Abstract
The persistently infected carrier stallion is the critical natural reservoir of equine arteritis virus (EAV), as venereal infection of mares frequently occurs after breeding to such stallions. Two Thoroughbred stallions that were infected during the 1984 outbreak of equine viral arteritis in central Kentucky subsequently became long-term EAV carriers. EAV genomes amplified from the semen of these two stallions were compared by sequence analysis of the six 3′ open reading frames (ORFs 2 through 7), which encode the four known structural proteins and two uncharacterized glycoproteins. The major variants of the EAV population that sequentially arose within the reproductive tract of each carrier stallion varied by approximately 1% per year, and the heterogeneity of the viral quasispecies increased during the course of long-term persistent infection. The various ORFs of the dominant EAV variants evolved independently, and there was apparently strong selective pressure on the uncharacterized GP3 protein during persistent infection. Amino acid changes also occurred in the V1 variable region of the GL protein. This region has been previously identified as a crucial neutralization domain, and selective pressures exerted on the V1 region during persistent EAV infection led to the emergence of virus variants with distinct neutralization properties. Thus, evolution of the EAV quasispecies that occurs during persistent infection of the stallion clearly can influence viral phenotypic properties such as neutralization and perhaps virulence.
Equine arteritis virus (EAV) is the cause of equine viral arteritis (EVA), a reproductive and respiratory disease of equids (61). EAV is transmitted either horizontally by aerosol during acute respiratory infection or venereally by natural or artificial breeding of mares to persistently infected carrier stallions (62). Up to 60% of stallions that acquire EAV by the respiratory route can subsequently become persistently infected. The carrier state can last from months to several years, during which time the virus is present solely in the reproductive tract, principally in the ampulla of the vas deferens (61). The carrier state is testosterone dependent and thus occurs only in stallions (41, 49). Susceptible mares bred to carrier stallions almost always become infected with EAV (61). While EAV infection is usually subclinical, there has recently been an apparent increase in the occurrence of clinical EVA (28). Many outbreaks of EVA are precipitated by venereal infection of a seronegative mare after breeding to a carrier stallion, with subsequent aerosol transmission to susceptible cohorts (2, 3, 61).
EAV has a single-stranded, positive-sense RNA genome of approximately 12.7 kb and is the prototype member of the genus Arterivirus in the family Arteriviridae (13). The EAV genome includes at least eight open reading frames (ORFs) (from 5′ to 3′, ORFs 1a, 1b, 2, 3, 4, 5, 6, and 7 [12]). The 9 kb located at the 5′ end of the genome contain ORFs 1a and 1b, which encode the viral replicase. The structural protein genes are overlapping, occupy 3 kb at the 3′ end of the genome, and are translated from a nested set of mRNAs (64). Generation of a 3′-coterminal set of mRNAs is a characteristic feature of the order Nidovirales, which includes the families Coronaviridae and Arteriviridae (8). ORFs 2 and 5 of EAV encode minor and major structural membrane glycoproteins, (GS and GL [13]), respectively. The GL protein is the most variable of the four known structural proteins and contains epitopes critical for virus neutralization (4, 6, 7, 31). ORF 6 encodes a structural membrane protein (M), and ORF 7 encodes the nucleocapsid protein (N) (13). The putative EAV GP3 and GP4 glycoproteins, encoded by ORFs 3 and 4 respectively, are uncharacterized.
RNA virus replication is characterized by high mutation rates, short generation times, and high yields (16). Therefore, RNA viruses exist not as a single genotype rather as a heterogeneous mixture of related genomes known as a viral quasispecies (8, 15, 33). Genetic variation has been repeatedly demonstrated among field isolates and laboratory strains of EAV and indirectly by the selection of neutralization resistant variants in vitro (4, 7, 10, 31). The carrier stallion is clearly central to the epidemiology of EAV infection, but the evolution of the EAV quasispecies that occurs during persistent infection and the potential emergence of novel variants with divergent phenotypic properties are yet to be characterized. Oligonucleotide fingerprinting of EAV strains isolated in cell culture from semen collected sequentially from two carrier stallions revealed ongoing nucleotide variation (47). It was not, however, determined which regions of the virus genome were principally affected, nor were the potential effects on virus phenotype investigated. To characterize the EAV quasispecies during persistent infection, detailed sequence analysis of the structural protein genes has been performed with viral RNA purified directly from semen collected sequentially over a 10-year period from two Thoroughbred carrier stallions that were initially infected during an EVA outbreak in Kentucky in 1984 (59). Phenotypic assays were performed with selected virus isolates. The results suggest that the EAV quasispecies evolves significantly in the individual carrier stallion, in distinct contrast to its relative genetic stability during EVA epizootics involving aerosol transmission of the virus (2, 3), and that variants with distinct phenotypes emerge during persistent infection.
MATERIALS AND METHODS
Disease outbreak, stallions, and semen collection.
An extensive outbreak of EVA occurred in Kentucky in 1984 (59). This outbreak was the first recorded occurrence of EVA in the North American Thoroughbred population, and subsequent investigation established that stallions could be chronically infected with EAV and that these carrier stallions are critical to the epidemiology of EAV infection (60, 62, 63). Two stallions from the same farm (designated D and E) were infected during this outbreak and subsequently became long-term carriers. Semen was collected from the two stallions at regular intervals (for D, 6/84 [month/year], 9/85, 12/86, 9/87, 7/88, 1/89, 1/91, 9/92, and 8/94; for E, 6/84, 9/85, 11/86, 2/88, 1/89, and 1/91) following initial infection of the stallions, which occurred in May 1984. Semen was collected until the stallions ceased shedding EAV, as determined by virus isolation as previously described (62). The field strain KY84 was originally isolated from pooled blood collected during the outbreak (5/84 [45]) from three stallions acutely infected with EAV (stallion E and two other stallions) and was serially passaged three times in horses and subsequently three times in rabbit kidney cells (RK-13; ATCC CCL37) before amplification of virus stock in RK-13 cells.
RNA extraction and PCR amplification.
Viral RNA was isolated directly from seminal plasma with a QIAamp viral RNA isolation kit (sequences and viruses are identified by stallion and year of isolation, e.g., D84). RNA was also isolated from the cell culture propagated KY84 strain as previously described (31). Viral RNA was reverse transcribed into cDNA with Superscript II and oligo(dT) primers (Gibco BRL). The first-strand cDNA was purified with GlassMAX DNA isolation system after RNase digestion (Gibco BRL). The entire 3-kb segment containing ORFs 2 through 7 was PCR amplified in two pieces by using Pfu Turbo DNA polymerase (Stratagene) (ORFs 2 through 4 were amplified with the primers located at positions 9705 to 9727 and 11218 to 11240; ORFs 5 through 7 were amplified with those at 11080 to 11101 and 12631 to 12651 [12]). Pfu polymerase was used to minimize artifactual substitutions (55). The resulting PCR products encompassed 2822 nucleotides at the 3′ end of the genome and included ORFs encoding the four known EAV structural proteins (GS, GL, M, and N) and two uncharacterized glycoproteins (GP3 and GP4). Fourteen different PCRs (100 μl/reaction) were carried out with each RNA sample, and the reaction products were pooled, concentrated (Centricon-30; Amicon), and purified by using a commercial kit (Qiaquick; Qiagen). A viral quasispecies is not accurately represented by a single sequence; thus, pooling of multiple reverse transcription-PCR (RT-PCR) products was done to obtain sequence data that most accurately represent the master sequence of the viral quasispecies present at the time of collection (16).
Cloning.
The PCR products that included ORFs 5 through 7 of the virus population present in the semen of stallion D in 1984 and 1994 were cloned into the plasmid pT7Blue-3 by using a Perfectly Blunt cloning kit (Novagen) according to the manufacturer’s protocol. Individual colonies were grown overnight in LB broth, and plasmid DNA was purified by using a Qiaprep kit (Qiagen). After restriction digestion, plasmid DNA was electrophoresed in 1% agarose gels and stained for confirmation of the insert. ORF 5 (nucleotides 11129 to 11896) was sequenced from 17 different clones from the D84 virus and 16 clones from the D94 virus.
Automatic sequencing.
Purified cDNA from PCR amplification products and plasmid DNA was sequenced by using a PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems). Approximately 100 ng of PCR-amplified cDNA or 1 μg of plasmid DNA and 10 pmol of primer were used in each reaction, and cycle sequencing was performed as previously described (31). Sequence data were collected with an ABI 377 automatic sequencer (Applied Biosystems) according to the manufacturer’s instructions. Sense and nonsense strands were each sequenced with a large library of primers (Genosys) (7, 31). The sequence of the KY84 strain was included in the analysis as the closest approximation of the original outbreak strain.
Sequence and phylogenetic analysis.
Computer analyses of DNA sequences were done with a Macintosh PowerPC and the MacDNASIS Pro version 3.5 (Hitachi) and Sequencher 3.0 (Gene Codes Corp.) programs. The TOPIR program of the Wisconsin package (version 8.0; Genetics Computer Group Inc.) and Clustal W version 1.7 (58) were used for multiple sequence alignment. Genetic distance, phylogenetic, and bootstrap analyses were done with PHYLIP version 3.5c for the Macintosh PowerPC (22, 23). Genetic distances (expected substitutions per site) were calculated for aligned sequences by using the DNADist program based on the Kimura two-parameter model (23, 37) with a transition/transversion ratio of 2.0. The distance matrices generated with DNADist were also used in the FITCH program (least-square method [24]) to generate phylogenetic trees. The FITCH program was carried out with randomized input order. The resulting phylogenetic trees are rooted with the corresponding ORFs of lactate dehydrogenase-elevating virus (29). Bootstrap sampling (22) was carried out on 1,000 replicate data sets with the SEQBOOT program to assess the confidence limits of the branch pattern. A value of ≥70% was considered significant (32). Estimates of the number of synonymous substitutions per synonymous site (Ks) and nonsynonymous substitutions per nonsynonymous site (Ka) were calculated with the MEGA program (version 1.01 [38]) by the method of Nei and Gojobori (48).
Virus isolation and neutralization assays.
Virus was isolated on RK-13 cells from semen collected from stallions D (6/84, 9/85, and 8/94) and E (6/84, 11/86, and 1/91) as previously described (62, 63). Stocks of each virus isolate were titrated by the method of Reed and Muench (52) after serial passage in RK-13 cells. Neutralization assays were performed as previously described with a panel of neutralizing monoclonal antibodies (MAbs) raised against the EAVUCD laboratory strain of EAV (5G11, 6D10, 7E5, 9F2, 10F11, and 10H4) or a neutralization-resistant variant derived from EAVUCD (1H9, 6A2, and 8D4) (4, 5). A nonneutralizing, N-protein-specific MAb (7B9) was used as a control (5). Neutralization assays were also done with equine polyclonal sera. Three of the sera were collected from stallions D and E (D86 serum, D98 serum, and E86 serum). One serum was collected after the second horse passage of the KY84 strain of EAV, 7 months postinfection (KY84 serum). Another serum was produced after nonlethal experimental infection of a horse with the well-characterized virulent Bucyrus strain of EAV (VBS53 [5, 7]). Serum from an uninfected horse was included as a control. Antibody titers were reported as the reciprocal of the highest final dilution that provided at least 50% protection of the monolayer against 1,000 50% tissue culture infective doses of virus.
Nucleotide sequence accession numbers.
The sequences analyzed were submitted to GenBank and have been assigned accession no. AF107266 to AF107274 (stallion D), AF107275 to AF107278 (stallion E), and AF107279 (KY84).
RESULTS
Sequences of the major EAV variants present in sequential semen samples from carrier stallions.
Two Thoroughbred stallions (D and E) were infected on the same farm during an outbreak of EVA in Kentucky in 1984. Recent studies indicate that EAV strains isolated from clinical samples obtained during the same outbreak and after limited cell culture passage are genetically stable (2, 3). Thus, we assume that these two stallions were infected with viruses that were very similar or identical to the original KY84 strain that was isolated during the outbreak.
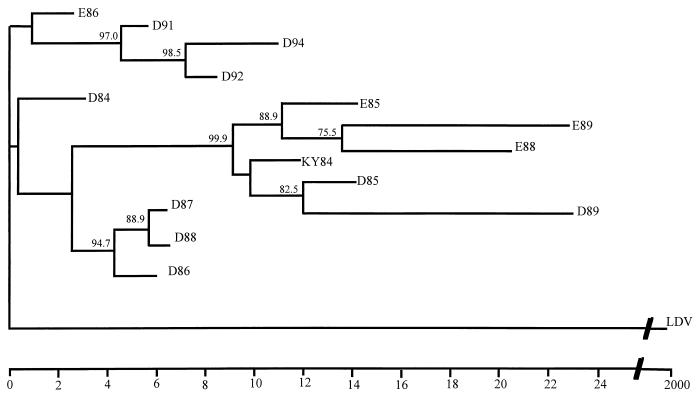
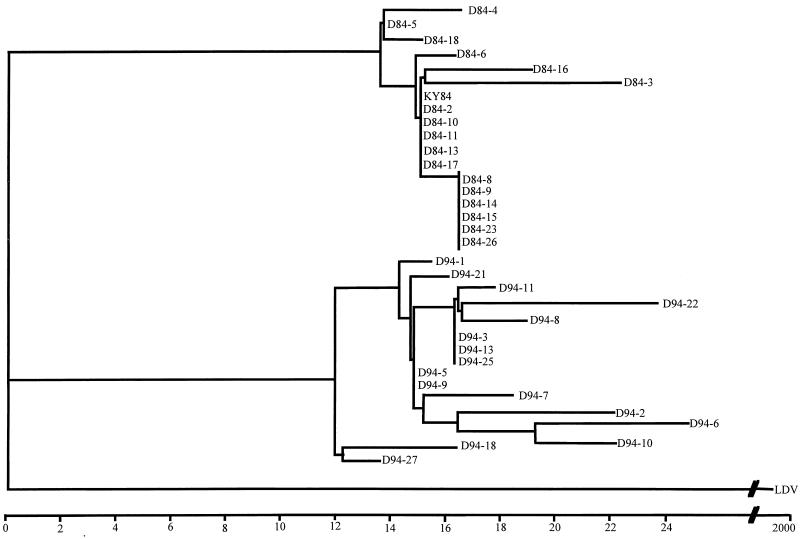
Viral RNA was isolated directly from the semen of the two carrier stallions, and a 3-kb portion was RT-PCR amplified and sequenced. To describe the relationships between the major EAV variants that sequentially arose during persistent infection of the stallions, sequences representing the major variant of the quasispecies from the ORF 2 start codon at position 9807 to the ORF 7 stop codon at 12628 were used to generate a phylogenetic tree (Fig. 1). The sequenced portions of viral RNA in semen were clearly distinct from year to year. The sequences of viruses from stallion D in the years 1991, 1992, and 1994 group phylogenetically with that of the E86 strain and are distinct from the branch containing the original outbreak strain KY84. These data indicate that EAV strains in the two stallions evolved down similar lineages and, in general, genetic distances increased over time relative to KY84. Some dominant species present earlier in infection of the two carrier stallions (D84, D86, D87, and D88 and E86) were more distant from KY84 than some later viral species (D89 and E89), suggesting that nucleotide reversions also occurred. Interestingly, many of the same mutations occurred in variants of EAV that evolved during persistent infection of the two stallions. Structural requirements likely exist at both the protein and RNA levels; thus, the changes that are fixed during persistent EAV infection of the carrier stallion are not random.
FIG. 1.
Phylogenetic tree describing the relationships between EAV genomes amplified from the semen of two persistently infected stallions (D and E) in the years 1984 through 1994. Horizontal branch lengths are scaled by a factor of 1,000 to Kimura two-parameter distances. Vertical lengths are not significant. Sequences from the corresponding ORFs of lactate dehydrogenase-elevating virus (LDV) are used as an outgroup to root the tree (29). Bootstrap values (shown when ≥70%) represent the percent occurrence of that clade per 1,000 bootstrap replicates.
The major variant of the viral quasispecies present in the semen of the carrier stallions D and E experienced mean nucleotide variation of 0.9 and 1.3%, respectively, per year throughout the entire 2,822-nucleotide segment that was sequenced (Table 1). The first and last dominant EAV species identified in semen from each stallion (D84-D94 and E85-E89) differed from each other by only 1.7%, indicating that new mutations as well as frequent reversions occur to generate new and unique quasispecies populations during persistent EAV infection. These results confirm those previously reported from two-dimensional oligonucleotide fingerprinting of viruses isolated in cell culture from stallions D and E during persistent infection (47) and are consistent with similar studies characterizing evolution during persistent infections with other RNA viruses (53, 57).
TABLE 1.
Percentage differences between EAV sequences (ORFs 2 through 7) amplified from semen collected from stallion D in 1984 through 1994 and stallion E in 1985 through 1989
| Sequences compared | No. of nucleotide changes | % Difference |
|---|---|---|
| D84 and D85 | 43 | 1.5 |
| D85 and D86 | 41 | 1.5 |
| D86 and D87 | 9 | 0.3 |
| D87 and D88 | 4 | 0.1 |
| D88 and D89 | 60 | 2.1 |
| D89 and D91 | 77 | 2.7 |
| D91 and D92 | 13 | 0.5 |
| D92 and D94 | 14 | 0.5 |
| Mean | 26.1 nta/yr | 0.9 |
| E85 and E86 | 47 | 1.7 |
| E86 and E88 | 62 | 2.2 |
| E88 and E89 | 42 | 1.5 |
| Mean | 37.8 nt/yr | 1.3 |
nt, nucleotides.
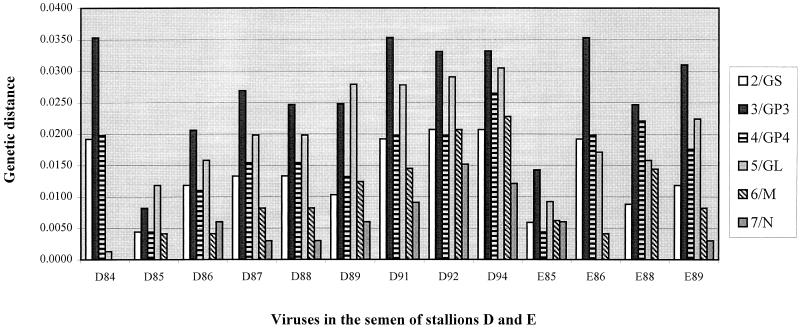
To investigate potential differences in the extent of evolution between the various ORFs of the dominant EAV variants during persistent infection, the genetic distances of the individual ORFs amplified from the semen of the stallions were calculated with the DNADist program of the PHYLIP package and compared to the outbreak strain KY84 (Fig. 2). Genetic distance is a value that reflects the number of expected nucleotide substitutions per site, accounting for differences in the number of transitions and transversions (23, 37). The data show that individual ORFs of the major EAV variants present in the semen of the two carrier stallions evolved independently of each other. ORFs 2, 3, and 4 of the D84 strain were more distinct from the outbreak strain than ORFs 5, 6, and 7, which were very closely related to KY84. The ORFs 2, 3, and 4 of the D85 strain (one year later) reverted to be more closely related to KY84, whereas genetic distances of ORF 5 increased linearly over time. ORF 3 was the most variable ORF during EAV persistence in these two stallions. Overall, ORFs 3 and 5 evolved most rapidly, ORFs 2 and 4 changed moderately, and ORFs 6 and 7 were substantially more conserved during persistent EAV infection. The relative rates of change of the various ORFs are consistent with results found after genetic characterization of various field isolates and laboratory strains of EAV (1, 7, 10, 31). ORF 6 encodes the M protein, which is a structural, protein with three transmembrane domains, and ORF 7 encodes the N protein (13). Structural requirements likely restrict amino acid alterations in these two proteins, and their genes are highly conserved, as few synonymous nucleotide changes occurred. ORFs 6 and 7 are located on the extreme 3′ end of the EAV genome and thus may be conserved due to structural restrictions imposed on nucleotides in this region from their potential function in genome packaging during assembly or the complex transcription process (16).
FIG. 2.
Genetic distances from the outbreak strain KY84 of individual EAV ORFs amplified from semen collected during persistent infection of the two stallions (D and E) were calculated with the DNADist program of the PHYLIP package.
Synonymous and nonsynonymous nucleotide changes.
The Ka/Ks ratio provides an estimate of the degree of selection responsible for amino acid substitutions. Fixation of nonsynonymous mutations can be accelerated by positive selection; therefore, the larger the Ka/Ks ratio, the stronger the selective pressure (38, 40). To examine the extent of selective pressure applied to individual EAV proteins during persistent infection of stallions, the Ka/Ks ratios were calculated for each ORF by using the MEGA program and are reported relative to the value for KY84. Other than ORF 3, the ORFs of major variant viruses in the semen of persistently infected stallions experienced little selective pressure on their entire encoded proteins (Table 2). The GL protein of the initial dominant variant in the semen of stallion D (D84) was identical to that of the KY84 strain, suggesting that GL protein-specific immune escape is not necessary for initial establishment of persistent EAV infection in the reproductive tract of the Thoroughbred stallion following aerosol acquisition of the virus. The Ka/Ks value was markedly greater for ORF 3, most notably in the two samples taken 1 year after the onset of the carrier state. D85, D89, and E85 ORF 3 sequences differed from that of KY84 by 4, 12, and 7 nucleotides and by 4, 10, and 5 amino acids, respectively. Thus, these Ka/Ks ratios realistically reflect an unusually large proportion of nonsynonymous mutations and not just a low synonymous value (denominator).
TABLE 2.
Ka/Ks ratios for ORFs 2 through 7 of EAV amplified from the semen of carrier stallions
| Stallion |
Ka/Ks
|
|||||
|---|---|---|---|---|---|---|
| 2/GSb | 3/GP3 | 4/GP4 | 5/GL | 6/M | 7/N | |
| D84 | 0.273 | 0.288 | 0.036 | 0.000 | 0.000 | 0.000 |
| D85 | 0.000 | 0.011/0c | 0.000 | 0.265 | 0.000 | 0.000 |
| D86 | 0.194 | 0.369 | 0.077 | 0.233 | 0.367 | 0.000 |
| D87 | 0.161 | 0.409 | 0.121 | 0.215 | 0.120 | 0.000 |
| D88 | 0.161 | 0.410 | 0.050 | 0.215 | 0.120 | 0.000 |
| D89 | 0.000 | 1.556 | 0.153 | 0.095 | 0.070 | 0.346 |
| D91 | 0.273 | 0.288 | 0.036 | 0.191 | 0.143 | 0.171 |
| D92 | 0.175 | 0.328 | 0.036 | 0.215 | 0.087 | 0.112 |
| D94 | 0.175 | 0.330 | 0.025 | 0.198 | 0.033 | 0.112 |
| E85 | 0.111 | 4.036 | 0.000 | 0.250 | 0.000 | 0.000 |
| E86 | 0.273 | 0.288 | 0.036 | 0.279 | 0.000 | 0.000 |
| E88 | 0.066 | 0.744 | 0.126 | 0.161 | 0.000 | 0.000 |
| E89 | 0.046 | 0.518 | 0.042 | 0.173 | 0.000 | 0.000 |
Calculated with the MEGA program and reported relative to KY84.
ORF/encoded protein.
All nucleotide changes were nonsynonymous.
Variable regions of the GP3 protein.
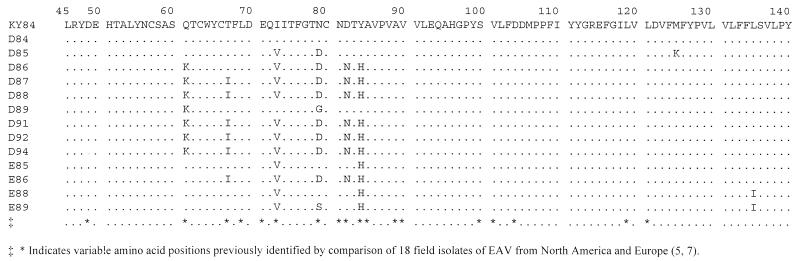
The uncharacterized GP3 protein, encoded by ORF 3, varied more than the other EAV proteins during persistent infection of the two carrier stallions. The variable regions of the GP3 protein are shown in Fig. 3A. Two highly variable regions, from amino acids 17 to 30 and 116 to 121, were identified in the GP3 protein. These regions are subsets of the variable regions previously identified in the GP3 protein of six field isolates of EAV (1). The EAV GP3 protein is predicted to be extensively glycosylated. The existing glycosylation sites were conserved; however, there were two amino acid changes that added potential glycosylation sites within the variable regions. Compared to KY84, most semen variants had a new potential glycosylation site at amino acid 28 (Fig. 4), and strain D89 had an additional potential glycosylation site at amino acid 118 (data not shown). The two variable amino acid regions of the GP3 protein corresponded to nucleotides 10337 to 10377 and 10634 to 10650, which varied, on average, 3.9 and 8.5%, respectively, per year, for carrier stallion D and 16.4 and 20.6%, respectively, per year for carrier stallion E (Fig. 3A). Amino acid variation that occurred in limited regions of the GP3 protein greatly exceeded the rate that would be expected by random variation of structurally insignificant domains. Altogether, our data indicate that strong selective pressure for amino acid change is likely exerted on the EAV GP3 protein during establishment and throughout persistent EAV infection of the Thoroughbred carrier stallion.
FIG. 3.
Regions of the GP3 and GL proteins had highly variable amino acid sequences during persistent EAV infection of the two Thoroughbred stallions. The amino acid ranges are shown on the left, variable sites are indicated with asterisks, and specific nucleotide and amino acid regions where variation occurred are shown in boxes on the right.
FIG. 4.
Variation in the region from nucleotides 10337 to 10379 that encodes the depicted regions of the GP3 and GS proteins, which are translated out of frame from each other.
The EAV genome is organized such that every internal ORF shares its 3′ and 5′ ends with the neighboring ORF; thus, the same nucleotides at the ends of the ORFs encode different proteins that are out of frame from each other (13). The highly variable nucleotide region 10337 to 10377 was located at the section where ORFs 2 and 3 overlap in different reading frames (Fig. 4). Most mutations occurred in the first position of the codon of the GP3 protein reading frame, resulting in an amino-terminal region of the GP3 protein that was highly variable during persistent infection. The GP3 amino terminus may act as a hydrophobic signal sequence, variation of which might explain the inconsistent inclusion in the virion of the equivalent GP3 protein in a related arterivirus (17, 43). The nucleotide changes in the region 10337 to 10377 predominantly occurred in the third position of the codon in the +1 reading frame from which the GS protein is read (Fig. 4). Thus, in distinct contrast to the amino acid plasticity that occurs in the GP3 protein, the carboxy-terminal region of the GS protein was apparently subjected to structural constraints, and variants that deviated from this requirement did not survive. This finding is consistent with results of a study comparing ORF2 of laboratory strains and field isolates of EAV (31).
Neutralization phenotype of sequential virus variants from the semen of carrier stallions.
The GL protein, encoded by ORF 5, contains the known neutralization determinants of EAV (4, 6). Specific regions of the GL protein important for EAV neutralization have been identified, as have regions that varied among field isolates and laboratory strains of EAV (5, 7). During persistent infection of stallions, variation in the GL protein occurred primarily within a specific section of the V1 variable region (amino acids 61 to 84; Fig. 3B and 5). This region corresponds to nucleotides 11308 to 11378, which evolved at average rates of 3.4 and 5.0% per year for stallions D and E, respectively. The V1 region is critical for neutralization by some murine MAbs and varied significantly among field strains of EAV from North America and Europe (2, 5, 7). Amino acid differences in this region correlated with differences in neutralization phenotype of various EAV field strains (2, 7). Many of these same critical amino acids also varied during long-term EAV persistence in the reproductive tracts of the two carrier stallions (Fig. 5).
FIG. 5.
Variation of the V1 variable region of the GL protein between sequential amplicons of EAV in the semen of the stallions D and E.
Neutralization assays with a large panel of well-characterized MAbs and polyclonal equine sera were done to determine whether the observed nucleotide variation in ORF 5 of EAV isolates from the semen of carrier stallions correlated with phenotypic differences in the corresponding viruses isolated from semen. A fourfold or greater difference in neutralization titer between viruses was interpreted as a significant change in neutralization phenotype (Table 3). The virus isolated from stallion D in 1994 had a notably lower titer with MAb 10H4 than the viruses isolated from the same stallion in 1984 and 1985. Virus isolate E91 was neutralized to a lower titer by MAb 7E5 and was not neutralized by MAb 6A2, which neutralized viruses isolated earlier from stallion E. Neutralization titers with the D94 virus and MAbs 7E5 and 9F2 and with the E91 virus and MAb 9F2 were lower, but not significantly so. MAb 10H4 recognizes an epitope that is distinct but interactive with that recognized by MAb 6D10 (4), yet MAb 6D10 neutralized all viruses isolated from the semen of the carrier stallions to a consistently high titer. MAbs 7E5 and 6A2 recognize distinct conformational epitopes that involve amino acids 69 to 99 and 102 (5). Specific mutations generated in vitro in neutralization-resistant variants (4, 5) were not duplicated in the variants isolated from the carrier stallions, but the MAbs recognize conformational epitopes that are apparently altered during persistent infection of stallions.
TABLE 3.
Neutralization titers of antibodies against selected EAV isolates from the semen of carrier stallions D and E
| Isolate | Neutralization titersa
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Monoclonal
|
Polyclonal
|
|||||||||||||||
| 5G11 | 6D10 | 7E5 | 9F2 | 10F11 | 10H4 | 1H9 | 6A2 | 8D4 | 7B9 | D86 | D98 | E86 | KY84 | VBS53 | NegEq | |
| KY84 | >4,096 | >4,096 | 1,024 | 512 | >4,096 | >4,096 | <32 | 256 | <32 | <32 | 1,024 | 512 | 512 | 512 | 256 | <32 |
| D84 | >4,096 | >4,096 | 512 | 256 | >4,096 | >4,096 | <32 | 256 | <32 | <32 | 1,024 | 512 | 512 | 512 | 512 | <32 |
| D85 | >4,096 | >4,096 | 1,024 | 256 | >4,096 | >4,096 | <32 | 256 | <32 | <32 | 1,024 | 256 | 1,024 | 512 | 512 | <32 |
| D94 | >4,096 | >4,096 | 256 | 128 | >4,096 | 512 | <32 | 256 | <32 | <32 | 256 | 128 | 512 | 128 | 128 | <32 |
| E84 | >4,096 | >4,096 | 1,024 | 512 | >4,096 | >4,096 | <32 | 256 | <32 | <32 | 1,024 | 512 | 1,024 | 512 | 256 | <32 |
| E86 | >4,096 | >4,096 | 512 | 512 | >4,096 | 2,048 | <32 | 512 | <32 | <32 | 1,024 | 512 | 1,024 | 512 | 256 | <32 |
| E91 | >4,096 | >4,096 | 128 | 256 | >4,096 | 1,024 | <32 | <32 | <32 | <32 | 512 | 128 | 256 | 256 | 64 | <32 |
Expressed as the inverse of the highest final dilution that provided at least 50% protection of RK-13 monolayers against 1,000 50% tissue culture infective doses of virus.
Subtle variation in the neutralization phenotype of individual viruses isolated from semen was also detected with the polyclonal equine sera; viruses D94 and E91 were neutralized to lower titers with equine polyclonal sera than viruses isolated earlier from the same stallions (Table 3). Differences in neutralization titers obtained with the VBS53 serum were significant for viruses isolated from both stallions. Titers were also lower for the D94 virus with the KY84 serum, and for the E91 virus with the E86 and D98 sera, than those for the earlier isolates from the same respective stallions. The results indicate that EAV variants with altered neutralization phenotypes emerged during the course of persistent infection of the two carrier stallions. Together these data imply that selective pressures exerted during the course of persistent EAV infection of stallions significantly influence the evolution of specific regions of the GL protein.
Phylogenetic analysis of cloned sequences.
To better characterize the EAV quasispecies present during persistent infection of stallions, ORF 5 was RT-PCR amplified and cloned from viral RNA isolated directly from semen collected in 1984 and 1994 from stallion D. Multiple clones from the viral quasispecies present in 1984 (17 clones) and 1994 (16 clones) were sequenced. The mean intrasample genetic distance almost doubled from 1984 to 1994 (from 0.0035 in 1984 to 0.0065 in 1994) but was low compared to the intersample genetic distances. One variant cloned from the 1994 sample had a deletion of approximately 100 bp that included the V1 region; thus, defective interfering particles may influence viral evolution during persistence, although their detection was not a goal of this study.
Phylogenetic analysis of the cloned ORF 5 sequences is shown in Fig. 6. Most ORF 5 clones are distinct, indicating that EAV exists as a mixture of related genomes; together with the data in Fig. 1, this finding suggests that different variants of this quasispecies emerge to become the majority variant during establishment and maintenance of persistent infection of the reproductive tract of the carrier stallion. Figure 6 shows that clones derived from the 1984 sample are less genetically diverse than those from the semen of the same stallion 10 years later. Phylogenetic analysis of the sequences of the various clones clearly shows that the heterogeneity of the quasispecies increased during the course of long-term persistent EAV infection of this stallion. We conclude that significant genetic and phenotypic evolution of the EAV quasispecies occurs in the reproductive tract of the Thoroughbred carrier stallion.
FIG. 6.
Phylogenetic tree showing the relationships between ORF 5 clones amplified from semen collected from stallion D in 1984 and 1994. The tree is rooted and scaled as for Fig. 1.
DISCUSSION
Genetic analyses of sequences derived from viral RNA in semen indicate that the EAV quasispecies evolves considerably during long-term persistent infection of the carrier stallion. The immune response exerts a major selective force during persistent infection with a variety of RNA viruses (25, 39, 42, 53). Our data also imply that selective pressures act on EAV during persistence in stallions, as illustrated by high rates of nonsynonymous changes in specific regions of the GP3 and GL proteins. The precise cell population that harbors EAV during persistent infection is not well defined, but testosterone is clearly essential for maintenance of persistence (41, 44). Androgens exert on reproductive tissues a variety of effects that could facilitate EAV persistence, such as maintenance of the susceptible host cell population(s) or immune suppression (46, 54). Other studies have implied that there is ongoing interaction between the host immune system and the EAV population that persists in the reproductive tract of the carrier stallion (30, 34, 49). Amino acid changes in the GL protein that occur during persistent infection of stallions are largely restricted to the critical V1 neutralization region; thus, neutralizing antibodies likely exert a selective pressure during EAV persistence, although other mechanisms are not excluded (5, 14, 57). The GP3 protein is also apparently subject to very strong selective pressure during establishment and maintenance of persistent EAV infection of Thoroughbred stallions. Preliminary studies using the EAV infectious cDNA clone (65) indicate that ORF 3 is essential for the production of infectious progeny virus in culture, but the structural role of the GP3 protein is uncertain (56). The equivalent protein in other arteriviruses is highly variable and antigenic, and antibodies directed toward it are apparently protective during infection with porcine reproductive and respiratory syndrome virus (17, 21, 36, 51). Further characterization of the specific virus protein domains that are subject to selection during persistent infection will be crucial for definition of the mechanisms of EAV persistence in the carrier stallion. Selective pressures might also differ during persistent EAV infection of other equine breeds (3).
We describe alteration in the neutralization phenotype of variants that emerged during persistent EAV infection of two Thoroughbred stallions. A previous study that described the test breeding of carrier stallions D and E, as well as seven others originally infected during the Kentucky 1984 outbreak, clearly demonstrated that clinical EVA disease was more common in susceptible mares bred to certain carrier stallions than to others in this group (63). Thus, genetic variation of EAV during persistent infection of stallions results not only in variants with altered neutralization phenotype but also in emergence of variants with distinct virulence characteristics.
When Eigen first introduced the concept of viral quasispecies, he emphasized that selection is applied to the entire heterogeneous population, not to individual variants (19). During EVA epizootics and early in persistence, the EAV quasispecies is relatively limited (2, 3), whereas later in persistent infection of the reproductive tract of the carrier stallion the quasispecies expands to better fill the sequence space. The Red Queen hypothesis proposes that all virus variants would also gain fitness as they compete for limited resources (15). The greater diversity of EAV variants generated during the course of persistent infection would result in a virus population that is better able to adapt to selective pressures inevitably encountered during venereal transfer of this large population to the recipient mare.
The high mutation frequency of RNA viruses generates a diverse quasispecies that facilitates their adaptability and survival. This mutability, however, also has inherent disadvantages such as the generation of variants with deleterious mutations (15). The transfer of a very small subset of a quasispecies (a genetic bottleneck) can isolate a less fit variant (11), and there is little opportunity for the bottlenecked variant to recover fitness when this occurs in RNA virus populations that do not recombine (15). The subsequent decrease in fitness is described in the process known as Muller’s ratchet, which has been demonstrated in vitro for three RNA viruses (9, 11, 18, 20). Bottleneck passages of viruses in culture result in reduced fitness that is recoverable with successive passage of large virus populations (11, 18). Transmission of aerosol droplets that contain few virions and adaptation to a new host or cell population are the principal means for in vivo occurrence of such a genetic bottleneck of RNA viruses (16, 26).
EAV is naturally transmitted either by aerosol droplet or infective semen (27, 61). The evolution and heterogeneity of the EAV quasispecies that occurs during persistent infection of the carrier stallion contrasts with its marked genetic stability during EVA epizootics involving horizontal aerosol spread of the virus (2, 3). The genetic stability of the virus population during EVA outbreaks results in a less diverse and therefore potentially less adaptable quasispecies (3). Indeed, widespread EVA epizootics are uncommon, as many field strains of EAV result only in seroconversion and subclinical infection in susceptible horses (35, 50, 61). Selective pressures exerted during the course of persistent infection of the reproductive tract of the carrier stallion clearly can be responsible for genotypic divergence and emergence of phenotypically novel EAV variants and likely compensate for the relatively limited virus population diversity that occurs during EVA outbreaks.
ACKNOWLEDGMENTS
We thank John Patton, Jenni Boonjakuaku, and Dave Pettigrew for valuable PCR and sequencing assistance and Dustin Lee for computer support.
This study was supported by the Grayson-Jockey Club Research Foundation; the Center for Equine Health at the University of California-Davis with funds provided by the Oak Tree Racing Association, the State of California Satellite Wagering Fund, and contributions by private donors; and USDA National Research Initiative Competitive Grant 97-35204-4736.
REFERENCES
- 1.Archambault D, Laganiere G, Carman S, St-Laurent G. Comparison of nucleic and amino acid sequences and phylogenetic analysis of open reading frames 3 and 4 of various equine arteritis virus isolates. Vet Res. 1997;28:505–516. [PubMed] [Google Scholar]
- 2.Balasuriya U B R, Evermann J F, Hedges J F, McKeirnan A J, Mitten J Q, Beyer J C, McCollum W H, Timoney P J, MacLachlan N J. Serologic and molecular characterization of an abortigenic strain of equine arteritis virus derived from infective frozen semen and an aborted equine fetus. J Am Vet Med Assoc. 1998;213:1586–1589. [PubMed] [Google Scholar]
- 3.Balasuriya, U. B. R., J. F. Hedges, P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. Genetic stability of equine arteritis virus during horizontal and vertical transmission in an outbreak of equine viral arteritis. Submitted for publication. [DOI] [PubMed]
- 4.Balasuriya U B R, MacLachlan N J, de Vries A A F, Rossitto P V, Rottier P J M. Identification of a neutralization site in the major envelope glycoprotein (GL) of equine arteritis virus. Virology. 1995;207:518–527. doi: 10.1006/viro.1995.1112. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya U B R, Patton J F, Rossitto P V, Timoney P J, McCollum W H, MacLachlan N J. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the GL envelope glycoprotein. Virology. 1997;232:114–128. doi: 10.1006/viro.1997.8551. [DOI] [PubMed] [Google Scholar]
- 6.Balasuriya U B R, Rossitto P V, DeMaula C D, MacLachlan N J. A 29K envelope glycoprotein of equine arteritis virus expresses neutralization determinants recognized by murine monoclonal antibodies. J Gen Virol. 1993;74:2525–2529. doi: 10.1099/0022-1317-74-11-2525. [DOI] [PubMed] [Google Scholar]
- 7.Balasuriya U B R, Timoney P J, McCollum W H, MacLachlan N J. Phylogenetic analysis of open reading frame 5 of field isolates of equine arteritis virus and identification of conserved and nonconserved regions in the GL envelope glycoprotein. Virology. 1995;214:690–697. doi: 10.1006/viro.1995.0087. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 9.Chao L. Fitness of RNA virus decreased by Muller’s ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 10.Chirnside E D, Wearing C M, Binns M M, Mumford J A. Comparison of M and N gene sequences distinguishes variation amongst equine arteritis virus isolates. J Gen Virol. 1994;75:1491–1497. doi: 10.1099/0022-1317-75-6-1491. [DOI] [PubMed] [Google Scholar]
- 11.Clarke D K, Duarte E A, Moya A, Elena S F, Domingo E, Holland J J. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo E, Diez J, Martinez M A, Hernandez J, Holguin A, Borrego B, Mateu M G. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J Gen Virol. 1993;74:2039–2045. doi: 10.1099/0022-1317-74-10-2039. . (Erratum, 75:949, 1994.) [DOI] [PubMed] [Google Scholar]
- 15.Domingo E, Escarmis C, Sevilla N, Moya A, Elena S F, Quer J, Novella I S, Holland J J. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 16.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 17.Drew T W, Lowings J P, Yapp F. Variation in open reading frames 3, 4 and 7 among porcine reproductive and respiratory syndrome virus isolates in the UK. Vet Microbiol. 1997;55:209–221. doi: 10.1016/s0378-1135(96)01328-4. [DOI] [PubMed] [Google Scholar]
- 18.Duarte E A, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. Many-trillionfold amplification of single RNA virus particles fails to overcome the Muller’s ratchet effect. J Virol. 1993;67:3620–3623. doi: 10.1128/jvi.67.6.3620-3623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 20.Escarmis C, Davila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 21.Faaberg K S, Plagemann P G W. ORF3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology. 1997;227:245–251. doi: 10.1006/viro.1996.8310. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. PHYLIP (Phylogeny Inference Package) 3.5c manual. Seattle, Wash: University of Washington; 1993. [Google Scholar]
- 24.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–384. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 25.Gebauer F, de la Torre J C, Gomes I, Mateu M G, Barahona H, Tiraboschi B, Bergmann I, Auge de Mello P, Domingo E. Rapid selection of genetic and antigenic variants of foot-and-mouth disease virus during persistence in cattle. J Virol. 1988;62:2041–2049. doi: 10.1128/jvi.62.6.2041-2049.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerone P J, Couch R B, Keefer G V, Douglas R G, Derrenbacher E B, Knight V. Assessment of experimental and natural viral aerosols. Bacteriol Rev. 1966;30:576–584. doi: 10.1128/br.30.3.576-588.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaser A L, Chirnside E D, Horzinek M C, de Vries A A F. Equine arteritis virus. Theriogenology. 1997;47:1275–1295. doi: 10.1016/S0093-691X(97)00107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaser A L, Rottier P J M, Horzinek M C, Colenbrander B. Equine arteritis virus: a review of clinical features and management aspects. Vet Q. 1996;18:95–99. doi: 10.1080/01652176.1996.9694625. [DOI] [PubMed] [Google Scholar]
- 29.Godeny E K, Chen L, Kumar S N, Methven S L, Koonin E V, Brinton M A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedges J F, Balasuriya U B R, Ahmad S, Timoney P J, McCollum W H, Yilma T D, MacLachlan N J. Detection of antibodies to equine arteritis virus by enzyme linked immunosorbant assays utilizing GL, M and N proteins expressed from recombinant baculoviruses. J Virol Methods. 1998;76:127–137. doi: 10.1016/s0166-0934(98)00131-1. [DOI] [PubMed] [Google Scholar]
- 31.Hedges J F, Balasuriya U B R, Timoney P J, McCollum W H, MacLachlan N J. Genetic variation in open reading frame 2 of field isolates and laboratory strains of equine arteritis virus. Virus Res. 1996;42:41–52. doi: 10.1016/0168-1702(96)01294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 33.Holland J J, de la Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 34.Holyoak G R, Giles R C, McCollum W H, Little T V, Timoney P J. Pathological changes associated with equine arteritis virus infection of the reproductive tract in prepubertal and peripubertal colts. J Comp Pathol. 1993;109:281–293. doi: 10.1016/s0021-9975(08)80253-8. [DOI] [PubMed] [Google Scholar]
- 35.Hullinger P J, Wilson W D, Rossitto P V, Patton J F, Thurmond M C, MacLachlan N J. Passive transfer, decay and protein specificity of equine arteritis virus antibodies in an endemically-infected standardbred herd. J Am Vet Med Assoc. 1998;213:839–842. [PubMed] [Google Scholar]
- 36.Katz J B, Shafer A L, Eernisse K A, Landgraf J G, Nelson E A. Antigenic differences between European and American isolates of porcine reproductive and respiratory syndrome virus (PRRSV) are encoded by the carboxy terminal portion of viral open reading frame 3. Vet Microbiol. 1995;44:65–76. doi: 10.1016/0378-1135(94)00113-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 39.Leroux C, Issel C J, Montelaro R C. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W H. Molecular evolution. Sunderland, Mass: Sinauer Associates Inc.; 1997. [Google Scholar]
- 41.Little T V, Holyoak G R, McCollum W H, Timoney P J. Output of equine arteritis virus from persistently infected stallions is testosterone dependent. In: Plowright W, Rossdale P D, Wade J F, editors. Proceedings of the 6th International Conference on Equine Infectious Diseases, Cambridge, 1991. Newmarket, England: R & W Publications; 1992. pp. 225–229. [Google Scholar]
- 42.Manzin A, Solforosi L, Petrelli E, Macarri G, Tosone G, Piazza M, Clementi M. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J Virol. 1998;72:6271–6276. doi: 10.1128/jvi.72.7.6271-6276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mardassi H, Gonin P, Gagnon C A, Massie B, Dea S. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J Virol. 1998;72:6298–6306. doi: 10.1128/jvi.72.8.6298-6306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCollum W H, Little T V, Timoney P J, Swerczek T W. Resistance of castrated male horses to attempted establishment of the carrier state with equine arteritis virus. J Comp Pathol. 1994;111:383–388. doi: 10.1016/s0021-9975(05)80096-9. [DOI] [PubMed] [Google Scholar]
- 45.McCollum W H, Timoney P J. Proceedings of the Grayson Foundation International Conference of Thoroughbred Breeders Organizations, Ireland 1984. 1984. The pathogenic qualities of the 1984 strain of equine arteritis virus; pp. 34–84. [Google Scholar]
- 46.McDonald L E, Pineda N H. Veterinary endocrinology and reproduction. 1989. p. 5. , 272–280. Lea & Febiger, Philadelphia, Pa. [Google Scholar]
- 47.Murphy T W, McCollum W H, Timoney P J. Variation in the genomic RNA of equine arteritis virus during long term persistence in the stallion. In: Plowright W, Rossdale P D, Wade J F, editors. Proceedings of the 6th International Conference on Equine Infectious Diseases, Cambridge 1991. Newmarket, England: R & W Publications; 1992. p. 327. [Google Scholar]
- 48.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and non-synonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 49.Neu S M, Timoney P J, McCollum W H. Persistent infection of the reproductive tract in stallions persistently infected with equine arteritis virus. In: Powell D G, editor. Proceedings of the 5th International Conference of Equine Infectious Diseases, Lexington 1987. Lexington, Ky: The University Press of Kentucky; 1988. pp. 149–154. [Google Scholar]
- 50.Patton, J. F., U. B. R. Balasuriya, J. F. Hedges, T. M. Schweidler, P. J. Hullinger, and N. J. MacLachlan. Phylogenetic characterization of a highly attenuated strain of equine arteritis virus from the semen of a persistently infected standardbred stallion. Arch. Virol., in press. [DOI] [PubMed]
- 51.Plana-Duran J, Climent I, Sarraseca A, Urniza E, Cortes E, Vela C, Casal J I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olo/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes. 1998;14:19–29. doi: 10.1023/a:1007931322271. [DOI] [PubMed] [Google Scholar]
- 52.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 53.Sevilla N, Domingo E. Evolution of a persistent aphthovirus in cytolytic infections: partial reversion of phenotypic traits accompanied by genetic diversification. J Virol. 1996;70:6617–6624. doi: 10.1128/jvi.70.10.6617-6624.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shivaji S, Scheit K H, Bhargava P M. Proteins of seminal plasma and secretions of the male reproductive tract. New York, N.Y: John Wiley & Sons; 1990. Immunosupressive factors in seminal plasma; pp. 375–389. [Google Scholar]
- 55.Smith D B, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 56.Snijder, E. J., and H. van Tol. 1998. Personal communication.
- 57.Spindler K, Horodyski F, Grabau E, Nichol S, Vandepol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timoney P J. Proceedings of the Grayson Foundation International Conference of Thoroughbred Breeders Organizations, Ireland 1984. 1984. Clinical, virological, and epidemiological features of the 1984 outbreak of equine viral arteritis in the Thoroughbred population in Kentucky, USA; pp. 24–33. [Google Scholar]
- 60.Timoney P J, McCollum W H. Equine viral arteritis: epidemiology and control. J Equine Vet Sci. 1988;8:54–59. [Google Scholar]
- 61.Timoney P J, McCollum W H. Equine viral arteritis. Vet Clin North Am Equine Pract. 1993;9:295–309. doi: 10.1016/S0749-0739(17)30397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timoney P J, McCollum W H, Murphy T W, Roberts A W, Willard J G, Carswell G D. The carrier state in equine arteritis virus infection in the stallion with specific emphasis on the venereal mode of virus transmission. J Reprod Fertil Suppl. 1987;35:95–102. [PubMed] [Google Scholar]
- 63.Timoney P J, McCollum W H, Roberts A W, Murphy T W. Demonstration of the carrier state in naturally acquired equine arteritis virus infection in the stallion. Res Vet Sci. 1986;41:279–280. [PubMed] [Google Scholar]
- 64.van Berlo M F, Rottier P J M, Horzinek M C, van der Zeijst B A M. Intracellular equine arteritis virus (EAV)-specific RNAs contain common sequences. Virology. 1986;152:492–496. doi: 10.1016/0042-6822(86)90154-6. [DOI] [PubMed] [Google Scholar]
- 65.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]