Abstract
Wastewater monitoring can provide insights into respiratory disease occurrence in communities that contribute to the wastewater system. Using daily measurements of RNA of influenza A (IAV), respiratory syncytial virus (RSV), and human metapneumovirus (HMPV), as well as SARS-CoV-2 in wastewater solids from eight publicly owned treatment works in the Greater San Francisco Bay Area of California between July 2022 and early July 2023, we identify a “tripledemic” when concentrations of IAV, RSV, and SARS-CoV-2 peaked at approximately the same time. HMPV was also widely circulating. We designed novel hydrolysis probe RT-PCR assays for different IAV subtype markers to discern that the dominant circulating IAV subtype was H3N2. We show that wastewater data can be used to identify the onset and offset of wastewater disease occurrence events. This information can provide insight into disease epidemiology and timely, localized information to inform hospital staffing and clinical decision making to respond to circulating viruses. Whereas RSV and IAV wastewater events were mostly regionally coherent, HMPV events displayed localized occurrence patterns.
Keywords: wastewater solids, respiratory viruses, tripledemic, wastewater-based epidemiology, influenza A, RSV, metapneumovirus, SARS-CoV-2
Introduction
Acute respiratory illness (ARI) accounts for a large burden of infectious disease1,2 and is a leading cause of death for children under 5 globally.3 ARI surveillance in much of the world is passive, relying on institutions to identify specific diseases through clinical specimens and regularly reporting results to government agencies. As a result, ARI surveillance is biased toward identifying infections in individuals with comorbidities and/or where symptoms are severe.4 Resultant lack of knowledge of occurrence and trends in circulating respiratory disease dynamics limits institutional awareness to guide public health response and clinical decision making. Additionally, the lack of robust, unbiased data on disease occurrence limits efforts to understand disease epidemiology.
During the COVID-19 pandemic, the circulation of respiratory viruses changed dramatically according to both clinical5,6 and wastewater7 For example, in the US, there was limited influenza or respiratory syncytial virus (RSV) circulation in the northern hemisphere winter of 2021–2022.5,8 During winter 2022–2023, the US experienced a “tripledemic” of respiratory viruses as clinics and hospitals recorded large numbers SARS-CoV-2, influenza, and RSV infections, at times overwhelming hospital capacity particularly in pediatric units.9 Nearly 40% of US households reported being infected by SARS-CoV-2, influenza, or RSV during the tripledemic period.10
In the present study, we explore the potential for wastewater monitoring, also referred to as wastewater-based epidemiology, to inform the onset and progression of important respiratory viruses during this tripledemic period in the US. We measured concentrations of genomic nucleic acids of influenza A (IAV), RSV, and SARS-CoV-2, as well as human metapneumovirus (HMPV), in wastewater solids daily in eight publicly owned treatment works (POTWs) in the Greater San Francisco Bay Area, California, USA during winter 2022–2023. We find that the co-occurrence of the onset of these outbreaks is identifiable using wastewater monitoring, and moreover, HMPV was also circulating. Whereas clinical testing data are affected by individuals’ test seeking behaviors and test availability,11−13 as well as institutional reporting delays which can be weeks in duration,14,15 wastewater data are available within 24 h of sample collection and provide information about the entire community contributing to the wastewater system. We posit that wastewater can be used to help identify the onset and offset of respiratory virus transmission events as well as the peak of such events. This in turn can inform clinical decision making as well as institutional public health messaging and individual behaviors.
Methods
Sample Collection
Eight POTWs located in the greater San Francisco Bay Area (extending to Sacramento) in California, USA contributed daily samples of wastewater settled solids for the prospective study (Figure S1, additional POTW details are in Wolfe et al.16). Approximately 50 mL of settled solids were collected daily between 7/1/22 (month/day/year format) and 7/5/23. Additional details of sample collection are in the Supporting Information (SI). Samples were immediately stored at 4 °C and transported to the laboratory, where processing began within 6 h of collection. 2938 samples were collected.
Sample Preparation and RNA Extraction
Details of sample preparation and nucleic-acid extraction have been described elsewhere7,16−18 and are provided in the SI. Samples were spiked with bovine coronavirus (BCoV) as a recovery control. RNA was extracted from 10 replicate aliquots per sample and then subjected to an inhibitor removal step. Extraction-negative controls (water) and extraction-positive controls were extracted using the same protocol as for the samples. The positive controls consisted of SARS-CoV-2 genomic RNA (ATCC VR-1986D), Twist Synthetic Influenza A H3N2 RNA Control (Twist 103002), Intact RSV B virus (Zepto NATFVP-NNS), and a gene block (double-stranded DNA [dsDNA] purchased from IDT) for the HMPV target suspended in a BCoV-s-spiked DNA/RNA shield solution (see SI).
Droplet Digital PCR
RNA extracts were used as a template in digital droplet RT-PCR assays for PMMoV, BCoV, SARS-CoV-2 N, IAV M, RSV N, and HMPV L gene targets in previously published and validated multiplex assays.7,19 Each of the 10 replicate nucleic-acid extracts were run in their own well so that there were 10 replicates per sample. Assays were multiplexed using the probe mixing approach (SI and Table S1). PMMoV is highly abundant in wastewater20 and is used here as an internal recovery and fecal strength control.21 Undiluted extract was used for the human viral assay template, and a 1:100 dilution of the extract was used for the BCoV/PMMoV assay template. Details of the digital RT-PCR have been published and are included in the SI.
Concentrations of RNA targets were converted to concentrations per gram of dry weight of solids (cp/g) using dimensional analysis. The total errors are reported as standard deviations and include errors associated with the Poisson distribution and variability among replicates. BCoV recovery was determined by normalizing the BCoV concentration to the expected concentration given the value measured in the spiked DNA/RNA shield. BCoV recovery was used as a process control and not used in the calculation of concentrations; samples were rerun in cases in which the recovery of BCoV was less than 1%.
Influenza Subtypes
We designed and tested novel RT-PCR primers and internal hydrolysis probes targeting the H1 subtype of the hemagglutinin (HA) gene, H3 subtype of the HA gene, N1 subtype of the neuraminidase (NA) gene, and N2 subtype of the NA gene of IAV. Primers and probes were then screened for specificity in silico and in vitro against other respiratory and enteric viruses (see SI, Table S2).
We retrospectively tested two samples per week between 7/1/22 and 3/31/23 from SJ and Ocean POTWs for the four influenza A subtype markers (N1, N2, H1, H3). We also reanalyzed the samples to quantify the IAV M gene in order to report the ratio of the subtype markers to M gene (see SI).
Clinical Data
We used state-aggregated weekly clinical sample positivity rates from sentinel laboratories for IAV, RSV, and HMPV; data were available through the last week of May 2023. Influenza, RSV, and a fraction of HMPV data are publicly available,22 and the remaining HMPV data were provided by California Department of Public Health (CDPH). State-aggregated daily positivity rates for COVID-19 are publicly available (https://data.chhs.ca.gov/dataset/covid-19-time-series-metrics-by-county-and-state).
Data Analysis
Human viral RNA concentrations were normalized by PMMoV RNA concentrations; 5-day (d) trimmed averages were used to visualize data and identify the date of peak maximum concentrations. The onset dates of IAV, RSV, and HMPV wastewater events were identified as the first day for which all samples in a 14 day look back period had concentrations higher than 2000 copies/g, which is approximately twice the lowest detectable concentration. The offset dates of IAV, RSV, and HMPV wastewater events were identified as the first day after an onset event for which only 7 samples during a 14-d look back period had concentrations over 2000 copies/g.
We tested whether PMMoV-normalized viral RNA concentrations at each POTW were correlated to the same measurements at the other eight POTWs (28 × 4 = 112 hypothesis tests), and whether PMMoV-normalized viral RNA concentrations were correlated within POTWs (48 tests) using Kendall’s tau as these data tend to not be normally distributed. We also tested hypotheses that PMMoV-normalized viral RNA correlated with state-aggregated positivity rates (32 tests). Since influenza RSV, and HMPV positivity rates are aggregated weekly, we used weekly median PMMoV-normalized viral RNA concentrations to test the hypothesis. As clinical data were available only through 5/20/23, we only test the associations through that date. We used a p value of 0.00026 (0.05/192) which corresponds to alpha = 0.05 with a Bonferroni correction. Results were similar when analyses were carried out with variables that were not PMMoV-normalized.
This study was reviewed by the State of California Health and Human Services Agency Committee for the Protection of Human Subjects and determined to be Exempt from oversight. Wastewater data are publicly available (https://purl.stanford.edu/gp563rt4747). Data collected between 7/1/22 and 12/31/22, and expanded methods, are publicly available through a Data Descriptor.23
Results
QA/QC
Results are reported as suggested in the Environmental Microbiology Minimal Information guidelines24 (Figure S2). Extraction and PCR negative and positive controls performed as expected (negative and positive, respectively). Median BCoV recovery in these samples was 1.3 (interquartile range = 0.96–1.81, n = 2938); values greater than 1 are likely a result of uncertainty in quantifying the amount of BCoV spiked in the DNA/RNA shield.
Novel IAV Subtype Assay Sensitivity and Specificity
In silico analysis indicated no cross reactivity of the novel RT-PCR probe-based assays (Table S3) with sequences deposited in NCBI. The novel IAV N1, N2, H1, and H3 assays were tested in vitro against nontarget viral gRNA as well as target gRNA. No cross reactivity was observed.
Viral RNA in Wastewater Solids
Viral concentrations were dynamic over the project duration, with most showing elevated concentrations in the winter (Figure 1). PMMoV-normalized HMPV, IAV, RSV, and SARS-CoV-2 RNA concentrations were each positively and significantly associated between the eight POTWs, respectively (IAV tau: 0.40 to 0.64, RSV tau: 0.56 to 0.69, HMPV tau: 0.37 to 0.55, SARS-CoV-2 tau: 0.25 to 0.40, all p < 10–12, Figure S3) suggesting coarse temporal coherence across the region.
Figure 1.
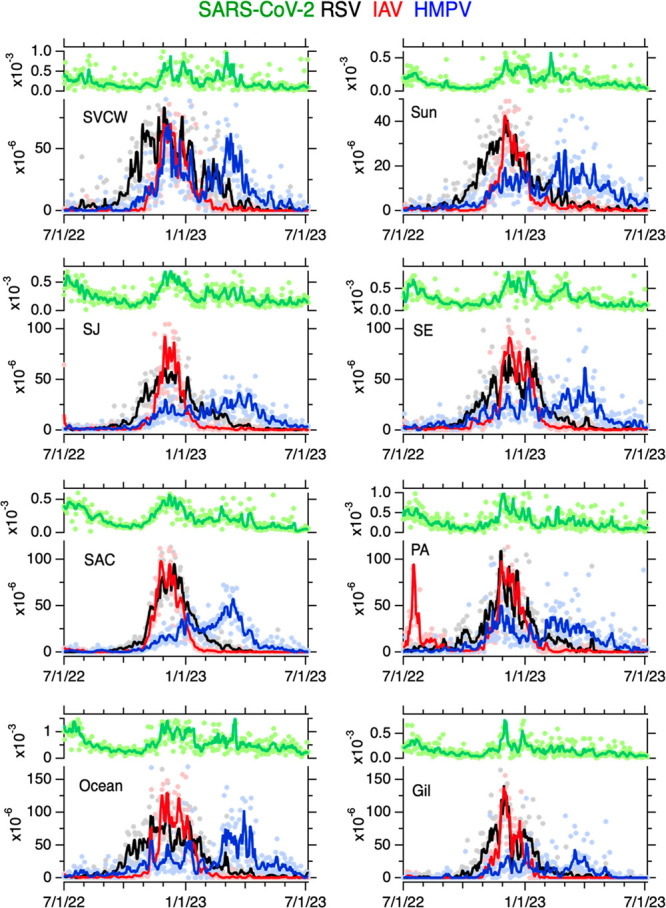
Time series of concentrations of SARS-CoV-2, RSV, IAV, and HMPV normalized by PMMoV in wastewater solids at eight POTWs between 7/1/22 and 7/5/23; the ratio is unitless. Raw data are shown as filled circles; lines are 5-d trimmed averages. Note the difference in scale between the SARS-CoV-2 (upper left axis of each panel) and the other viruses (lower left axis of each panel). The acronym for each POTW is provided in the top left corner.
Within each POTW, PMMoV-normalized IAV, RSV, HMPV, and SARS-CoV-2 RNA concentrations were generally positively associated with each other suggesting coherence within a POTW (tau: 0.14 to 0.58, all p < 0.00026, depending on POTW and specific measurements, Figure S4). An exception is at PA where HMPV was not associated with IAV. Taus were generally highest between IAV and RSV, and lowest between HMPV and both IAV and SARS-CoV-2.
State-aggregated clinical specimen positivity rates for influenza, RSV, HMPV, and SARS-CoV-2 show peaks in the winter months (Figure S5). Weekly median PMMoV-normalized concentrations of all viruses were significantly, positively correlated with the associated clinical positivity rate (see SI, all p < 10–6).
The peak wastewater concentrations varied across POTWs for different viral targets (Table 1). Peak RSV occurred between 11/25/22 and 12/14/22, peak IAV occurred between 11/26/22 and 12/13/22, and peak HMPV occurred between 11/26/22 and 3/30/23, depending on POTW. For context, peak SARS-CoV-2 occurred between 11/30/22 and 3/16/23.
Table 1. Date of Onset and Offset, As Well As Peak of Wastewater Events for Each Human Viral Target Measured in Wastewater Solids for Each of 8 POTWsa.
| HMPV |
IAV |
RSV |
SC2 | ||||
|---|---|---|---|---|---|---|---|
| POTW | onset/offset | peak | onset/offset | peak | onset/offset | peak | peak |
| Gil | 3 Dec - 18 Mar | 3 Jan | 13 Nov - 25 Jan | 30 Nov | 24 Oct - 20 Mar | 30 Nov | 1 Dec |
| SJ | 30 Oct - ? | 22 March | 14 July - 23 July | 30 Nov | 17 Sept - 18 April | 20 Dec | 1 Dec |
| 8 Nov - 3 Mar | |||||||
| PA | 1 Nov - 21 June | 26 Nov | 14 July - 21 Sept | 26 Nov | 11 Oct - 2 April | 25 Nov | 30 Nov |
| 11 Nov - 13 Feb | |||||||
| SVCW | 12 Nov - 21 June | 6 Dec | 4 Nov - 24 Feb | 9 Dec | 23 Sept - 25 April | 13 Dec | 4 Mar |
| SE | 1 Sept - 24 Sept | 1 April | 26 Oct - 25 Feb | 8 Dec | 6 Oct - 25 Mar | 29 Nov | 5 Jan |
| 13 Oct - 3 July | |||||||
| Ocean | 6 Aug - 3 July | 30 March | 18 Nov - 22 Feb | 13 Dec | 23 Sept - 17 April | 4 Dec | 16 Mar |
| SAC | 10 Nov - 4 June | 13 March | 4 Nov - 30 Jan | 24 Nov | 19 Oct - 27 Mar | 14 Dec | 7 Dec |
| Sun | 4 Nov - ? | 2 March | 10 Nov - 1 Mar | 2 Dec | 26 Sept - 29 April | 3 Dec | 9 Feb |
A “?” indicates that the offset had not yet occurred during the study period. Dates for two events are provided if two wastewater events were identified. SC2 is SARS-CoV-2, only the peak is provided for SC2.
We identified dates of wastewater event onsets and offsets for RSV, IAV, and HMPV (Table 1), but not for SARS-CoV-2 as its levels were such that the entire period of the study would be defined as a wastewater event. We observed wintertime RSV wastewater events at all 8 POTWs with onsets between 9/17/22 and 10/24/22, and offsets between 3/20/23 and 4/29/23, depending on POTW. The period of time between onset and offset varied between 147 and 215 days (median = 190 days).
We observed a summer IAV wastewater event at two POTWs (SJ and PA); both onset on 7/14/22 and offset on 7/23/22 (SJ) and 9/21/22 (PA). We observed wintertime wastewater events at all 8 POTWs with onsets between 10/26/22 and 11/18/22 and offsets between 1/25/23 and 3/3/23. The duration of the wintertime events ranged from 73 and 122 days (median of 104 days). IAV subtype analysis detected N2, H1, and H3 at both SJ and Ocean, and N1 at Ocean. When detected, the ratio of N1/IAV ranged from 0.05 to 0.8 (median = 0.2), N2/IAV ranged from 0.1 to over 1 (median = 0.6), H1/IAV ranged from from 0.03 to 0.8 (median = 0.1), and H3/IAV ranged from from 0.1 to 0.6 (median = 0.3) (Figure S6).
We observed a fall HMPV wastwater event at one POTW (SE) with onset on 9/1/22 and offset on 9/24/22. We observed wintertime HMPV wastewater events for all 8 POTWs with onsets between 8/6/22 and 12/3/22 and offsets between 5/18/23 and 6/21/23; note that, as of 7/5/23, HMPV had not offset at two POTWs. HMPV wastewater event durations were (assuming offset occurred on 7/5, for the two POTW not yet offset) between 166 and 333 days (median of 245 days).
Discussion
Influenza A, RSV, and SARS-CoV-2 RNA in wastewater solids in the Greater San Francisco Bay Area of California are reflective of the winter 2022–2023 “tripledemic” when cases of influenza, RSV, and COVID-19 increased dramatically with similar onset periods, sometimes overwhelming hospital capacity.9 Wastewater monitoring of human metapneumovirus (HMPV) RNA suggests that HMPV was also circulating in communities during the tripledemic but with slightly different outbreak timing and continued activity after IAV and RSV offset in most areas. HMPV was discovered in 2001,25 and clinical presentation can be indistinguishable from influenza and RSV.26 Hospitalization rates associated with HMPV for children are similar to those for influenza.27 Although there are vaccines available for IAV28 and the first vaccine was approved by the FDA for RSV in 2023 (although only for adults age >60 yo),29,30 there is no HMPV vaccine currently available.31 IAV subtype analysis suggested that H3N2 was more common than H1N1, consistent with subtyping of clinical samples.22
Wastewater data was able to resolve information on occurrence patterns at higher resolution than clinical data, showing differences in occurrence patterns of IAV, RSV, SARS-CoV-2, and HMPV community infections. For IAV, we identified two localized summertime events at two POTWs located 15 km apart in Santa Clara County and a late localized fall HMPV event was identified at one POTW in San Francisco County, neither of which were reflected in their respective state-aggregated positivity rate data.
Wintertime wastewater events were evident for IAV, RSV, and HMPV, but their characteristics differed. IAV events were temporally coherent throughout the region with onsets, peaks, and offsets occurring at all POTWs within the same three, three, and five week windows, respectively. RSV events were also regionally temporally coherent; all POTWs had onsets, peaks, and offsets within the same five-, three-, and five week windows, respectively. However, wintertime RSV onset and offsets occurred one month earlier and later, respectively, than those of IAV across all POTWs, a pattern also discernible in the state-aggregated clinical data. RSV peak concentrations occurred at the same time as the peak IAV concentrations. SARS-CoV-2 concentrations also had local peaks at the same time as RSV and IAV suggesting community infections reach their height in unison, consistent with reports of the tripledemic overwhelming some hospital capacities.9
HMPV wastewater events differed widely in the timing of onset and peak among the eight POTWs, suggesting localized dynamics; onset dates varied by up to 4 months, and the timing of the peak varied by up to 5 months. Localized HMPV dynamics are further supported by the relatively weak correlations between HMPV concentrations and concentrations of the other viruses that showed patterns of regional coherence. Although HMPV concentrations were positively associated with the state-aggregated positivity rate data, there are clearly localized dynamics in the wastewater events that are not reflected in the state-aggregated clinical data. More localized data on HMPV circulation are not available as testing is extremely limited.7 Previous studies of clinical HMPV infections suggest variable seasonal infection from year-to-year in contrast to typical patterns in seasonal RSV and IAV infections.27
A global systematic review32 indicates that seasonal IAV, RSV, and HMPV epidemics typically overlap. RSV epidemics typically start earlier than IAV epidemics by 0.3 months in temperate regions.32 In our study, RSV wastewater events onset 1.2 months (median) before IAV. The same review indicates that IAV and RSV epidemics in temperate areas are 3.8 and 4.6 months in duration, respectively. Wastewater events for IAV and RSV were 3.5 and 4.8 months in duration (medians). Temperate HMPV epidemics began 1.7 months after RSV and were 4.8 months in duration. In the present study, HMPV wastewater events began 1 month after RSV (median) and were 8.2 months (median) in duration.
While the clinical test positivity data used here provide insight into disease circulation during the study time period, there is no data on community incidence or prevalence of IAV, RSV, or HMPV in the study area. Additionally, data on COVID-19 incidence and prevalence degraded during this time period owing to the wide availability of at home rapid tests,12,33 the results of which are not reportable to public health agencies in our study area. IAV, RSV, HMPV, and COVID-19 test positivity rates serve as proxies for disease occurrence, but it is important to acknowledge that the rates reflect those of severely symptomatic cases. There is also a significant delay in those data being available, usually with results available and updated 2–4 weeks after specimen collection. Wastewater represents composite biological samples from an entire community, including those who are asymptomatic or mildly symptomatic. Wastewater therefore compliments data on clinical test positivity. Given the delays associated with the positivity rate data and the sparseness of localized positivity rate data, wastewater can serve as an indicator of the first onsets of localized, community infections. While spatial and temporal coherence was observed for the IAV and RSV events, HMPV showed more localized dynamics. This has been shown previously for other diseases; localized dynamics of mpox were also evident from wastewater monitoring.34 We explored other methods of identifying wastewater events including using different concentration thresholds and look-back periods. All gave similar relative results among the viruses; a public health organization could decide on data analysis methods for identifying wastewater events that were fit for their purposes.
Acknowledgments
We acknowledge the numerous people who contributed to wastewater sample collection, Allegra Koch for support with the literature review, and Alexander Yu for suggestions on the draft manuscript. TOC art was created with BioRender.com. This research was performed on the ancestral and unceded lands of the Muwekma Ohlone people. We pay our respects to them and their Elders, past and present, and are grateful for the opportunity to live and work here. This work is supported by gifts from the CDC Foundation and the Sergey Brin Family Foundation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.estlett.3c00385.
Additional methodological details as well as Tables S1–S3, and Figures S1–S6. (PDF)
The authors declare the following competing financial interest(s): B.W., B.H., and D.D. are employees of Verily Life Sciences, LLC.
Supplementary Material
References
- Jin X.; Ren J.; Li R.; Gao Y.; Zhang H.; Li J.; Zhang J.; Wang X.; Wang G. Global Burden of Upper Respiratory Infections in 204 Countries and Territories, from 1990 to 2019. eClinicalMedicine 2021, 37, 100986. 10.1016/j.eclinm.2021.100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos T.; Lim S. S.; Abbafati C.; Abbas K. M.; Abbasi M.; Abbasifard M.; Abbasi-Kangevari M.; Abbastabar H.; Abd-Allah F.; Abdelalim A.; Abdollahi M.; Abdollahpour I.; Abolhassani H.; Aboyans V.; Abrams E. M.; Abreu L. G.; Abrigo M. R. M.; Abu-Raddad L. J.; Abushouk A. I.; Acebedo A.; et al. Lancet 2020, 396 (10258), 1204–1222. 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Bhutta Z. A.; Coates M. M.; Coggeshall M.; Dandona L.; Diallo K.; Franca E. B.; Fraser M.; Fullman N.; Gething P. W.; Hay S. I.; Kinfu Y.; Kita M.; Kulikoff X. R.; Larson H. J.; Liang J.; Liang X.; Lim S. S.; Lind M.; Lopez A. D.; et al. Lancet 2016, 388 (10053), 1725–1774. 10.1016/S0140-6736(16)31575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M. E.; Biggs H. M.; Haynes A.; Dahl R. M.; Mustaquim D.; Gerber S. I.; Watson J. T. Human Coronavirus Circulation in the United States 2014–2017. Journal of Clinical Virology 2018, 101, 52–56. 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S. J.; Winn A. K.; Budd A. P.; Prill M. M.; Steel J.; Midgley C. M.; Kniss K.; Burns E.; Rowe T.; Foust A.; Jasso G.; Merced-Morales A.; Davis C. T.; Jang Y.; Jones J.; Daly P.; Gubareva L.; Barnes J.; Kondor R.; Sessions W.; Smith C.; Wentworth D. E.; Garg S.; Havers F. P.; Fry A. M.; Hall A. J.; Brammer L.; Silk B. J. Changes in Influenza and Other Respiratory Virus Activity During the COVID-19 Pandemic - United States, 2020–2021. MMWR Morb Mortal Wkly Rep 2021, 70 (29), 1013–1019. 10.15585/mmwr.mm7029a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E. J.; Uyeki T. M.; Chu H. Y. The Effects of the COVID-19 Pandemic on Community Respiratory Virus Activity. Nature Reviews Microbiology 2022, 21 (3), 195–210. 10.1038/s41579-022-00807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A. B.; Hughes B.; Duong D.; Chan-Herur V.; Buchman A.; Wolfe M. K.; White B. J. Wastewater Concentrations of Human Influenza, Metapneumovirus, Parainfluenza, Respiratory Syncytial Virus, Rhinovirus, and Seasonal Coronavirus Nucleic-Acids during the COVID-19 Pandemic: A Surveillance Study. Lancet Microbe 2023, 4 (5), e340–e348. 10.1016/S2666-5247(22)00386-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid S.; Winn A.; Parikh R.; Jones J.; McMorrow M.; Prill M.; Silk B. J.; Scobie H. M.; Hall A. J. Seasonality of Respiratory Syncytial Virus — United States, 2017–2023. MMWR Morb Mortal Wkly Rep 2023, 72, 355–361. 10.15585/mmwr.mm7214a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlow B. Triple-Demic Overwhelms Paediatric Units in US Hospitals. Lancet Child & Adolescent Health 2023, 7 (2), 86. 10.1016/S2352-4642(22)00372-8. [DOI] [PubMed] [Google Scholar]
- Weixel N. ‘Tripledemic’ Infected Nearly 40% of Households, Survey Finds. Hill. February 7, 2023. https://thehill.com/policy/healthcare/3846161-tripledemic-infected-nearly-40-percent-of-households-survey-finds/ (accessed 2023-05-22). [Google Scholar]
- Chow Y. P.; Chin B. H. Z.; Loo J. M.; Moorthy L. R.; Jairaman J.; Tan L. H.; Tay W. W. Y. Clinical and Epidemiological Characteristics of Patients Seeking COVID-19 Testing in a Private Centre in Malaysia: Is There a Role for Private Healthcare in Battling the Outbreak?. PLoS One 2021, 16 (10), e0258671 10.1371/journal.pone.0258671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader B.; Gertz A.; Iuliano A. D.; Gilmer M.; Wronski L.; Astley C. M.; Sewalk K.; Varrelman T. J.; Cohen J.; Parikh R.; Reese H. E.; Reed C.; Brownstein J. S. Use of At-Home COVID-19 Tests — United States, August 23, 2021–March 12, 2022. MMWR Morb Mortal Wkly Rep 2022, 71, 489–494. 10.15585/mmwr.mm7113e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElfish P. A.; Purvis R.; James L. P.; Willis D. E.; Andersen J. A. Perceived Barriers to COVID-19 Testing. International Journal of Environmental Research and Public Health 2021, 18 (5), 2278. 10.3390/ijerph18052278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jajosky R. A.; Groseclose S. L. Evaluation of Reporting Timeliness of Public Health Surveillance Systems for Infectious Diseases. BMC Public Health 2004, 4 (1), 29. 10.1186/1471-2458-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L.; Gog J. R.; Olson D.; Viboud C. Infectious Disease Surveillance in the Big Data Era: Towards Faster and Locally Relevant Systems. Journal of Infectious Diseases 2016, 214 (suppl_4), S380–S385. 10.1093/infdis/jiw376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. K.; Topol A.; Knudson A.; Simpson A.; White B.; Vugia D. J.; Yu A. T.; Li L.; Balliet M.; Stoddard P.; Han G. S.; Wigginton K. R.; Boehm A. B. High-Frequency, High-Throughput Quantification of SARS-CoV-2 RNA in Wastewater Settled Solids at Eight Publicly Owned Treatment Works in Northern California Shows Strong Association with COVID-19 Incidence. mSystems 2021, 6 (5), e00829–21. 10.1128/mSystems.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A.; Wolfe M.; White B.; Wigginton K.; Boehm A.. High Throughput Pre-Analytical Processing of Wastewater Settled Solids for SARS-CoV-2 RNA Analyses. protocols.io 2021. DOI: dx.doi.org/10.17504/protocols.io.btyqnpvw. [Google Scholar]
- Topol A.; Wolfe M.; Wigginton K.; White B.; Boehm A.. High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA. protocols.io 2021. DOI: dx.doi.org/10.17504/protocols.io.btyrnpv6. [Google Scholar]
- Huisman J. S.; Scire J.; Caduff L.; Fernandez-Cassi X.; Ganesanandamoorthy P.; Kull A.; Scheidegger A.; Stachler E.; Boehm A. B.; Hughes B.; Knudson A.; Topol A.; Wigginton K. R.; Wolfe M. K.; Kohn T.; Ort C.; Stadler T.; Julian T. R. Wastewater-Based Estimation of the Effective Reproductive Number of SARS-CoV-2. Environ. Health Perspect. 2022, 130 (5), 057011–1. 10.1289/EHP10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E. M.; Nguyen K. H.; Harwood V. J.; Breitbart M. Pepper Mild Mottle Virus: A Plant Pathogen with a Greater Purpose in (Waste)Water Treatment Development and Public Health Management. Water Res. 2018, 144, 1–12. 10.1016/j.watres.2018.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A.; Topol A.; White B.; Wolfe M. K.; Wigginton K.; Boehm A. B. Effect of Storage Conditions on SARS-CoV-2 RNA Quantification in Wastewater Solids. PeerJ. 2021, 9, e11933 10.7717/peerj.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- California Influenza Surveillance Program. Influenza and Other Respiratory Viruses Weekly Report; 2023. https://www.cdph.ca.gov/Programs/CID/DCDC/Pages/Immunization/Influenza.aspx (accessed 2023-06-30).
- Boehm A. B.; Wolfe M. K.; Wigginton K. R.; Bidwell A.; White B. J.; Hughes B.; Duong; Chan-Herur V.; Bischel H. N.; Naughton C. C. Human Viral Nucleic Acids Concentrations in Wastewater Solids from Central and Coastal California USA. Scientific Data 2023, 10, 396. 10.1038/s41597-023-02297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M. A.; Boehm A. B.; Salit M.; Spencer S. K.; Wigginton K. R.; Noble R. T. The Environmental Microbiology Minimum Information (EMMI) Guidelines: QPCR and DPCR Quality and Reporting for Environmental Microbiology. Environ. Sci. Technol. 2021, 55 (15), 10210–10223. 10.1021/acs.est.1c01767. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B. G.; de Jong J. C.; Groen J.; Kuiken T.; de Groot R.; Fouchier R. A.; Osterhaus A. D. A Newly Discovered Human Pneumovirus Isolated from Young Children with Respiratory Tract Disease. Nat. Med. 2001, 7 (6), 719–724. 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B. G.; Osterhaus D. M. E.; Fouchier R. A. M. Clinical Impact and Diagnosis of Human Metapneumovirus Infection. Pediatric Infectious Disease Journal 2004, 23 (1), S25. 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- Edwards K. M.; Zhu Y.; Griffin M. R.; Weinberg G. A.; Hall C. B.; Szilagyi P. G.; Staat M. A.; Iwane M.; Prill M. M.; Williams J. V. Burden of Human Metapneumovirus Infection in Young Children. N Engl J. Med. 2013, 368 (7), 633–643. 10.1056/NEJMoa1204630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.-J.; Crank M. C.; Shiver J.; Graham B. S.; Mascola J. R.; Nabel G. J. Next-Generation Influenza Vaccines: Opportunities and Challenges. Nat. Rev. Drug Discovery 2020, 19 (4), 239–252. 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias A.; Rodríguez-Fernández R.; Oliva S.; Peeples M. E.; Ramilo O. The Journey to a Respiratory Syncytial Virus Vaccine. Annals of Allergy, Asthma & Immunology 2020, 125 (1), 36–46. 10.1016/j.anai.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US FDA. FDA Approves First Respiratory Syncytial Virus (RSV) Vaccine. https://www.fda.gov/news-events/press-announcements/fda-approves-first-respiratory-syncytial-virus-rsv-vaccine (accessed 2023-06-08).
- Center for Disease Control. Human metapneumovirus. https://www.cdc.gov/ncird/human-metapneumovirus.html (accessed 2023-05-23).
- Li Y.; Reeves R. M.; Wang X.; Bassat Q.; Brooks W. A.; Cohen C.; Moore D. P.; Nunes M.; Rath B.; Campbell H.; Nair H.; Acacio S.; Alonso W. J.; Antonio M.; Ayora Talavera G.; Badarch D.; Baillie V. L.; Barrera-Badillo G.; Bigogo G.; Broor S.; et al. Global Patterns in Monthly Activity of Influenza Virus, Respiratory Syncytial Virus, Parainfluenza Virus, and Metapneumovirus: A Systematic Analysis. Lancet Global Health 2019, 7 (8), e1031–e1045. 10.1016/S2214-109X(19)30264-5. [DOI] [PubMed] [Google Scholar]
- Boehm A. B.; Wolfe M. K.; White B. J.; Hughes B.; Duong D. Divergence of Wastewater SARS-CoV-2 and Reported Laboratory-Confirmed COVID-19 Incident Case Data Coincident with Wide-Spread Availability of at-Home COVID-19 Antigen Tests. PeerJ. 2023, 11, e15631 10.7717/peerj.15631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. K.; Yu A. T.; Duong D.; Rane M. S.; Hughes B.; Chan-Herur V.; Donnelly M.; Chai S.; White B. J.; Vugia D. J.; Boehm A. B. Use of Wastewater for Mpox Outbreak Surveillance in California. N Engl J. Med. 2023, 388, 570. 10.1056/NEJMc2213882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


