PURPOSE
Myelodysplastic syndromes (MDS) are heterogeneous myeloid neoplasms in which a risk-adapted treatment strategy is needed. Recently, a new clinical-molecular prognostic model, the Molecular International Prognostic Scoring System (IPSS-M) was proposed to improve the prediction of clinical outcome of the currently available tool (Revised International Prognostic Scoring System [IPSS-R]). We aimed to provide an extensive validation of IPSS-M.
METHODS
A total of 2,876 patients with primary MDS from the GenoMed4All consortium were retrospectively analyzed.
RESULTS
IPSS-M improved prognostic discrimination across all clinical end points with respect to IPSS-R (concordance was 0.81 v 0.74 for overall survival and 0.89 v 0.76 for leukemia-free survival, respectively). This was true even in those patients without detectable gene mutations. Compared with the IPSS-R based stratification, the IPSS-M risk group changed in 46% of patients (23.6% and 22.4% of subjects were upstaged and downstaged, respectively).
In patients treated with hematopoietic stem cell transplantation (HSCT), IPSS-M significantly improved the prediction of the risk of disease relapse and the probability of post-transplantation survival versus IPSS-R (concordance was 0.76 v 0.60 for overall survival and 0.89 v 0.70 for probability of relapse, respectively). In high-risk patients treated with hypomethylating agents (HMA), IPSS-M failed to stratify individual probability of response; response duration and probability of survival were inversely related to IPSS-M risk.
Finally, we tested the accuracy in predicting IPSS-M when molecular information was missed and we defined a minimum set of 15 relevant genes associated with high performance of the score.
CONCLUSION
IPSS-M improves MDS prognostication and might result in a more effective selection of candidates to HSCT. Additional factors other than gene mutations can be involved in determining HMA sensitivity. The definition of a minimum set of relevant genes may facilitate the clinical implementation of the score.
INTRODUCTION
Myelodysplastic syndromes (MDS) are heterogeneous neoplasms ranging from indolent conditions to cases rapidly progressing into acute myeloid leukemia and therefore a risk-adapted treatment strategy is needed.1 Disease-related risk is currently assessed by the Revised International Prognostic Scoring System (IPSS-R), on the basis of bone marrow blasts, blood cytopenias, and cytogenetic abnormalities.2 Although IPSS-R is an excellent tool for clinical decision making, this scoring system has its weaknesses and may fail to capture reliable prognostic information at individual patient level.3,4
CONTEXT
Key Objective
Are gene mutations relevant to improve the prediction of clinical outcomes in patients with myelodysplastic syndromes and to define the probability of response to currently available treatments?
Knowledge Generated
Molecular International Prognostic Scoring System (IPSS-M, including both clinical and genomic features) improves prognostic discrimination across all clinical end points compared with the currently available scores. In patients treated with allogeneic stem-cell transplantation, IPSS-M significantly improves the prediction of the risk of disease relapse and the probability of post-transplantation survival. In patients treated with hypomethylating agents, IPSS-M fails to stratify individual probability of response.
Relevance (C.F. Craddock)
The IPSS-M represents an important new prognostic model in patients with myelodysplastic syndromes and informs the rational selection of treatment strategies including stem-cell transplantation.*
*Relevance section written by JCO Associate Editor Charles F. Craddock, MD.
In MDS, conventional prognostic tools on the basis of clinical and hematologic features are being complemented by introducing somatic gene mutations that were shown to be valuable prognostic markers.4-8 Recently, the International Working Group for Prognosis in MDS (IWG-PM) proposed a clinical-molecular prognostic model (Molecular IPSS [IPSS-M]) that was developed using hematologic parameters, cytogenetic abnormalities, and mutations of 31 MDS-related genes.9 IPSS-M improved prognostic discrimination across all clinical end points compared with IPSS-R.
In this study, we aimed to address the issue of clinical implementability of IPSS-M by (i) providing an extensive validation of its prognostic value (also focusing on patients without detectable mutations); (ii) investigating the predictive and prognostic power of IPSS-M in patients receiving disease-modifying treatment (hypomethylating agents [HMA] and hematopoietic stem cell transplantation [HSCT]); and (iii) testing the accuracy in predicting IPSS-M when molecular information was missed to define a minimum set of relevant genes associated with high performance of the score.
Study Populations and Procedures
The study was conducted by GenoMed4All consortium10 and supported by EuroBloodNET, the European Reference Network on rare hematologic diseases.11 The Humanitas Ethics Committee approved the study. Written informed consent was obtained from each participant. This study was registered at ClinicalTrials.gov (ClinicalTrials.gov identifier: NCT04889729).
Inclusion criteria were age ≥ 18 years, a diagnosis of primary MDS according to WHO 2016 criteria,12 and available information on IPSS-M related variables collected at diagnosis and before starting disease-modifying treatments (if any). Patients affected with therapy-related myeloid neoplasms or incomplete information on IPSS-M variables were excluded. A total of 2,876 patients matched study criteria (Data Supplement, online only).
Karyotypes were classified using the International System for Cytogenetic Nomenclature Criteria. Mutation screening of MDS-related genes was performed on DNA bone marrow mononuclear cells or peripheral blood granulocytes (Data Supplement).
Patients were reclassified according to WHO 2022 and International Consensus Classification of Myeloid Neoplasms criteria.13,14 IPSS-M score was calculated according to the original publication.9
Statistical Analysis
Survival curves were estimated with the Kaplan-Meier method and differences among groups were evaluated by log-rank test. Overall survival (OS) and leukemia-free survival (LFS) were defined as the time between diagnosis and death (from any cause) or last follow-up (for censored observations) and the time between diagnosis and acute myeloid leukemia evolution (if any) or last follow-up (for censored observations), respectively. When focusing on patient populations receiving a specific treatment, OS was calculated as the time between start of treatment and death/last follow-up. The probability of relapse after treatment was estimated according to standardized criteria.15 For patients treated with HSCT, when estimating nonrelapse mortality (NRM), any death in the absence of disease relapse was considered an event. The cumulative incidence of relapse and NRM was estimated by competing risk approach.16
Multivariable survival analyses were performed by Cox's proportional hazards regression models (IPSS-M was incorporated as ordinal variable in the models). The discriminatory power of the models and the relative goodness of fit for the predictive score were evaluated using Harrell's concordance index.17 To compare different statistical models, we used in addition the Akaike18 information criterion (AIC), which allows the evaluation of a model by combining goodness of fit and complexity, with a lower AIC indicating a better trade-off between fit and complexity.
The impact of single IPSS-M factors on the prediction of clinical outcomes was evaluated by fitting a random-effects Cox's model.19,20 The percentage of variation of the logarithmic hazard explained by each set of variables was estimated (Data Supplement).
The accuracy of IPSS-M in predicting the probability of survival in the presence of missing molecular data was calculated as the number of correctly classified patients divided by the size of patient's cohort. The accuracy loss was calculated as the fraction of wrongly classified patients divided by the population size.
RESULTS
Clinical Characteristics of Patients and Gene Mutations
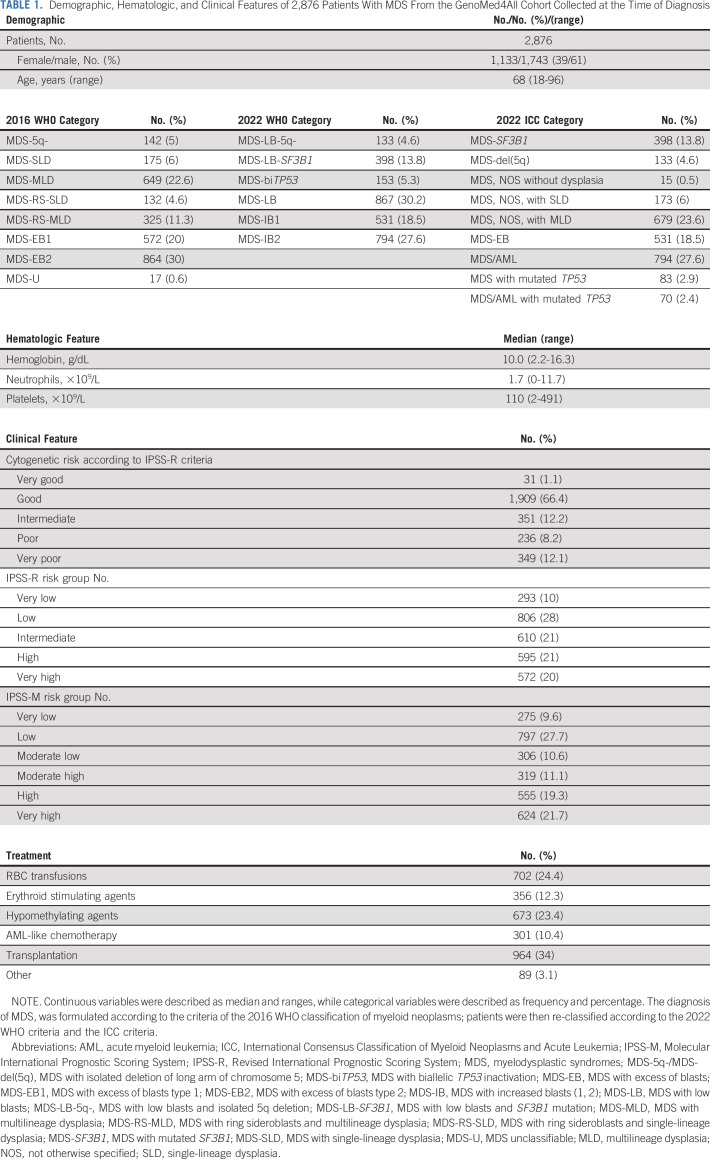
Clinical features at diagnosis of the 2,876 patients with MDS enrolled in the study are reported in Table 1. Study participants included 1,743 men (61%) and 1,133 women (39%). Date range of diagnosis was from 1999 to 2018. Median age at diagnosis was 68 years (range, 18-96 years). Follow-up was updated on December, 2020. Median duration of follow-up was 37.5 months (95% CI, 36.2 to 38.8 months).
TABLE 1.
Demographic, Hematologic, and Clinical Features of 2,876 Patients With MDS From the GenoMed4All Cohort Collected at the Time of Diagnosis
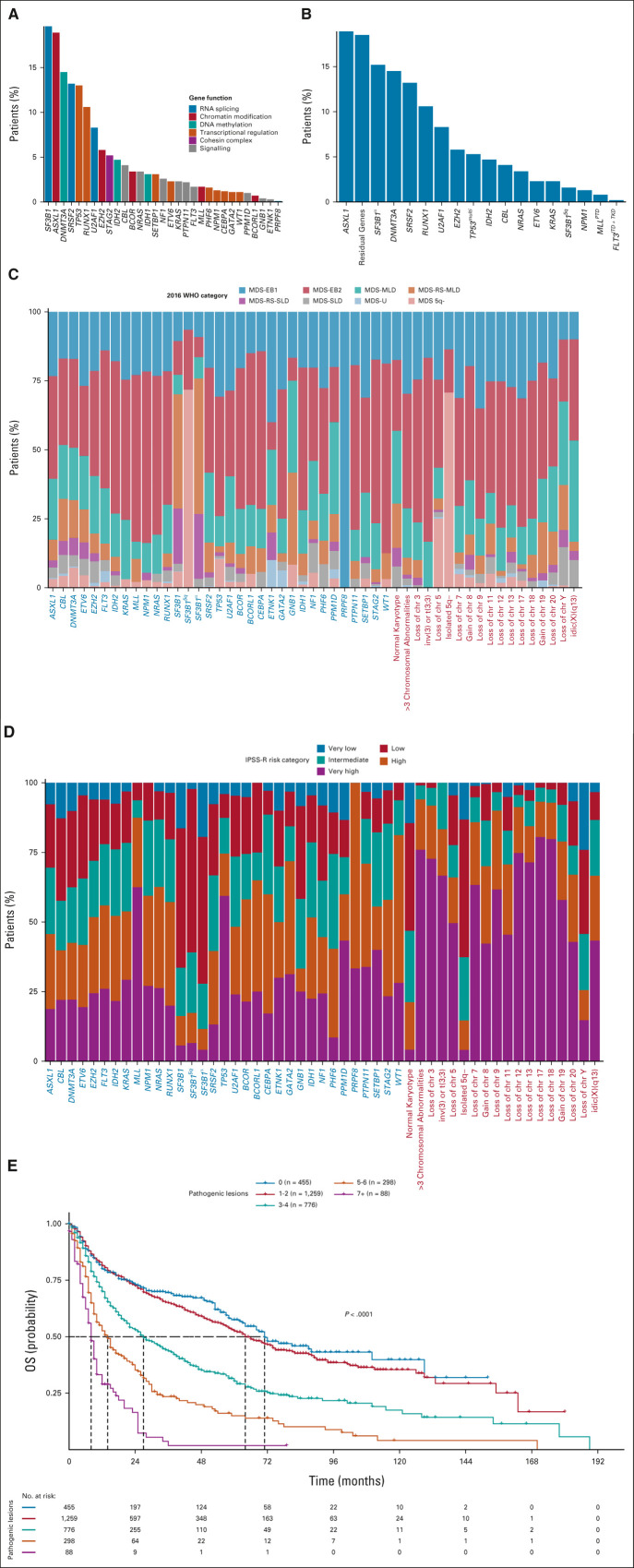
Considering IPSS-M–related genomic features, we identified 6,749 genomic lesions at diagnosis (median, 3; range, 0-12). 2,421 patients (84.1%) presented one or more genomic alterations (mutations and/or chromosomal abnormalities). 2,369 patients (82.4%) had one or more somatic mutations on 31 IPSS-M–related genes, whereas 1,297 showed abnormal karyotype (45%; Fig 1; Data Supplement). Probability of OS and LFS according to the mutational status of IPSS-M–related genes was reported in the Data Supplement.
FIG 1.
Molecular landscape of patients with MDS from the GenoMed4all cohort. (A) Frequency of mutations of the 31 genes included into IPSS-M score in 2,876 patients from the GenoMed4All cohort. Colors linked to the bars represent the gene function. (B) Frequency of mutations on genomic features grouped according to IPSS-M criteria. (C and D) Frequency of gene mutations and chromosomal abnormalities broken down by MDS subtypes according to 2016 WHO criteria and IPSS-R risk category, respectively. Mutations on genes are grouped according to IPSS-M criteria as main effect genes (gene labels are highlighted in blue) and residual genes (gene labels are highlighted in dark green). (E) Kaplan-Meier probability estimates of OS across numbers of oncogenic alterations per patient (gene mutations and cytogenetic abnormalities). P value is from log-rank test. Frequency of IPSS-M–related gene mutations and chromosomal abnormalities broken down by MDS subtypes according to 2022 WHO criteria and ICC criteria is available in the Data Supplement. ICC, International Consensus Classification of Myeloid Neoplasms and Acute Leukemia; MDS, myelodysplastic syndromes; MDS 5q-, MDS with isolated deletion of long arm of chromosome five; MDS-EB1, MDS with excess of blasts, type 1; MDS-EB2, MDS with excess of blasts, type 2; MDS-MLD, MDS with multilineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts and multilineage dysplasia; MDS-RS-SLD, MDS with ring sideroblasts and single-lineage dysplasia; MDS-SLD, MDS with single-lineage dysplasia; MDS-U, MDS unclassifiable; IPSS-R, Revised International Prognostic Scoring System; IPSS-M, Molecular International Prognostic Scoring System; OS, overall survival.
Validation of the Prognostic Power of IPSS-M and Comparison With IPSS-R
We calculated IPSS-M in the study cohort at diagnosis.9 Cytogenetic abnormalities were classified according the IPSS-R criteria.2 Gene mutations were considered as binary variables with the exception of TP53 (not mutated, monoallelic mutation, multihit mutations) and SF3B1 (SF3B15q [SF3B1 mutation in the presence of isolated del(5q) only or with one additional aberration excluding -7/del(7q)], and SF3B1α [SF3B1 mutation without comutations in BCOR, BCORL1, RUNX1, NRAS, STAG2, SRSF2, and del(5q)]9).
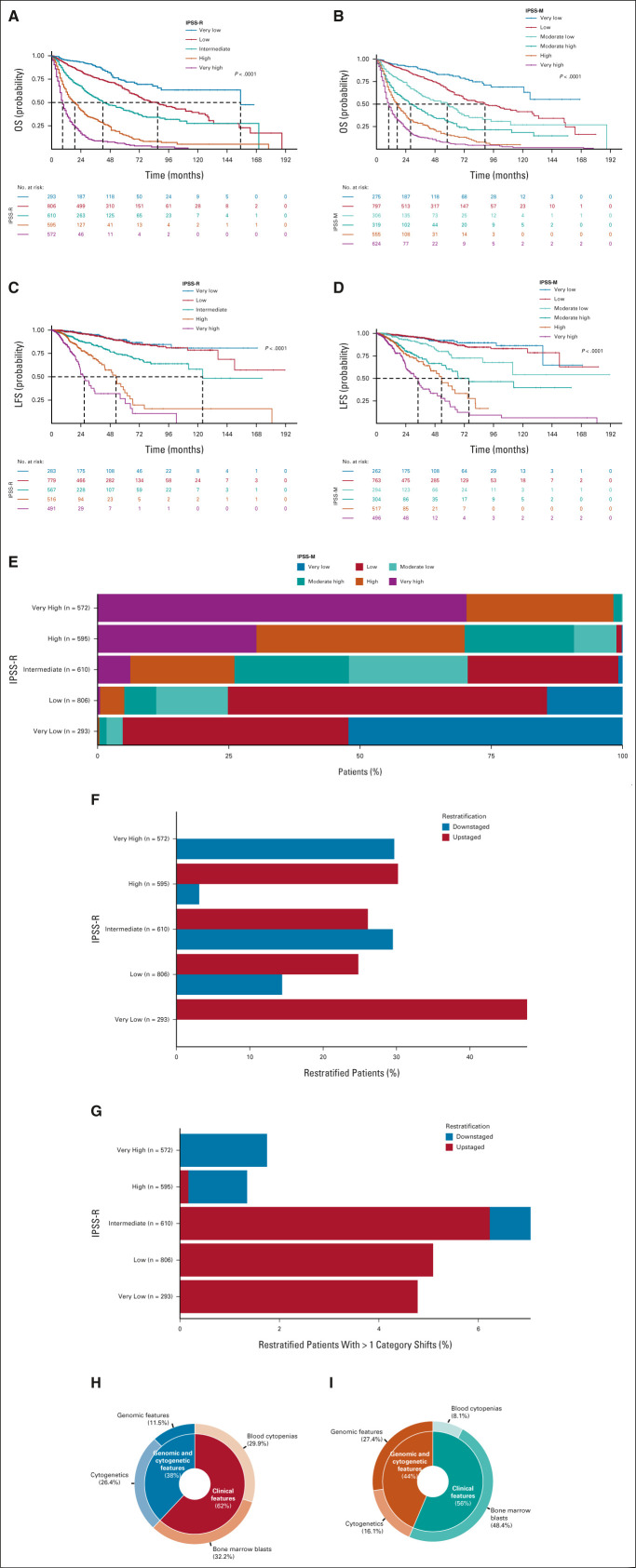
Accordingly, 9.6% of patients (n = 275) were classified as very low risk, 27.7% (n = 797) as low risk, 10.6% (n = 306) as moderate low risk, 11.1% (n = 319) as moderate high, 19.3% (n = 555) as high risk, and 21.7% (n = 624) as very high risk.
We analyzed the probability of OS and LFS for all IPSS-M categories. Patients who received HSCT were censored at the time of the procedure. IPSS-M categories showed significantly different probabilities of both OS and LFS (both P < .001; Fig 2). The independent effect of IPSS-M on clinical outcome was maintained in a multivariable model including age and sex as covariates (HR, 1.67; 95% CI, 1.61 to 1.73; P < .001 for OS; and HR, 1.79; 95% CI, 1.73 to 1.86; P < .001 for LFS).
FIG 2.
Clinical assessment of IPSS-R and IPSS-M in the GenoMed4All MDS cohort. Kaplan-Meier probability estimates of (A and B) OS and (C and D) LFS for 2,876 patients with MDS from the GenoMed4All cohort stratified by IPSS-R and IPSS-M risk categories, respectively. P values are from log-rank test. (E) Restratification of IPSS-R to IPSS-M risk groups in the MDS cohort. Each bar represents an IPSS-R category and shows the percentage of patients that is restratified in the IPSS-M categories (indicated with different colors). (F) Distribution of the restratified patients in each IPSS-R category, counting the proportion of patients who are downstaged (highlighted in blue) and upstaged (highlighted in red) with the IPSS-M classification. (G) Distribution of the restratified patients in each IPSS-R category, counting the proportion of patients with more than one shift in IPSS-M risk category (downstaged and upstaged cases are highlighted in blue and in red, respectively). (H-I) Fraction of explained variation related to the IPSS-M prognostic factors for OS and LFS, respectively. IPSS-R, Revised International Prognostic Scoring System; IPSS-M, Molecular International Prognostic Scoring System; LFS, leukemia-free survival; MDS, myelodysplastic syndromes; OS, overall survival.
IPSS-M showed superior performance with respect to conventional IPSS-R scoring system: concordance was 0.81 (95% CI, 0.79 to 0.82) versus 0.74 (95% CI, 0.73 to 0.76) for OS and 0.89 (95% CI, 0.87 to 0.91) versus 0.76 (95% CI, 0.73 to 0.79) for LFS, respectively. In addition, to evaluate the effect of IPSS-M versus IPSS-R, we fitted two separate multivariable Cox's models including age and sex as covariates, comparing them by the AIC. AIC for the model with IPSS-M versus IPSS-R was 17,455.43 versus 17,469.33 for OS and 3,973.26 versus 4,011.64 for LFS, thus confirming the importance of accounting for gene mutations in the prognostic model.
The five-to-five comparison of IPSS-R and IPSS-M patients' distribution (in which we merged moderate low and moderate high to moderate in IPSS-M) resulted in the restratification of 46% of patients (1,324 of 2,876). Of these, 23.6% (n = 679) were upstaged and 22.4% (n = 645) were downstaged (Fig 2). A total of 115 patients (4%) were reclassified by more than one risk strata. Highlighting the implications of this restratification, marked differences in survival were observed between IPSS-M categories within each IPSS-R risk category; by contrast, the IPSS-R did not stratify patient outcomes within IPSS-M risk strata (Data Supplement).
We specifically studied the prognostic impact of gene mutations on main effect IPSS-M genes that were associated with adverse prognosis9 and their contribution on patients restratification from IPSS-R to IPSS-M risk categories (Data Supplement). Among restratified patients, 193 (26%) had one mutated adverse IPSS-M main effect gene, whereas 275 (37%) had two or more mutated genes. In details, in the very low + low IPSS-R category (n = 1,099), 214 patients (19.5%) were upstaged, of which 198 (93%) have more than one mutated IPSS-M genes. Considering patients classified in the intermediate IPSS-R category (n = 610), 180 (29%) were downstaged, the majority of them had no mutations (67%); by contrast, 159 subjects (26%) were upstaged, and 69% of these patients carried two or more main effect IPSS-M mutated genes. In the very high + high IPSS-R category (n = 1,167), instead, 189 patients (16%) were reclassified in lower-risk classes and only 33% of these presented more than one mutated gene. Thus, patient restratification was not a single gene effect, but the cumulative contribution of the prognostic mutations for each subject.
Then, we addressed the issue of the prognostic value of IPSS-M in those patients without detectable mutations in the 31 IPSS-M–related genes. 507 subjects entered the analysis. IPSS-M categories maintained a significant effect on probability of both OS and LFS (both P < .001; Data Supplement). IPSS-M maintained superior performance to conventional IPSS-R scoring system in this patient setting: concordance was 0.89 [0.86-0.91] versus 0.73 [0.69-0.77] for OS and 0.91 [0.90-0.92] versus 0.81 [0.75-0.87] for LFS, respectively. By comparing two multivariable models including IPSS-M versus IPSS-R, AIC was 1,573.04 versus 1,590.11 for OS, respectively, and 491.91 versus 498.61 for LFS, respectively, thus confirming the best prognostic performance of IPSS-M in this population.
Finally, we evaluated the prognostic impact of IPSS-M–related variables in terms of percentage of explained variation for clinical outcomes (OS and LFS; Fig 2). Clinical features had a high predictive power for both OS (bone marrow blasts and cytopenias) and LFS (bone marrow blasts). IPSS-M–related genomic variables had a strong predictive power, that is increased for the LFS outcome, highlighting the impact of genomic landscape on the prediction of the risk of disease evolution.
Predictive and Prognostic Effect of IPSS-M in Patients Receiving Specific Treatments
In MDS, an increasing proportion of patients undergo to disease-modifying therapies, including HSCT and HMA (for high-risk subjects who are not eligible to HSCT). Therefore, it is relevant to know if IPSS-M may provide information on the probability of response to specific treatments (predictive value) and the probability of survival after treatment (prognostic value).
We therefore analyzed the predictive/prognostic value of IPSS-M in two populations treated with HSCT and HMA according to currently available guidelines on the basis of IPSS-R, age, performance stats, and donor avaialbility.1 To investigate the predictive value of IPSS-M, the risk of disease relapse in patients treated with HSCT and the overall response rate (including the achievement of complete response (CR), partial response, marrow CR, and stable disease with hematologic improvement according to 2006 IWG criteria)15 for patients treated with HMA were used as primary end points, while the prognostic value of IPSS-M was tested on the probability of OS since the start of treatment in both cases.
Nine hundred sixty-four patients receiving HSCT entered the analysis, in which clinical and genomic information for IPSS-M calculation was available at the time of transplant in patients who were transplanted upfront and before chemotherapy/HMA in those receiving treatment before transplantation (Data Supplement). Patients receiving HSCT were reclassified according to IPSS-M criteria: 126 (13.1%) patients were classified as low-risk, 108 (11.2%) patients as moderate low, 136 (14.1%) as moderate high, and 290 (30.1%) and 304 (31.5%) as high and very high risk, respectively. As illustrated in Figure 3, the 5-year OS probability was 61% in low-, 55% in moderate low-, 46% in moderate high-, 33% in high-, and 27% in very high-risk patients (P < .0001). In these risk groups, by competing risk analysis, the 5-year cumulative incidence of relapse was 14%, 14%, 15%, 20% and 29%, respectively (P < .001; Fig 3). A five-to-five mapping between the IPSS-R and IPSS-M categories resulted in the restratification of 45% (n = 433) of the patients. Of these, 21% (n = 204) were upstaged and 24% (n = 229) were downstaged (Fig 3).
FIG 3.
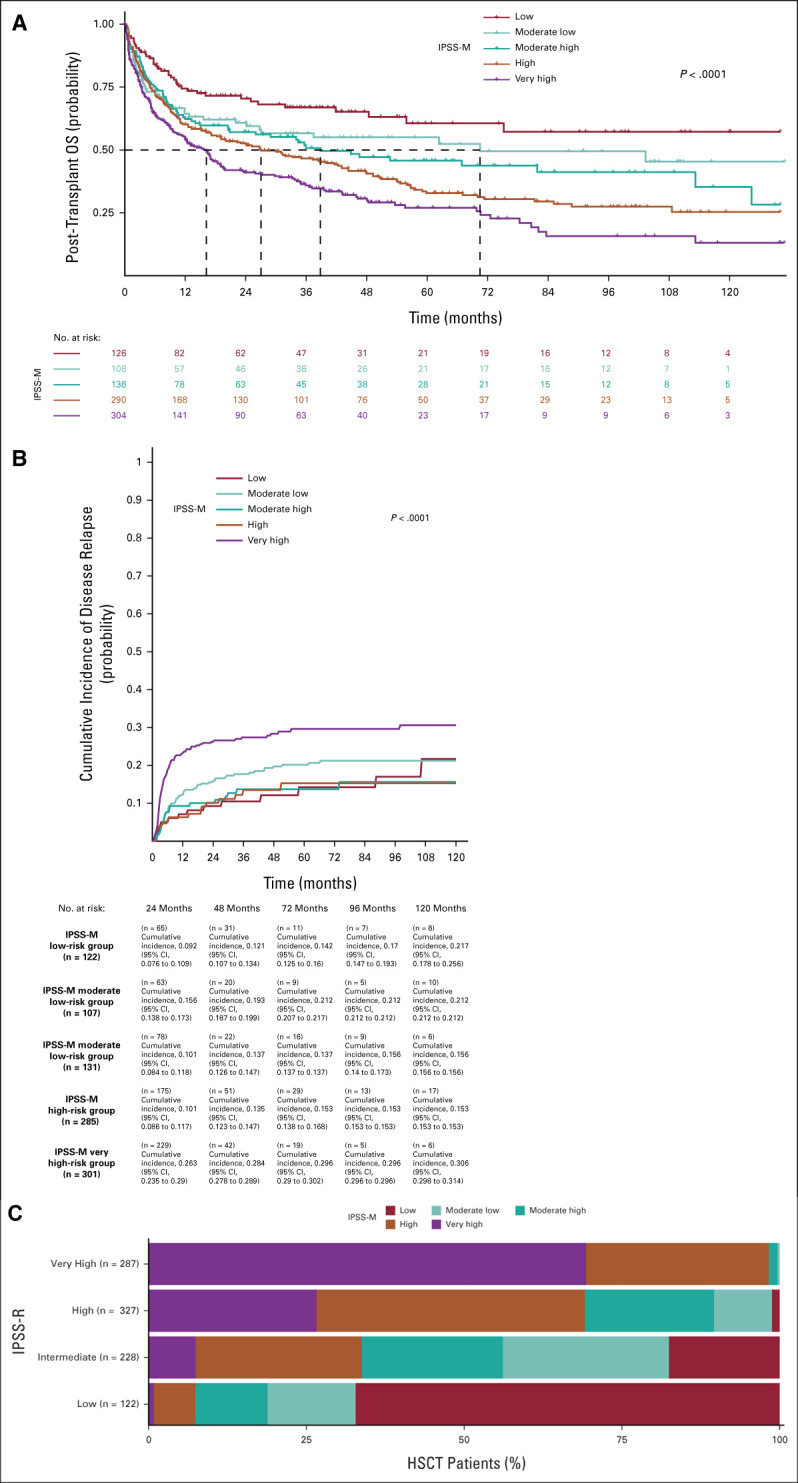
Clinical assessment of IPSS-R and IPSS-M in 964 MDS patients from the GenoMed4All cohort who received allogeneic stem-cell transplantation (HSCT). (A) Kaplan-Meier probability estimates of OS and (B) cumulative incidence of disease relapse (estimated with a competing risk approach including NRM) for 964 patients from the GenoMed4All cohort who received HSCT, stratified by IPSS-M risk categories. P values are from log-rank test. (C) Restratification of IPSS-R to IPSS-M risk groups in the MDS cohort. Each bar represents an IPSS-R category and shows the percentage of patients that is restratified in the IPSS-M categories (indicated with different colors). HSCT, hematopoietic stem-cell transplantation; IPSS-M, Molecular International Prognostic Scoring System; IPSS-R, Revised International Prognostic Scoring System; LFS, leukemia-free survival; MDS, myelodysplastic syndromes; NRM, nonrelapse mortality; OS, overall survival.
We analyzed the prognostic effect of the IPSS-M score by a multivariable model, including recipient age and sex, time from diagnosis to transplantation, source of hematopoietic stem cells, type of donor, disease status at transplant (active/progressive disease v complete remission), and conditioning regimen (reduced-intensity v standard conditioning). The IPSS-M score was significantly associated with OS (HR, 1.18 [95% CI, 1.08 to 1.27]; P < .001) and probability of relapse (HR, 1.38 [95% CI, 1.21 to 1.56]; P < .001)
IPSS-M showed superior performance to conventional IPSS-R in predicting both OS and probability of relapse after HSCT (concordance was 0.76 [95% CI, 0.73 to 0.78] v 0.60 [95% CI, 0.57 to 0.64] for OS, and 0.89 [95% CI, 0.87 to 0.91] v 0.70 [95% CI, 0.65 to 0.74] for probability of relapse, respectively). By comparing two multivariable models including IPSS-M versus IPSS-R, AIC was 6,545.87 versus 6,559.96 for OS, respectively, and 2,404.97 versus 2,416.78 for probability of relapse, respectively, thus confirming the best prognostic performance of IPSS-M in predicting post-transplantation outcomes.
Recipient age was a significant risk factor for OS and NRM (HR, 1.01 [95% CI, 1.00 to 1.02]; P = .028, and HR, 1.01 [95% CI, 1.01 to 1.02]; P < .001, respectively). Lack of complete remission after pretransplantation treatment (induction chemotherapy/HMA) showed an independent effect on relapse (HR, 1.78 [95% CI, 1.32 to 2.41]; P < .001). Patients receiving standard conditioning regimens showed a reduced probability of relapse (HR, 0.63 [95% CI, 0.49 to 0.82]; P < .001). With respect to donor-recipient HLA match, patients receiving transplant from mismatched unrelated donors showed a significantly reduced OS (HR, 1.2 [95% CI, 1.082 to 1.33]; P = .012) and a significantly increased NRM (HR, 1.33 [95% CI, 1.08 to 1.63]; P = .007) than those transplanted from a HLA-matched donor.
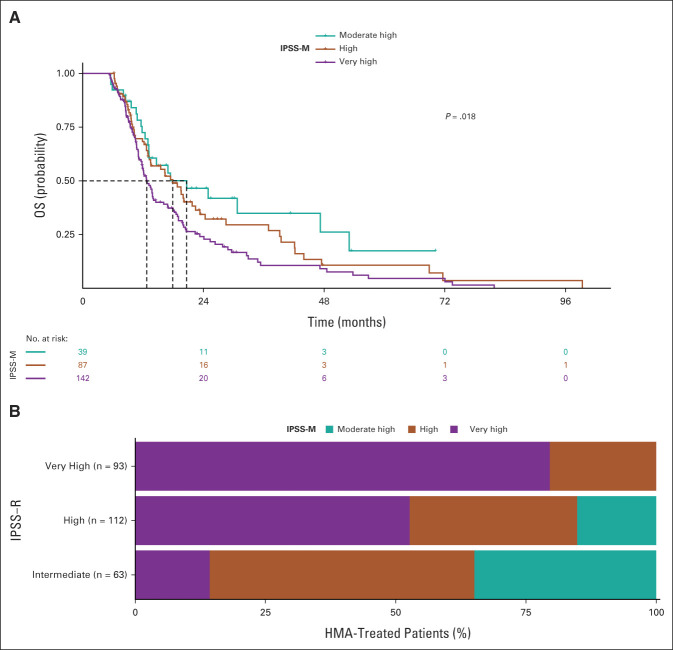
We then investigated the predictive/prognostic effect of IPSS-M in a cohort of high-risk patients with MDS ineligible for HSCT who received HMA. Inclusion criteria were bone marrow blasts ≥ 10% and availability of clinical and genomic information before starting treatment. 268 patients entered the analysis.
Patients were reclassified according to IPSS-M criteria: 39 patients (15%) had moderate high, 87 (32%) had high, and 142 (53%) had very high risk (Data Supplement). Median duration of MDS before the onset of HMA was 5 months (range, 1-11 months). Patients received HMA for a median of six cycles (range, 1-32 cycles) without significant difference among IPSS-M categories (P = .41). The probability of overall response (CR, marrow CR, partial response, and stable disease with hematologic improvement) evaluated after 4-6 cycles of treatment was 42%, without significant difference among IPSS-M categories (P = .19).
Median OS in the whole population treated by HMA was 13.9 months. As illustrated in Figure 4, the estimated median OS was 20.7 months in moderate high-, 17.9 months in high-, and 12.7 in very high-risk patients (P < .001; HR, 1.34 [95% CI, 1.08 to 1.65]; P = .006 in a multivariable model adjusted by age and sex).
FIG 4.
Clinical assessment of IPSS-R and IPSS-M in patients with MDS from the GenoMed4All cohort who received HMA. (A) Kaplan-Meier probability estimates of OS of patients with MDS from the GenoMed4All cohort who received HMA (n = 268) stratified by IPSS-M risk categories. P values are from log-rank test. (B) Restratification of IPSS-R to IPSS-M risk groups in the HMA-treated MDS patients. Each bar represents an IPSS-R category and shows the percentage of patients that is restratified in the IPSS-M categories (indicated with different colors). HMA, hypomethylating agents; IPSS-M, Molecular International Prognostic Scoring System; IPSS-R, Revised International Prognostic Scoring System; LFS, leukemia-free survival; MDS, myelodysplastic syndromes; OS, overall survival.
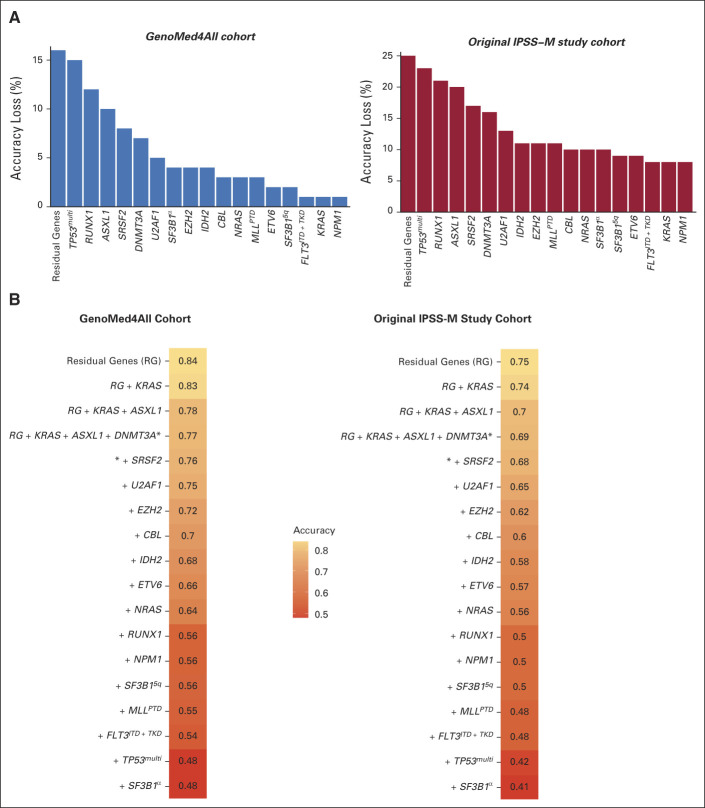
Accuracy of IPSS-M Prediction when Molecular Information was Missed
We analyzed the loss of accuracy of IPSS-M prediction when one or more IPSS-M–related molecular features are missing.
GenoMed4all and IWG-PM9 populations were used as learning and validation cohorts, respectively. We first evaluated the impact of a missing information from each of the IPSS-M genomic features.9 Figure 5 shows the accuracy loss of IPSS-M prediction for each missing genomic variable. Then, we evaluated the IPSS-M prediction accuracy in the presence of a combination of missing genomic features (starting from missing information on residual genes and then considering the main effect genes ordered by their prognostic weights estimated on probability of LFS; Fig 5).12 Information on mutational status of a set of 15 genes (ASXL1, CBL, DNMT3A, ETV6, EZH2, FLT3, IDH2, MLLPTD, NPM1, NRAS, RUNX1, SF3B1, SRSF2, TP53multihit, and U2AF1) was required to have an accuracy of IPSS-M prediction of 80% and 70% in the GenoMed4all and IWG-PM cohorts, respectively, while considering a set of 10 and seven genes, the accuracy of IPSS-M prediction decreased to < 70% versus < 60%, respectively, and to < 60% versus < 50%, respectively (Fig 5).
FIG 5.
Accuracy of IPSS-M prediction when molecular information was missed. (A) Impact of a missing information from each of the IPSS-M genomic feature on the accuracy of IPSS-M risk prediction in GenoMed4all (n = 2,876) and IWG-PM cohorts (n = 2,957). The height of the bar is proportional to the accuracy loss in IPSS-M prediction in the presence of a missing genomic feature, estimated as the percentage of patients classified in the wrong risk category. (B) Prediction of IPSS-M accuracy in the presence of a combination of missing genomic features in the GenoMed4all and IWG-PM cohorts (starting from missing information on residual genes and then considering the main effect genes ordered by their prognostic weights estimated on probability of LFS). The accuracy was estimated as the number of patients classified in the correct risk category divided by the patient population's size. IPSS-M, Molecular International Prognostic Scoring System; IPSS-R, Revised International Prognostic Scoring System; IWG-PM, International Working Group for Prognosis in MDS; LFS, leukemia-free survival.
DISCUSSION
A more precise risk score is essential to improve precision medicine strategies for patients with MDS. These in turn can identify patient groups that may respond better versus do not benefit from current treatment approaches.1-9 In this study, we provided an extensive validation of the recently developed IPSS-M9 and we confirmed that the molecular score performed better than the conventional IPSS-R. This was also true in patients without detectable mutations, thus suggesting that the statistical model used to develop IPSS-M is more efficient per se in capturing prognostic information with respect to conventional Cox's model.9
The precise definition of the probability of leukemic evolution is particularly important in the lower-risk groups, which represent the majority of patients with MDS, and in whom new treatment approaches, including HSCT, may be addressed in a refined manner.1,21 In this context, our findings confirmed that in the very low-low intermediate IPSS-R risk group, 20% of patients were reclassified into a less favorable prognostic category, > 90% of which had one or more mutated main effect IPSS-M genes. Thus, the clinical implementation of IPSS-M is expected to result in a more effective selection of candidates to disease-modifying therapies (including HSCT) among patients with early-stage disesase.21-23 Transplantation performed early after the diagnosis is associated with the most favorable outcome,21 and therefore, patients with higher risk according to IPSS-M should be considered to receive a transplant procedure earlier than the conventional scoring system (IPSS-R) would dictate.24,25 We observed in addition that, in patients with MDS treated with HSCT, IPSS-M significantly improved the prediction of the probability of OS with respect to IPSS-R. In particular, IPSS-M was able to efficiently capture the probability of relapse, thus potentially refining the choice of the optimal conditioning regimen at individual patient level26 (a myeloablative conditioning should be preferable in eligible subjects who are at higher risk of relapse according to genomic features) and improving the identification of patients with high risk of transplantation failure that can be considered for preemptive treatments of disease recurrence.22,23
HMA are the only class of drugs approved for the treatment of higher-risk MDS not eligible for HSCT. However, only 40%-50% of patients experience hematologic improvement, and CR occurs in 10%-15% of cases.1,27 Effective methods for identifying patients who are most likely to respond to HMA would be of immediate clinical utility. Models on the basis of clinical features are not sufficiently conclusive to deny eligible patients a trial of HMA based on their predictions alone.28,29
In our study, IPSS-M failed to stratify individual probability of response; however, response duration and probability of OS were inversely related to IPSS-M risk. This is in line with observation that the IPSS-M is a very good tool to reflect the disease biology and the aggressiveness of MDS subtypes.28-30 Additional factors other than gene mutations can be involved in determining sensitivity to HMA.31,32
Molecular testing is not yet routine globally because of cost, infrastructure, and reimbursement considerations.1,9 We analyzed the accuracy of IPSS-M prediction in both GenoMed4All and IWG-PM cohorts when one or more molecular features are missing. Considering a minimum data set of 15 relevant genes, the accuracy of IPSS-M prediction was 80% and 70%, respectively, while reducing the number of available genes to 10 or less, the accuracy of IPSS-M prediction was significantly lower in both cohorts. These findings may facilitate the clinical implementation of the score into a real-world clinical setting and may help clinicians to define the robustness of the prognosis prediction according to the amount of available information.
Our study may present some limitations, mainly because of the retrospective nature of the data. However, we were able to analyze a large population of patients with MDS, and the collection of DNA for genomic screening was provided independently from disease diagnosis, risk category, and treatment, thus limiting the risk of a selection bias effect and improving the generalizability of the results.
Despite the improved prognostication provided by IPSS-M, we observed that demographic features have a high predictive prognostic power, and clinical parameters (bone marrow blasts and anemia) still retain a strong predictive effect on survival, suggesting that these variables reflect important features of the disease state that are not captured by genomic landscape.4 Accordingly, including sex and age information and combining gene mutation with gene expression data33 might further improve outcome prediction in MDS in next future.
Marie Robin
Research Funding: AbbVie (Inst), Medac (Inst)
Travel, Accommodations, Expenses: Medac
Manja Meggendorfer
Employment: MLL Munich Leukemia Laboratory
Juan Carlos Caballero Berrocal
Honoraria: Takeda
Massimo Bernardi
Honoraria: Celgene, Interlabo, Janssen, AbbVie, Astellas Pharma
Consulting or Advisory Role: Pfizer
Travel, Accommodations, Expenses: Medac, Amgen, Sanofi, Jazz Pharmaceuticals, Biotest, AbbVie, Takeda
Lorenza Borin
Consulting or Advisory Role: Celgene
Speakers' Bureau: Genzyme
Travel, Accommodations, Expenses: Genzyme
Antonio Russo
Travel, Accommodations, Expenses: Pfizer, Novartis
Patrizia Chiusolo
Speakers' Bureau: Astellas Pharma, Novartis, Bristol Myers Squibb/Medarex
Luisa Giaccone
Honoraria: MSD, Novartis
Travel, Accommodations, Expenses: Novartis Italy, Janssen
Maria Teresa Voso
Honoraria: AbbVie, Astellas Pharma, Jazz Pharmaceuticals, Celene/BMS
Speakers' Bureau: Celgene
Research Funding: Celgene (Inst), Novartis (Inst)
Esther N. Oliva
Honoraria: Celgene, Novartis, Amgen, Alexion Pharmaceuticals, Daiichi Sankyo Europe GmbH, Sobi, SERVIER, Janssen
Consulting or Advisory Role: Amgen, Celgene, Novartis, Janssen, Daiichi Sankyo, Alexion Pharmaceuticals
Speakers' Bureau: Celgene, Novartis, Amgen
Patents, Royalties, Other Intellectual Property: Royalties for QOL-E instrument, Royalties for HM-PRO
Olivier Nibourel
Honoraria: Novartis, Incyte
Consulting or Advisory Role: Novartis, Incyte
Travel, Accommodations, Expenses: Incyte
Niccolo’ Bolli
Honoraria: Pfizer, Janssen Oncology, Amgen, GlaxoSmithKline, Jazz Pharmaceuticals, Takeda
Consulting or Advisory Role: Janssen
Speakers' Bureau: Celgene, Amgen
Alessandro Rambaldi
Consulting or Advisory Role: Amgen, Omeros, Novartis, Astellas Pharma, Jazz Pharmaceuticals, Roche, AbbVie, Janssen, Pfizer, Incyte, Kite/Gilead
Francesco Passamonti
Consulting or Advisory Role: ROCHE, AMOMED, Novartis, Celgene/Bristol Myers Squibb, Sandoz, Sierra Oncology, AbbVie, Karyiopharma, Sumitomo Dainippon Pharma Oncology
Speakers' Bureau: Novartis, AOP Orphan Pharmaceuticals, Celgene/Bristol Myers Squibb, AbbVie
Armando Santoro
Consulting or Advisory Role: Bristol Myers Squibb, SERVIER, Gilead Sciences, Pfizer, Eisai, Bayer, MSD, Sanofi, ARQULE, Incyte
Speakers' Bureau: Takeda, Roche, AbbVie, Amgen, Celgene, AstraZeneca, ArQule, Lilly, Sandoz, Novartis, BMS, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD
Ulrich Germing
Honoraria: Celgene, Novartis, Jazz Pharmaceuticals
Consulting or Advisory Role: Celgene
Research Funding: Celgene (Inst), Novartis (Inst)
Shahram Kordasti
Honoraria: Beckman Coulter, GWT-TUD, Alexion Pharmaceuticals
Consulting or Advisory Role: Syneos Health, Novartis, Pfizer
Speakers' Bureau: Pfizer
Research Funding: Celgene, Novartis, MorphoSys
Valeria Santini
Honoraria: Celgene/Bristol Myers Squibb, Novartis
Consulting or Advisory Role: Celgene/Bristol Myers Squibb, Novartis, Menarini, Takeda, Gilead Sciences, AbbVie, Syros Pharmaceuticals, SERVIER
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Janssen-Cilag, Celgene
Maria Diez-Campelo
Honoraria: Celgene, Novartis
Consulting or Advisory Role: Celgene, Novartis, GlaxoSmithKline, Blueprint Medicines
Travel, Accommodations, Expenses: Gilead Sciences
Guillermo Sanz
Honoraria: Celgene, Roche
Consulting or Advisory Role: AbbVie, Celgene, Novartis, Takeda, ExCellThera
Speakers' Bureau: Takeda, Novartis, Bristol Myers Squibb/Celgene
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Celgene, BeiGene
Wolfgang Kern
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Stock and Other Ownership Interests: MLL Munich Leukemia Laboratory
Uwe Platzbecker
Honoraria: Celgene/Jazz, AbbVie, Curis, Geron, Janssen
Consulting or Advisory Role: Celgene/Jazz, Novartis, BMS GmbH & Co KG
Research Funding: Amgen (Inst), Janssen (Inst), Novartis (Inst), BerGenBio (Inst), Celgene (Inst), Chris (Inst)
Patents, Royalties, Other Intellectual Property: part of a patent for a TFR-2 antibody (Rauner et al. Nature Metabolics 2019)
Travel, Accommodations, Expenses: Celgene
Lionel Ades
Honoraria: Celgene, AbbVie, Jazz Pharmaceuticals, BerGenBio, Silence Therapeutics, Novartis
Research Funding: Celgene (Inst)
Pierre Fenaux
Honoraria: Celgene
Research Funding: Celgene (Inst)
Torsten Haferlach
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Consulting or Advisory Role: Illumina
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented as oral communication to the 2022 ASH meeting (December 11, 2022).
SUPPORT
The study was conducted by GenoMed4all consortium (https://genomed4all.eu/) and supported by EuroBloodNET, the European Reference Network on rare hematologic diseases. (https://eurobloodnet.eu/)
European Union (GenoMed4All project No. 101017549 to M.G.D.P., C.D.G., T.H., U.P., P.F., M.D.C.; Transcan 7 Horizon 2020—EuroMDS project No. 20180424 to M.G.D.P., F.S., U.P., P.F.; HARMONY project No. 116026 to GC); AIRC Foundation (Associazione Italiana per la Ricerca contro il Cancro, Milan Italy—Project No. 22053 to M.G.D.P. and No. 26216 to G.C.); PRIN 2017 (Ministry of University & Research, Italy—Project 2017WXR7ZT to M.G.D.P.); Ricerca Finalizzata 2016 and 2018 (Italian Ministry of Health, Italy—Project RF2016-02364918 to M.G.D.P. and Project NET-2018-12,365,935 to M.G.D.P., F.P., M.T.V.); Cariplo Foundation (Milan Italy—Project No. 2016-0860 to M.G.D.P.); Beat Leukemia Foundation, Milan Italy (to M.G.D.P.)
CLINICAL TRIAL INFORMATION
G.C. and M.G.D.P. contributed equally as last senior authors.
DATA SHARING STATEMENT
Requests for access to data from the study should be addressed to GenoMed4All scientific committee (please contact Matteo G Della Porta at matteo.della_porta@hunimed.eu). All proposals requesting data access will need to specify how the data will be used, and all proposals will need the approval of the GenoMed4All scientific committee before data release.
AUTHOR CONTRIBUTIONS
Conception and design: Elisabetta Sauta, Francesco Passamonti, Armando Santoro, Shahram Kordasti, Valeria Santini, Pierre Fenaux, Torsten Haferlach, Gastone Castellani, Matteo Giovanni Della Porta
Financial support: Gastone Castellani
Administrative support: Marilena Bicchieri
Provision of study materials or patients: Marie Robin, Lin-Pierre Zhao, Massimo Bernardi, Giulia Rivoli, Lorenza Borin, Cristina Astrid Tentori, Luisa Giaccone, Maria Teresa Voso, Esther Natalie Oliva, Alessandro Rambaldi, Francesco Passamonti, Armando Santoro, Ulrich Germing, Valeria Santini, Maria Diez-Campelo, Francesc Sole, Uwe Platzbecker, Lionel Ades, Torsten Haferlach
Collection and assembly of data: Elisabetta Sauta, Marie Robin, Erica Travaglino, Manja Meggendorfer, Lin-Pierre Zhao, Juan Carlos Caballero Berrocal, Giulia Maggioni, Massimo Bernardi, Carmen Di Grazia, Luca Vago, Giulia Rivoli, Lorenza Borin, Cristina Astrid Tentori, Marta Ubezio, Alessia Campagna, Antonio Russo, Daniele Mannina, Luca Lanino, Patrizia Chiusolo, Luisa Giaccone, Maria Teresa Voso, Marta Riva, Esther Natalie Oliva, Elena Riva, Marilena Bicchieri, Alessandro Rambaldi, Francesco Passamonti, Ulrich Germing, Valeria Santini, Maria Diez-Campelo, Guillermo Sanz, Wolfgang Kern, Uwe Platzbecker, Lionel Ades, Pierre Fenaux, Matteo Giovanni Della Porta
Data analysis and interpretation: Elisabetta Sauta, Marie Robin, Matteo Bersanelli, Erica Travaglino, Claudia Sala, Giulia Maggioni, Saverio D'Amico, Luca Lanino, Matteo Zampini, Olivier Nibourel, Niccolo’ Bolli,Francesco Passamonti, Victor Savevski, Armando Santoro, Shahram Kordasti, Valeria Santini, Maria Diez-Campelo, Francesc Sole, Wolfgang Kern, Uwe Platzbecker, Gastone Castellani, Matteo Giovanni Della Porta
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Real-World Validation of Molecular International Prognostic Scoring System for Myelodysplastic Syndromes
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Marie Robin
Research Funding: AbbVie (Inst), Medac (Inst)
Travel, Accommodations, Expenses: Medac
Manja Meggendorfer
Employment: MLL Munich Leukemia Laboratory
Juan Carlos Caballero Berrocal
Honoraria: Takeda
Massimo Bernardi
Honoraria: Celgene, Interlabo, Janssen, AbbVie, Astellas Pharma
Consulting or Advisory Role: Pfizer
Travel, Accommodations, Expenses: Medac, Amgen, Sanofi, Jazz Pharmaceuticals, Biotest, AbbVie, Takeda
Lorenza Borin
Consulting or Advisory Role: Celgene
Speakers' Bureau: Genzyme
Travel, Accommodations, Expenses: Genzyme
Antonio Russo
Travel, Accommodations, Expenses: Pfizer, Novartis
Patrizia Chiusolo
Speakers' Bureau: Astellas Pharma, Novartis, Bristol Myers Squibb/Medarex
Luisa Giaccone
Honoraria: MSD, Novartis
Travel, Accommodations, Expenses: Novartis Italy, Janssen
Maria Teresa Voso
Honoraria: AbbVie, Astellas Pharma, Jazz Pharmaceuticals, Celene/BMS
Speakers' Bureau: Celgene
Research Funding: Celgene (Inst), Novartis (Inst)
Esther N. Oliva
Honoraria: Celgene, Novartis, Amgen, Alexion Pharmaceuticals, Daiichi Sankyo Europe GmbH, Sobi, SERVIER, Janssen
Consulting or Advisory Role: Amgen, Celgene, Novartis, Janssen, Daiichi Sankyo, Alexion Pharmaceuticals
Speakers' Bureau: Celgene, Novartis, Amgen
Patents, Royalties, Other Intellectual Property: Royalties for QOL-E instrument, Royalties for HM-PRO
Olivier Nibourel
Honoraria: Novartis, Incyte
Consulting or Advisory Role: Novartis, Incyte
Travel, Accommodations, Expenses: Incyte
Niccolo’ Bolli
Honoraria: Pfizer, Janssen Oncology, Amgen, GlaxoSmithKline, Jazz Pharmaceuticals, Takeda
Consulting or Advisory Role: Janssen
Speakers' Bureau: Celgene, Amgen
Alessandro Rambaldi
Consulting or Advisory Role: Amgen, Omeros, Novartis, Astellas Pharma, Jazz Pharmaceuticals, Roche, AbbVie, Janssen, Pfizer, Incyte, Kite/Gilead
Francesco Passamonti
Consulting or Advisory Role: ROCHE, AMOMED, Novartis, Celgene/Bristol Myers Squibb, Sandoz, Sierra Oncology, AbbVie, Karyiopharma, Sumitomo Dainippon Pharma Oncology
Speakers' Bureau: Novartis, AOP Orphan Pharmaceuticals, Celgene/Bristol Myers Squibb, AbbVie
Armando Santoro
Consulting or Advisory Role: Bristol Myers Squibb, SERVIER, Gilead Sciences, Pfizer, Eisai, Bayer, MSD, Sanofi, ARQULE, Incyte
Speakers' Bureau: Takeda, Roche, AbbVie, Amgen, Celgene, AstraZeneca, ArQule, Lilly, Sandoz, Novartis, BMS, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD
Ulrich Germing
Honoraria: Celgene, Novartis, Jazz Pharmaceuticals
Consulting or Advisory Role: Celgene
Research Funding: Celgene (Inst), Novartis (Inst)
Shahram Kordasti
Honoraria: Beckman Coulter, GWT-TUD, Alexion Pharmaceuticals
Consulting or Advisory Role: Syneos Health, Novartis, Pfizer
Speakers' Bureau: Pfizer
Research Funding: Celgene, Novartis, MorphoSys
Valeria Santini
Honoraria: Celgene/Bristol Myers Squibb, Novartis
Consulting or Advisory Role: Celgene/Bristol Myers Squibb, Novartis, Menarini, Takeda, Gilead Sciences, AbbVie, Syros Pharmaceuticals, SERVIER
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Janssen-Cilag, Celgene
Maria Diez-Campelo
Honoraria: Celgene, Novartis
Consulting or Advisory Role: Celgene, Novartis, GlaxoSmithKline, Blueprint Medicines
Travel, Accommodations, Expenses: Gilead Sciences
Guillermo Sanz
Honoraria: Celgene, Roche
Consulting or Advisory Role: AbbVie, Celgene, Novartis, Takeda, ExCellThera
Speakers' Bureau: Takeda, Novartis, Bristol Myers Squibb/Celgene
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Celgene, BeiGene
Wolfgang Kern
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Stock and Other Ownership Interests: MLL Munich Leukemia Laboratory
Uwe Platzbecker
Honoraria: Celgene/Jazz, AbbVie, Curis, Geron, Janssen
Consulting or Advisory Role: Celgene/Jazz, Novartis, BMS GmbH & Co KG
Research Funding: Amgen (Inst), Janssen (Inst), Novartis (Inst), BerGenBio (Inst), Celgene (Inst), Chris (Inst)
Patents, Royalties, Other Intellectual Property: part of a patent for a TFR-2 antibody (Rauner et al. Nature Metabolics 2019)
Travel, Accommodations, Expenses: Celgene
Lionel Ades
Honoraria: Celgene, AbbVie, Jazz Pharmaceuticals, BerGenBio, Silence Therapeutics, Novartis
Research Funding: Celgene (Inst)
Pierre Fenaux
Honoraria: Celgene
Research Funding: Celgene (Inst)
Torsten Haferlach
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Consulting or Advisory Role: Illumina
No other potential conflicts of interest were reported.
REFERENCES
- 1.Malcovati L, Hellström-Lindberg E, Bowen D, et al. : Diagnosis and treatment of primary myelodysplastic syndromes in adults: Recommendations from the European LeukemiaNet. Blood 122:2943-2964, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg PL, Tuechler H, Schanz J, et al. : Revised international prognostic scoring system for myelodysplastic syndromes. Blood 120:2454-2465, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Della Porta MG, Tuechler H, Malcovati L, et al. : Validation of WHO classification-based Prognostic Scoring System (WPSS) for myelodysplastic syndromes and comparison with the revised International Prognostic Scoring System (IPSS-R). A study of the International Working Group for Prognosis in Myelodysplasia (IWG-PM). Leukemia 29:1502-1513, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Bersanelli M, Travaglino E, Meggendorfer M, et al. : Classification and personalized prognostic assessment on the basis of clinical and genomic features in myelodysplastic syndromes. J Clin Oncol 39:1223-1233, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazzola M, Della Porta MG, Malcovati L: The genetic basis of myelodysplasia and its clinical relevance. Blood 122:4021-4034, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Gerstung M, Malcovati L, et al. : Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122:3616-3627, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard E, Nannya Y, Hasserjian RP, et al. : Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 26:1549-1556, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazha A, Komrokji R, Meggendorfer M, et al. : Personalized prediction model to risk stratify patients with myelodysplastic syndromes. J Clin Oncol 39:3737-3746, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard E, Tuechler H, Greenberg PL, et al. : Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid 1:2022, 2022 [DOI] [PubMed] [Google Scholar]
- 10.GenoMed4All : https://GenoMed4All.eu/
- 11.EuroBloodNet : https://eurobloodnet.eu/
- 12.Arber DA, Orazi A, Hasserjian R, et al. : The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391-2405, 2016 [DOI] [PubMed] [Google Scholar]
- 13.Khoury JD, Solary E, Abla O, et al. : The 5th edition of the World Health Organization classification of haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia 36:1703-1719, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arber DA, Orazi A, Hasserjian RP, et al. : International consensus classification of myeloid neoplasms and acute leukemias: Integrating morphologic, clinical, and genomic data. Blood 140:1200-1228, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheson BD, Greenberg PL, Bennett JM, et al. : Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood 108:419-425, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Chen BE, Kramer JL, Greene MH, et al. : Competing risks analysis of correlated failure time data. Biometrics 64:172-179, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrell FE, Jr, Califf RM, Pryor DB, et al. : Evaluating the yield of medical tests. JAMA 247:2543-2546, 1982 [PubMed] [Google Scholar]
- 18.Akaike H: A new look at the statistical model identification. IEEE Trans Automat Contr 19:716-723, 1974 [Google Scholar]
- 19.Gerstung M, Papaemmanuil E, Martincorena I, et al. : Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet 49:332-340, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GitHub : http://github.com/mg14/CoxHD
- 21.de Witte T, Bowen D, Robin M, et al. : Allogeneic hematopoietic stem cell transplantation for MDS and CMML: Recommendations from an international expert panel. Blood 129:1753-1762, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Della Porta MG, Gallì A, Bacigalupo A, et al. : Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol 34:3627-3637, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsley RC, Saber W, Mar BG, et al. : Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med 376:536-547, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cutler CS, Lee SJ, Greenberg P, et al. : A decision analysis of allogeneic bone marrow transplantation for the myelodysplastic syndromes: Delayed transplantation for low-risk myelodysplasia is associated with improved outcome. Blood 104:579-585, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Della Porta MG, Jackson CH, Alessandrino EP, et al. : Decision analysis of allogeneic hematopoietic stem cell transplantation for patients with myelodysplastic syndrome stratified according to the revised International Prognostic Scoring System. Leukemia 31:2449-2457, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kröger N, Iacobelli S, Franke GN, et al. : Dose-reduced versus standard conditioning followed by allogeneic stem-cell transplantation for patients with myelodysplastic syndrome: A prospective randomized phase III study of the EBMT (RICMAC trial). J Clin Oncol 35:2157-2164, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. : Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol 10:223-232, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeidan AM, Sekeres MA, Garcia-Manero G, et al. : Comparison of risk stratification tools in predicting outcomes of patients with higher-risk myelodysplastic syndromes treated with azanucleosides. Leukemia 30:649-657, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuendgen A, Müller-Thomas C, Lauseker M, et al. : Efficacy of azacitidine is independent of molecular and clinical characteristics—An analysis of 128 patients with myelodysplastic syndromes or acute myeloid leukemia and a review of the literature. Oncotarget 9:27882-27894, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch JS, Petti AA, Miller CA, et al. : TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med 375:2023-2036, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Follo MY, Finelli C, Mongiorgi S, et al. : Reduction of phosphoinositide-phospholipase C beta1 methylation predicts the responsiveness to azacitidine in high-risk MDS. Proc Natl Acad Sci USA 106:16811-16816, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YC, Kwon J, Fabiani E, et al. : Demethylation and up-regulation of an oncogene after hypomethylating therapy. N Engl J Med 386:19982022-19992010, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerstung M, Pellagatti A, Malcovati L, et al. : Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun 6:5901, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Requests for access to data from the study should be addressed to GenoMed4All scientific committee (please contact Matteo G Della Porta at matteo.della_porta@hunimed.eu). All proposals requesting data access will need to specify how the data will be used, and all proposals will need the approval of the GenoMed4All scientific committee before data release.