1 to 15, identical experiments carried out on BHK cells previously transfected with the different mutant DI9c derivatives (Fig. 5). (B) Calculation of the replication abilities of the different 3′ UTR DI9c derivatives. Five independent RNase protection experiments were performed with each of the mutant DI9c derivatives and the major protected bands (indicated in panel A, lane 9) quantified with a phosphorimager (see Materials and Methods). The relative ability of replication was thus calculated for each of the mutants (mu) with respect to the wild-type (wt) DI9c RNA (estimated as 100% replication competent) and depicted as a column diagram. PSL, photostimulated luminescence. The error bars indicate standard deviations.
These results clearly revealed the importance of an overall intact secondary structure of the SL I motif. They further underlined the relevance of certain sequence elements within the SS region and the SL I hexaloop region, as well as within the SL I stem region, for the RNA replication process (see Discussion).
DISCUSSION
We recently described the surprising observation that a subgenomic BVDV RNA, DI9c, supports the entire RNA replication pathway in vivo (see above) (Fig. 1B). This finding opened up the opportunity to examine the functional role of the proteins as well as the role of cis-acting elements encoded by this viral RNA by using a straightforward reverse-genetic approach, i.e., site-directed mutagenesis of the cDNA construct, in vitro transcription of the cRNA derivative, and, after its transfection into host cells, monitoring of RNA replication in vivo (7). The work presented here was aimed at determining characteristic structural features of the RNA 3′ terminus (3′ UTR) and at gaining some evidence as to its role during the RNA replication process.
Since it was suggested that all pestivirus 3′ UTRs exhibit rather extensive RNA folding (6, 22), it was important to establish the RNA secondary structure of the DI9c 3′ UTR by experimental means prior to commencing a functional study. As shown by the enzymatic- and chemical-probing data, no distinct motifs could be recognized for the 5′ terminus of the 3′ UTR (Fig. 2 and 3). This part of the sequence, which in all pestivirus genomes is rich in repeating UA stretches, has been previously denoted variable region 3′V of the 3′ UTR (22) (Fig. 1A) and displays a remarkable heterogeneity in size, ranging between 83 (BVDV Osloss) and 170 (BDV X818) nucleotides. We concluded that the structural detail of this region was less pronounced and/or displayed multiple alternatives which cannot be recorded with the applied experimental procedures. In remarkable contrast, the 3′-terminal part of the 3′ UTR was demonstrated to comprise a number of characteristic RNA motifs. Moreover, these were found to be subject to stringent constraints, since variant transcript RNAs (Fig. 1B) yielded congruent structure-probing results and no dramatic differences were observed under various experimental conditions (data not shown). The most obvious features were located within a stretch of approximately 70 nucleotides immediately upstream of the 3′ terminus of the viral RNA; the latter was confirmed to consist of four single-stranded C residues (Fig. 2, 3, and 4). Largely in accordance with the predictions made by the FOLD program (Fig. 4A), this 70-nucleotide element is composed of SL I, a huge, stable stem-loop structure folded by 56 nucleotides (Fig. 4A, positions 133 to 188; calculated ΔGmax, ∼−21.7 kcal/mol at 37°C; melting temperature in 1 M NaCl, 75.5°C) and of a stretch of single-stranded nucleotides further upstream, the SS region, of which nine residues (AGCACUUUA [124 to 132]) are identical within all pestivirus genomes known so far (6) (Fig. 1A and 4A). However, contrary to all predictions, it was evident that SL I does not contain a tetraloop but that it contains a hexaloop made up of the nucleotide sequence AUAGUU (159 to 164), varying among the different pestivirus members only at the second or third position (AUAGUU [BVDV NADL and BVDV SD-1], ACAGUU [CSFV Alfort], AGAGUU [BDV X818], and AUGGUU [BVDV Osloss]) (variations are boldface italic). Another invariant region is located in direct proximity to the SL I loop: following the model (Fig. 4), nucleotides GACGUC (152 to 157) and GACUA (166 to 170) form a bulge and stem region that, among all pestiviruses, differs only in strain BDV X818 (GAAGUC/GACUA [6]). The structure-probing data further demonstrated a second stem-loop structure (SL II), formed by the RNA sequence upstream of the SS region, which may, however, exhibit alternative loop structures, as indicated by an incompatibility of the enzymatic digestion and chemical modification profiles (Fig. 4) (see Results). SL II exhibits a thermodynamic stability significantly lower (ΔGmax, ∼−9.5 kcal/mol at 37°C; melting temperature in 1 M NaCl, 58.6°C) than that of SL I.
Intramolecular cross talk of RNA sequences yielding pseudoknot structures, or even long-distance base pairing between 5′ and 3′ UTR sequences, has been described as being an important functional feature in a number of single-stranded RNA viruses for the modulation of translation, replication, and encapsidation processes (4, 5, 33, 49, 50, 62, 63, 72). Such interactions were presumed for flaviviruses and pestiviruses as well (29, 46). Our data on the 137 3′-terminally located nucleotides of the BVDV genome suggest that most of these residues are not involved in stable base-pairing events other than those forming the stem structures of SL I and SL II (Fig. 4). Certain compounds of the conserved SL I bulge and stem region (see above) or the loop of SL II, however, were neither hydrolyzed by ribonucleases nor chemically modified (Fig. 4) and thus may represent candidates for involvement in the formation of sealed tertiary structure, noncanonical base pairing (28), or, indeed, intramolecular interactions. In this context it should be noted that tertiary “kissing” interactions, such as those found within the 3′ UTRs of poliovirus (52) and coxsackie B virus (42), are difficult to monitor under the experimental conditions of our analysis. It is also noteworthy that RNA-RNA interactions may also be mediated and/or stabilized in trans by binding of host cellular and/or viral proteins, as suggested for a potential interaction of the genomic 5′ and 3′ termini of HCV (32, 68). As soon as factors are identified that specifically associate with the pestivirus UTRs (see below), a detailed investigation of this interesting topic will be an important task.
Distinctive conserved RNA motifs comprising up to 100 nucleotides, which are located near or even encompass the 3′ terminus of the RNA genome, have now been predicted and/or experimentally proven for all members of the family Flaviviridae (6, 11, 13, 22, 32, 37, 55, 65, 72). In the second part of this work, our main interest was to investigate their relevance as cis-acting elements during the amplification process of the BVDV replicon. During mutagenesis of the DI9c 3′ UTR, we focused on those RNA elements which during structure probing turned out to be the most striking features, i.e., the overall structure of SL I and particularly exposed nucleotide residues within the SL I hexaloop and the SS region (Fig. 5). First of all, the capability of the RNA molecule for autonomous replication was found to be essentially dependent on the presence of a complete set of intact backbone stems of SL I (mutations 8, 10, and 12). Since RNA replication could be restored by compensatory base changes (mutations 9, 11, and 13), although in one case (mutant 11) only back to wild-type level (Fig. 6), we concluded that the overall RNA structure of SL I as well as sequence determinants within the stems accounted for the importance of this motif as a replication signal. By far the lowest compensatory effect obtained with the mutant 13 derivative may be similarly explained, since almost the entire stem structure consists of conserved nucleotide residues (Fig. 1A and 4A) and the adjoining nucleotides, A (159) and U (164), previously shown to be part of the SL I hexaloop, were also changed.
The interpretation of the data obtained by mutagenesis of residues located within the conserved single-stranded stretches of the SS region and the SL I hexaloop was found to be more complex. In summary, all mutations yielded a clear negative effect on the replication efficiency of the replicon (Fig. 6), thus further validating the importance of these sequence elements. In keeping with the phylogenetic data mentioned above (Fig. 4A), RNA molecules where the variable UA (160 and 161) residues within the SL I hexaloop had been replaced by CG were found to be replication competent at a low level (Fig. 6) whereas replacement of the 100%-conserved GU (162 and 163) nucleotides led, as expected, to RNAs which were unable to amplify (Fig. 1A, 5, and 6). Along the same lines, residues GCAC (125 to 128), constituents of the invariant portion of SS, were found to be essential for the replication process. Up to this point, data could be explained by different stringencies of selective constraints displayed by the respective sequence elements. More surprising, however, were the results showing that RNAs containing nucleotide substitutions at positions 122 to 124 (ACA; mutant 1) and 129, 130, 131, and 132 (UUUA; mutants 4 to 7) are apparently replication competent, irrespective of the fact that these residues also comprise components of the conserved stretch within the SS motif. Moreover, substitutions of neighboring nucleotides revealed remarkable differences (compare, for instance, Fig. 5 and 6, mutants 3 and 4, C [128] and U [129]). Of course, our genetic approach has been limited by the fact that all variations of nucleotides were not tested at each position; certain differences concerning the effect of mutagenesis may thus be caused by the nature of the particular substituting nucleotide. However, the reasons for the observation that replication could be detected even after replacement of residues that are conserved within all pestivirus genomes remain to be established. Most likely, these residues perform multiple functions, in replication as well as during other stages of the viral life cycle, such as packaging of the RNA genome. Experiments that may substantiate this hypothesis, e.g., testing these mutations in the context of a full-length BVDV genome, are in progress.
The remarkable stability and conservation of structure as well as sequence characteristics of the SL I motif suggest that this element is involved in RNA-protein interactions similar to quite a number of examples known from other virus systems. Whereas, in several cases, the secondary or tertiary structures of the RNA motifs were shown to represent the only essential constraint that defines the specificity of interaction with a protein (9, 15, 23, 26, 33, 39, 40, 48, 60), in other examples protein binding was proposed to yield a conformational change within the RNA, thus generating a novel recognition signal for the functional interaction of other factors (3, 58). Recognition of RNA by proteins was described as also occurring independently of structure in a sequence-specific manner (36, 41). In accordance with our functional data, it seems likely that nucleotides GU (162 and 163) of the SL I hexaloop and GCAC (125 to 128) of the SS intervening sequence may represent well-defined interaction sites for viral and/or cellular proteins during RNA replication. For West Nile encephalitis virus and HCV, cellular proteins that interact with 3′-terminally encoded RNA elements have been described (10, 32, 68). Interaction of the viral proteins NS3 (viral protease and RNA helicase) and NS5B (RNA-dependent RNA polymerase) has been described in Japanese encephalitis virus and Dengue virus (16, 18), respectively. This work represents an essential starting point in the search for viral and host proteins capable of selective binding to the defined RNA motifs of the pestivirus 3′ UTR and to further determine the functional aspects of these interactions in the RNA replication process.
ACKNOWLEDGMENTS
This study was supported by the SFB 535 Invasionsmechanismen und Replikationsstrategien von Krankheitserregern (C.W.G.) and the Graduiertenkolleg Biochemie von Nukleoproteinkomplexen (H.Y.) from the Deutsche Forschungsgemeinschaft at the Justus-Liebig-Universität Giessen. S.-E.B. is supported by the Infektionsforschung-Stipendienprogramm (2131) of the BMBF (Bundesministerium Bildung und Forschung) administrated by the Deutsches Krebsforschungszentrum (DKFZ).
We thank Norbert Tautz and Mary Kromeier for critically reading the manuscript and H.-J. Thiel for support.
REFERENCES
- 1.Andino R, Rieckhoff G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′-end of poliovirus RNA. Cell. 1990;63:269–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 3.Baer M L, Houser F, Loesch-Fries L S, Gehrke L. Specific RNA binding by aminoterminal peptides of alfalfa mosaic virus coat protein. EMBO J. 1994;13:727–735. doi: 10.1002/j.1460-2075.1994.tb06312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball L A. Requirements for the self-directed replication of flock-house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball L A, Li Y. cis-acting requirements for the replication of flock-house virus RNA 2. J Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becher P, Orlich M, Thiel H-J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black D L, Pinto A L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blight K J, Rice C M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumenthal T, Carmichael G G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 13.Brinton M A, Fernandez A V, Dispoto J H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 14.Brown E A, Zhang H, Ping L, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang M F, Sun C Y, Chen C J, Chang S C. Functional motifs of delta antigen essential for RNA binding and replication of hepatitis delta virus. J Virol. 1993;67:2529–2536. doi: 10.1128/jvi.67.5.2529-2536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christiansen J, Garrett R. Enzymatic and chemical probing of ribosomal RNA-protein interactions. Methods Enzymol. 1988;164:456–468. doi: 10.1016/s0076-6879(88)64061-4. [DOI] [PubMed] [Google Scholar]
- 18.Cui T, Sugrue R J, Xu Q, Lee A K W, Chan Y-C, Fu J. Recombinant Dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 19.David C, Gargouri-Bouzid R, Haenni A-L. RNA replication of plant viruses containing an RNA genome. Prog Nucleic Acid Res Mol Biol. 1992;42:157–227. doi: 10.1016/s0079-6603(08)60576-0. [DOI] [PubMed] [Google Scholar]
- 20.Deiman B A, Kortlever R M, Pleij C W. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–879. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 22.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dildine S L, Semler B L. Conservation of RNA-protein interactions among picornaviruses. J Virol. 1992;66:4364–4376. doi: 10.1128/jvi.66.7.4364-4376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.England T E, Uhlenbeck O C. 3′-terminal labelling of RNA with T4 RNA ligase. Nature. 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 25.Francki R I, Fauquet D L, Knudson D L, Brown F. Classification and nomenclature of viruses. Arch Virol Suppl. 1991;2:223–233. [Google Scholar]
- 26.Furuya T, Lai M M C. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Y, Trowbridge R, Macnaughton T B, Westaway E G, Shannon A D, Gowans E J. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhea virus. J Gen Virol. 1996;77:2729–2736. doi: 10.1099/0022-1317-77-11-2729. [DOI] [PubMed] [Google Scholar]
- 28.Gutell R R, Woese C R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci USA. 1990;87:663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn C S, Hahn Y S, Rice C M, Lee E, Dalgarno L, Strauss E G, Strauss J H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 30.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry site within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 31.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 1035–1058. [Google Scholar]
- 32.Ito T, Lai M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang S K, Pestova T V, Hellen C U T, Witherell G W, Wimmer E. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- 36.Karn J, Gait M J, Churcher M J, Mann D A, Mikaelian I, Pritchard C. Control of human deficiency virus gene expression by the RNA-binding proteins tat and rev. In: Nagai K, Mattaj I W, editors. RNA-protein interactions. Oxford, England: IRL Press; 1994. pp. 192–220. [Google Scholar]
- 37.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le S Y, Siddiqui A, Maizel J V. A common structural core in the internal ribosome entry sites of picornavirus, hepatitis C virus and pestivirus. Virus Genes. 1996;12:135–147. doi: 10.1007/BF00572952. [DOI] [PubMed] [Google Scholar]
- 39.Leopardi R, Hukkanen V, Vanionpää R, Salmi A A. Cell proteins bind to the sites within the 3′ noncoding region and the positive-strand leader sequence of measles virus RNA. J Virol. 1993;67:785–790. doi: 10.1128/jvi.67.2.785-790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J H, Chang M F, Baker S C, Govindarajan S, Lai M M C. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J Virol. 1990;64:4051–4058. doi: 10.1128/jvi.64.9.4051-4058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattaj I W. RNA recognition: a family matter? Cell. 1993;73:837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- 42.Melchers W J G, Hoenderop J G J, Bruins Slot H J, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyers G, Thiel H-J, Rümenapf T. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol. 1996;70:1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moormann R J, Warmerdam P A, van-der-Meer B, Schaaper W M, Wensvoort G, Hulst M M. Molecular cloning and nucleotide sequence of hog cholera virus strain Brescia and mapping of the genomic region encoding envelope protein E1. Virology. 1990;177:184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- 47.Moormann R J M, van Gennip H G P, Miedema G K L, Hulst M M, van Rijn P A. Infectious RNA transcribed from an engineered full-length cDNA template of the genome of a pestivirus. J Virol. 1996;70:763–770. doi: 10.1128/jvi.70.2.763-770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakhasi H L, Cao X-Q, Rouault T A, Liu T Y. Specific binding of host cell proteins to the 3′-terminal stem-loop structure of rubella virus negative-strand RNA. J Virol. 1991;65:5961–5967. doi: 10.1128/jvi.65.11.5961-5967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Neill R E, Palese P. cis-acting signals and trans-acting factors involved in influenza virus RNA synthesis. Infect Agents Dis. 1994;3:77–84. [PubMed] [Google Scholar]
- 50.Pattnaik A K, Ball A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed from a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 52.Pilipenko E V, Poperechny K V, Maslova S V, Melchers W J G, Bruins Slot H J, Agol V I. cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 53.Pleij C W A. RNA pseudoknots. Curr Opin Struct Biol. 1994;4:337–344. [Google Scholar]
- 54.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 55.Rauscher S, Flamm C, Mandl C W, Heinz F X, Stadler P F. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA. 1997;3:779–791. [PMC free article] [PubMed] [Google Scholar]
- 56.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 931–960. [Google Scholar]
- 57.Ruggli N, Tratschin J D, Mittelholzer C, Hofmann M A. Nucleotide sequence of classical swine fever strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi P-Y, Li W, Brinton M A. Cell proteins bind specifically to West Nile minus-strand 3′ stem-loop RNA. J Virol. 1996;70:6278–6287. doi: 10.1128/jvi.70.9.6278-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi P-Y, Brinton M A, Veal J M, Zhong Y Y, Wilson W D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 60.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stern S, Moazed D, Noller H F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 62.Strauss J H, Strauss E G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- 63.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka T, Kato N, Cho M-J, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H-J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 1059–1074. [Google Scholar]
- 68.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wengler G, Castle E. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J Gen Virol. 1986;67:1183–1188. doi: 10.1099/0022-1317-67-6-1183. [DOI] [PubMed] [Google Scholar]
- 72.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 73.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]